Abstract
One of the common methods for assessing energy functions of proteins is selection of native or near-native structures from decoys. This is an efficient but indirect test of the energy functions because decoy structures are typically generated either by sampling procedures or by a separate energy function. As a result, these decoys may not contain the global minimum structure that reflects the true folding accuracy of the energy functions. This paper proposes to assess energy functions by ab initio refolding of fully unfolded terminal segments with secondary structures while keeping the rest of the proteins fixed in their native conformations. Global energy minimization of these short unfolded segments, a challenging yet tractable problem, is a direct test of the energy functions. As an illustrative example, refolding terminal segments is employed to assess two closely related all-atom statistical energy functions, DFIRE (distance-scaled, finite, ideal-gas reference state) and DOPE (discrete optimized protein energy). We found that a simple sequence-position dependence contained in the DOPE energy function leads to an intrinsic bias toward the formation of helical structures. Meanwhile, a finer statistical treatment of short-range interactions yields a significant improvement in the accuracy of segment refolding by DFIRE. The updated DFIRE energy function yields success rates of 100% and 67%, respectively, for its ability to sample and fold fully unfolded terminal segments of 15 proteins to within 3.5 Å global root-mean-squared distance from the corresponding native structures. The updated DFIRE energy function is available as DFIRE 2.0 upon request.
Keywords: new methods, protein structure prediction, statistical energy function
Energy functions of proteins are developed to quantitatively capture the physical interaction that determines how proteins fold and interact with other biologically active molecules. Existing energy functions of proteins are obtained through a physical-based approach (Brooks et al. 1983; Weiner et al. 1986; Ponder and Richards 1987; Jorgensen et al. 1996; Scott et al. 1999), a statistical (knowledge-based) approach (Tanaka and Scheraga 1976), or their combination (empirical approach). A knowledge-based or statistical energy function is obtained directly from statistical analysis of known protein structures (Tanaka and Scheraga 1976).
Different statistical energy functions differ only in how their reference states are defined. We have introduced a physical reference state of uniformly distributed ideal gas points in finite protein-size spheres and assumed that the number of pairs of ideal gas points is proportional to the constant fractional power of the distance between two ideal gas points (r α) with α < 2 to account for the finite-size effect. This reference state together with a distance-scaling approximation yields the DFIRE (distance-scaled, finite, ideal-gas reference state) statistical energy function (Zhou and Zhou 2002). Recently, Shen and Sali introduced the theory of the discrete optimized protein energy (DOPE) based on the same finite ideal-gas reference state but with an analytical, protein-size dependent α parameter and a different scaling scheme (Shen and Sali 2006). A large-scale comparison among DFIRE, DOPE, and more than 20 other scoring functions (Eramian et al. 2006) suggests that DFIRE and DOPE perform superiorly over other scoring functions in near-native selections, whereas the difference between the two is small.
Decoy selection is only an indirect test of an energy function because decoys are often generated by a separate energy function or by sampling only. Here, we propose a more direct test of energy functions through ab initio refolding of completely unfolded segments with secondary structures while the rest of the protein remains folded. We hypothesize that a more direct test could detect the hidden difference between two closely related energy functions. In addition to assessing energy functions, terminal-segment refolding itself is biologically relevant. The folding of many proteins is assisted by a prefolded domain (pro-domain) (Llinas and Marqusee 1998; Atwell and Wells 1999).
Refolding the terminal segment while fixing the rest of a protein, however, is challenging because terminal regions are more flexible and often exposed (Jacob and Unger 2007) and, thus, searching backbone and side chain conformations at the same time is nontrivial. In fact, Zhu et al. (2006) showed that several physical-based and knowledge-based energy functions have difficulty folding even partially unfolded segments with secondary structures, and the best performance is given by the DFIRE energy function. Here, in order to separate testing energy functions from testing sampling techniques, we limit the maximum size of unfolded regions to less than 15 residues for a strand-containing segment, and less than 25 residues for a helix-containing segment.
