Abstract
During systematic analysis of nonbonded contacts in protein–ligand complexes derived from crystal structures in the Protein Data Bank, Cl–π interactions have been found, not only in the well-documented serine proteases but also, to a lesser extent, in other proteins. From geometric analysis of such Cl–π interactions in the crystal structures, two distinct geometries were found: the “edge-on” approach of a Cl atom to a ring atom or C–C bond and the “face-on” approach toward the ring centroid with an average interatomic distance of 3.6 Å. High-level ab initio calculations using benzene–chlorohydrocarbon model systems elucidated that the calculated Cl–π interaction energy is −2.01 kcal/mol, and the dispersion force is the major source of attraction. We also discussed the geometric flexibility in Cl–π interactions and a relationship between the intensity of the π density in an aromatic ring and the interaction position of the Cl atom.
Keywords: protein–ligand interaction, Protein Data Bank (PDB), Cl–π, interaction, ab initio calculation, dispersion interaction
Recent increase in the amount of crystal structures for protein–ligand complexes has helped us understand the contributions of noncanonical interactions, such as CH–O (Pierce et al. 2002), CH–π (Umezawa and Nishio 1998a,b), π–π (Boehr et al. 2002), and cation–π interactions (Kryger et al. 1999), to molecular recognition in protein–ligand interactions. However, these interactions are rarely included in the force fields used in structure-based drug design, because precise information about them is not available. We believe that an exhaustive analysis of crystal-structure databases will provide us with new insight into noncanonical interactions and aid the development of new scoring functions in docking studies. Therefore, we carried out a systematic analysis of nonbonded contacts for 14 polar and aromatic amino acid side chains in the crystal structures of protein–ligand complexes contained in the Protein Data Bank (PDB) and have reported the preferences and propensities of the patterns and types of weak interactions for each amino acid side chain (Imai et al. 2007). Our analysis revealed that the propensities for noncanonical interactions are not always consistent among side chains with similar characteristics, and there still remain attractive contacts to be investigated. In particular, π-related interactions such as halogen–π and S–π interactions are of interest.
Halogen substitution is an important approach for drug design, because halogenation may result in altered physico-chemical properties. However, explanation of the structure–activity effects of halogens has been limited to considerations of electron-withdrawing effects and the steric effects of halogen substituents on aromatic rings. During our recent analysis mentioned above, halogen atoms were sometimes found lying within 4 Å from the centroid of aromatic rings in side chains, which was suggestive of some form of interaction. In particular, cases of chloro (Cl) groups playing a key role in ligand binding were recognized for several serine proteases, such as trypsin (Stubbs et al. 2002), Factor Xa (Adler et al. 2002; Choi-Sledeski et al. 2003; Maignan et al. 2003; Nazare et al. 2005; Roehrig et al. 2005), and thrombin (Tucker et al. 1998), where a Cl group in high-affinity ligands has been found to bind to the aromatic ring of Tyr in the active site, replacing a water molecule. These interactions have been well documented, and Saraogi et al. (2003) reported a database study for halogen–π interactions in the PDB, but there has yet to be a systematic analysis and theoretical study. To further elucidate the nature of these interactions, we describe here precise analyses of the Cl–π interactions of protein–ligand complexes in crystal structures from the PDB and evaluate the Cl–π interaction for the preferred geometries. The interaction energies of model structures, using ab initio computational methods, are shown.
Results and Discussion
Database search of Cl–π interactions in protein–ligand complexes of the PDB
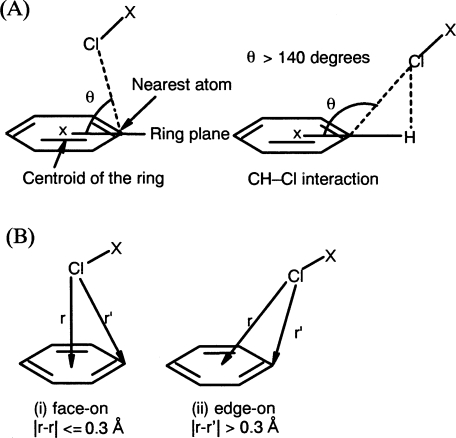
Searches of protein–ligand complex structures in the PDB showed more than 200 ligand data (338 interaction data), where a Cl atom lies close to an aromatic ring centroid of a side chain at a shorter distance than 4.5 Å (see Materials and Methods). However, the results include structures outside of the distance criteria, such as those better classified as CH–Cl interactions. To efficiently extract only Cl–π interactions, the geometries of the interactions shown in Figure 1A were calculated for each datum, and almost all the CH–Cl interactions in the results were found to have a θ angle >140°. Accordingly, θ < 140° was adopted as an additional criterion for Cl–π interactions. After removing CH–Cl interactions by final visual inspection, a total of 59 interaction data were selected. Most of the Cl–π interactions were found to occur with chlorophenyl or chloronaphthyl groups of the ligands, and only two cases were found with nonaromatic moieties, (R)-halothane (PDBID:1XZ1) and chloramphenicol (PDBID:1USQ).
