Abstract
Background
Variants in numerous genes are thought to affect the success or failure of cancer chemotherapy. Interindividual variability can result from genes involved in drug metabolism and transport, drug targets (receptors, enzymes, etc), and proteins relevant to cell survival (e.g., cell cycle, DNA repair, and apoptosis). The purpose of the current study is to establish a flexible, cost-effective, high-throughput genotyping platform for candidate genes involved in chemoresistance and -sensitivity, and treatment outcomes.
Methods
We have adopted SNPlex for genotyping 432 single nucleotide polymorphisms (SNPs) in 160 candidate genes implicated in response to anticancer chemotherapy.
Results
The genotyping panels were applied to 39 patients with chronic lymphocytic leukemia undergoing flavopiridol chemotherapy, and 90 patients with colorectal cancer. 408 SNPs (94%) produced successful genotyping results. Additional genotyping methods were established for polymorphisms undetectable by SNPlex, including multiplexed SNaPshot for CYP2D6 SNPs, and PCR amplification with fluorescently labeled primers for the UGT1A1 promoter (TA)nTAA repeat polymorphism.
Conclusion
This genotyping panel is useful for supporting clinical anticancer drug trials to identify polymorphisms that contribute to interindividual variability in drug response. Availability of population genetic data across multiple studies has the potential to yield genetic biomarkers for optimizing anticancer therapy.
Background
Pharmacogenetic studies have shown that polymorphisms in genes related to drug metabolism, transport, and drug targets contribute to interindividual variability in drug efficacy and adverse effects. Hence, pharmacogenetic biomarkers have the potential of optimizing chemotherapy for individual patients [1,2]. This is exemplified with genotyping of thiopurine S-methyltransferase (TPMT), which inactivates thioguanine, to avoid serious toxicity in childhood leukemias [1,3]. Homozygous carriers of defective TPMT alleles experience drastically slowed thioguanine inactivation and are at high risk unless the thioguanine dose is reduced more than tenfold. Similarly, deficiency of dihydropyrimidine dehydrogenase (DPYD) activity predisposes to 5-fluorouracil toxicity [4]. Many more examples begin to emerge, with a number of genomic biomarkers listed on the US FDA website for use in guiding drug efficacy and/or safety [5]. In addition, the US FDA has issued "Guidance for Industry Pharmacogenomic Data Submissions" in 2005 for drugs in clinical trials [6]. The purpose is to identify potential biomarkers of interindividual variability in drug response for personalized drug treatment achieving maximum benefit and minimum toxicity. However, the relationship between genotype and phenotype (drug levels, but more importantly, therapy outcome) is confounded by numerous factors, such as age, sex, body weight, nutrition, organ function and comedications, involvement of multiple genes, and population admixture [7]. In this study we have established genotyping panels of relevant candidate genes that could interfere with response to chemotherapy and clinical outcomes; the genotyping panels are flexibly designed so that new candidate genes can be added as needed.
To exploit genetic information in cancer treatment, we must adopt a comprehensive approach, assessing which genes play critical roles in the response to any given drug. For example, irinotecan has become standard in the treatment of intestinal carcinomas. The following genes/proteins could play a role in the response of individual patients: carboxyesterases that activate irinotecan to SN38, CYP3A4 which inactivates irinotecan, UDP-glucuronosyltransferase 1A1 (UGT1A1) which inactivates SN38, and several transporters involved in shuttling irinotecan and SN38 in and out of cells. Among these, (TA)nTAA repeats in the promoter region of UGT1A1 appear to have a significant impact on irinotecan response and toxicity [8]. This information has been added to the package insert of irinotecan as a warning, and the US FDA has approved a prospective genetic biomarker assay to support individualized dosing [9]. However, given the complexity of the metabolic pathway, the UGT1A1 polymorphisms account for only a portion of observed phenotypic variability (e.g., toxicity) [9]. A more comprehensive view of polymorphisms in multiple genes may improve the predictive accuracy of genotype information – even a relatively small increase in predictive power could translate into clinical benefits. In this study we have developed large-scale genotyping methods to provide information on genetic variants of candidate genes involved in drug metabolism and transport.
Drug response is further affected by genes involved in apoptosis, DNA repair, redox cycling, and cell cycle progression. These factors appear to function as main determinants of drug resistance, the principal problem for successful cancer chemotherapy. For example, the DNA-repair enzyme O6-methylguanine-DNA methyltransferase (MGMT) is implicated in resistance to alkylating agents [10]. We adopt here a candidate gene approach to determine genetic factors in cancer biology that are likely relevant to an individual's response to chemotherapy. On the other hand, genome-wide SNP analyses are now available using very large-scale array genotyping methods, a trend that might eventually replace candidate gene panels. However, our knowledge of genetic variants in even the most intensely studied candidate genes remains fragmentary, and we expect that long-term, genotyping panels containing only a few strong biomarker genes with complete information on genetic variants will prove valuable clinically.
A second critical factor is the selection of polymorphisms for genotyping within the candidate genes. This involves known functional polymorphisms, polymorphisms of relative frequencies (> 5%) that are likely to affect function (gene regulation, mRNA processing and splicing, translation, and protein functions), and haplotype-tag SNPs providing maximum information on haplotype structures. Numerous Web tools are available to optimize the SNP selection. We summarize here details of the genotyping panels specifically developed for cancer chemotherapy. Similar panels have been proposed elsewhere [11] but the present study extend these panels with further candidate genes to maximize its utility.
Because the polymorphisms/variants differ at the molecular level (SNPs, insertions/deletions, repeats, translocations, LOH, and gene/chromosomal duplications), no single method can readily detect all genotypes. Rather, we first select a versatile method capable of covering a majority of polymorphisms at low cost. The remainder must be completed with a set of varying technologies, at a smaller scale. The aim of this project is to establish a platform for genotyping single nucleotide polymorphisms (SNPs, representing a majority of genetic variants) of genes involved in drug metabolism, transport, and targets, and DNA repair, cell signaling, cell cycle, apoptosis [11-14]. Various high-throughput genotyping platforms are available, each with advantages and disadvantages [15-17]. For example, Affymetrix SNP array is a practical platform for genome-wide genotyping. However, the SNP set is not readily adaptable to include a few newly emerging candidate genes, and the cost for genotyping is relatively high if one wishes to focus on select candidate genes.
In our study, several hundred SNPs need to be genotyped in various numbers of samples. In addition, the SNP set needs to be flexible for different research designs. To establish a flexible, cost-effective, high-throughput genotyping method, we adapted SNPlex genotyping established and systemically validated by Applied Biosystems to have high precision [18,19]. The method can detect 48 SNPs in one single well for each patient sample, adapted here to a 96-well plate format covering more than 400 SNPs for cancer chemotherapy. SNPlex is designed to detect single nucleotide polymorphisms, but not other genetic changes including insertions/deletions and variable number tandem repeat (VNTR) polymorphisms. Additional genotyping strategies, such as multiplexed SNaPshot for CYP2D6 and PCR using fluorescently labeled primers to detect the UGT1A1 promoter dinucleotide repeat polymorphism, serve as examples of complementary methods. The goal is to generate a common set of genotyping data for cancer treatment trials, thereby, growing the patient and control cohorts for retrospective and prospective analyses. The panel described here can be expanded while new functional polymorphisms are being discovered, and it is suitable for relatively small trials to large cohorts, economically covering up to 1,000 SNPs.
To illustrate potential applications, we show here genotyping results obtained with our SNP panels related to genes involved in cancer biology. For this, we have genotyped a cohort of colorectal cancer patients. In addition, we have applied the drug metabolism and transport gene panels to a Phase I leukemia trial, of which detailed results will be reported elsewhere.
Methods
Selection of genes and polymorphisms
The objective was to include genes likely to be involved in therapy outcome. For many of these main candidate genes, genetic studies have already suggested or confirmed functional polymorphisms, but we also include other potential candidate genes/polymorphisms. The main focus of the current study was to include known functional polymorphisms candidate genes based on available literature. The genotyping panels have not been geared primarily to cover all main haplotypes for each gene, but rather to focus on functional SNPs as much as they are known. The purpose therefore is not primarily the discovery of new functional polymorphisms, but rather the assessment of the clinical impact of known ones. We anticipate that in the future we will be able to focus the genotyping panels even more on known functional SNPs, in an effort to develop clinically relevant biomarker panels. The approach takes into consideration that new candidate genes and polymorphisms continue to emerge [20] that need to be flexibly included in the genotyping panels.