Several variants of the DFIRE energy function are tested in this paper with a genetic algorithm for global energy minimization. We first update the DFIRE energy function with 30 distance bins that allow finer statistics to extract short-range repulsive and long-range interactions (uDFIRE). We then obtain an asymmetric DFIRE (aDFIRE) energy function that depends on the relative sequence positions of the interacting residues, an approach adopted in the DOPE energy function. Segment refolding by these energy functions suggests that both aDFIRE and DOPE have stronger local interactions and intrinsically bias toward the formation of longer helical segments than uDFIRE does, while uDFIRE appears to be more balanced in reproducing helical and strand segments. The most significant improvement, resulting from the change of the binning method from DFIRE to uDFIRE, highlights the importance of fine treatment of repulsive interactions for shape complementarity in segment refolding.
Results
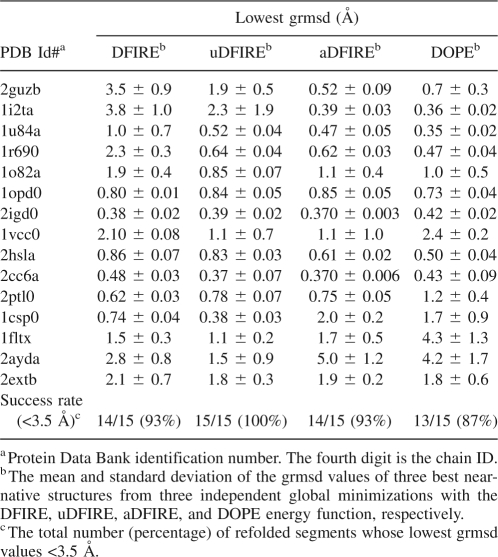
Table 1 shows the refolding results of four single helices, two two-helix bundles, seven single strands, one mixed helix/strand segment, and one β hairpin that were unfolded and refolded in the presence of a fixed folded region. The accuracy of refolded segments is described by a local root-mean-squared distance (lrmsd) that is calculated by superposing the unfolded and native segments or by a global rmsd (grmsd) between the refolded and the native segment conformations that are calculated after the superposition of the fixed regions. The values of lrmsd and grmsd are calculated from the positions of Cα atoms. The former measures the accuracy of the segment structure only, while the latter describes both the structure and its orientation relative to the rest of the protein. We report the results of four energy functions: DFIRE based on the original 20 bins (2 Å for the first distance bin, 0.5 Å per bin up to 8 Å, and 1 Å per bin from 8 Å to 15 Å), updated DFIRE (uDFIRE) based on the 30 equal-distance bins (0.5 Å per bin for the same distance range), asymmetric DFIRE (aDFIRE), where the interaction between two atoms depends on the difference (positive or negative) in the two sequence positions of their respective residues, and DOPE, which is also an asymmetric energy function. Three independent global energy minimizations by a genetic algorithm (see Materials and Methods) are performed for each segment in the presence of the fixed, folded region, and only one global minimum structure from each minimization is employed to produce the results in Table 1.
Table 1.
Restoration of unfolded terminal regions by a genetic algorithm with the original DFIRE, updated DFIRE (uDFIRE) with 30 bins, asymmetric DFIRE (aDFIRE) with a simple sequence dependence, and the DOPE energy function
It is clear from Table 1 that the uDFIRE energy makes a significant improvement over the DFIRE energy function with a smaller number of bins. Large reductions in rmsd values of refolded structures are observed for 1r690, 1o82a, and 2ayda, and smaller improvements are achieved for 1u84a, 1opd0, and 1csp0. Only three segments (1i2ta, 1vcc0, and 2extb) are refolded with significantly larger grmsd values by uDFIRE than by DFIRE (difference >0.5 Å). All three are misfolded by uDFIRE and DFIRE (grmsd > 6 Å). That is, the increases of grmsd values due to the use of uDFIRE for these segments are not that important. The overall success rates for refolding to within a 3.5 Å grmsd from the native conformation are 47% by DFIRE and 67% by uDFIRE, respectively, while aDFIRE and DOPE have the same success rate as uDFIRE.