Figure 1.
(A) Schematic of Cl–π interactions. The interaction is defined by the θ angle of the Cl atom relative to the plane of the ring. (B) Two geometries found in Cl–π interactions: (i) face-on and (ii) edge-on geometries. The geometry is defined by the difference between two distances, which are between the Cl atom and the centroid or the nearest atom of the aromatic ring.
Frequency and geometries of Cl–π interactions
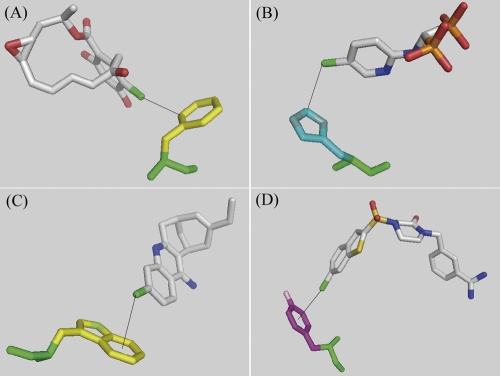
Analysis of Cl–π interactions based on the type of the aromatic side chain revealed that there are 21, 15, 15, and eight data with the Cl atom pointing toward Phe, Tyr, Trp, and His aromatic rings, respectively, and no distinguishable propensity for any aromatic amino acid could be observed except for His. For the His-related interactions, an additional four interactions with a Cl atom pointing toward a N atom rather than the aromatic ring of the His side chain were found, and they were excluded from Cl–π interactions because of the possibility of Cl–N(H) interactions. The distances from the Cl atom to the ring centroid were shorter in Trp and Tyr than in Phe and His (the average distances were 3.9 Å, 3.9 Å, 4.3 Å, and 4.0 Å, respectively). Two geometries were found to be dominant in Cl–π interactions from visual inspection, namely, where the Cl atom approaches toward the aromatic “atoms” and “bonds,” or toward the aromatic “center.” We classified these geometries as “edge-on” and “face-on” as shown in Figure 1B. To describe this difference in the geometries, several parameters were examined such as the θ angles or distances, and the value of the difference between the two distances (center and nearest) seemed to match best with the results of visual inspection: Interactions with a difference value of 0.3 Å or less belong to “face-on” geometry, and the rest are “edge-on” (geometric data for 59 Cl–π interactions are summarized in the Supplemental material). Similar geometries were proposed on the basis of spectroscopic data for OH–π and NH–π interactions (Joris et al. 1968) and by crystallographic analysis for CH–π interactions (Allen et al. 1996) of cyclopropane. Although the edge-on geometry (25 interactions) was found more often than face-on (10 interactions) as a whole, the edge-on geometry was preferred for Phe (14 vs. 0) (Fig. 2A, PDBID:1BGQ) and His (4 vs. 0) (Fig. 2B, PDBID:1TLS), while the face-on geometry was preferred in Trp (3 vs. 5) (Fig. 2C, PDBID:1E66) and Tyr (4 vs. 5) (Fig. 2D, PDBID:1NFU). These results could be due to limitations on the ligand allocation by the shape of the binding pocket of the specific proteins; however, the π densities of phenol and indole rings are considered to be higher than that of benzene; it suggests that the close proximity of the Cl atom to the ring centroid being found more frequently in Tyr or Trp than in Phe could be related to the intensity of the π density in the aromatic ring.
Figure 2.
Proximal ring atoms to the Cl atom. (A) Phe (PDBID: 1BGQ), (B) His (PDBID: 1TLS), (C) Trp (PDBID: 1E66), (D) Tyr (PDBID: 1NFU). (Black solid line) Cl–π interaction. Figures were prepared using the program PyMOL (DeLano Scientific).
Furthermore, most of the interatomic distances between the Cl atom and its nearest aromatic “atoms” were within 4 Å, and there were 21 out of 59 data with shorter interatomic distances than the sum of each van der Waals radius (the sum is 3.45 Å, with 1.75 Å and 1.7 Å for the Cl and C atoms, respectively); the shortest was 3.0 Å, and the average was 3.6 Å.