We chose candidate SNPs that for the most part have been implicated in cancer biology and chemotherapy in more than one study. For the selected genes, we first surveyed recent reviews for known polymorphisms reported to be related to cancer risk or drug metabolism [12-14], and a lung cancer risk study targeting 250 SNPs in 101 genes [11]. We further searched PubMed for additional polymorphisms associated with cancer risk, revealing genes that are also likely to affect treatment outcome [21]. Lastly, we searched the NCBI dbSNP database for SNPs in the transcribed regions with > 5% minor allele frequency to capture the main haplotypes in genes where only 1–2 SNPs had been selected by the other methods. SNPs from dbSNP were frequent and fully validated by different research projects, such as HapMap project [22] and the NCI SNP500 Cancer project [23]. SNPs in high LD (D'>0.7) with another SNP already in the panel were generally excluded, although in some case we added such SNPs for the assays design, to assure that either one was represented in the panel design. For cytochrome P450 genes, we included the known functional polymorphisms from human allele nomenclature database for cytochrome P450 enzymes [24]. We also searched the UDP-glucuronosyltransferase (UGT) alleles nomenclature database [25] and NAT nomenclature database [26]. We also consulted various drug transporter databases, including the human membrane transporter database [27] and PharmGKB [28].
SNPlex probe pools and reagents
For the selected SNPs [see Additional file 1], either NCBI SNP reference cluster IDs (rs numbers) or SNP sequences were submitted to Applied Biosystems for the design of SNPlex panels following their proprietary selection algorithms. We separated the genes into different groups: drug metabolism and transport, DNA repair/apoptosis and cell cycle/cell growth/drug targets. DNA sequence surrounding a specific polymorphism must meet specific requirements for probe design, including but not limited to: A. genomic screening; the DNA sequence flanking the target SNP must be unique and not have more than 1 genomic alignment hit with 21 or more contiguous bases to ensure annealing specificity, and there is no second SNP nearby. B. The target sequences should have appropriate features for annealing efficiency. C. Pooling rules: stringent pooling rules are used to determine optimal multiplex composition. SNPlex panels and reagents were synthesized by Applied Biosystems.
DNA samples
Thirty nine blood DNA samples from chronic lymphocytic leukemia patients were collected by Dr. John Byrd following the institutional review board (IRB) protocol at the Ohio State University for a flavopiridol phase I clinical trials at The Ohio State University Comprehensive Cancer Center. In addition, 90 colorectal cancer samples were chosen from a series of 1262 consecutively accrued patients with colorectal carcinoma diagnosed in the main hospitals of Metropolitan Columbus, whose tumors did not show microsatellite instability, as described previously [29]. Control groups were obtained from previously genotyped cohorts where the same SNPs are accessible (HapMap and other datasets as indicated). The research protocol and consent form were approved by the institutional review board at each participating hospital, and all patients provided written informed consent.
SNPlex genotyping
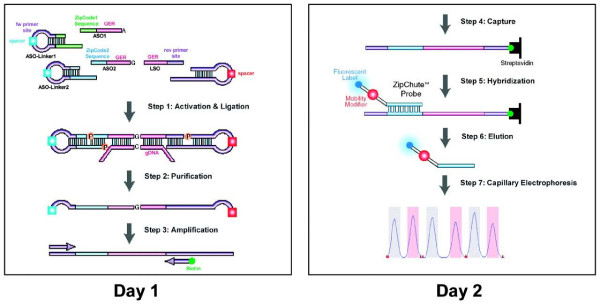
SNPlex genotyping was carried out according to the manufacturer's suggested protocol with slight modifications to accommodate a manual procedure using 96-well plate (90 testing DNAs plus positive and no DNA template controls, and 4 wells for allelic ladders). The multi-step procedure has been previously described (Figure 1) [18,19]. Step 1, for DNA fragmentation, 40 nanogram genomic DNA (2 μl) was fragmented at 99°C for 10 min. Step 2 involved phosphorylation and ligation of allele-specific oligonucleotide (ASO, 2 for each SNP) ligation probes, locus-specific oligonucleotide (LSO, one for each SNP) ligation probes and linkers. SNPlex ASO and LSO ligation probes, universal linkers, dATP and oligonucleotide ligation assay (OLA) master mix containing DNA kinase and ligase were mixed and 3 μl were added to the fragmented DNA. The ligation probes and universal linkers were phosphorylated and then ligated based on the sequence specificity complementary to the genomic DNA template. Step 3, exonuclease digestion served to purify the ligation product. Lambda Exonuclease and Exonuclease I were used to remove unligated probes, universal linkers and genomic DNA. In Step 4, the purified ligation product was PCR amplified using one pair of universal primers, with one primer labeled by biotin. Step 5, the biotinylated PCR products are bound to 96-well streptavidin-coated plate (Sigma). The PCR products were then denatured and the unbounded strands were washed off. Step 6 consisted of hybridization with fluorescence labeled universal ZipChute probe. For 48 SNPs, 96 unique ZipChute probes were hybridized to the complimentary sequence within each allele-specific oligonucleotide (ASO) probes. Each ZipChute probe migrates differently due to a mobility modifier. Step 7, the bound ZipChute probes were eluted and analyzed on Applied Biosystems 3730 DNA Analyzer. Step 8, the elution profiles were analyzed by GeneMapper software (Applied Biosystems) to determine genotypes. With 48 SNPs per run, 48 capillaries in the Applied Biosystems 3730 DNA Analyzer, and up to three injections per hour, the throughput is 5–10,000 SNPs per hour.
Figure 1.
SNPlex genotyping system assay protocol. On the first day, the OLA reaction, exonuclease purification, and PCR amplification are performed. On the second day, the amplicons are immobilized on streptavidin-coated microtiter plates. ZipChute probes are hybridized to complementary ZipCode sequences, and non-hybridized ZipChute probes are washed away. The bound ZipChute probes are eluted and analyzed by capillary electrophoresis using Applied Biosystems 3730. Reproduced from Journal of Biomolecular Techniques (Reference [18]) with permission from Andreas R Tobler at Applied Biosystems.
Multiplex SNaPshot genotyping
SNaPshot was performed following a previously published procedure based on single nucleotide primer extension that has been successfully adapted to the Applied Biosystems 3730 DNA Analyzer [30]. A stretch of genomic DNA (50 to 150 base pairs) was amplified by PCR, and the genotype was measured by primer extension using fluorescently labeled terminator nucleotides. Three single nucleotide polymorphisms, rs42427 in the APC gene, rs1800392 in WRN, and rs2228000 in XPC, were multiplexed for the study. Three pairs of PCR primers were amplified simultaneously in 15 μl reactions using 2× ReadyMix™ Taq PCR Reaction Mix with MgCl2 (Sigma, St. Louis, MO). For each SNP, 0.15 μl PCR forward and reverse primers (10 μM) were added to the PCR reactions. The amplification was carried out for 30 cycles starting with denaturation at 95°C for 30 s, and then primer annealing at 60°C for 1 min, followed by extension at 72°C for 1 min. The forward and reverse primers were as follows: for rs42427 in APC, 5'-CCCTCCAAATGAGTTAGCTGCT-3' and 5'-GCCTTCTGTAGGAATGGTATCTCG-3'; for rs222800 in XPC, 5'-GGAGCCATCGTAAGGACCCA-3' and 5'-TGCCTCTTTTACTGCTTGAAGAGC-3'; for SNP rs1800392 in WRN, 5'-GGTCCAACAATCATCTACTGTCCTT and 5'-TGATGAATGTCTTTCCTTGTGCTAAA-3'. After PCR amplification, the reactions were treated with Exonuclease I and Bacterial Antarctic Alkaline Phosphatase (New England Biolabs, Beverly, MA). For the primer extension, a gene-specific primer was designed with its 3'-end one base from the SNP position. The forward extension primers were as follows: for rs42427 in APC, 5'-TTTTTTTTTTTTTTTTTTCTGGAGAAGGAGTTAGAGGAGG (40 mer); for rs222800 in XPC, 5'-TAAGGACCCAAGCTTGCCAG-3' (20 mer), for SNP rs1800392 in WRN, 5'-TTTTTTTTCAAGTTACAGGTGAACTTAGGAAACT-3' (34 mer). SNaPshot reagent from Applied Biosystems was used to incorporate a single fluorescently labeled dideoxynucleotide into the 3' end of the primer directed by the DNA template. The primer extension reactions were analyzed using an Applied Biosystems 3730 capillary electrophoresis DNA instrument, and analyzed with GeneMapper 3.0 software (Applied Biosystems), with a throughput of 150 to 750 per hour (if multiplexed to 5 reactions). For CYP2D6, the multiplexed SNaPshot was carried out following a previously published protocol with slight modifications [31]. The forward PCR primer (5'-ATGGCAGCTGCCATACAATCCACCTG-3') was redesigned to analyze the promoter SNP rs1080985. The SNaPshot extension primer for rs1080985 was 5'-(T)58CCTGGACAACTTGGAAGAACC-3'. A total of 12 polymorphims were analyzed in parallel, by designing extension primers that are separable by capillary electrophoresis.
Genotyping of UGT1A1 promoter (TA)nTAA dinucleotide repeat polymorphism
The UGT1A1 dinucleotide repeat was genotyped according to previously designed PCR sequences and PCR conditions [32]. The forward primer sequence was 5'-GTCACGTGACACAGTCAAAC-3'. The reverse primer sequence was 5'-TTTGCTCCTGCCAGAGGTT-3' and FAM-labeled. The PCR products were analyzed using an Applied Biosystems 3730 DNA Analyzer.
