Abstract
Mutations in 11 genes that encode ion channels or their associated proteins cause inherited long QT syndrome (LQTS) and account for ≈75–80% of cases (LQT1–11). Direct sequencing of SNTA1, the gene encoding α1-syntrophin, was performed in a cohort of LQTS patients that were negative for mutations in the 11 known LQTS-susceptibility genes. A missense mutation (A390V-SNTA1) was found in a patient with recurrent syncope and markedly prolonged QT interval (QTc, 530 ms). SNTA1 links neuronal nitric oxide synthase (nNOS) to the nNOS inhibitor plasma membrane Ca-ATPase subtype 4b (PMCA4b); SNTA1 also is known to associate with the cardiac sodium channel SCN5A. By using a GST-fusion protein of the C terminus of SCN5A, we showed that WT-SNTA1 interacted with SCN5A, nNOS, and PMCA4b. In contrast, A390V-SNTA1 selectively disrupted association of PMCA4b with this complex and increased direct nitrosylation of SCN5A. A390V-SNTA1 expressed with SCN5A, nNOS, and PMCA4b in heterologous cells increased peak and late sodium current compared with WT-SNTA1, and the increase was partially inhibited by NOS blockers. Expression of A390V-SNTA1 in cardiac myocytes also increased late sodium current. We conclude that the A390V mutation disrupted binding with PMCA4b, released inhibition of nNOS, caused S-nitrosylation of SCN5A, and was associated with increased late sodium current, which is the characteristic biophysical dysfunction for sodium-channel-mediated LQTS (LQT3). These results establish an SNTA1-based nNOS complex attached to SCN5A as a key regulator of sodium current and suggest that SNTA1 be considered a rare LQTS-susceptibility gene.
Keywords: cardiac arrhythmia, ion channel, nitrosylation, plasma membrane Ca-ATPase, sodium current
Long QT Syndrome (LQTS) is characterized by QT prolongation and syncope or death from arrhythmia. Mutations in 11 genes have been linked to inherited LQTS; 5 of these genes encode cardiac ion channels (KCNQ1, KCNH2, SCN5A, KCNJ2, and CACNA1C), and 6 encode ion channel subunits or channel-interacting proteins (ChIPs) (ANKB, KCNE1, KCNE2, CAV3, SCN4B, and AKAP9) (1). The three most common LQTS-susceptibility genes, KCNQ1 (LQT1), KCNH2 (LQT2), and SCN5A (LQT3), account for ≈75% of patients with LQTS, whereas each of the remaining LQTS-susceptibility genes are uncommon, accounting for <1% each, leaving 20% of LQTS patients with a yet-to-be-discovered genotype. Genes encoding ChIPs that affect the ion channels behind LQT1–3 (2) represent logical candidates for LQTS-susceptibility genes. Notably, this strategy has yielded caveolin-3 and LQT9 (3), SCN4B and LQT10 (4), and the most recent LQTS disease gene discovery, yotiao and LQT11 (5). When a mutation is found in a candidate gene in an LQTS patient, the demonstration of altered biophysical function provides a plausible mechanism for arrhythmogenesis and supports the pathogenicity of the gene and mutation. Characterization of the molecular mechanism by which the mutation causes altered channel function provides additional insight into basic channel function and regulation.
Nitric oxide (NO) is an endogenous signaling molecule synthesized from l-arginine by nitric oxide synthases: neural NOS (nNOS or NOS1), inducible NOS (iNOS or NOS2), and endothelial NOS (eNOS or NOS3) (6). Both nNOS and eNOS are expressed constitutively throughout the heart, and iNOS is expressed under pathophysiological conditions. In the heart, NO has many effects, including those on contractility (7), but importantly NO also increased late sodium current (INa) amplitude (8), the signature biophysical mechanism associated with SCN5A-mediated LQT3. Also, genome-wide association studies suggest an impact of polymorphisms in the nNOS regulator, NOS1AP or CAPON, on the QT interval of the electrocardiogram (9, 10). These data support the hypothesis that genes encoding proteins interacting with nNOS are candidate LQTS-susceptibility genes.