The effect of the ligand π electron system in Cl–π interaction
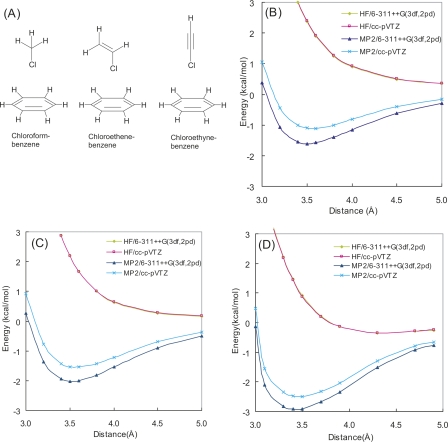
Among the 59 Cl–π interactions found in the PDB search, all cases except two were found to involve Cl groups bound to aromatic rings, which may mean that such Cl atoms have a greater propensity for Cl–π interactions, or that the existence of other cooperative aromatic interactions, such as CH–π interactions, are required to form the weaker Cl–π interactions. To elucidate whether a Cl group bound to an aromatic ring has a greater propensity for Cl–π interaction than one bound to an aliphatic group, we used a simple model system, which consists of chloroform, chloroethene, or chloroethyne as a prototypical Cl-containing ligand, and benzene as an aromatic side chain (Fig. 3A).
Figure 3.
(A) Model molecules of chloroform–benzene, chloroethene–benzene, and chloroethyne–benzene complexes considered in this study. HF and HF + MP2 potential energy curves of (B) chloroform–benzene, (C) chloroethene–benzene, and (D) chloroethyne–benzene complexes.
For each optimized complex structure using MP2/6-311++G(3df, 2pd) (the atomic coordinates of the complexes are given in the Supplemental material), with variable interatomic distance between the Cl atom and the nearest benzene atom, single-point energy calculations were performed at HF and MP2 levels using 6-311++G(3df, 2pd) and cc-pVTZ basis sets to generate interaction energy potential maps (Fig. 3B–D). The potential maps demonstrated that the energy minima were around the intermolecular distance of 3.5 Å in each complex and agreed with the observed geometry in Factor Xa crystal structures (Fig. 2D, PDBID: 1NFU) (Maignan et al. 2003). Figure 3B–D, also shows that the interaction energies of Cl atoms bound to π-electron systems are greater than those of Cl atoms bound to non-π systems and suggests the advantage of Cl atoms bound to aromatic rings for Cl–π interaction. Since the interaction energy of the chloroethyne complex is greater than that of the chloroethene complex, which will have additional CH–π interactions, it suggests that the Cl atom can be affected by the π-electron system of a ligand aromatic ring and further contribute to the stabilization of Cl–π interactions.
The essential source of Cl–π interaction
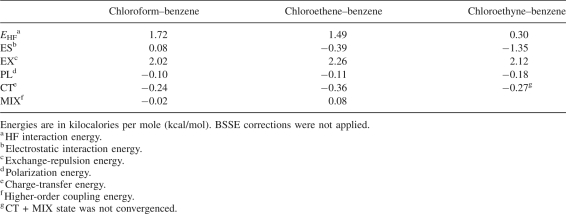
The calculated interaction potentials demonstrate that the Cl–π interaction is a greatly attractive force even if the interaction is distant. This means that the major source of attraction in Cl–π interaction is not orbital overlap such as for charge transfer, but interaction such as for electrostatic or dispersion interactions. As shown in Figure 3B–D, the HF interaction energies were repulsive, whereas HF + MP2 interaction energies were attractive. As the difference between HF and MP2 interaction energies is mainly an attractive dispersion interaction (Jaffe and Smith 1996), the dispersion effect is suggested to greatly contribute to Cl–π interaction. To estimate the contribution of electrostatic energies in the Cl–π interaction, the energy decomposition analysis (EDA) by Kitaura and Morokuma (1976) was applied to the HF interaction energies of three model complexes calculated with the cc-pVTZ basis sets (Table 1). The electrostatic interaction is repulsive in the chloroform–benzene complex and attractive in chloroethene–benzene and chloroethyne–benzene complexes. Although the electrostatic interaction (ES) is attractive in the two complexes, its magnitude is weaker than the exchange-repulsion (EX) interaction. Other attractive polarization (PL) and charge transfer (CT) interactions are also weak, and the sum of these attractive interaction energies does not overcome the exchange-repulsion energy. This fact indicates that the dispersion interaction is essential to make these complexes stable. Since small basis sets were reported to considerably underestimate the attractive dispersion interaction (Tsuzuki et al. 1994), our choice of basis sets appears to be large enough to discuss Cl–π interaction. To exclude the contribution of CH–π interactions, the chloroethyne–benzene complex was used for the following calculations.