Data analysis
Hardy-Weinberg equilibrium for each SNP was analyzed using HelixTree according to the manufacture's manual (Golden Helix, Inc. Bozeman, MT, USA).
Results
Genes and polymorphisms selected for genotyping by SNPlex
We have designed cancer genotyping SNPlex panels, selecting genes involved in drug metabolism and transport, DNA repair and apoptosis, cell cycle/cell growth/drug targets. We have selected polymorphisms for genotyping along the following criteria: polymorphisms known to affect enzyme/transporter functions, and SNPs in transcribed genic regions and htSNPs with high abundance obtained from HapMap and other databases. We have selected 560 SNPs for 160 genes, ordered into different categories:
Transporters: ABCA1, ABCA2, ABCA3, ABCA9, ABCA10, MDR1/ABCB1, ABCB4, ABCB11, ABCC1, ABCC2, ABCC3, ABCC4, ABCC5, ABCC6, ABCG2/BCRP, ABCG5, ABCG8, SLC19A1 (RFC) and SLC21A6.
Phase I metabolism enzymes: CYP1A1, 1A2, 1B1, 2A6, 2B6, 2C8, 2C9, 2C18, 2C19, 2D6, CYP2E1, 3A4, 3A5, 17A1, DIA4/NQO1, EPHX1/EH, MPO and SOD2.
Phase II metabolism enzymes: GSTA1 GSTA2, GSTA4, GSTM1, GSTM3, GSTP1, GSTT1, GSTT2, NAT1, NAT2, SULT1A1, SULT1A2, TPMT, COMT, UGT1A1, UGT1A6, UGT1A7, UGT1A9 and UGT2B7.
DNA repair genes: ADPRT/PARP, ADPRTL1, APEX1/APE1, ATM, ATR, BARD1, BLM, BRCA1, BRCA2, CHEK2, ERCC2/XPD, ERCC4/XPF, ERCC5/XPG, FANCD2, LIG1, LIG3, LIG4, MGMT/AGT, MLH1, MPG, MSH2, MSH3, MSH6, MYH/MUTYH, NBS1, NT5E, OGG1, PCNA, PMS2, POLB, RAD23A, RAD51, RAD52, RAD54B, RAD9A, RECQL, WRN, XPA, XPC, XRCC1, XRCC2, XRCC3, XRCC4, XRCC5 and XRCC9/FANCG.
Drug targets, cell signaling, cell cycle and apoptosis related genes: DHFR, DPYD, TYMS, VKORC1, EGFR, ERBB2, FLT1 (VEGFR1), KDR (VEGFR2), FLT4 (VEGFR3), PDGFRA, PDGFRB, KIT, RET, CDA, BAX, CASP3, CASP8, CASP9, CASP10, CCND1, CCNH, CDK7, CDKN1A/p21, CDKN1B/p27, CDKN2A/p16, CDKN2B/p15, GADD45A, IRS2, MDM2, RB1, TERC/hTR, TERT, TP53, TP53BP1, TP53BP2, TP73, APC, NF1, NF2, HPC1, VHL, ECRG1, WT1, MEN1, SMAD2, SMAD4, TNFRSF10A, PTCH and CDH1.
Among the 560 SNPs, 432 SNPs (77%) were successfully designed to be included in the SNPlex panels [see Additional file 1]. The SNPs were divided into several groups so that a subset of the SNPlex panels might be sufficient for a specific research project.
• Drug metabolism and transports: 4 panels, 189 SNPs.
• DNA repair: 3 panels, 148 SNPs.
• Cell cycle/growth/apoptosis: 2 panels, 95 SNPs.
The selection of polymorphisms for this study included some redundancy to account for limitation of the SNPlex approach. Any polymorphisms that could not be included with the SNPlex panels were omitted, or if thought to be critical, targeted by alternative methods. For example, a majority of the SNPs that are not suitable for SNPlex genotyping can be genotyped by multiplexed SNaPshot assay (see multiplexed SNaPshot for CYP2D6 in this manuscript as an example). Similarly, small insertions/deletions and repeats can be amplified by PCR and the variants determined by PCR product size difference based on gel electrophoresis or capillary electrophoresis using fluorescent-labeled primers (see UGT1A1 promoter dinucleotide repeat polymorphism in this manuscript). Based on the sequence information and literature search, possible alternative methods for detection of these genetic variants are listed in Additional file 1.
Validation of SNPlex results using SNaPshot
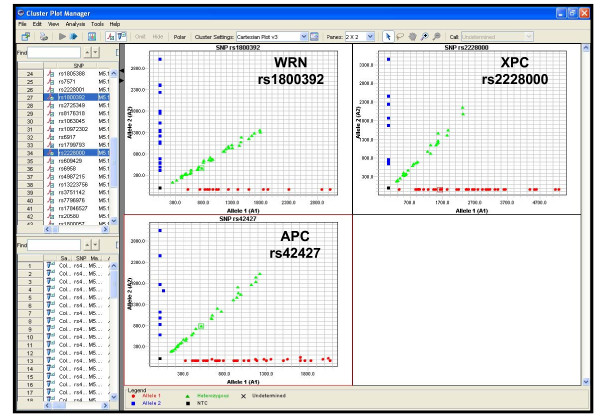
We selected SNaPshot (Applied Biosystems), based on single base-pair extension, as a reliable reference genotyping method [33,34]. Three SNPs, rs1800392 in WRN gene, rs2228000 in XPC, and rs42427 in APC, were genotyped using multiplexed SNaPshot. For 74 colorectal cancer samples, the two methods produced identical results for all three SNPs. We monitored the quality of SNPlex genotyping for the three SNPs (Figure 2) to identify the output features indicative of obtaining robust results. The homozygous and heterozygous alleles were clearly separated into different clusters on the Cartesian plots (Figure 2). Similar precision of genotype calls was observed in other projects performed in our laboratory comparing the genotyping results generated from SNPlex, SNaPshot, or TaqMan real-time PCR genotyping (unpublished data). This is consistent with the precision rate that was reported by Applied Biosystems [18].
Figure 2.
SNPlex genotyping results of three SNPs in colorectal cancer patients were identical to those measured by multiplexed SNaPshot.
Examples of single nucleotide polymorphisms known to be associated with cancer risk or drug treatment response
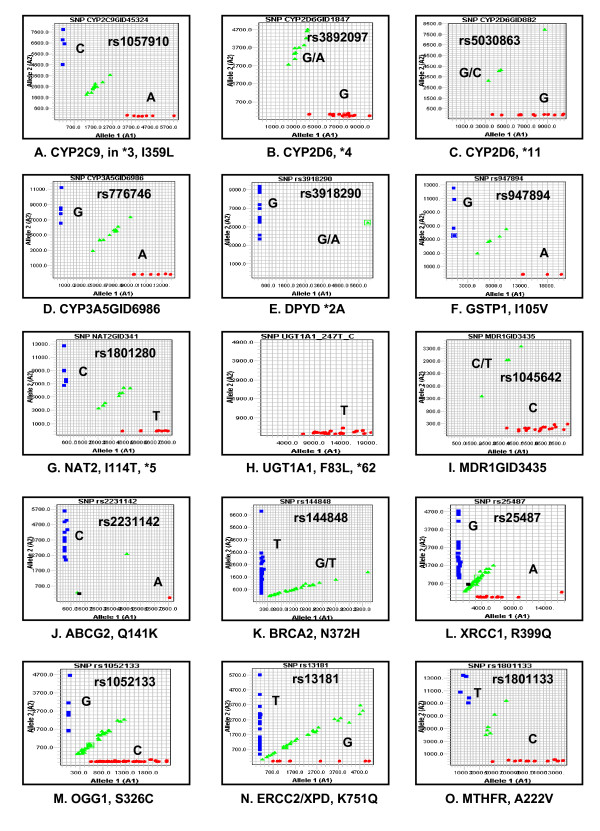
Examples of the most extensively studied polymorphisms with clinical relevance, such as cancer risk and cancer therapeutic response are summarized in Table 1, which includes 66 SNPs for 24 genes that can be genotyped by our SNPlex panels. We selected 15 SNPs based on their potential significance to cancer treatment response and cancer risk, to illustrate the SNPlex genotyping results (Figure 3). In each case, homozygous and heterozygous genotypes were clearly separated, and therefore, readily assigned.
Table 1.