α1-Syntrophin (SNTA1) is a member of the family of dystrophin-associated proteins containing multiple protein interaction motifs that act as molecular scaffolds for nNOS and plasma membrane Ca-ATPase (PMCA) (11). The cardiac isoform of PMCA, PMCA4b, participates in the nNOS complex and inhibits NO synthesis (12). SNTA1 also interacts with the pore-forming α subunit (SCN5A, also known as NaV1.5) of the cardiac sodium channel macromolecular complex at the C terminus (13), but whether or not SCN5A interacts with this entire complex is unclear, as is the nature of the functional consequences and implications for LQTS. In this study, we show that SNTA1 connects SCN5A to the nNOS–PMCA complex in heart and that a perturbation in SNTA1, stemming from a rare missense mutation found in an LQTS patient, selectively disrupted association with PMCA4b, increased SCN5A nitrosylation, and increased late sodium current (INa) in both a noncardiomyocyte heterologous expression system and in native cardiomyocytes.
Results
Identification of a Mutation (A390V) in SNTA1 from a Cohort of Patients with Previously Genotype-Negative LQTS.
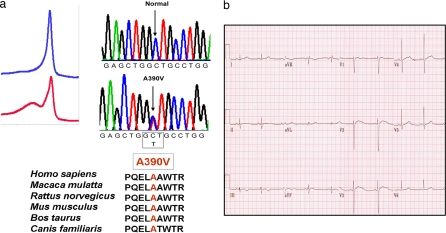
We performed comprehensive ORF/splice mutational analysis of SNTA1 by using PCR, denaturing HPLC, and direct DNA sequencing for a cohort of 50 unrelated patients (34 females; average age at diagnosis, 26 ± 16 years; average QTc, 531 ± 60 ms; QTc range, 480–759 ms) with genotype-negative LQTS that is lacking mutations in genes for LQT1–11. In addition, these patients had a “strong clinical phenotype,” defined as symptomatic cases with QTc ≥ 480 ms. A male, diagnosed with LQTS at 18 years of age after syncope episodes hosted the A390V-SNTA1 mutation (Fig. 1a Upper). A390V-SNTA1 was a rare missense mutation that occurred in a highly conserved region across species (Fig. 1a Lower) and was absent in 600 reference alleles. The electrocardiogram showed marked QT interval prolongation with a heart-rate-corrected QT interval (QTc) of 529 ms (Fig. 1b). The proband had no other symptoms of cardiac or skeletal muscle disease.
Fig. 1.
An LQTS patient with the mutation A390V-SNTA1. (a) Sequence chromatogram of the proband and the amino acid conservation of A390 across species. (b) A 12-lead electrocardiogram from the LQTS index case with the A390V-SNTA1 missense mutation. The QTc exceeds 520 ms.
SCN5A C Terminus Is Linked to nNOS and PMCA4b by SNTA1.
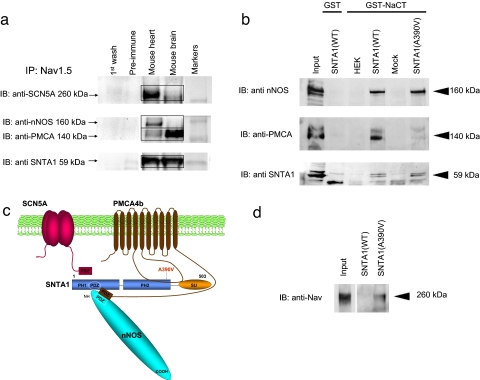
In mouse cardiac homogenates, anti-SCN5A antibodies coimmunoprecipitated nNOS, PMCA4b, and SNTA1 (Fig. 2a). A GST-labeled C terminus of SCN5A (GST-NaCT) pulled down the complex of nNOS, PMCA4b, and WT-SNTA1 (Fig. 2b) when coexpressed in HEK293 cells. Together these results support the hypothesis that SCN5A interacts with the nNOS complex through the C terminus (Fig. 2c). The mutation A390V-SNTA1 is located near the linker between the PH2 and SU domains of syntrophin, where there is a strong binding region for SNTA1 with PMCA4b (Fig. 2c). A second weaker interaction occurs between nNOS and PMCA4b via PDZ domains (11).
Fig. 2.