Table 1.
Interaction energy components of three model complexes obtained by Kitaura–Morokuma energy decomposition analysis at the HF/cc-pVTZ level
Verification of geometry preference
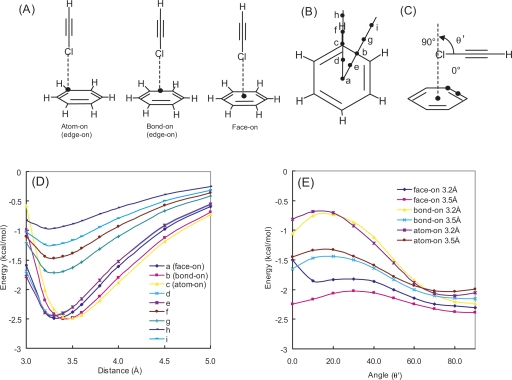
Two geometries for the Cl–π interaction, edge-on and face-on, were found in the PDB search, and edge-on was found more often than face-on. This preference was verified by comparison of the interaction energies for various geometric models including atom-on, bond-on, and face-on, as shown in Figure 4A. The interaction energy potential maps were generated using MP2/cc-pVTZ single-point calculations at points over the benzene ring, considering the symmetry shown in Figure 4B. As shown in Figure 4D, except for the cases in which the intermolecular distance is 3.0 Å, the potential energy surfaces are very flat within the benzene ring (positions a–e), and the interaction energy of chloroethyne weakens with the distance from the benzene ring, which suggests that the attractive force arises from the recognition of an aromatic ring by the Cl atom. Relatively unstable energies of atom-on (position c) and bond-on (position b) geometries, with an intermolecular distance of 3.0 Å, would be caused by the unfavorable repulsion between aromatic carbon atoms and the Cl atom.
Figure 4.
(A) Three geometries of chloroethyne–benzene complex. (B) Approaching points for Cl atom of chloroethyne. (C) Definition of relative angle (θ′) between chloroethyne molecule and benzene ring. Potential energy curves of chloroethyne–benzene complex. Plot of HF + MP2 interaction energies calculated with cc-pVTZ basis set (D) along various approaches and (E) along angle variation.
Potential energy maps were also generated for the relative angles of a chloroethyne molecule against a benzene, as shown in Figure 4C for the intermolecular distances of 3.2 Å and 3.5 Å with MP2/cc-pVTZ single-point calculations. Figure 4E demonstrates that for the θ′ angle of 60°–90°, the potential energy surfaces are very flat, within 0.4 kcal/mol of the energy difference for all distances and geometries; while for θ′ angles where chloroethyne is oriented nearly parallel with benzene, large energy differences were observed, both among geometries and between the distances. Energy maxima for the θ′ angle of 10°–20°, found in both atom-on and bond-on geometries, may be caused by the unfavorable repulsion between aromatic carbon atoms and the Cl atom. Meanwhile, the face-on geometry has no preference for the θ′ angle, with the only exception being that the interaction for θ′ = 0° at 3.2 Å was unstable. In this conformation, chloroethyne would overlap with the benzene much more than for atom-on or bond-on geometry, and it may cause the relative destabilization.
These results demonstrate that the Cl–π interaction has no geometry preference when the Cl atom approaches toward a benzene ring at a nearly perpendicular direction, but otherwise, face-on geometry is preferred, since it avoids unfavorable molecular contacts.
Calculation of Cl–π interaction energy
Tsuzuki et al. (2000) has reported that MP2 level calculation overestimates the interaction energy for CH–π interaction. Thus, the interaction energy for the chloroethyne–benzene complex was calculated by the CCSD(T) method to evaluate the amount that the correlation energy is overestimated by MP2. An edge-on geometric model derived from the crystal structure of a factor Xa–inhibitor complex was used because the Cl–π interaction energy was very similar to such a model, as mentioned above. Table 2 shows that the interaction energy of Cl–π interaction is also overestimated with MP2 calculation. Tsuzuki et al. (2006) also reported that weak intermolecular interaction energy can be precisely calculated using extrapolated MP2 interaction energies at the basis set limit [E MP2(limit)] and CCSD(T) correction terms. The extrapolation method for the estimation of E MP2(limit) proposed by Helgaker et al. (1997) was reported to give a good fit, especially when the electron correlation energy has a large contribution. Moreover, Helgaker's formula E = a + bX −3 (X is 2 for aug-cc-pVDZ, 3 for aug-cc-pVTZ, etc.) can be fitted to the interaction energies calculated using only two parameters, the energies calculated by aug-cc-pVXZ (X = D and T). Therefore, the E MP2(limit) was estimated by Helgaker's method as a correlation energy, and a value of −4.21 kcal/mol was obtained. From this value, the correlation term of the CCSD(T) interaction energy at basis limit [E CCSD(T)(limit)] was calculated as −2.93 kcal/mol. To estimate the Cl–π interaction energy (E int) as a sum of the correlation term of E CCSD(T)(limit) and converged HF interaction energy, HF interaction energy was calculated by HF/cc-pVQZ as 0.92 kcal/mol. Thus, E int was estimated to be −2.01 kcal/mol, as summarized in Table 3. This value is greater than the CH–π interaction energy (−1.454 kcal/mol) (Ringer et al. 2006).