Select examples of SNPs with clinical significance.
|
Phase I metabolism enzymes, Allele nomenclature for Cytochrome P450 enzymes [24]: | ||||
| Gene | rs# | location | Function | SNPlex |
| CYP2C9 | rs1799853 | *2, R144C | PM 0.25% in Caucasians, life-threatening bleeding after given warfarin | No |
| rs1057910 | *3, I359L | Yes | ||
| CYP2C19 | rs4244285 | *2, 681G>A, exon 5, splicing defect | PM phenotype 2–5% in Caucasians, 18–23% in Asians, > 87% PM in Caucasians is *2 and *3; > 99% PM in Asians has *2 and *3. CYP2C19*2 homozygotes did not respond to antiangiogenic drug thalidomide treatment | No |
| rs4986893 | *3, 17948G>A, exon 4 premature stop | Yes | ||
| rs28399504 | *4, transcription ablation | Failed | ||
| 90033C>T, R433W, *5A, *5B | No enzymatic activity | Yes | ||
| *7, 19294T>A | Splicing defect, no enzymatic activity | Yes | ||
| CYP2D6 | rs16947 | *2, 2851C>T, R296C | Normal, nucleotide position corrected according to [47] | Yes |
| rs3892097 or rs1800716 | *4, 1847G>A, splicing defect | The CYP2D6 PM is about 5–10% of Caucasians. 99% PM has *3, *4, *5, *6, *7, *8 and *11. *3, *5 and *6 are deletions | Yes | |
| rs28371704 | 983A>G, H94R | In *4A, *4B, *4F, *4G, *4H and *4J | Failed | |
| rs5030867 | *7, 2936A>C, H324P | No enzymatic activity | Yes | |
| rs5030865 | *8, 1759G>T | Stop codon, no enzymatic activity | Yes | |
| rs1065852 | *10, 100C>T, P34S | Decrease enzymatic activity | Yes | |
| rs5030863 | *11, 882G>C | Splicing defect, no enzymatic activity | Yes | |
| rs28371706 | *17, 1022C>T, T107I | Decrease enzymatic activity | Yes | |
| rs28371717 | *33, 2484G>T, A237S | Normal | Yes | |
| *44, 2951G>C | Splicing defect, no enzymatic activity | Yes | ||
| CYP3A4 | rs11773597 | *1F, m747C>G | Trans-regulation of gene expression is important. Overall, no major pharmacokinetic consequences for the identified CYP3A4 SNPs have been observed for the metabolism of anti-cancer drugs [12] | Yes |
| rs2740574 | *1B, m392A>G | Yes | ||
| Yes | ||||
| *4, 13989A>G, | In AF209389 | Yes | ||
| *8, 14026G>A | In AF209389, R130Q | Yes | ||
| CYP3A5 | rs28365083 | *2, 27289C>A, T398N | Failed | |
| rs776746 | *3, 6986A>G, splicing inclusion | *3 is the most frequent polymorphism (about 90% in Caucasians). Splicing defect, severely decrease of enzymatic activity [12] | Yes | |
| rs28365085 | *3d, 31551T>C, I488T | Yes | ||
| *5, 12952T>C | Splicing defect | Yes | ||
| *8, 3699C>T, R28C | Decreased enzymatic activity | Yes | ||
| rs28383479 | *9, 19386G>A, A337T | Decreased enzymatic activity | Failed | |
| rs15524 | *10, 31611C>T | Decreasde enzymatic activity | Yes | |
| DPYD | rs3918290 | splice variant IVS14+1G>A | *2A, Skipping exon 14, ↑ 5FU neurotoxicity [12] | Yes |
| NQO1 | rs1800566 | *2, C609T, R187S | *2 and *3 have reduced protein level and enzymatic activity. NQO1 is needed for the activation of mitomycin C, 17AAG (HSP90 inhibitor) and inactivation of benzene-like leukemogenic agents [13] | Yes |
| rs4986998 | *3, C465T, R139W | Yes | ||
|
Phase II metabolism enzymes NAT allele nomenclature [26]: UGT allele nomenclature [25]: | ||||
| Gene | rs# | location | Function | SNPlex |
| NAT2 | rs1801280 | 341T>C, I114T, *5A to*5J, *14C and *14F | Alleles with decreased activity include NAT2*5B, NAT2*6A, NAT*7A or B, NAT2*10, NAT2*14A or B, NAT2*17, NAT2*18 and NAT2*19 [12, 14] Low NAT2 activity is related to the increased risk of isoniazid hepatotoxicity |
Yes |
| rs1799929 | 481C>T, L161L, *5A, *5B, *5F, *5G, *5H, *5I, *6E, *11A, *11B, *12C and *14C | Yes | ||
| rs1208 | 803A>G, K268R,*5B, *5C, *5F, *5G, *5H, *5I, *6C, *12A, *12B, *12C, *12D, *14E and *14F | Yes | ||
| rs1041983 | 282C>T, Y94Y, *13, *5G, *5J, *6A, *6C, *6D, *7B, *12B, *14B, *14D, *14G | Yes | ||
| rs1799930 | 590G>A, R197Q *5E, *5J, *6A, *6B to *6E, *14D | Yes | ||
| rs1799931 | , 857G>A, G286E *7A, *7B | Yes | ||
| 499G>A in sequence X14672, E167K, *10 | Yes | |||
| rs1801279 | 191G>A, R64Q *14A to *14G, | Yes | ||
| 434A>C A in sequence X14672, Q145P, *17 | Yes | |||
| 845A>C A in sequence X14672, K282T, *18 | Yes | |||
| rs1805158 | 190C>T, R64W, *19 | Yes | ||
| TPMT | rs1800462 | *2, 238G>C | Null genotype associated with hematopoietic thiopurine toxicity, homozygous frequency 1/300 [4] | No |
| rs1800460 | *3A, 460G>A | No | ||
| rs1142345 | *3C, 719A>G | No | ||
| UGT1A1 | TA (5–8) TAA | UGT1A1 *28 (7 TAs) associated with increased irinotecan toxicity. Caucasians ~32% | No | |
| rs4148323 | 211G>A, G71R, *6 | Reduced enzymatic activity | Yes | |
| rs34993780 | 1456T>G, Y486D, *7 | Yes | ||
| rs35350960 | 686C>A, P229Q, *27 | Yes | ||
| 247T>C, F83L, *62 | Causing Gilbert's syndrome | Yes | ||
| GSTT1 | Deletion causing null genotype | Null allele has been associated with better or poorer survival in leukemia patients following chemotherapy [12] | No | |
| GSTP1 | rs947894 | 313A>G I105V | Val associated with decreased enzyme activity and increased survival after 5FU/oxaliplatin treatment of colorectal cancer patients [54] | Yes |
| GSTM1 | Deletion causing null genotype | Null allele is associated with increased survival after chemotherapy for multiple cancers [13, 14] | No | |
| SULT1A1 | rs9282861 | *2, R213H, HaeII | His/His has lower enzymatic activity and is associated with poor survival following tamoxifen therapy [55] | No |
| Transporter Genes | ||||
| Gene | rs# | location | Function | SNPlex |
| ABCB1 | rs1045642 | 3435C>T | C3435 associated with higher drug transport activity | Yes |
| rs1128503 | 1236T>C | Yes | ||
| rs2229109 | 1199G>A | Yes | ||
| ABCC2 | rs2273697 | 1249G>A, Val417Ile | 1249AA associated with decreased mRNA [56] | Yes |
| ABCG2 | rs2231142 | 421C>A, Q141K | Minor alleles with lower BRCP expression, enhanced drug sensitivity [12] | Yes |
| rs2231137 | G34 G>A V12M | No | ||
| 944–949 deletion | No | |||
| SLC19A1 | rs1051266 | 80G>A Arg27His | Patients with the 80AA genotype had higher plasma MTX levels, suggesting decreased cellular uptake of MTX | Yes |
| SLCO1B1/SCL21A6 | rs4149056 | T521C, Val174Ala, *5 | *5 and *15 are associated with decreased transport activity [57] | Yes |
| rs2306283 | Asp130Asn, *15 | Yes | ||
| DNA repair genes | ||||
| Gene | rs# | location | Function | SNPlex |
| BRCA2 | rs144848 | N372H | Cancer risk [51] | Yes |
| OGG1 | rs1052133 | S326C | Cancer risk [51] | Yes |
| XRCC1 | rs1799782 | R194W | Cancer risk [51] | Yes |
| rs25487 | R399Q | Gln399 associated with oxaliplatin/5-FU resistance | Yes | |
| rs25489 | R280H | Yes | ||
| ERCC2/XPD | rs13181 | K751Q | Lys751 associated with improved oxaliplatin/5-FU treatment outcome [52] | Yes |
| TP53 | rs1042522 | R72P | Cancer risk | Failed |
| MGMT | rs12917 | 262C>T, L84F | Decreased repair of DNA damage [58] | Yes |
| CHEK2 | 1100delC | Protein truncation, cancer risk [59] | No | |
| Drug target, pathway genes | ||||
| Gene | rs# | location | Function | SNPlex |
| DHFR | rs5030762 | 829T>C | SNP 829T>C located in the untranslated region of the DHFR, associated with ↑ of DHFR mRNA, ↓ responsiveness to methotrexate | No |
| MTHFR | rs1801133 | 677C>T, A222V | minor allele frequency 24–46%% in Caucasians, T allele is associated with reduced enzyme activity, increased toxicity to methotrexate [13, 53] | Yes |
| rs1801131 | 1298A>C, E429A | Reduced MTHFR enzyme activity [13, 53] | Yes | |
| TYMS | 2–9 28 bp repeats in the 5' promoter enhancer | 3 repeats ↑ RNA, TSER*3 associated with drug resistance of 5FU and methotrexate | No | |
| CDA | rs2072671 | 79A>C, K27Q | Minor allele has lower activity to inactivate gemcitabine than the wild-type [60] | Yes |
| 208G>A, A70T | 70TT has lower activity to inactive cytidine and ara-C than the wild-type [61] | Yes | ||
| Cell cycle genes | ||||
| CCND1 | rs603965 | 870A>G | Alternative transcript encodes a protein with enhanced cell transformation activity, and modifies caner risk [62] | Yes |
Figure 3.