While the overall success rates given by uDFIRE, aDFIRE, or DOPE are the same, there are some intrinsic differences between the three energy functions. For example, the uDFIRE energy function fails to fold the terminal helix of 2guzb and 1i2ta (grmsd > 9 Å) while DOPE misfolds the terminal strand in 1csp0 (grmsd = 5 Å) and 2ayda (grmsd = 16 Å). A close examination of 2guzb and 1i2ta indicates that their C-terminal helices are long and partially exposed (1i2ta is shown in Fig. 1A). uDFIRE does not make an accurate prediction of the C-terminal segments of the two proteins because it attempts to tightly pack the terminal segment with the rest of the protein. Both aDFIRE and DOPE must have stronger local interactions than uDFIRE so that the partially exposed helix can be stabilized in the absence of strong interaction with the rest of proteins. We further discovered that slightly more accurate refolding of mixed α-helical and β-strand terminal regions of 2hsla by aDFIRE and DOPE than by uDFIRE is due to a more accurate prediction of the helical portion of the refolded segment. However, bias toward a long helical structure has a negative impact on folding of some other segments. For example, DOPE predicts helical segments for the terminal strands of 1csp0, 1fltx, and 2ayda (1csp0 is shown in Fig. 1C). Similar behavior is observed for the terminal segments in 1csp0 and 2ayda predicted by aDFIRE. Stronger local interactions are likely the source for the misfolded single helix by DOPE and aDFIRE rather than native two helices in the two-helix terminal segment of 1o82a (Fig. 1B).
Figure 1.
The segment structures (red) refolded by updated DFIRE (uDFIRE, center left), asymmetric DFIRE (aDFIRE, center right), DOPE (right) for five proteins—(A) 1i2ta, (B) 1o82a, (C) 1csp0, (D) 2ptl0, and (E) 1vcc0—are compared with their respective native conformations (left). The fixed portion of each protein is colored in light green. For segments whose three independent runs yield quite different structures, only the lowest energy structures are shown. In the first three cases, aDFIRE and DOPE have a stronger ability to form helices correctly (1i2ta) or incorrectly (1o82a and 1csp0) than uDFIRE.
While the difference between the three energy functions (uDFIRE, aDFIRE, and DOPE) is obvious for the seven above-mentioned segments, their similarity prevails for the eight remaining segments. All make high-resolution predictions of the terminal segments of 1u84a (1α), 1r690 (2α), 1opd0 (1α), 2igd0 (1β), and 2cc6a (1β) to within 1.5 Å grmsd. They also refold the terminal strand of 2ptl0 at 3 Å grmsd. An example (2ptl0) is shown in Figure 1D. All three predict a slightly curved strand, likely due to the lack of orientation-dependent polar interactions. The successful folding of these segments suggests that shape complementary is sufficient for accurate refolding of some segments in the presence of a prefolded region. All three energy functions, however, misfold the strands in 1vcc0 and 2extb. An example (1vcc0) is shown in Figure 1E. All three energy functions yield a helical structure because there is space available for helical formation and a helix has a better interaction with the rest of protein. Thus, a strand conformation, if not confined by shape complementary, requires an orientation-dependent interstrand interaction for its stabilization. This interaction is absent in all three energy functions.
The analysis above is limited to the average grmsd values of the global minimum structures. However, in some cases, very different global minimum structures are obtained from independent runs. This is reflected from several large standard deviations in grmsd values of the three global minimum structures from three independent runs (Table 1). Interestingly, only one standard deviation of three independent runs is >1 Å for uDFIRE. The corresponding number is three for either DOPE or aDFIRE. More tests are required to be certain whether or not uDFIRE is more specific than DOPE or aDFIRE in folding.
Different structures from independent runs indicate the limitation of the global minimization technique employed in this study. This raises the question of whether misfolding some segments is caused by the failure of sampling. Figure 2 shows two segment conformations (1o82a and 1vcc0, respectively) sampled by DOPE, aDFIRE, and uDFIRE. In the first case, uDFIRE is more specific (producing single stable structure). In the second case (1vcc0), different energy functions have different abilities to sample near-native structures. uDFIRE and aDFIRE can sample near-native conformations of <2 Å grmsd while DOPE cannot, although all three make wrong predictions. Thus, near-native structures can be successfully sampled even when their energy values are not competitive.
