Abstract
Background
Hepatitis C virus (HCV) infection is a major global public health problem in both developed and developing countries. The prevalence and genetic diversity of HCV in pregnant women in Gabon, central Africa, is not known. We therefore evaluated the prevalence and the circulating genotypes of HCV in a large population cohort of pregnant women.
Methods
Blood samples (947) were collected from pregnant women in the five main cities of the country. The prevalence was evaluated by two ELISA tests, and the circulating genotypes were characterized by sequencing and phylogenetic analysis.
Results
Twenty pregnant women (2.1%) were infected with HCV. The seroprevalence differed significantly by region (p = 0.004) and increased significantly with age (p = 0.05), being 1.3% at 14–20 years, 1.1% at 21–25 years, 1.9% at 26–30 years, 4.1% at 31–35 years and 6.0% at > 35 years. Sequencing in the 5'-UTR and NS5B regions showed that the circulating strains belonged to genotypes 4 (4e and 4c).
Conclusion
We found that the HCV seroprevalence in pregnant women in Gabon is almost as high as that in other African countries and increases with age. Furthermore, only genotype 4 (4e and 4c) was found. More extensive studies aiming to evaluate the prevalence and heterogeneity of HCV genotypes circulating in the general population of the country are needed.
Background
Hepatitis C virus (HCV) infection is a major global public health problem in both developed and developing countries. More than 170 million people are chronically infected with HCV worldwide, and the infection results in the development of chronic liver diseases, including liver cirrhosis and hepatocellular carcinoma. HCV has been clustered into six distinct genotypes and numerous subtypes on the basis of nucleic acid sequence [1]. Genotyping of HCV strains is an important tool for epidemiological and clinical purposes; in particular, examination of sequence diversity can aid understanding of different patterns of serological reactivity, virulence and response to treatment [2,3].
Genotypes 1, 2 and 3 are distributed widely throughout western countries, whereas types 5 and 6 are largely confined to South Africa and southeast Asia, respectively, and type 4 was first found predominantly in the Middle East [4] and then in central Africa [5-7]. Data on HCV prevalence and molecular epidemiology are available for only a few African countries. In Gabon, central Africa, the seroprevalence was previously reported to be up to 6.5% in the general population of the eastern part of the country [8], and genotype 4 was found by screening a few sequences [7]. These data are based on small groups of individuals and third-generation enzyme-linked immunosorbent assays (ELISAs), molecular (reverse transcriptase polymerase chain reaction (RT-PCR)) assays having rarely been used. Furthermore, no data are available on the presence of circulating HCV or the genotypes in pregnant women in Gabon. A serological and molecular study was therefore undertaken on a large population cohort of pregnant women in the five main cities of the country.
Methods
Area and study population
Gabon occupies 270 000 km2 in the Gulf of Guinea near the equator, with tropical forest covering three-quarters of the territory. The population was estimated in 2003 to be 1 273 000 persons, belonging to more than 40 ethnic groups.
To determine the prevalence and the circulating genotypes of HCV in pregnant women in Gabon, five sentinel sites were selected: Libreville, the capital, in the north-west; Port-Gentil, the port and economic capital in the west; Lambaréné, in the centre of the country; Franceville in the south-east; and Oyem in the north-east (Figure 1). Between January and March 2005, blood samples were collected from all pregnant women at their first antenatal examination, after full informed consent had been obtained. The samples were anonymous, but age and geographic origin were retained. The study obtained ethical clearance from the public health authorities (Ministry of Public Health and the "Programme National de Lutte contre le Sida" in Gabon).
Figure 1.
Map of Gabon in central Africa with the selected sentinel sites at which blood was collected from pregnant women.