The nNOS complex is linked to SCN5A and disrupted by A390V-SNTA1. (a) SCN5A is associated with nNOS, PMCA, and SNTA1 in mouse heart homogenates. Homogenates were subjected to immunoprecipitation with anti-SCN5A antibody, and the precipitates were analyzed by immunoblotting. PMCA, nNOS, and SNTA1 were detected in the immunoprecipitates, whereas control preimmune serum did not immunoprecipitate the proteins. (b) The GST-labeled C terminus of SCN5A was incubated with lysates obtained from cells transiently expressing nNOS and PMCA4b and either WT-SNTA1 or A390V-SNTA1. Bound proteins were analyzed by SDS/PAGE and immunoblotted with anti-nNOS, anti-PMCA, and anti-SNTA antibodies. With WT-SNTA1, all three proteins were associated, but A390V-SNTA1 caused dissociation of PMCA4b. (c) Schematic representation of the sodium channel macromolecular complex with SCN5A, SNTA1, nNOS, and PMCA4b showing the location of A390V-SNTA1 in the PH2 domain. The PDZ domains of SCN5A, nNOS, SNTA1, and PMCA4b are indicated by boxes. The long intracellular linker between transmembrane domains 4–5 of PMCA4b interacts with the PH2 and SU domains of SNTA1. (d) A390V caused increased S-nitrosylation of SCN5A relative to WT by using the nitrosylation biotin switch assay. Cell lysates were prepared from cells stably expressing SCN5A and transiently expressing nNOS and PMCA4b and either WT-SNTA1 or A390V-SNTA1. Samples were biotinylated and subjected to SDS/PAGE and Western blot with the anti-SCN5A channel antibody.
A390V-SNTA1 Disrupted Association of PMCA4b with the nNOS Complex.
A390V-SNTA1 selectively disrupted participation of PMCA4b in the nNOS complex as indicated by the disappearance of the band for PMCA4b (Fig. 2b), but the C terminus continued to pull down both SNTA1 and nNOS (Fig. 2b). We hypothesized that dissociation of PMCA4b caused by A390V-SNTA1 would release the PMCA4b-mediated inhibition of nNOS and increase local concentrations of NO. By using a biotin switch assay (14), we found that direct S-nitrosylation of SCN5A was minimal with WT-SNTA1 but that S-nitrosylation was clearly evident with A390V-SNTA1 (Fig. 2d).
A390V-SNTA1 Increased Peak and Late Sodium Current Through an nNOS-Dependent Mechanism.
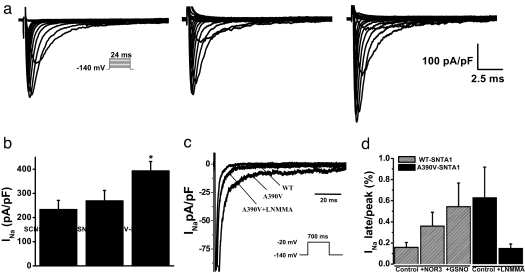
We performed functional characterization of A390V-SNTA1 with HEK cells coexpressing SCN5A, nNOS, PMCA4b, and either WT-SNTA1 or A390V-SNTA1. A390V caused a significant increase in peak INa (Fig. 3 a and b and Table 1) and in late INa (Fig. 3 c and d). When NG-monomethyl-l-arginine (l-NMMA), an NOS inhibitor, was administered into the culture medium for 24 h, the late INa caused by A390V-SNTA1 was abolished, and the increase in peak INa was partially reversed (Fig. 3 c and d and Table 1). The NO donors NOR3 (30 μM) and GSNO (100 μM) caused increased peak INa (Table 1) and increased late INa (Fig. 3d and Table 1) with WT-SNTA1, showing that NO was associated with increased peak and late INa like that found with the A390V mutation. Moreover, NOS inhibition restored the INa amplitudes found with the A390V mutation to WT levels (Fig. 3d). Together with the macromolecular complex association data (Fig. 2b) and the SCN5A direct nitrosylation data (Fig. 2d), these functional data support the hypothesis that A390V-SNTA1 disrupted the PMCA4b-mediated inhibition of nNOS and caused an NO-mediated accentuation of late INa, perhaps by direct S-nitrosylation of the channel.