Figure 2.
The energies of the segment conformations are sampled for proteins 1o82a (A) and 1vcc0 (B) by the genetic algorithm coupled with either DOPE (top), aDFIRE (middle), or uDFIRE (bottom). They are plotted as a function of their global rmsd values from their respective native structures. The results of three independent global minimizations are shown in three different colors.
To further examine the ability of different energy functions to sample near-native conformations, Table 2 shows the lowest global rmsd values (average over three independent runs) sampled by DFIRE, uDFIRE, aDFIRE, and DOPE. Although the success rate of folding to within 3.5 Å grmsd is only 67%, the success rate of reaching (sampling) near-native structures (within 3.5 Å grmsd) is significantly higher. uDFIRE has the highest success rate of 100%, followed by aDFIRE and DFIRE (93%) and DOPE (87%). However, the actual difference among the four is small (two segments). Thus, more studies are needed to be certain if the uDFIRE energy function is best suitable for sampling both helical- and strand-containing fragments.
Table 2.
The lowest global rmsd (grmsd) values for the structures sampled by the original DFIRE, updated DFIRE (uDFIRE) with 30 bins, asymmetric DFIRE (aDFIRE) with a simple sequence dependence, and the DOPE energy function
Discussion
In this paper, we demonstrate that terminal-segment refolding allows the revelation of the fine difference between the energy functions that are otherwise difficult to detect by commonly used decoy selections. We find that DOPE and aDFIRE, because of their dependence of relative sequence positions, favor helices over sheets (2ayda, 1fltx, and 1csp0) and longer helices over short ones (2guzb, 1i2ta, and 1o82a). This hidden bias produces more accurate structures for some segments (2guzb, 1i2ta, 2hsla) while it misfolds others (2ayda, 1csp0, 1o82a). Stronger local interactions for helical formation likely result from the simple two-state dependence on the sequence positions of two residues I and J (I > J or J < I), which overestimates the effect of the dependence on sequence positions. It overestimates because the dependence on sequence positions should be limited to neighboring residues (|I − J| within a small number), rather than for all residues. More sophisticated sequence-position dependence has been proposed (Melo et al. 2002; Zhang and Skolnick 2004; Bradley et al. 2005; Ferrada and Melo 2007). We are currently testing various approaches to account for covalent bonding (Melo et al. 2002; Cheng et al. 2007; Ferrada and Melo 2007).
The effect caused by the protein-size-dependent α (the difference between DOPE and aDFIRE) is small. This was observed previously in decoy selections (Eramian et al. 2006; Shen and Sali 2006). For example, the relative occurrence of the most accurate 20% models among the 20% best scoring models compared with that for the entire decoy set is 3.85 for DFIRE and 3.92 for DOPE (Shen and Sali 2006) in the moulder decoy set of 20 proteins. For the same decoy set, the success rate of selecting best near-native structures is 25.4% by DFIRE and 24.7% by DOPE (Eramian et al. 2006). We compare the energy parameters of DOPE and aDFIRE by calculating the correlation coefficients for the two sets of parameters for all atomic pairs at each distance bin. The correlation coefficients are all >0.9 from the bin of 0.5–1 Å to the bin of 9.5–10 Å. For example, at the distance bin of 4–4.5 Å, the correlation coefficient between the two sets of data is 0.9888 with a slope of 0.996. The difference between the two sets of parameters is even smaller for attractive interactions (negative values) and larger for repulsive interactions. Repulsive interactions typically involve fewer occurrences, and, thus, the larger difference for repulsive interactions is in part due to different statistical information from different databases for DOPE (1472 proteins) and aDFIRE (3572 proteins). Using the same database might further increase the similarity between aDFIRE and DOPE. For segment refolding, aDFIRE and DOPE yield similarly accurate structures for 10 out of 15 segments (grmsd difference <0.5 Å). aDFIRE produces a more accurate terminal β segment in 1csp0, while DOPE folds the terminal helix of 2guzb more accurately. We found that DOPE is more likely than aDFIRE to form helical structures.