Serological tests
All samples were screened for anti-HCV antibodies with a commercial third-generation ELISA (Monolisa, anti-HCV Plus version 2, Bio-Rad, Marne-La-Coquette, France). The results of the assay were expressed quantitatively as the ratio of the optical density of the test sample to the calculated cut-off absorbance, as recommended by the manufacturer. Sera with ratios > 1.1 were considered to be positive, sera with R values of 0.9–1.1 were recorded as indeterminate, whereas those with R values < 0.9 were considered negative. Positive and indeterminate sera were re-tested with another third-generation ELISA assay, Innotest HCV Ab IV (Innogenetics NV HCV, Gand, Belgium). Samples were considered HCV positive when they were positive in the two ELISA tests.
HCV amplification and molecular and phylogenetic analysis
Viral RNA was extracted from 140 μl of plasma with a Qiamp® viral mini kit according to the manufacturer's protocol (Qiagen, Courtaboeuf, France). The extracted RNA was used as a template and amplified by nested RT-PCR with primers specific to the 5'UTR (5' non-coding region), as described previously [9,10]. Reverse transcription and first amplifications were performed with a Titan One Tube RT-PCR kit (Roche Diagnostics, Mannheim, Germany), according to the manufacturer's protocol. A portion of the NS5B region was also amplified with degenerated primers, as described previously [11], and then sequenced according to standard procedures. The HCV sequences and phylogenetic results were studied as reported previously [11]. The GenBank accession numbers of the new NS5b sequences are EU0334530 to EU0334534, and those of the 5'UTR sequences are EU0334535 to EU0334545.
Statistical analysis
Statistical analysis was performed by the chi-squared test with Yates' correction, and the prevalences and odds ratios were calculated. The corresponding 95% confidence intervals were reported as measures of statistical significance. Analyses were performed with Epi-Info (version 6.04dfr, ENSP-Epiconcept-InUS, 2001).
Results
HCV prevalence in pregnant women
Between January and March 2005, sera from all 947 pregnant women recruited at the five sentinel sites were screened for HCV antibodies with the two ELISA tests; 20 pregnant women (2.1%) were found to be seropositives for HCV (Table 1). The seroprevalence differed significantly by region (p = 0.004), with the highest prevalence (4.9%) in Franceville (Haut Ogooué region in the south-east) and no cases in Port Gentil. There was a significant increase in HCV seroprevalence with age (p = 0.05), being 1.3% at 14–20 years, 1.1% at 21–25 years, 1.9% at 26–30 years, 4.1% at 31–35 years and 6.0% at > 35 years (Table 1).
Table 1.
Prevalence of antibodies to hepatitis C virus (HCV) in pregnant women in Gabon (central Africa) by geographical area and age group
| Variable | HCV antibodies | |||
| No. positive/no. tested | % | Odds ratio | 95% CI | |
| Sentinel site | ||||
| Port Gentil | 0/168 | 0 | ||
| Lambaréné | 3/331 | 0.9 | 0.33 | 0.1–1.13 |
| Libreville | 5/199 | 2.5 | 1.29 | 0.46–3.59 |
| Oyem | 3/68 | 4.4 | 2.39 | 0.68–8.37 |
| Franceville | 9/181 | 4.9 | 3.64 | 1.49–8.92 |
| Age range (years) | ||||
| 14–20 | 4/293 | 1.3 | 0.57 | 0.19–1.72 |
| 21–25 | 3/266 | 1.1 | 0.46 | 0.13–1.58 |
| 26–30 | 4/202 | 1.9 | 0.94 | 0.31–2.84 |
| 31–35 | 5/120 | 4.1 | 2.4 | 0.86–6.73 |
| > 35 | 4/66 | 6.0 | 3.5 | 1.15–10.94 |
| All | 20/947 | 2.1 | ||
HCV RNA detection and molecular analysis of circulating genotypes
HCV RNA targeting 5'-UTR was found in 13 of the 20 positive samples, showing that 1.3% of the pregnant women were viraemic. Sequencing in the 5'-UTR region was successful for 11 RNA-positive isolates (Table 2). BLAST searches of the 5'-UTR sequences classified the 11 strains as genotype 4. Of these, nine belonged to genotype 4e, with 99–100% homology with the reference sequences GAB809 (L29625) and CAM73 (L29591) from Gabon and Cameroon, respectively [12]. The other two sequences belonged to genotype 4c, with 98% homology with the reference sequences Z6 (M84862) and GB358 (L29608) from Zaire and Gabon, respectively [13] (Table 2).