Fig. 3.
Biophysical properties of SCN5A coexpressed with PMCA4b and nNOS without SNTA1 (labeled as SCN5A), with WT-SNTA1, or with A390V-SNTA1. (a) Whole-cell current traces from representative cells show increased peak INa associated with A390V-SNTA1. (Inset) INa was elicited by test depolarization to 24 ms from a holding potential of −140 mV. (b) Summary data of peak INa densities from each group (n = 9–16). (c) Representative traces showing increased late INa due to A390V-SNTA1 compared with WT-SNTA1 and A390V-SNTA1 plus l-NMMA. (Inset) Currents were elicited by a test depolarization pulse from −140 mV to −20 mV for 700 ms. (d) Summary data for late INa normalized to peak INa after leak subtraction. Late INa was measured as the mean between 600 and 700 ms after the initiation of the depolarization (n = 4–18). Symbols represent means, and bars represent the SEM. *, P < 0.05 versus WT-SNTA1.
Table 1.
Peak INa density, the percentage of late normalized to peak, and sodium channel kinetics from n experiments averaged and reported as mean ± SEM
| Samples | Peak INa |
Activation |
Inactivation |
INaL |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| pA/pF | n | V1/2, mV | K | n | V1/2, mV | K | n | % | n | |
| SCN5A | −233 ± 38 | 9 | −40 ± 2 | 4 | 9 | −77 ± 2 | 5 | 9 | 0.34 ± 0.09 | 5 |
| SNTA-WT | −238 ± 33 | 10 | −44 ± 2 | 4 | 7 | −76 ± 2 | 5 | 9 | 0.16 ± 0.04 | 9 |
| SNTA-A390V | −393 ± 39* | 16 | −41 ± 2 | 4 | 9 | −69 ± 2* | 6 | 15 | 0.62 ± 0.29 | 18 |
| SNTA-A390V + l-NMMA | −336 ± 37* | 6 | −44 ± 2 | 4 | 6 | −75 ± 2 | 5 | 6 | 0.15 ± 0.04 | 4 |
| SNTA-WT + NOR3 | −400 ± 67* | 12 | −47 ± 1 | 4 | 11 | −78 ± 1 | 5 | 12 | 0.31 ± 0.07 | 12 |
| SNTA-WT + GSNO | −427 ± 151* | 5 | −39 ± 2 | 4 | 3 | −77 ± 3 | 5 | 5 | 0.54 ± 0.22* | 5 |
The parameter fits were obtained from individual experiments with Boltzmann or exponential fits (see Materials and Methods).
*P < 0.05 versus SNTA-WT.
A390V-SNTA1 Altered Sodium Channel Gating Properties Through an nNOS-Dependent Mechanism.
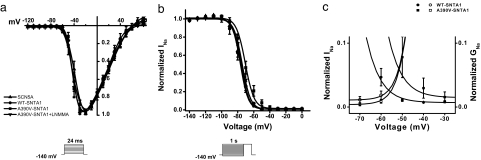
The voltage dependence of activation and inactivation was measured in HEK cells coexpressing SCN5A, nNOS, PMCA4b, and either WT-SNTA1 or A390V-SNTA1. A390V-SNTA1 caused no change in activation (Fig. 4a) but caused a significant depolarizing shift of 6 mV in channel inactivation (Fig. 4b and Table 1), which increased the overlap of the activation and inactivation curves (Fig. 4c) to increase the “window current” (15). The inactivation curve for the A390V-SNTA1 was normalized after exposing the cells to l-NMMA for 24 h (Fig. 4b), showing this gating effect of A390V also was mediated by NO. We also found that the time course of INa current decay was slower in A390V-SNTA1 compared with that of WT-SNTA1 (data not shown). The difference in INa decay for the A390V-SNTA1 compared with that of WT-SNTA1 reached significance at more positive test potentials.
Fig. 4.