SNPlex genotyping plots for selected functional SNPs. Known functional consequences are listed in Table 1 for each SNP.
Additional methods to accommodate polymorphisms not suitable for SNPlex
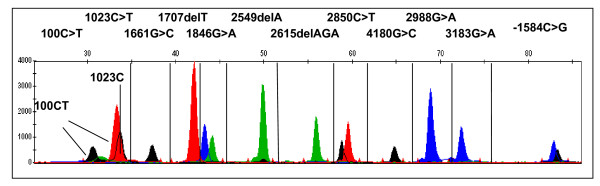
SNPs excluded from SNPlex panels for genomic sequence and other types of polymorphisms, such as insertions/deletions and variable number tandem repeat (VNTR), can be genotyped by different approaches, including multiplexed SNaPshot, TaqMan PCR, sequencing and SYBR Green melting curve assays [35], to detect all genetic variants of interest for a specific project. Mutiplexed SNaPshot is a primer extension assay with fluorescent terminator dyes, followed by capillary electrophoresis (Applied Biosystems 3730 DNA Aanalyzer). For faster throughput, we multiplex SNaPshot (up to 12 SNPs per reaction) for CYP2D6, by using extension primers of varying length (Fig. 4), yielding a throughput of between 150 (single SNP) to 1,800 (multiplex) per hour. Moreover, base pair deletions or insertions can be detected (Fig 4, 1707 del T, 2549 del A, 2615 del AGA).
Figure 4.
Multiplex SNaPshot genotyping assay for CYP2D6.
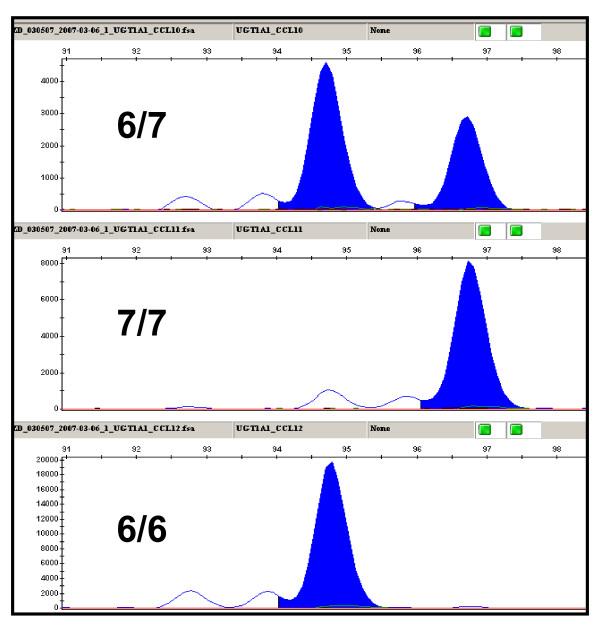
The UGT1A1 promoter dinucleotide repeat polymorphism, (TA)nTAA, was analyzed by capillary electrophoresis following PCR amplification. The repeats were amplified with unique flanking sequences on each side. The variation of dinucleotide repeat number leads to PCR products of different sizes that can be distinguished by capillary electrophoresis (Applied Biosystems 3730 DNA Analyzer, Figure 5).
Figure 5.
Genotyping for UGT1A1 promoter (TA)nTAA dinucleotide repeat polymorphism. The three samples are (TA)6/7TAA heterozygous, and (TA)7/7TAA and (TA)6/6TAA homozygous.
Pilot genotyping study for peripheral blood DNA from leukemia and colorectal cancer patients to evaluate the SNPlex panels
A. Genes for drug metabolism and transport in chronic lymphocytic leukemia patients
Flavopiridol, a broad inhibitor of cyclin-dependent kinases, is metabolized by UGT1A9 and UGT1A1, and interacts with a number of transporters, including MRP2, and BCRP (but apparently not with MDR1) [36-40]. Polymorphisms in each of these genes could affect the pharmacokinetic profile, and hence, treatment outcome. Our goals are to genotype and integrate the data with pharmacoanalytical and clinical results. In a pilot project, the 4 drug metabolism and transporter panels (189 SNPs) were applied to 39 DNA samples obtained from peripheral blood of chronic lymphocytic leukemia patients enrolled in flavopiridol clinical trials ongoing at The Ohio State University. To ensure the genotyping quality, the samples with signal intensity for the majority of SNP peaks below 1000 RFU (relative fluorescent units) were eliminated. For the 189 SNPs related to drug metabolizing and transport, 177 (94%) produced successful genotyping information. SNPs with low genotyping quality or that failed entirely in the SNPlex assays are listed in Table 2. The criteria for successful genotyping were based on the information from the manufacture's manual and our validation process (see below). Over 95% (136 out of 146) of the SNPs follow Hardy-Weinberg equilibrium (Chi-squared test, P > 0.01).
Table 2.
SNPs showing low genotyping quality or failing in the SNPlex analysis.
| Panels | DME | Cell cycle | DNA repair | |
| SNPs* | rs6413432 (CYP2E1) rs28371704 (CYP2D6) rs7439366 (UGT2B7) CYP1A2_m730C_T NAT1GID97 CYP2C19GID1 | rs4987138 (CYP2D7P1) rs28383479 (CYP3A5) CYP3A5GID27289 CYP1A2GID3534 NAT1GID560 CYP2C19GID80161 | rs2066827 (CDKN1B/p27) rs1799939 (RET) rs1042522 (TP53) rs17882155 (TP53) | rs1801321 (RAD51) rs3219489 (MUTYH) rs4986940 (XRCC9) rs3218384 (XRCC2) |
*For sequence information, refer to the column in Additional file 1 titled "Unique name in the panel".
B. Cancer-related genes in colorectal cancer patients
We genotyped 90 blood samples from Caucasian colorectal cancer patients using the 5 SNPlex panels for polymorphisms in DNA repair and cell cycle/growth/apoptosis. This is a pilot study to identify polymorphisms that contribute to colorectal cancer risk, and possibly treatment outcomes. For the 2 SNPlex panels related to cell cycle/drug target/apoptosis, 91 out of 95 (96%) SNPs were successful. For the three DNA repair panels, 140 out of 144 (97%) were successful (See Table 2 for SNPs showing low genotyping quality or that failed in the SNPlex analyses). Chi-squared test indicates all SNPs follow Hardy-Weinberg equilibrium (P > 0.01).
Discussion
We have adapted SNPlex as a platform for genotyping 432 SNPs in 160 genes related to the efficacy and toxicity of anticancer chemotherapy, and cancer risk. Stringent quality control criteria were used to attain optimal results. For example, DNA samples with the majority of signal peaks lower than 1000 RFU (relative fluorescence units) were discarded. In addition, DNA quality is a key factor for successful genotyping. Genomic DNA from blood samples and cell lines yielded high success rates. Our pilot studies indicate that 408 SNPs (94%) produced successful genotyping results. This is consistent with a previous study, where 19,779 nonsynonymous SNPs were genotyped by SNPlex in more than 1000 samples for a genome-wide association study [41]. The system allows 48 SNPs/panel to be genotyped simultaneously in each well for one DNA sample in 96-well or 384-well plate format, with a throughput of 5–10,000 SNPs per hour.
The goal is to develop genotyping panels containing polymorphisms shown to be relevant to disease and drug therapy. Therefore, the genotyping platform needs to be flexible to accommodate new findings, while the number of pertinent SNPs remains rather modest at present. In contrast, for discovery of new candidate genes and polymorphisms, very large SNP panels are beginning to be the norm. The SNPlex platform is designed for genotyping assays involving an intermediate number of SNPs (30–500). As each panel is multiplexed to maximally 48 SNPs, multiple panels need to run for larger SNP panel genotyping. As reagent cost is ~$5.00 per run ($0.10/SNP), the method is cost-effective for targeted genotyping of up to 500 to maximally 1000 candidate SNPs. Use of multiple panels permits flexibility in genotyping for specific applications, involving just a few samples or large cohorts. From our experience, for genotyping more than 500–1000 SNPs in any given project, alternative methods such as bead arrays may be more practical because of the increasing number of SNPlex panels needed. However, the optimal method will change rapidly on a yearly basis.