It is somewhat surprising that uDFIRE makes a significant improvement over DFIRE. uDFIRE produces more accurate structures for six segments and similarly accurate structures for all other proteins (either the grmsd difference is <0.5 Å or both grmsd values are ≥6 Å, i.e., misfolded structures). The only major difference between uDFIRE and DFIRE is that the former has more bins within the same interaction range of 15 Å. We believe that the improvement is mostly contributed by a fine treatment of short-range interactions from a single 2 Å bin to four 0.5 Å bins, rather than a fine treatment of long-range interactions from seven 1 Å bins to 14 0.5 Å bins between 8 and 15 Å. Indeed, changing four 0.5 Å bins back to a single 2 Å bin makes uDFIRE fold 1o82a at the same level of accuracy (4.5 Å) as DFIRE does (5.2 Å). This highlights the importance of an accurate description of repulsive interactions for induced fitting in segment refolding. It is interesting to note that DFIRE and DOPE are able to form near perfect helices and reasonable strands in the absence of hydrogen-bonding interactions. This is mostly due to induced fitting of the unfolded segment to prefolded domains. It remains to be seen if DFIRE or any other distance-dependent only potential can make an ab initio folding for some helical proteins or even β proteins. We have implemented orientation-dependent dipolar interactions into the DFIRE energy functions. A test of refolding by the new dipolar DFIRE energy function to the same 15 terminal fragments yields a folding rate of 87% (13/15) to within 2 Å grmsd from the corresponding native conformations (Yang and Zhou 2008). The successful refolding (including the β hairpin in 2extb) highlights the importance of orientation-dependent polar interactions in the formation of secondary and tertiary structures of proteins.
The high success rate in refolding fully unfolded fragments by the all-atom statistical energy function highlights its potential for structural refinement. By comparison, it is known that molecular dynamics simulations often fail to refine homology (Baker and Sali 2001; Summa and Levitt 2007) models or partially unfolded structures (Zhu et al. 2006). One possible cause is that the entropy of packing is not included in a typical physical-based force field and it cannot be described adequately by short nanosecond dynamics simulations. This is supported by the fact that a more accurate description of the free energy of packing leads to 20% improvement in the success rate of fragment refolding from DFIRE to uDFIRE. Certainly, one cannot discount other equally plausible causes such as inadequate conformational sampling and shortcomings of the force field and/or solvent model.
Materials and Methods
The genetic algorithm and the sampling method are the same as those described elsewhere (Yang and Zhou 2008). We include them here briefly for completeness.
Genetic algorithm for global minimization of unfolded terminal segments
The selected terminal segment of a given protein is described by internal coordinates: the bond lengths, bond angles, planar torsion angles, backbone φ/ψ torsion angles, and side-chain χ angles. The bond lengths, bond angles, and planar torsion angles of the segment are fixed with standard values from the AMBER 99 force field (Weiner et al. 1986). The initial conformation of the segment is obtained by randomly assigned backbone φ/ψ torsion angles according to the observed residue-specific probability in the backbone-dependent rotamer library (Dunbrack Jr. and Karplus 1993) (http://www.fccc.edu/research/labs/dunbrack/bbdep.html; version as of May 2002). The side-chain χ angles are then randomly assigned according to rotamer probability based on the previously assigned main-chain φ/ψ angles. The initial conformations are generated 16 Å–20 Å away from the native conformation in terms of global rmsd values (see Table 1). A given conformation is locally minimized by randomly choosing a new φ/ψ angle of a selected residue from its own bin or its nearest-neighboring 24 angle bins. The side-chain χ angles are then chosen based on the new backbone angles as above. The new conformation is accepted if it has a lower energy than the current conformation and rejected if not. This procedure repeats until reaching either 100 successive rejections of new conformations or a total of 1000 attempted angle changes.