Table 2.
Interaction energies of chloroethyne–benzene complex calculated with electron correlation correction by several methods
Table 3.
Estimated MP2 and CCSD(T) basis set limit interaction energies of chloroethyne–benzene complex
Effect of aromatic ring π density on the Cl–π interaction geometries
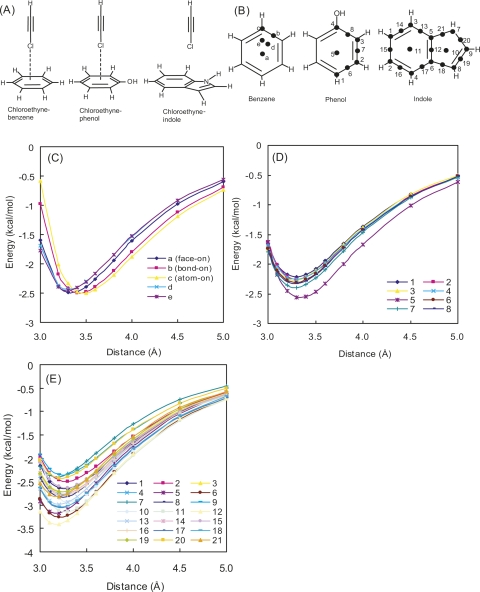
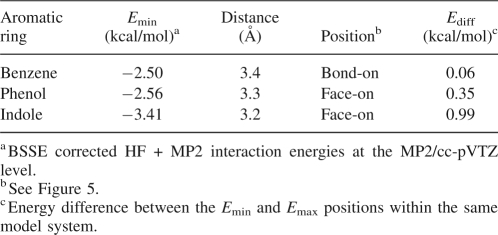
In the PDB search, Phe and His preferred edge-on geometry, but in the case of Tyr and Trp, face-on geometry was also found. As π densities of aromatic rings are estimated to increase in the order benzene < phenol < indole (Mecozzi et al. 1996), our observation suggested that the face-on geometry is preferred by π electron-rich aromatic rings. To better understand the effect of π density on the geometries, the interaction energy potential maps were generated for chloroethyne–benzene, chloroethyne–phenol, and chloroethyne–indole complexes (Fig. 5A,B), with single-point calculations using MP2/cc-pVTZ (Fig. 5C–E). Table 4 shows that the stability of the interactions and a tendency to shorten the intermolecular distance increased in proportion to the intensity of the π density. The differences of interaction energies among the interaction positions were also affected by the intensity of π density: The chloroethyne–benzene complex possesses similar interaction energies at any position, while chloroethyne–phenol and chloroethyne–indole complexes possess their minimum energies for face-on geometries. We also found that the interaction position providing the minimum interaction energy for the chloroethyne–indole complex was at the bond in the center of the indole ring. These results support the hypothesis that the face-on geometry is preferred by π electron-rich aromatic rings.
Figure 5.
(A) Model molecules of chloroethyne–benzene, chloroethyne–phenol, and chloroethyne–indole complexes. (B) In single-point energy calculations for drawing potential energy curves, the Cl atom was approached along a straight line perpendicular to the aromatic ring plane, whose crossing point is shown by the filled circle for the benzene, phenol, and indole complexes. Potential energy curves of HF + MP2 interaction energies calculated with the cc-pVTZ basis set. (C) Chloroethyne–benzene, (D) chloroethyne–phenol, (E) chloroethyne–indole complexes. Distance is measured from the aromatic ring plane.
Table 4.