SNPlex is based on DNA ligation; therefore, its specificity is based on the characteristics of DNA sequence. The method can only be used to detect single nucleotide polymorphisms but commonly fails for genotyping repetitive sequences, insertion or deletions, and duplications. In addition, DNA sequence surrounding a specific polymorphism must meet specific criteria for probe design. As a result, the design process will disqualify a number of SNPs for SNPlex genotyping. Approximately 77% of selected SNPs were admissible for SNPlex analysis. Different genotyping strategies, such as multiplexed SNaPshot, TaqMan real-time PCR or sequencing, are complementary for genotyping all types of genetics variants.
An important aspect of this study is the careful selection of candidate genes and SNPs for genotyping. One limitation of the targeted SNP approach is that the panels fall short of covering all functional SNPs. Novel genetic polymorphisms associated with complex diseases, such as cancer, are identified in an increasing pace. For example, results from genome-wide association studies (GWAS) continue to reveal new polymorphisms that suggest the presence of functional variants in candidate genes [42,43]. However, the odds ratios of implicated polymorphisms in these case control studies usually range at or below 1.5, insufficient for inclusion with the intended genotyping panels that are eventually geared towards establishing clinical biomarkers for therapy. Yet, we expect that functional polymorphisms with high odds ratios with respect to specific phenotypes (e.g., treatment outcomes) will emerge from GWAS and its follow-up studies, then to be incorporated into the SNPlex panels. Since the SNPlex platform is flexible and expandable, a small subset of genetic polymorphisms, especially SNPs, could be easily added to the panel established in the current study. In addition, the selection of SNPs was not designed to optimize haplotype tagging – commonly used to survey variation in a gene, and this may represent a limitation of the present panels. Rather, the intent was to genotype a maximum number of SNPs either known or suspected of being functionally relevant – with newly discovered functional variants to be added in additional panels in the future. Moreover, most of the SNPs are in the transcribed regions. We can use these SNPs as markers for analysis of allelic mRNA expression imbalance, a powerful means for discovering regulatory SNPs that alter gene expression and RNA stability. It is estimated that regulatory SNPs are more abundant than nonsynonymous polymorphisms that alter the amino acids [33,34,44,45].
The selected functional SNPs in genes related to drug response and cancer risk are readily detectable using the methods established in the current study (Table 1 and Figure 3). Below, we briefly discuss polymorphisms of clinical significance (see reviews for detailed information [12,13]), to illustrate potential clinical applications of the genotyping panels we have established.
Phase I metabolizing enzymes
Cytochrome P450's are Phase I drug metabolizing enzymes harboring numerous mutations. For example, the two most important allele variants of CYP2C9, CYP2C9*2 and CYP2C9*3, cause a poor metabolizer phenotype associated with adverse warfarin effects [12]. Figure 3A shows the genotyping results with rs1057910 (Fig. 3A) in CYP2C9*3.
CYP2D6 metabolizes many commonly used drugs and is one of the best studied cytochrome P450 enzymes, with numerous variant alleles designated *1 to *61. The incidence of CYP2D6 poor metabolizers, carrying two null alleles, is 5–10% of Caucasians, imparting increased risk of adverse reactions from drugs requiring 2D6 metabolism for elimination. Nearly 99% of poor metabolizers have any two of the following alleles: *3, *4, *5, *6, *7 *8 or *11 [12,46]. Our SNPlex panels include key polymorphisms for alleles *4, *7, *8 and *11. For example, SNP rs3892097 (1847G>A, Fig. 3B), a common SNP in CYP2D6*4A to *4N, causes splicing defect [47], and it accounts for more than 75% of poor metabolizers in Caucasian [46]. CYP2D6*5 is a deletion of the entire functional CYP2D6 gene. In alleles *3 and *6, single nucleotide deletions causing CYP2D6 protein reading frame shift are undetectable by SNPlex genotyping. Multiplexed SNaPshot is complementary to SNPlex and can detect alleles *3 and *6 [31]. In addition, 4 SNPs, rs1065852, rs28371706, rs3892097 and rs16947, overlap in SNPlex panels and multiplexed SNaPshot, serving as a quality control. CYP2D6 catalyzes the conversion of tamoxifen to more potent metabolites, and poor CYP2D6 enzymatic activity has been associated with tamoxifen treatment outcome [12].
SNP rs776746 in CYP3A5 (6986A>G, CYP3A5*3, Fig. 3D) causes aberrantly spliced mRNA that is unstable, resulting in severely decreases protein level in the liver. The CYP3A5*3 allele frequency is approximately 90% in Caucasians [12].
Dihydropyrimidine dehydrogenase (DPYD) is a rate-limiting phase I metabolizing enzyme for 5-FU inactivation in the liver. SNP rs3918290 (Fig. 3E) is located at an RNA splicing donor site, causing DPYD exon 14 skipping (deletion) and leading to inactive enzyme. DPYD deficiency conveys risk for severe, life-threatening 5-FU toxicity [13].
Phase II metabolizing enzymes
For GSTP1 I105V (rs947894, Fig. 3F), Val/Val homozygotes express lower enzyme activity and decreased clearance rate of chemotherapeutic compounds, which leads to an increased survival following 5FU/oxaliplatin treatment of colorectal cancer patients. A better survival was also observed for breast cancer patients following treatment [12].
Polymorphisms affecting acetylator phenotype are common genetic variants for the biotransformation of drugs and carcinogens. N-acetyltransferase 2 (NAT2) polymorphisms are among the best studied examples in pharmacogenetics (Table 1 and Fig. 3G, rs1801280). These polymorphisms affect enzyme activity and are associated with drug toxicity and increased risk to develop certain cancers [48].
Uridine diphosphate glucuronosyltransferase 1A1 (UGT1A1) mediates glucocuronidation of bilirubin and anticancer drugs, such as SN38 (active irinotecan metabolite with antitumor activity). The UGT1A1*28 (promoter (TA)6TAA to (TA)7TAA) is a common genetic variant reducing UGT1A1 activity associated with irinotecan toxicity and hyperbilirubinaemia. Since this is a dinucleotide repeat variation, it is not suitable for detection with SNPlex. Fluorescently labeled PCR was designed to amplify the repeat and flanking DNA region. The repeat number was determined by the PCR product length (Figure 5). The SNPs in UGT1A1*6, *7, *27 and *62 are in the SNPlex panels (Table 1 and Fig. 3H).
Transporters
ABCB1/Multidrug resistance (MDR1) transporter is an efflux pump. High expression of MDR1 conveys resistance to a number of chemotherapeutic agents, including paclitaxel, doxorubicin and irinotecan [49]. C3435T (rs1045642, Fig. 3I) is a synonymous SNP without amino acid change. However, the T allele has been reported to affect RNA stability [33] and possibly translation [50] and lead to decreased protein expression. Nevertheless, varying results have been reported about the effects of MDR1 polymorphisms on pharmacokinetics and pharmacodynamics [12,13]; possibly, the functional polymorphism(s) behave differently in different tissues.
ABCG2 is another extrusion transporter that renders chemoresistance to a variety of anticancer drugs, such as mitoxantrone, methotrexate, doxorubicin and camptothecin-based anticancer drugs [49]. The minor allele of rs2231142 (Q141K, Fig. 3J) is associated with decreased protein expression and results in hypersensitivity to anticancer drugs in caner cell lines [12].
DNA repair genes
BRCA2 N372H (rs144848, Fig. 3K), XRCC1 R399Q (rs25487, Fig. 3L), and OGG1 S326C (rs1052133, Fig. 3M) are three SNPs in DNA repair genes consistently associated with cancer risk, supported by thirty studies [51]. In addition, the ERCC2/XPD variant Lys751Gln (rs13181, Fig. 3N) was associated with the response to treatment with 5-fluorouracil and oxaliplatin in colorectal cancer patients. Lys/Lys homozygotes responded better and had longer survival time [52]. However, contradictory results were observed for cisplatin treatment of non-small cell lung cancer patients [13].
Drug target/pathway genes
5,10-methylenetrtrahydrofolate reductase (MTHFR), a key enzyme in folate metabolism, catalyzes the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, which is involved in DNA and protein synthesis as a methyl donor [1]. SNP rs1801133 (C677T, A222V, Fig. 3O) in MTHFR is a functional variant associated with reduced MTHFR enzyme activity in TT homozygotes compared with heterozygots. As a result, the polymorphism increases toxicity to methotrexate [53].
In summary, the selected SNPs have broad applications for cancer research. Furthermore, the SNP panels are not limited to genes involved in cancer treatment outcomes with current drugs in clinical use. Hence, the developed SNPlex panels are not only applicable to studying pharmacogenomics/genetics of novel anticancer compounds under development, but also any drugs for the treatment of other diseases that are metabolized and/or transported by these gene products.
SNPlex has the advantage of being flexible and expandable for different studies, critical for translational research applications, including clinical drug trials. With the implementation of this platform, we have established a pharmacogenomics core with specific application to cancer chemotherapy. We hypothesize that genotyping on a large scale, both with respect to number of polymorphisms and subject populations, will yield valuable information on treatment outcomes. This concept will be applied to Phase I and II clinical trials with novel drugs or drug combinations, in comparison to pharmacokinetic analyses. Availability of population data across all subjects, collected over several years, will support multiple studies and has the potential to reveal novel mechanisms affecting drug response.