The minimized initial conformations serve as the first parent generation. The parent conformations are ranked with the normalized fitness function and the standard fitness proportionate selection procedure (Goldberg and Smith 1987). We define the fitness function of each conformation in generation l, relative to other conformations in the same generation, as
 |
where the conformation density ρi = ∑j,j ≠ i Sij, Sij is the number of residues in identical conformational states between conformations i and j, ei is the energy of conformation i (including the fixed portion of the protein) based on either the DFIRE or DOPE energy function, el –1 min and ▵el –1 are the lowest energy and the rmsd of the energies in the parent generation l − 1, respectively, with Tenv set to 1.5. Two residues are considered in an identical conformation state if |φi − φj| < 10°, |ψi − ψj| < 10°, and |φi − φj| + |ψi − ψj| < 15°. The sequentially ranked conformation pair is chosen to be the parent to breed two new conformations, first by a two-point crossover and then by mutation operations. This evolution process continues until the global minimum conformation is not changed for 100 successive generations or a total number of 400 generations is reached. A population of 120 conformations (i.e., Nc = 120) was used in this study.
Segment refolding
Table 3 lists 15 small globular proteins (<100 residues) with diverse structural topologies, both as a whole and in their terminal regions. Each refolding was performed three times, with different random initial conformations for the unfolded segments. Multiple global minimizations were used to check the robustness of the final folded structures. Because this paper focuses on evaluating the proposed energy function rather than testing sampling techniques, we limited the maximum size of unfolded regions to be <15 residues for a strand-containing segment, and <25 residues for a helix-containing segment. Each refolding event takes ∼40 h for the DFIRE energy function on a single CPU in an AMD Dual-Core Opteron Processor (2.4 GHz).
Table 3.
Fifteen unfolded terminal regions
The DOPE, aDFIRE, and uDFIRE energy functions
All DFIRE variants were extracted from a database of 3574 nonredundant (<30% homology) high-resolution proteins (resolution <2.0 Å and R % factor <0.25) from Hobohm et al. (1992).
uDFIRE is an updated DFIRE energy function by increasing the number of bins from 20 to 30 without changing the cutoff distance of 15 Å. uDFIRE uses a constant 0.5 Å for each bin and a total of 30 bins, 158 residue-specific atom types with identical atom types merged (e.g., NH1 and NH2 in Arg), while the original DFIRE used a variable bin width (2 Å for the first 2 Å, 0.5 Å from 2 Å and 8 Å, and 1 Å from 8 to 15 Å) and 167 residue-specific atom types (Zhou and Zhou 2002). Moreover, uDFIRE is a continuous potential based on a linear interpolation. No interpolation was used in DFIRE (Zhou and Zhou 2002). The same bin procedure and linear interpolation are used in DOPE.
We further introduce aDFIRE for examining the effect of the dependence on relative sequence positions adopted in DOPE. The pair interaction energy between two atoms i and j of residues I and J, uij, is different from u ji. The former is exacted from the database when the sequence position of residue I is greater than that of J, and the latter is used when the opposite is true.
The latest version of DOPE was kindly provided by the authors. It uses 30 distance bins with 0.5 Å per bin, asymmetric pairwise interaction, and linear interpolation between bins. We have integrated their energy function into our genetic algorithm for global energy minimization.
Acknowledgment
This work was supported by the NIH (R01 GM 966049 and R01 GM 068530).
Footnotes
Reprint requests to: Yaoqi Zhou, Indiana University School of Informatics, Indiana University–Purdue University Indianapolis, and Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, 719 Indiana Avenue, Walker Plaza Building Suite 319, Indianapolis, IN 46202, USA; e-mail: yqzhou@iupui.edu; fax: (317) 278-9201.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.033480.107.