Comparison of interaction energy for three chloroethyne–aromatic ring complex models
In this study, we performed precise analyses of Cl–π interactions in the PDB, defined as “edge-on” and “face-on” structures. Based on several observations in the search, we performed theoretical studies using ab initio calculations and found the following points:
Cl–π interaction is clearly an attractive interaction, where the major source of attraction is the dispersion force, and the calculated Cl–π interaction energy is −2.01 kcal/mol, which is greater than that of the CH–π interaction;
the edge-on geometry is predominant in crystallographic observations, while Cl–π interaction has no geometry preference regarding the approach of the Cl atom toward benzene rings at a nearly perpendicular direction, but otherwise, face-on geometry is preferred to avoid unfavorable molecular contacts;
the intensity of the π density in the aromatic ring could determine the position of the Cl atom interaction, at the center, the atoms, or the bonds of the ring;
Cl atoms bound to an aromatic ring have a greater propensity for stable Cl–π interaction compared to those bound to non-aromatic moieties in a ligand.
We concluded that the Cl–π interaction is both experimentally and theoretically reliable and an attractive interaction that is a means for ligand recognition by proteins.
Materials and Methods
Cl–π interaction search of the PDB
Cl–π interactions were searched for protein–ligand complex structures, using a subset of the PDB structures in Relibase+ version 2.1.1 (Hendlich et al. 2003) (38,672 PDB sets, 367,044 ligand data, up to August 2006). Throughout this work, a Cl–π interaction was defined as any contact where the interatomic distance between the centroid of the aromatic plane of a side chain and the Cl atom of a ligand is shorter than 4.5 Å. Details of the searching procedure have been reported in a previous article (Imai et al. 2007). In summary, data for 219 ligands were selected by the search via the graphical user interface of Relibase+, and with in-house perl scripts and python scripts implemented by Reliscript (a Relibase library of python scripts); they were categorized according to the amino acid to give data for 338 interactions, since Cl atoms were often surrounded by two or three different amino acids. To determine the geometries of the interactions, the θ angle of the Cl atom relative to the plane of the ring was measured, as shown in Figure 1A. Finally, each complex was checked by visual inspection, and data for 59 interactions were selected in total.
Ab initio calculations
The Gaussian 03 program (Frisch et al. 2004) was used for ab initio molecular orbital calculations. The 6-311++G(3df, 2pd) and cc-pVXZ (X = D, T, and Q) basis sets were used for the calculations, which were run on the Fujitsu Primepower HPC2500 of the Academic Center for Computing and Media Studies, Kyoto University. Electron correlation correction was accounted for using the second-order Møller-Plesset perturbation method (MP2) and by coupled cluster theory using single and double substitutions with noniterative triple excitations [CCSD(T)]. The geometries of isolated molecules and complex models were optimized at MP2/6-311++G(3df, 2pd) levels. The basis set superposition error (BSSE) (Ransil 1961) was corrected using the counterpoise method (Boys and Bernardi 1970). MP2 correlation interaction energy at the basis set limit was estimated by the method proposed by Helgaker et al. (1997). To estimate the magnitude of electrostatic energies in the Cl–π interaction, the energy decomposition analysis (EDA) of Kitaura and Morokuma (1976) was applied to the HF interaction energies of the model complexes calculated with the cc-pVTZ basis set. The EDA calculations were performed with the GAMESS program package (Schmidt et al. 1993).
Electronic supplemental material
A summary for the Cl–π interactions in Figure 2; geometric data for 59 Cl–π interactions; the optimized geometries of chloroform–benzene, chloroethene–benzene, and chloroethyne–benzene complexes at the MP2/6-311++G(3df, 2pd); and the complete reference of Gaussian03 are available in the Supplemental material.
Acknowledgments
The authors gratefully acknowledge Kaneyoshi Kato and Hiroyuki Kimura for their suggestions and encouragement. We thank David Cork for critical reading of the manuscript. I.N. and K.K. are grateful to Astellas Pharma, Inc. for financial support.
Footnotes
Reprint requests to: Yumi N. Imai, Takeda Pharmaceutical Company, Ltd., 2-17-85, Juso-honmachi, Yodogawa-ku, Osaka 532-8686, Japan; e-mail: imai_yumi@takeda.co.jp; fax: 81-6-6308-9010.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.033910.107.