Table 2.
Epidemiological status and results of molecular screening and 5'UTR sequence analysis for the 20 serologically HCV-positive pregnant women in Gabon, central Africa.
| Sample | Age (years) | Region | PCR 5'UTR | BLAST % homology with accession number | Subtype | Accession number | |
| Gab104 | 32 | Lambaréné | Positive | 98 | L29608 | 4c | EU334535 |
| Gab121 | 18 | Lambaréné | Negative | - | - | - | |
| Gab977 | 26 | Lambaréné | Positive | 99 | L29625 | 4e | EU334536 |
| Gab652 | 16 | Libreville | Positive | 100 | L29625 | 4e | EU334537 |
| Gab704 | 28 | Libreville | Positive | 99 | L29625 | 4e | EU334538 |
| Gab705 | 21 | Libreville | Negative | - | - | - | |
| Gab757 | 21 | Libreville | Negative | - | - | - | |
| Gab770 | 33 | Libreville | Positive | 100 | L29591 | 4e | EU334539 |
| Gab1089 | 23 | Franceville | Positive | 100 | L29625 | 4e | EU334540 |
| Gab1115 | 40 | Franceville | Negative | - | - | - | |
| Gab1139 | 26 | Franceville | Positive | 99 | L29625 | 4e | EU334541 |
| Gab1148 | 19 | Franceville | Positive | 98 | M84862 | 4c | EU334542 |
| Gab1174 | 29 | Franceville | Positive | 100 | L29591 | 4e | EU334543 |
| Gab1192 | 30 | Franceville | Negative | - | - | - | |
| Gab1213 | 34 | Franceville | Negative | - | - | - | |
| Gab1228 | 15 | Franceville | Positive | 99 | L29625 | 4e | EU334544 |
| Gab1229 | 31 | Franceville | Positive | NA | NA | NA | |
| Gab1664 | 25 | Oyem | Negative | - | - | - | |
| Gab1669 | 19 | Oyem | Positive | NA | NA | NA | |
| Gab1670 | 19 | Oyem | Positive | 99 | L29625 | 4e | EU334545 |
NA, not available; only partial sequence of the amplicon could be obtained.
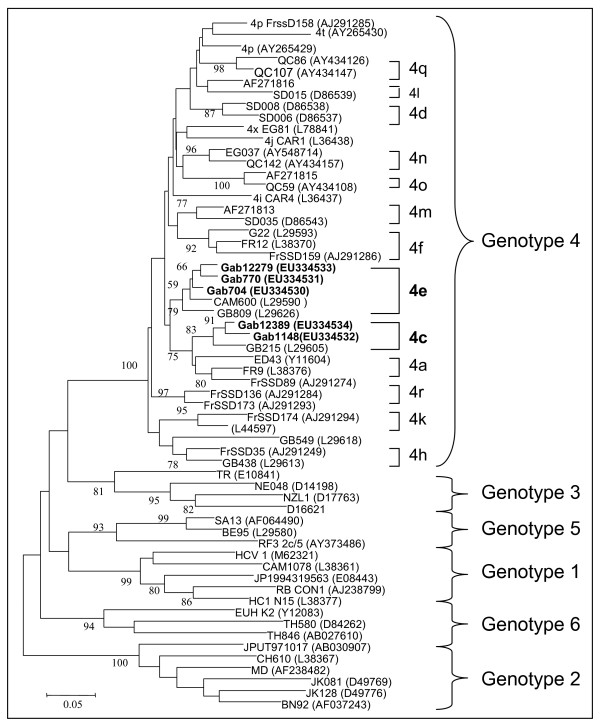
To confirm the circulating genotypes obtained with 5'-UTR sequences and for phylogenetic analysis, the NS5B regions of three strains were amplified and sequenced. We also amplified and sequenced two new positive samples obtained from routinely screened pregnant women in the region with the highest HCV prevalence (Franceville). These two samples were also included in the phylogenetic analysis. Sequence analysis of the NS5B region confirmed that the strains from pregnant women were closely related to genotypes 4e and 4c, with 94.5% and 93.7% homology with other strains from Gabon, as described previously [12]. Phylogenetic analysis of the NS5B region confirmed the sequence homology analysis, showing that the strains from pregnant women in Gabon belonged to genotypes 4c and 4e (Figure 2). Within genotype 4e, our strains clustered with strains from Cameroon and with other strains from Gabon described previously [12]. In genotype 4c, the strains from Gabon were closely related to other strains identified previously by Stuyver et al. (1994).