References
- Atwell, S., Wells, J.A. Selection for improved subtiligases by phage display. Proc. Natl. Acad. Sci. 1999;96:9497–9502. doi: 10.1073/pnas.96.17.9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, D., Sali, A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- Bradley, P., Misura, K.M.S., Baker, D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- Brooks, B.R., Bruccoleri, R.E., Olafson, B.D., States, D.J., Swaminathan, S., Karplus, M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- Cheng, J., Pei, J., Lai, L. A free-rotating and self-avoiding chain model for deriving statistical potentials based on protein structures. Biophys. J. 2007;92:3868–3877. doi: 10.1529/biophysj.106.102152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbrack R.L., Jr, Karplus, M. Backbone-dependent rotamer library for proteins: Application to side-chain prediction. J. Mol. Biol. 1993;230:543–574. doi: 10.1006/jmbi.1993.1170. [DOI] [PubMed] [Google Scholar]
- Eramian, D., Shen, M., Devos, D., Melo, F., Sali, A., Marti-Renom, M.A. A composite score for predicting errors in protein structure models. Protein Sci. 2006;15:1653–1666. doi: 10.1110/ps.062095806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrada, E., Melo, F. Nonbonded terms extrapolated from nonlocal knowledge-based energy functions improve error detection in near-native protein structure models. Protein Sci. 2007;16:1410–1421. doi: 10.1110/ps.062735907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, D.E., Smith, R.E. Nonstationary function optimization using genetic algorithm with dominance and diploidy. In: Grefenstette J.J., editor. Proceedings of the second international conference on genetic algorithms and their application. Lawrence Erlbaum Associates, Inc; Cambridge, MA, USA: 1987. pp. 59–68. [Google Scholar]
- Hobohm, U., Scharf, M., Schneider, R., Sander, C. Selection of representative protein data sets. Protein Sci. 1992;1:409–417. doi: 10.1002/pro.5560010313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, E., Unger, R. A tale of two tails: Why are terminal residues of proteins exposed? Bioinformatics. 2007;2:E225–E230. doi: 10.1093/bioinformatics/btl318. [DOI] [PubMed] [Google Scholar]
- Jorgensen, W.L., Maxwell, D.S., Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11225–11236. [Google Scholar]
- Llinas, M., Marqusee, S. Subdomain interactions as a determinant in the folding and stability of T4 lysozyme. Protein Sci. 1998;7:96–104. doi: 10.1002/pro.5560070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo, F., Sanchez, R., Sali, A. Statistical potentials for fold assessment. Protein Sci. 2002;430:430–448. doi: 10.1002/pro.110430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder, J.W., Richards, F.M. An efficient Newton-like method for molecular mechanics energy minimization of large molecules. J. Comput. Chem. 1987;8:1016–1026. [Google Scholar]
- Scott, W.R.P., Hünenberger, P.H., Tironi, I.G., Mark, A.E., Billeter, S.R., Fennen, J., Torda, A.E., Huber, T., Krüger, P., van Gunsteren, W.F. The GROMOS biomolecular simulation program package. J. Phys. Chem. A. 1999;103:3596–3607. [Google Scholar]
- Shen, M., Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summa, C.M., Levitt, M. Near-native structure refinement using in vacuo energy minimization. Proc. Natl. Acad. Sci. 2007;104:3177–3182. doi: 10.1073/pnas.0611593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, S., Scheraga, H.A. Medium- and long-range interaction parameters between amino acids for predicting three-dimensional structures of proteins. Macromolecules. 1976;9:945–950. doi: 10.1021/ma60054a013. [DOI] [PubMed] [Google Scholar]
- Weiner, S.J., Kollman, P., Nguyen, D., Case, D. An all atom force field for simulations of proteins and nucleic acids. J. Comput. Chem. 1986;7:230–252. doi: 10.1002/jcc.540070216. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Zhou, Y. Specific interactions for ab initio folding of protein terminal regions with secondary structures. Proteins. 2008 doi: 10.1002/prot.21968. (in press) [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Skolnick, J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc. Natl. Acad. Sci. 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H., Zhou, Y. Distance-scaled, finite, ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Xie, L., Honig, B. Structural refinement of protein segments containing secondary structure elements: Local sampling, knowledge-based potentials and clustering. Proteins. 2006;65:463–479. doi: 10.1002/prot.21085. [DOI] [PubMed] [Google Scholar]