Voltage-dependence relationships of steady-state activation, inactivation, and decay. INa was measured from cells transiently expressing SCN5A, PMCA4b, and nNOS without SNTA1 (squares), with WT-SNTA1 (circles), A390V-SNTA1 (triangles), and A390V-SNTA1 plus the nNOS blocker l-NMMA (inverted triangles). (a) A390V-SNTA1 did not affect steady-state activation. (b) A390V-SNTA1 shifted steady-state inactivation by +6 mV. (c) The peak-current activation data from a are replotted as a conductance curve with inactivation relationships from b to show that A390V-SNTA1 increases the overlap of these relationships. Lines represent fits to Boltzmann equations with parameters of the fit and n numbers in Table 1.
A390V-SNTA1 Increased Late Sodium Current in Native Cardiac Cells.
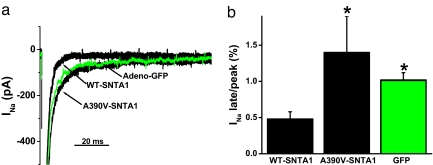
To assess the effects of A390V-SNTA1 in a more native myocardial cell environment, neonatal cardiomyocytes were transduced for 48 h with adenovirus containing the WT or the A390V-SNTA1/Ires-GFP recombinants. Only beating, GFP-positive cardiomyocytes were selected for study. Representative current traces (Fig. 5a) and summary data from five to six cells (Fig. 5b) show significantly increased late INa in the A390V-SNTA1 cells compared with that of the cardiac cells transduced with WT-SNTA1. Myocytes infected with GFP alone had increased late INa compared with overexpression of WT-SNTA1.
Fig. 5.
A390V-SNTA1 increased late INa in native cardiac cells compared with that of WT-SNTA1. (a) Whole-cell INa traces were elicited by step depolarization of 775 ms in duration to −20 mV from a holding potential of −140 mV and normalized to cell capacitance. Adenoviral recombinants of WT-SNTA1, A390V-SNTA1-Ires-GFP, and GFP alone were used to transduce neonatal cardiomyocytes for 48 h. (b) Summary data for the percentage of late INa at 775 ms normalized to peak current.
Discussion
SNTA1 — An LQTS-Susceptibility Gene.
SNTA1 is here proposed as a susceptibility gene for inherited LQTS. Although the proband is not part of a large family that would allow for classic linkage analysis, other evidence supports the pathogenic nature of this mutation. A390V-SNTA1 was found in a patient with a strong LQTS phenotype, all known susceptibility genes for LQTS were found to be negative, and this mutation was absent from 600 reference alleles. The mutation also occurred in a region of SNTA1 that is highly conserved and thought to be important for structure and function. Perhaps most persuasively, investigations in both heterologous cells and cardiac myocytes showed that the mutation caused a marked increase in late INa comparable to the increases seen in patients with LQT3 mutations (16). The data showing a plausible molecular mechanism for the effects of the A390V-SNTA1 mutation on the regulation and function of the cardiac sodium channel through the nNOS complex further support the view that the mutation in this gene is pathogenic. If the genotype-phenotype relationship is supported by further studies in additional patients, then SNTA1 would join the LQT9 gene CAV3 (17) and the LQT10 gene SCN4B (4) as a rare LQTS-susceptibility gene (LQT12) that together with CAV3 and SCN4B produce LQTS through actions on SCN5A to cause a net “gain of function” with increased late INa.
SCN5A Association with the nNOS Complex.
SNTA1 was reported previously to interact with nNOS and PMCA (11), and in a separate study SNTA1 was shown to interact with the C terminus of SCN5A (13). The cardiac isoform of PMCA is PMCA4b, and it was shown to participate in the nNOS complex in heart to inhibit NO synthesis (12). Although these individual associations were known previously, the coimmunopreciptation data from myocytes (Fig. 2a) indicate the complete complex. In addition, we used a GST-fusion protein of the C terminus of SCN5A as bait to show that the complex of nNOS, SNTA1, and PMCA4b associated with SCN5A when expressed in HEK cells. We also showed that the A390V-SNTA1 mutation selectively disrupted the association of PMCA4b but that nNOS and SNTA1 association with SCN5A was maintained. These results suggest that this nNOS complex may be important for modulation of the cardiac sodium channel through local levels of NO.
Direct Nitrosylation of SCN5A by NO.