Figure 2.
Phylogenetic analysis of HCV NS5B sequences. A neighbour-joining phylogenetic tree was constructed from Gabonese isolates from pregnant women, and published reference sequences for the various HCV genotypes were obtained from GenBank. The Kimura two-parameter method of estimating genetic distance was used. Numbers next to the nodes of the tree represent bootstrap values (1000 replicates). In genotype 4, sequences found in Gabon are indicated in bold. The GenBank accession numbers of the new NS5b sequences of HCV from Gabon are EU0334530 to EU0334534.
Discussion
We found an overall seroprevalence of HCV in pregnant women in Gabon of 2.1%, which is similar to that in other African countries, such as Cameroon (1.9%) [14] and Burkina Faso (2%) [15]. The prevalence in our study was lower than that described previously in the general population of eastern Gabon (6.5%) [8]. This difference could be related to age, as we observed an increasing prevalence with age and low prevalences in the youngest age groups (Table 1). Similar observations have been made in previous studies in several African countries, including Cameroon and the Democratic Republic of the Congo, in both pregnant women and the general population [14,16]. Although studies to evaluate the routes of HCV transmission in Africa are needed [17], ritual procedures, such as scarification and tattooing, which are common tribal customs in various areas of Gabon, may facilitate transmission [18]. Furthermore, in rural health centres in Gabon, drugs such as antibiotics are commonly given by intramuscular injection, and the sterilization procedures are often inadequate, increasing the risk for HCV transmission. People are therefore exposed to new infection throughout their lives. This might explain in part the increasing prevalence of HCV with age.
We also showed that HCV seroprevalence differed significantly by region. Many factors can influence the transmission of HCV in Africa, including environmental, ethnic, ritual (see above), economic and genetic factors. In Egypt, it was reported that transmission of HCV was related to extensive antischistosomiasis injection campaigns [19]. It is not known whether previous vaccination campaigns in Gabon had the same effects in various areas of the country. Further epidemiological, molecular and genetic studies are required to determine the factors responsible for the high prevalence of HCV in some regions of Gabon.
Of the 20 seropositive women, seven were ELISA-positive but RNA-negative. These seven samples also had a low ratio in the two ELISA tests. It has been reported previously that during the natural history of HCV infection, 20–30% of infected individuals eliminate the virus spontaneously [20,21]. Results similar to ours were obtained in studies conducted in Cameroon and Japan [14,22]. The presence of HCV antibodies in the pregnant women in our study might have been due to a previous infection.
We also showed that the HCV circulating in pregnant women in Gabon belongs to genotype 4 (4e and 4c). In a previous study, Xu et al. [7] published three sequences of HCV selected from 1172 persons in the general population of Gabon. Sequence comparisons showed that these three sequences were closely related to genotype 4, and the authors suggested that they might be related to genotype 4c or to 4d as described by Buck et al. [13]. In our study, the sequences of 11 samples were also found to be closely related to genotypes 4c and 4e. These two studies thus confirm that genotype 4 is the only genotype circulating in Gabon.