Supplemental material: see www.proteinscience.org
References
- Adler, M., Kochanny, M.J., Ye, B., Rumennik, G., Light, D.R., Biancalana, S., Whitlow, M. Crystal structures of two potent nonamidine inhibitors bound to factor Xa. Biochemistry. 2002;41:15514–15523. doi: 10.1021/bi0264061. [DOI] [PubMed] [Google Scholar]
- Allen, F.H., Lommerse, J.P.M., Hoy, V.J., Howard, J.A.K., Desiraju, G.R. The hydrogen-bond C–H donor and π–acceptor characteristics of three-membered rings. Acta Crystallogr. B. 1996;52:734–745. [Google Scholar]
- Boehr, D.D., Farley, A.R., Wright, G.D., Cox, J.R. Analysis of the π–π stacking interactions between the aminoglycoside antibiotic kinase APH(3′)-IIIa and its nucleotide ligands. Chem. Biol. 2002;9:1209–1217. doi: 10.1016/s1074-5521(02)00245-4. [DOI] [PubMed] [Google Scholar]
- Boys, S.F., Bernardi, F. The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol. Phys. 1970;19:553–566. [Google Scholar]
- Choi-Sledeski, Y.M., Kearney, R., Poli, G., Pauls, H., Gardner, C., Gong, Y., Becker, M., Davis, R., Spada, A., Liang, G., et al. Discovery of an orally efficacious inhibitor of coagulation factor Xa which incorporates a neutral P1 ligand. J. Med. Chem. 2003;46:681–684. doi: 10.1021/jm020384z. [DOI] [PubMed] [Google Scholar]
- Frisch, M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R., Montgomery J.A., Jr, Vreven, T., Kudin, K.N., Burant, J.C., et al. 2004 Gaussian 03; Revision C.02. Gaussian, Inc. Wallingford, CT. [Google Scholar]
- Helgaker, T., Klopper, W., Koch, H., Noga, J. Basis-set convergence of correlated calculations on water. J. Chem. Phys. 1997;106:9639–9646. [Google Scholar]
- Hendlich, M., Bergner, A., Gunther, J., Klebe, G. Relibase: Design and development of a database for comprehensive analysis of protein–ligand interactions. J. Mol. Biol. 2003;326:607–620. doi: 10.1016/s0022-2836(02)01408-0. [DOI] [PubMed] [Google Scholar]
- Imai, Y.N., Inoue, Y., Yamamoto, Y. Propensities of polar and aromatic amino acids in noncanonical interactions: Nonbonded contacts analysis of protein–ligand complexes in crystal structures. J. Med. Chem. 2007;50:1189–1196. doi: 10.1021/jm061038a. [DOI] [PubMed] [Google Scholar]
- Jaffe, R.L., Smith, G.D. A quantum chemistry study of benzene dimmer. J. Chem. Phys. 1996;105:2780–2788. [Google Scholar]
- Joris, L., Schleyer, P.R., Gleiter, R. Cyclopropane rings as proton–acceptor groups in hydrogen bonding. J. Am. Chem. Soc. 1968;90:327–336. [Google Scholar]
- Kitaura, K., Morokuma, K. A new energy decomposition scheme for molecular interactions within the Hartree-Fock approximation. Int. J. Quantum Chem. 1976;10:325–340. [Google Scholar]
- Kryger, G., Silman, I., Sussman, J.L. Structure of acetylcholinesterase complexed with E2020 (Aricept®): Implications for the design of new anti-Alzheimer drugs. Struct. Fold. Des. 1999;7:297–307. doi: 10.1016/s0969-2126(99)80040-9. [DOI] [PubMed] [Google Scholar]
- Maignan, S., Guilloteau, J.P., Choi-Sledeski, Y.M., Becker, M.R., Ewing, W.R., Pauls, H.W., Spada, A.P., Mikol, V. Molecular structures of human factor Xa complexed with ketopiperazine inhibitors: Preference for a neutral group in the S1 pocket. J. Med. Chem. 2003;46:685–690. doi: 10.1021/jm0203837. (PDBID: 1NFU) [DOI] [PubMed] [Google Scholar]
- Mecozzi, S., West A.P., Jr, Dougherty, D.A. Cation–π interactions in aromatics of biological and medicinal interest: Electrostatic potential surfaces as a useful qualitative guide. Proc. Natl. Acad. Sci. 1996;93:10566–10571. doi: 10.1073/pnas.93.20.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazare, M., Will, D.W., Matter, H., Schreuder, H., Ritter, K., Urmann, M., Essrich, M., Bauer, A., Wagner, M., Czech, J., et al. Probing the subpockets of factor Xa reveals two binding modes for inhibitors based on a 2-carboxyindole scaffold: A study combining structure–activity relationship and X-ray crystallography. J. Med. Chem. 2005;48:4511–4525. doi: 10.1021/jm0490540. [DOI] [PubMed] [Google Scholar]