Conclusion
We have established SNPlex as a platform for genotyping more than 400 SNPs in 160 genes related to the efficacy and toxicity of anticancer chemotherapy, and cancer risk. The selected SNPs have broad applications for cancer research to study pharmacogenomics/genetics of current drugs in clinical use and novel anticancer compounds under development. In addition, since the phase I and phase II metabolizing enzymes and transporters are common genes in the absorption and elimination of therapeutic agents for diseases other than cancer, the platform has broad applications for pharmacogenomics studies at large.
Abbreviations
SNP: single nucleotide polymorphism.
All human gene symbols (names) are approved by HUGO gene nomenclature committee.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
ZD, ACP and DW were involved in the study design, acquisition of data, and interpretation of data, and drafted the manuscript; HH collected and classified all colorectal cancer samples, and assisted in drafting the manuscript; WS conceived the study, was responsible for its design and coordination, helped in the evaluation of the results and revised the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Selected genetic polymorphisms in 160 genes for cancer pharmacogenomics. This table includes all selected genes, genetic polymorphisms, sequences or rs numbers for selected SNPs.
Acknowledgments
Acknowledgements
This study was in part supported by a grant "Plasma Membrane Transporters", GM61390, from the National Institute of Health, General Medical Sciences. We thank Dr. Albert de la Chapelle for providing the colorectal cancer samples. We thank Andreas R Tobler at Applied Biosystems for the permission to reproduce Figure 1 from Journal of Biomolecular Techniques (Reference [18]).
Contributor Information
Zunyan Dai, Email: dai.15@osu.edu.
Audrey C Papp, Email: papp.2@osu.edu.
Danxin Wang, Email: Wang.808@osu.edu.
Heather Hampel, Email: Heather.Hampel@osumc.edu.
Wolfgang Sadee, Email: Wolfgang.Sadee@osumc.edu.
References
- Cheok MH, Evans WE. Acute lymphoblastic leukaemia: a model for the pharmacogenomics of cancer therapy. Nat Rev Cancer. 2006;6:117–129. doi: 10.1038/nrc1800. [DOI] [PubMed] [Google Scholar]
- Marsh S, McLeod HL. Pharmacogenomics: from bedside to clinical practice. Hum Mol Genet. 2006;15 Spec No 1:R89–93. doi: 10.1093/hmg/ddl087. [DOI] [PubMed] [Google Scholar]
- Wang L, Nguyen TV, McLaughlin RW, Sikkink LA, Ramirez-Alvarado M, Weinshilboum RM. Human thiopurine S-methyltransferase pharmacogenetics: variant allozyme misfolding and aggresome formation. Proc Natl Acad Sci U S A. 2005;102:9394–9399. doi: 10.1073/pnas.0502352102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dervieux T, Meshkin B, Neri B. Pharmacogenetic testing: proofs of principle and pharmacoeconomic implications. Mutat Res. 2005;573:180–194. doi: 10.1016/j.mrfmmm.2004.07.025. [DOI] [PubMed] [Google Scholar]
- Table of Valid Genomic Biomarkers in the Context of Approved Drug Labels http://www.fda.gov:80/cder/genomics/genomic_biomarkers_table.htm
- Guidance for Industry Pharmacogenomic Data Submissions http://www.fda.gov/cder/guidance/6400fnl.pdf
- Sadee W, Dai Z. Pharmacogenetics/genomics and personalized medicine. Hum Mol Genet. 2005;14 Spec No. 2:R207–14. doi: 10.1093/hmg/ddi261. [DOI] [PubMed] [Google Scholar]
- de Jong FA, de Jonge MJ, Verweij J, Mathijssen RH. Role of pharmacogenetics in irinotecan therapy. Cancer Lett. 2006;234:90–106. doi: 10.1016/j.canlet.2005.04.040. [DOI] [PubMed] [Google Scholar]
- O'Dwyer PJ, Catalano RB. Uridine diphosphate glucuronosyltransferase (UGT) 1A1 and irinotecan: practical pharmacogenomics arrives in cancer therapy. J Clin Oncol. 2006;24:4534–4538. doi: 10.1200/JCO.2006.07.3031. [DOI] [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- Landi S, Gemignani F, Monnier S, Canzian F. A database of single-nucleotide polymorphisms and a genotyping microarray for genetic epidemiology of lung cancer. Exp Lung Res. 2005;31:223–258. doi: 10.1080/01902140490495624. [DOI] [PubMed] [Google Scholar]
- Bosch TM, Meijerman I, Beijnen JH, Schellens JH. Genetic polymorphisms of drug-metabolising enzymes and drug transporters in the chemotherapeutic treatment of cancer. Clin Pharmacokinet. 2006;45:253–285. doi: 10.2165/00003088-200645030-00003. [DOI] [PubMed] [Google Scholar]
- Efferth T, Volm M. Pharmacogenetics for individualized cancer chemotherapy. Pharmacol Ther. 2005;107:155–176. doi: 10.1016/j.pharmthera.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Tate SK, Sisodiya SM. Pharmacogenetics goes genomic. Nat Rev Genet. 2003;4:937–947. doi: 10.1038/nrg1229. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z, Dracopoli NC. Progress in high throughput SNP genotyping methods. Pharmacogenomics J. 2002;2:103–110. doi: 10.1038/sj.tpj.6500094. [DOI] [PubMed] [Google Scholar]
- Fan JB, Chee MS, Gunderson KL. Highly parallel genomic assays. Nat Rev Genet. 2006;7:632–644. doi: 10.1038/nrg1901. [DOI] [PubMed] [Google Scholar]
- Syvanen AC. Toward genome-wide SNP genotyping. Nat Genet. 2005;37 Suppl:S5–10. doi: 10.1038/ng1558. [DOI] [PubMed] [Google Scholar]
- Tobler AR, Short S, Andersen MR, Paner TM, Briggs JC, Lambert SM, Wu PP, Wang Y, Spoonde AY, Koehler RT, Peyret N, Chen C, Broomer AJ, Ridzon DA, Zhou H, Hoo BS, Hayashibara KC, Leong LN, Ma CN, Rosenblum BB, Day JP, Ziegle JS, De La Vega FM, Rhodes MD, Hennessy KM, Wenz HM. The SNPlex genotyping system: a flexible and scalable platform for SNP genotyping. J Biomol Tech. 2005;16:398–406. [PMC free article] [PubMed] [Google Scholar]
- De la Vega FM, Lazaruk KD, Rhodes MD, Wenz MH. Assessment of two flexible and compatible SNP genotyping platforms: TaqMan SNP Genotyping Assays and the SNPlex Genotyping System. Mutat Res. 2005;573:111–135. doi: 10.1016/j.mrfmmm.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Pennisi E. Breakthrough of the year. Human genetic variation. Science. 2007;318:1842–1843. doi: 10.1126/science.318.5858.1842. [DOI] [PubMed] [Google Scholar]
- Dai Z, Huang Y, Sadee W. Growth factor signaling and resistance to cancer chemotherapy. Curr Top Med Chem. 2004;4:1347–1356. doi: 10.2174/1568026043387746. [DOI] [PubMed] [Google Scholar]
- The HapMap project http://www.hapmap.org/cgi-perl/gbrowse/hapmap_B35
- The NCI SNP500Cancer project http://cgap.nci.nih.gov/Genes
- Human allele nomenclature database for cytochrome P450 enzymes http://www.cypalleles.ki.se/
- The UDP-glucuronosyltransferase (UGT) alleles nomenclature database http://galien.pha.ulaval.ca/alleles/alleles.html
- The NAT nomenclature database http://louisville.edu/medschool/pharmacology/NAT.html
- The human membrane transporter database http://lab.digibench.net/transporter
- PharmGKB http://www.pharmgkb.org/do/serve?id=home.welcome
- Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Nakagawa H, Sotamaa K, Prior TW, Westman J, Panescu J, Fix D, Lockman J, Comeras I, de la Chapelle A. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- Pinsonneault J, Nielsen CU, Sadee W. Genetic variants of the human H+/dipeptide transporter PEPT2: analysis of haplotype functions. J Pharmacol Exp Ther. 2004;311:1088–1096. doi: 10.1124/jpet.104.073098. [DOI] [PubMed] [Google Scholar]
- Sistonen J, Fuselli S, Levo A, Sajantila A. CYP2D6 genotyping by a multiplex primer extension reaction. Clin Chem. 2005;51:1291–1295. doi: 10.1373/clinchem.2004.046466. [DOI] [PubMed] [Google Scholar]
- Monaghan G, Ryan M, Seddon R, Hume R, Burchell B. Genetic variation in bilirubin UPD-glucuronosyltransferase gene promoter and Gilbert's syndrome. Lancet. 1996;347:578–581. doi: 10.1016/S0140-6736(96)91273-8. [DOI] [PubMed] [Google Scholar]