In our study, we also found that 4e predominates in five widely separated geographical regions of the country. Sub-Saharan Africa is known to have a wide diversity of genotypes 1, 2, and 4 and a remarkable diversity of subtypes [6,11,23]; genotypes 4 and 5 predominate in several African countries [24-26], and genotype 4f has been described in Cameroon. We found only genotypes 4e and 4c in pregnant women in Gabon. Indeed, the plasma samples used in the present study, obtained after screening for HIV, HTLV, HBV/HDV and HEV, were insufficient to perform more sequence analyses. However, the two strains obtained from further routinely screened pregnant women in the region of the highest HCV prevalence (Franceville) confirm, however, that the circulating strains in this population belong to genotype 4 (4c and 4e). A larger study in the general population is needed to better characterize the circulating genotypes.
Conclusion
Our study represents the first serological and molecular epidemiological investigation on HCV in a homogeneous population in Gabon. We showed that the HCV seroprevalence in pregnant women in Gabon is almost as high as that in other African countries and increases with age. Thus, preventive measures to decrease the spread and transmission of HCV in Gabon are warranted. These measures should include systematic HCV screening of pregnant women, in order to counsel them about the risk for HCV transmission and the development of associated diseases.
Only genotype 4 (4e and 4c) was found in these pregnant women. More extensive studies aiming to evaluate the heterogeneity of HCV genotypes in samples collected from the general population of the country are needed.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
G–RN-A, MM and MB carried out the serological and molecular studies. RN and DR participated in sequence alignments and phylogenetic analysis. AM collected the sera and compiled the epidemiological data. FB–V and MK participated in the design of the study, the statistical analysis and the drafting of the manuscript. All authors read and approved the final version of the manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
Guy-Roger Ndong-Atome is the recipient of a fellowship from the European Community. We thank Marie-Thérèse Bedjabaga and Patricia Keba (Programme National de Lutte contre le Sida, Libreville, Gabon) for technical help. The Centre International de Recherches Médicales de Franceville (CIRMF), Franceville, Gabon, is funded by the Gabonese Government, Total-Gabon and the French Foreign Ministry. This work was supported by funds from the Service de Coopération et d'Action Culturelle, French Embassy, Libreville, Gabon.
Contributor Information
Guy-Roger Ndong-Atome, Email: roger_ndong@yahoo.com.
Maria Makuwa, Email: mmakuwa@yahoo.com.
Richard Njouom, Email: njouom@pasteur-yaounde.org.
Michel Branger, Email: michel.branger@bch.ap-hop-paris.fr.
Francoise Brun-Vézinet, Email: francoise.brun-vezinet@bch.ap-hop-paris.fr.
Antoine Mahé, Email: maheant@yahoo.fr.
Dominique Rousset, Email: rousset@pasteur-yaounde.org.
Mirdad Kazanji, Email: m.kazanji@cirmf.org.