- Pierce, A.C., Sandretto, K.L., Bemis, G.W. Kinase inhibitors and the case for CH…O hydrogen bonds in protein–ligand binding. Proteins. 2002;49:567–576. doi: 10.1002/prot.10259. [DOI] [PubMed] [Google Scholar]
- Ransil, B.J. Studies in molecular structure. IV. Potential curve for the interaction of two helium atoms in single-configuration LCAO MO SCF approximation. J. Chem. Phys. 1961;34:2109–2118. [Google Scholar]
- Ringer, A.L., Figgs, M.S., Sinnokrot, M.O., Sherrill, C.D. Aliphatic C–H/π interactions: Methane–benzene, methane–phenol, and methane–indole complexes. J. Phys. Chem. A. 2006;110:10822–10828. doi: 10.1021/jp062740l. [DOI] [PubMed] [Google Scholar]
- Roehrig, S., Straub, A., Pohlmann, J., Lampe, T., Pernerstorfer, J., Schlemmer, K.H., Reinemer, P., Perzborn, E. Discovery of the novel antithrombotic agent 5-chloro-N-({(5S)-2-oxo-3- [4-(3-oxomorpholin-4-yl)phenyl]-1,3-oxazolidin-5-yl}methyl)thiophene- 2-carboxamide (BAY 59-7939): An oral, direct factor Xa inhibitor. J. Med. Chem. 2005;48:5900–5908. doi: 10.1021/jm050101d. [DOI] [PubMed] [Google Scholar]
- Saraogi, I., Vijay, V.G., Das, S., Sekar, K., Guru Row, T.N. C–halogen…π interactions in proteins: A database study. Cryst. Eng. 2003;6:69–77. [Google Scholar]
- Schmidt, M.W., Baldridge, K.K., Boatz, J.A., Elbert, S.T., Gordon, M.S., Jensen, J.H., Koseki, S., Matsunaga, N., Nguyen, K.A., Su, S., et al. General atomic and molecular electronic structure system. J. Comput. Chem. 1993;14:1347–1363. [Google Scholar]
- Stubbs, M.T., Reyda, S., Dullweber, F., Möller, M., Klebe, G., Dorsch, D., Mederski, W.W.K.R., Wurziger, H. pH-Dependent binding modes observed in trypsin crystals: Lessons for structure-based drug design. ChemBioChem. 2002;3:246–249. doi: 10.1002/1439-7633(20020301)3:2/3<246::aid-cbic246>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Tsuzuki, S., Uchimaru, T., Tanabe, K. Basis set effects on the intermolecular interaction of hydrocarbon molecules obtained by an ab initio molecular orbital method: Evaluation of dispersion energy. J. Mol. Struct. (Theochem.) 1994;307:107–118. [Google Scholar]
- Tsuzuki, S., Honda, K., Uchimaru, T., Mikami, M., Tanabe, K. The magnitude of the CH/π interaction between benzene and some model hydrocarbons. J. Am. Chem. Soc. 2000;122:3746–3753. [Google Scholar]
- Tsuzuki, S., Honda, K., Uchimaru, T., Mikami, M. Estimated MP2 and CCSD(T) interaction energies of n-alkane dimers at the basis set limit: Comparison of the methods of Helgaker et al. and Feller. J. Chem. Phys. 2006;124:114304–114310. doi: 10.1063/1.2178795. [DOI] [PubMed] [Google Scholar]
- Tucker, T.J., Brady, S.F., Lumma, W.C., Lewis, S.D., Gardell, S.J., Naylor-Olsen, A.M., Yan, Y., Sisko, J.T., Stauffer, K.J., Lucas, B.J., et al. Design and synthesis of a series of potent and orally bioavailable noncovalent thrombin inhibitors that utilize nonbasic groups in the P1 position. J. Med. Chem. 1998;41:3210–3219. doi: 10.1021/jm9801713. [DOI] [PubMed] [Google Scholar]
- Umezawa, Y., Nishio, M. CH/π interactions in the crystal structure of class I MHC antigens and their complexes with peptides. Bioorg. Med. Chem. 1998a;6:2507–2515. doi: 10.1016/s0968-0896(98)80024-2. [DOI] [PubMed] [Google Scholar]
- Umezawa, Y., Nishio, M. CH/π interactions as demonstrated in the crystal structure of guanine-nucleotide binding proteins, Src homology-2 domains and human growth hormone in complex with their specific ligands. Bioorg. Med. Chem. 1998b;6:493–504. doi: 10.1016/s0968-0896(98)00002-9. [DOI] [PubMed] [Google Scholar]