- Wang D, Johnson AD, Papp AC, Kroetz DL, Sadee W. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenet Genomics. 2005;15:693–704. doi: 10.1097/01.fpc.0000175600.26893.fa. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang D, Johnson AD, Papp AC, Sadee W. Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem. 2005;280:32618–32624. doi: 10.1074/jbc.M504942200. [DOI] [PubMed] [Google Scholar]
- Papp AC, Pinsonneault JK, Cooke G, Sadee W. Single nucleotide polymorphism genotyping using allele-specific PCR and fluorescence melting curves. Biotechniques. 2003;34:1068–1072. doi: 10.2144/03345dd03. [DOI] [PubMed] [Google Scholar]
- Hagenauer B, Salamon A, Thalhammer T, Kunert O, Haslinger E, Klingler P, Senderowicz AM, Sausville EA, Jager W. In vitro glucuronidation of the cyclin-dependent kinase inhibitor flavopiridol by rat and human liver microsomes: involvement of UDP-glucuronosyltransferases 1A1 and 1A9. Drug Metab Dispos. 2001;29:407–414. [PubMed] [Google Scholar]
- Villeneuve L, Girard H, Fortier LC, Gagne JF, Guillemette C. Novel functional polymorphisms in the UGT1A7 and UGT1A9 glucuronidating enzymes in Caucasian and African-American subjects and their impact on the metabolism of 7-ethyl-10-hydroxycamptothecin and flavopiridol anticancer drugs. J Pharmacol Exp Ther. 2003;307:117–128. doi: 10.1124/jpet.103.054072. [DOI] [PubMed] [Google Scholar]
- Nakanishi T, Karp JE, Tan M, Doyle LA, Peters T, Yang W, Wei D, Ross DD. Quantitative analysis of breast cancer resistance protein and cellular resistance to flavopiridol in acute leukemia patients. Clin Cancer Res. 2003;9:3320–3328. [PubMed] [Google Scholar]
- Boerner SA, Tourne ME, Kaufmann SH, Bible KC. Effect of P-glycoprotein on flavopiridol sensitivity. Br J Cancer. 2001;84:1391–1396. doi: 10.1054/bjoc.2000.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager W, Gehring E, Hagenauer B, Aust S, Senderowicz A, Thalhammer T. The role of hepatic Mrp2 in the interaction of flavopiridol and bilirubin: impact on therapy. Int J Clin Pharmacol Ther. 2003;41:610–611. doi: 10.5414/cpp41610. [DOI] [PubMed] [Google Scholar]
- Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, Gunther S, Prescott NJ, Onnie CM, Hasler R, Sipos B, Folsch UR, Lengauer T, Platzer M, Mathew CG, Krawczak M, Schreiber S. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai YY, Chen WV, Shete S, Spitz MR, Houlston RS. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, Spain S, Lubbe S, Walther A, Sullivan K, Jaeger E, Fielding S, Rowan A, Vijayakrishnan J, Domingo E, Chandler I, Kemp Z, Qureshi M, Farrington SM, Tenesa A, Prendergast JG, Barnetson RA, Penegar S, Barclay E, Wood W, Martin L, Gorman M, Thomas H, Peto J, Bishop DT, Gray R, Maher ER, Lucassen A, Kerr D, Evans DG, Schafmayer C, Buch S, Volzke H, Hampe J, Schreiber S, John U, Koessler T, Pharoah P, van Wezel T, Morreau H, Wijnen JT, Hopper JL, Southey MC, Giles GG, Severi G, Castellvi-Bel S, Ruiz-Ponte C, Carracedo A, Castells A, Forsti A, Hemminki K, Vodicka P, Naccarati A, Lipton L, Ho JW, Cheng KK, Sham PC, Luk J, Agundez JA, Ladero JM, de la Hoya M, Caldes T, Niittymaki I, Tuupanen S, Karhu A, Aaltonen L, Cazier JB, Campbell H, Dunlop MG, Houlston RS. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- Pastinen T, Hudson TJ. Cis-acting regulatory variation in the human genome. Science. 2004;306:647–650. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Wang D, Sadee W. Polymorphisms affecting gene regulation and mRNA processing: broad implications for pharmacogenetics. Pharmacol Ther. 2005;106:19–38. doi: 10.1016/j.pharmthera.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3:229–243. doi: 10.1517/14622416.3.2.229. [DOI] [PubMed] [Google Scholar]
- CYP2D6 allele nomenclature http://www.cypalleles.ki.se/cyp2d6.htm
- Blum M, Demierre A, Grant DM, Heim M, Meyer UA. Molecular mechanism of slow acetylation of drugs and carcinogens in humans. Proc Natl Acad Sci U S A. 1991;88:5237–5241. doi: 10.1073/pnas.88.12.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–1530. [PubMed] [Google Scholar]
- Park DJ, Stoehlmacher J, Zhang W, Tsao-Wei DD, Groshen S, Lenz HJ. A Xeroderma pigmentosum group D gene polymorphism predicts clinical outcome to platinum-based chemotherapy in patients with advanced colorectal cancer. Cancer Res. 2001;61:8654–8658. [PubMed] [Google Scholar]
- Yong WP, Innocenti F, Ratain MJ. The role of pharmacogenetics in cancer therapeutics. Br J Clin Pharmacol. 2006;62:35–46. doi: 10.1111/j.1365-2125.2006.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoehlmacher J, Park DJ, Zhang W, Groshen S, Tsao-Wei DD, Yu MC, Lenz HJ. Association between glutathione S-transferase P1, T1, and M1 genetic polymorphism and survival of patients with metastatic colorectal cancer. J Natl Cancer Inst. 2002;94:936–942. doi: 10.1093/jnci/94.12.936. [DOI] [PubMed] [Google Scholar]
- Nowell S, Sweeney C, Winters M, Stone A, Lang NP, Hutchins LF, Kadlubar FF, Ambrosone CB. Association between sulfotransferase 1A1 genotype and survival of breast cancer patients receiving tamoxifen therapy. J Natl Cancer Inst. 2002;94:1635–1640. doi: 10.1093/jnci/94.21.1635. [DOI] [PubMed] [Google Scholar]
- Meyer zu Schwabedissen HE, Jedlitschky G, Gratz M, Haenisch S, Linnemann K, Fusch C, Cascorbi I, Kroemer HK. Variable expression of MRP2 (ABCC2) in human placenta: influence of gestational age and cellular differentiation. Drug Metab Dispos. 2005;33:896–904. doi: 10.1124/dmd.104.003335. [DOI] [PubMed] [Google Scholar]
- Kameyama Y, Yamashita K, Kobayashi K, Hosokawa M, Chiba K. Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics. 2005;15:513–522. doi: 10.1097/01.fpc.0000170913.73780.5f. [DOI] [PubMed] [Google Scholar]
- Hill CE, Wickliffe JK, Wolfe KJ, Kinslow CJ, Lopez MS, Abdel-Rahman SZ. The L84F and the I143V polymorphisms in the O6-methylguanine-DNA-methyltransferase (MGMT) gene increase human sensitivity to the genotoxic effects of the tobacco-specific nitrosamine carcinogen NNK. Pharmacogenet Genomics. 2005;15:571–578. doi: 10.1097/01.fpc.0000167332.38528.a5. [DOI] [PubMed] [Google Scholar]
- Cybulski C, Gorski B, Huzarski T, Masojc B, Mierzejewski M, Debniak T, Teodorczyk U, Byrski T, Gronwald J, Matyjasik J, Zlowocka E, Lenner M, Grabowska E, Nej K, Castaneda J, Medrek K, Szymanska A, Szymanska J, Kurzawski G, Suchy J, Oszurek O, Witek A, Narod SA, Lubinski J. CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet. 2004;75:1131–1135. doi: 10.1086/426403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Salavaggione OE, Ji Y, Pelleymounter LL, Eckloff BW, Wieben ED, Ames MM, Weinshilboum RM. Gemcitabine pharmacogenomics: cytidine deaminase and deoxycytidylate deaminase gene resequencing and functional genomics. Clin Cancer Res. 2006;12:1794–1803. doi: 10.1158/1078-0432.CCR-05-1969. [DOI] [PubMed] [Google Scholar]
- Yue L, Saikawa Y, Ota K, Tanaka M, Nishimura R, Uehara T, Maeba H, Ito T, Sasaki T, Koizumi S. A functional single-nucleotide polymorphism in the human cytidine deaminase gene contributing to ara-C sensitivity. Pharmacogenetics. 2003;13:29–38. doi: 10.1097/00008571-200301000-00005. [DOI] [PubMed] [Google Scholar]
- Shu XO, Moore DB, Cai Q, Cheng J, Wen W, Pierce L, Cai H, Gao YT, Zheng W. Association of cyclin D1 genotype with breast cancer risk and survival. Cancer Epidemiol Biomarkers Prev. 2005;14:91–97. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Selected genetic polymorphisms in 160 genes for cancer pharmacogenomics. This table includes all selected genes, genetic polymorphisms, sequences or rs numbers for selected SNPs.