References
- Simmonds P, Holmes EC, Cha TA, Chan SW, McOmish F, Irvine B, Beall E, Yap PL, Kolberg J, Urdea MS. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74 ( Pt 11):2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- Fried MW. Viral factors affecting the outcome of therapy for chronic hepatitis C. Rev Gastroenterol Disord. 2004;4 Suppl 1:S8–S13. [PubMed] [Google Scholar]
- Halfon P, Neumann AU, Bourliere M, Rieu A, Chadapaud S, Khiri H, Ouzan D, Cacoub P. Slow viral dynamics of hepatitis C virus genotype 4. J Viral Hepat. 2003;10:351–353. doi: 10.1046/j.1365-2893.2003.00433.x. [DOI] [PubMed] [Google Scholar]
- Ray SC, Arthur RR, Carella A, Bukh J, Thomas DL. Genetic epidemiology of hepatitis C virus throughout egypt. J Infect Dis. 2000;182:698–707. doi: 10.1086/315786. [DOI] [PubMed] [Google Scholar]
- Fretz C, Jeannel D, Stuyver L, Herve V, Lunel F, Boudifa A, Mathiot C, de The G, Fournel JJ. HCV infection in a rural population of the Central African Republic (CAR): evidence for three additional subtypes of genotype 4. J Med Virol. 1995;47:435–437. doi: 10.1002/jmv.1890470423. [DOI] [PubMed] [Google Scholar]
- Njouom R, Pasquier C, Ayouba A, Gessain A, Froment A, Mfoupouendoun J, Pouillot R, Dubois M, Sandres-Saune K, Thonnon J, Izopet J, Nerrienet E. High rate of hepatitis C virus infection and predominance of genotype 4 among elderly inhabitants of a remote village of the rain forest of South Cameroon. J Med Virol. 2003;71:219–225. doi: 10.1002/jmv.10473. [DOI] [PubMed] [Google Scholar]
- Xu LZ, Larzul D, Delaporte E, Brechot C, Kremsdorf D. Hepatitis C virus genotype 4 is highly prevalent in central Africa (Gabon) J Gen Virol. 1994;75:2393–2398. doi: 10.1099/0022-1317-75-9-2393. [DOI] [PubMed] [Google Scholar]
- Delaporte E, Thiers V, Dazza MC, Romeo R, Mlika-Cabanne N, Aptel I, Schrijvers D, Brechot C, Larouze B. High level of hepatitis C endemicity in Gabon, equatorial Africa. Trans R Soc Trop Med Hyg. 1993;87:636–637. doi: 10.1016/0035-9203(93)90269-V. [DOI] [PubMed] [Google Scholar]
- Bukh J, Purcell RH, Miller RH. Importance of primer selection for the detection of hepatitis C virus RNA with the polymerase chain reaction assay. Proc Natl Acad Sci U S A. 1992;89:187–191. doi: 10.1073/pnas.89.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno O, Mizokami M, Wu RR, Saleh MG, Ohba K, Orito E, Mukaide M, Williams R, Lau JY. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J Clin Microbiol. 1997;35:201–207. doi: 10.1128/jcm.35.1.201-207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier C, Njouom R, Ayouba A, Dubois M, Sartre MT, Vessiere A, Timba I, Thonnon J, Izopet J, Nerrienet E. Distribution and heterogeneity of hepatitis C genotypes in hepatitis patients in Cameroon. J Med Virol. 2005;77:390–398. doi: 10.1002/jmv.20468. [DOI] [PubMed] [Google Scholar]
- Stuyver L, Rossau R, Wyseur A, Duhamel M, Vanderborght B, Van Heuverswyn H, Maertens G. Typing of hepatitis C virus isolates and characterization of new subtypes using a line probe assay. J Gen Virol. 1993 ;74:1093–1102. doi: 10.1099/0022-1317-74-6-1093. [DOI] [PubMed] [Google Scholar]
- Bukh J, Purcell RH, Miller RH. Sequence analysis of the 5' noncoding region of hepatitis C virus. Proc Natl Acad Sci U S A. 1992;89:4942–4946. doi: 10.1073/pnas.89.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njouom R, Pasquier C, Ayouba A, Sandres-Saune K, Mfoupouendoun J, Mony Lobe M, Tene G, Thonnon J, Izopet J, Nerrienet E. Hepatitis C virus infection among pregnant women in Yaounde, Cameroon: prevalence, viremia, and genotypes. J Med Virol. 2003;69:384–390. doi: 10.1002/jmv.10300. [DOI] [PubMed] [Google Scholar]