Two NO-associated pathways are used commonly for signaling (18). One is indirect through cGMP (19), a second messenger that tends to oppose the actions of cAMP. The other is direct modification via S-nitrosylation of target proteins where NO binds with sulfhydryl groups of specific cysteine residues and alters function directly through this posttranslational modification (20). NO is a short-lived diatomic free radical, and physiological effects are likely to depend on the local concentration of NO in the molecular neighborhood of NOS and downstream targets. We showed that A390V-SNTA1 was associated with increased direct nitrosylation of SCN5A (Fig. 2d). Although direct S-nitrosylation has been shown for other ion channels (21) and inferred indirectly for sodium channels (22), our study provides a demonstration of direct S-nitrosylation of the cardiac sodium channel. This finding supports the idea that the mutation was associated with alteration of the local NO compared with that of WT-SNTA1 and suggests that the observed modulation of INa may have been mediated directly by S-nitrosylation of the sodium channel.
A390V-SNTA1 Increased Late INa Consistent with NO Modulation.
A major effect of A390V-SNTA1, and the one important for LQTS pathogenesis, was the increased late INa observed in both heterologous cells cotransfected with SCN5A, A390V-SNTA1, nNOS, and PMCA4b (Fig. 3) and myocytes transduced with A390V-SNTA1 (Fig. 5). The increased late INa with A390V-SNTA was prevented by the nNOS inhibitor l-NMMA (Fig. 3 c and d), supporting the involvement of a NO mechanism for the increase. In nerve terminals and ventricular myocytes, the release of caged NO was shown previously to increase late INa (8) in a manner that was independent of guanylate cyclase, and the effect on late INa was blocked by N-ethylmaleimide treatment, suggesting direct involvement of the S-nitrosylation pathway. In addition to late INa, we also observed a depolarizing shift in steady-state inactivation (Fig. 4b) that was reversed by l-NMMA and a significant increase in peak INa density (Table 1) that was partially reversed by l-NMMA. In a study where NO was applied exogenously to cardiac myocytes, the major effect was actually a decrease in INa density that depended on cGMP and cAMP (23) without alterations in kinetics or late INa. Our results are more consistent with those of Ahern et al. (8); we speculate that the method of NO delivery may be important for the observed differences with endogenous NO generation in our study and caged NO release in the Ahern et al. study (8) giving different and perhaps more physiological results than exogenous application (23), although species differences may exist also. The data support our hypothesis that A390V-SNTA1 increased late INa and exerts other effects by direct S-nitrosylation of SCN5A but that A390V-SNTA1 possibly exerts action through direct S-nitrosylation of other regulatory proteins of the SCN5A complex or by cGMP-dependent phosphorylation of SCN5A or other proteins of the complex. The detailed molecular mechanisms of the possible role for direct nitrosylation versus the cGMP pathway in the SCN5A macromolecular complex will require further study.
Implications and Limitations for Clinical and Basic Science.
The clinical findings and functional data reported here support SNTA1 as an LQTS-susceptibility gene acting through SCN5A and a localized nNOS complex to increase late INa. It is important to note that mutant SNTA1 is presently limited to a single patient and the seeming rarity of SNTA1 mutations in LQTS awaits exploration of other LQTS patient cohorts. So far, mutated SNTA1 was present in just 1 of 50 (2%) patients with phenotype-positive but heretofore genotype-negative LQTS. Among our entire cohort of clinically definite LQTS, SNTA1-mediated LQTS accounts for <1% of LQTS and is as common/rare as LQT6, LQT8, LQT10, and LQT11 (data not shown). In addition, we have not excluded the possibility that A390V-SNTA1 may not be specific to SCN5A but also may interact with other ion channel complexes, producing additional effects. In particular, the disassociation of PMCA4b from the complex caused by A390V-SNTA1 may affect the transport function of that protein in ways that may be arrhythmogenic. The results that we present here, however, add to our increasing understanding of the SCN5A macromolecular complex and support a general mechanism for locally generated NO regulation of late INa amplitude in heart. This mechanism may account for previously observed effects of polymorphisms involving an nNOS regulator on the QT interval (9, 10), and it suggests that other ChIPs such as PMCA4b are logical candidates for the pathogenesis of LQTS.
Materials and Methods
LQTS-Negative Cohort.