- Serme AK, Ilboudo PD, Samandoulgou A, Simpore J, Bougouma A, Sombie AR. Prevalence of Hepatitis C virus infection in pregnant women and mother-child transmission in Ouagadougou, Burkina Faso. Bull Soc Pathol Exot. 2006;99:108–109. [PubMed] [Google Scholar]
- Laurent C, Henzel D, Mulanga-Kabeya C, Maertens G, Larouzé B, Delaporte E. Seroepidemiological survey of hepatitis C virus among commercial sex workers and pregnant women in Kinshasa, Democratic Republic of Congo. Int J Epidemiol. 2001;30:872–877. doi: 10.1093/ije/30.4.872. [DOI] [PubMed] [Google Scholar]
- Madhava V, Burgess C, Drucker E. Epidemiology of chronic hepatitis C virus infection in sub-Saharan Africa. Lancet Infect Dis. 2002;2:293–302. doi: 10.1016/S1473-3099(02)00264-5. [DOI] [PubMed] [Google Scholar]
- Abildgaard N, Peterslund NA. Hepatitis C virus transmitted by tattooing needle. Lancet. 1991;17:460. doi: 10.1016/0140-6736(91)91095-C. [DOI] [PubMed] [Google Scholar]
- Pybus OG, Drummond AJ, Nakano T, Robertson BH, Rambaut A. The epidemiology and iatrogenic transmission of hepatitis C virus in Egypt: a Bayesian coalescent approach. Mol Biol Evol. 2003;20:381–387. doi: 10.1093/molbev/msg043. [DOI] [PubMed] [Google Scholar]
- Uto H, Hayashi K, Kusumoto K, Hasuike S, Nagata K, Kodama M, Ido A, Kohara M, Stuver SO, Tsubouchi H. Spontaneous elimination of hepatitis C virus RNA in individuals with persistent infection in a hyperendemic area of Japan. Hepatol Res. 2006;34:28–34. doi: 10.1016/j.hepres.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Kondili LA, Chionne P, Costantino A, Villano U, Lo Noce C, Pannozzo F, Mele A, Giampaoli S, Rapicetta M. Infection rate and spontaneous seroreversion of anti-hepatitis C virus during the natural course of hepatitis C virus infection in the general population. Gut. 2002;50:693–696. doi: 10.1136/gut.50.5.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto E, Noguchi S, Taniai M, Hayashi N. Characteristics of patients sero-positive for hepatitis C virus (HCV) antibodies but negative for HCV-RNA by polymerase chain reaction. J Hepatol. 1998;29:856–857. doi: 10.1016/S0168-8278(98)80271-7. [DOI] [PubMed] [Google Scholar]
- Ndjomou J, Kupfer B, Kochan B, Zekeng L, Kaptue L, Matz B. Hepatitis C virus infection and genotypes among human immunodeficiency virus high-risk groups in Cameroon. J Med Virol. 2002;66:179–186. doi: 10.1002/jmv.2128. [DOI] [PubMed] [Google Scholar]
- Matera G, Lamberti A, Quirino A, Foca D, Giancotti A, Barreca GS, Guadagnino V, Liberto MC. Changes in the prevalence of hepatitis C virus (HCV) genotype 4 in Calabria, Southern Italy. Diagn Microbiol Infect Dis. 2002;42:169–173. doi: 10.1016/S0732-8893(01)00350-9. [DOI] [PubMed] [Google Scholar]
- Nicot F, Legrand-Abravanel F, Sandres-Saune K, Boulestin A, Dubois M, Alric L, Vinel JP, Pasquier C, Izopet J. Heterogeneity of hepatitis C virus genotype 4 strains circulating in south-western France. J Gen Virol. 2005;86:107–114. doi: 10.1099/vir.0.80409-0. [DOI] [PubMed] [Google Scholar]
- van Asten L, Verhaest I, Lamzira S, Hernandez-Aguado I, Zangerle R, Boufassa F, Rezza G, Broers B, Robertson JR, Brettle RP, McMenamin J, Prins M, Cochrane A, Simmonds P, Coutinho RA, Bruisten S. Spread of hepatitis C virus among European injection drug users infected with HIV: a phylogenetic analysis. J Infect Dis. 2004;189:292–302. doi: 10.1086/380821. [DOI] [PubMed] [Google Scholar]