Between August 1997 and 2004, 541 consecutive, unrelated patients (358 females) were referred to the Mayo Clinic Windland Smith Rice Sudden Death Genomics Laboratory in Rochester, MN, for LQTS genetic testing (24). Clinical data including physical examination, personal history of syncope, seizures, or aborted cardiac arrest, family history, and 12-lead electrocardiogram analysis were collected. Mutational analysis of the genes responsible for LQT1–10 was performed previously (3, 24, 25). Patients were characterized as either genotype-positive or -negative based on this primary analysis. This study focuses on the subset of 50 (34 females; average age at diagnosis, 26 ± 16 years; average QTc, 531 ± 60.4 ms; QTc range, 480–759 ms) unrelated patients negative for LQT1–10 with a clinically robust phenotype of LQTS (QTc ≥ 480 ms or Schwartz score ≥ 3.0) (26).
SNTA1 Mutational Analysis.
After written approval for this protocol by the Mayo Foundation Institutional Review Board, genomic DNA was extracted from peripheral blood lymphocytes with the Purgene DNA extraction kit (Gentra, Inc.). A mutational analysis of the entire coding region of SNTA1 (chromosome 20q11.2) was performed on genomic DNA with PCR, denaturing HPLC, and direct DNA sequencing as previously described (27). PCR primers and conditions are available upon request.
Reagents and Antibodies.
l-NMMA was obtained from Sigma–Aldrich. Nitrosoglutathione (GSNO) and 4-ethyl-2-hydroxyimino-5-nitro-3-hexenamide (NOR3) were from Cayman Chemical. Rabbit anti-α1-syntrophin antibody was purchased from Sigma–Aldrich, mouse anti-nNOS antibody was from Invitrogen, and mouse anti-calcium pump pan PMCA ATPase antibody was from Abcam. Rabbit anti-cardiac Na channel antibody and goat HRP-conjugated anti-mouse IgG antibody were from Upstate, and goat HRP-conjugated anti-rabbit IgG (H+L) antibody was from Bio-Rad.
Plasmid Construction.
The cDNA of the α1-syntrophin gene, SNTA1 (GenBank accession no. NM_003098), was subcloned into pIRES2EGFP (Clontech Laboratories). A mutation of A390V in the PH2 domain in SNTA1 was introduced into WT SNTA1 by the PCR-based overlap-extension method. The cDNAs of nNOS and PMCA4b were kind gifts from Solomon H. Snyder (Johns Hopkins University, Baltimore) and Emanuel E. Strehler (Mayo Clinic), respectively. To generate the GST-labeled C terminus of the SCN5A expression vector, the region corresponding to Ile-1771 through the termination was inserted in-frame into pGEX-5X-2 (Amersham Biosciences). All clones were sequenced to confirm integrity.
Plasmid Constructions on Bicistronic Adenovirus Shuttle Vectors.
For transduction into primary cardiac myocytes, we generated the bicistronic adenovirus shuttle vector of SNTA1 and GFP. SNTA1 cDNA was subcloned into an entry vector of pENTR1A-IRES2EGFP, which was created by incorporation of the multicloning sites, an internal ribosome entry site, and EGFP coding region from pIRES2EGFP into pENTR1A (Invitrogen). A recombination reaction between pAd/CMV/V5-DEST (Invitrogen) generated pAdSNTA1-IRES2EGFP. The A390V mutant was incorporated into the WT by site-directed mutagenesis. All clones were sequenced to confirm integrity.
Transfection into HEK Cells and Neonatal Cardiomyocytes.
Expressing vectors containing SCN5A (GenBank accession no. AB158469), nNOS (GenBank accession no. NM_052799), PMCA4b (GenBank accession no. AY560895), and either WT-SNTA1 or A390V-SNTA1 in pIRES2EGFP at the ratio of 1:1:1:0.25, respectively, were transfected into HEK cells with FuGENE6 reagent according to the manufacturer's procedure. Neonatal ventricular myocytes were isolated from the hearts of 1- to 2-day-old mice through collagenase digestion (Worthington Biochemical Corp.) in calcium- and magnesium-free Hank's balance salt solution (pH 7.4). The isolated cells were washed three times with PBS and plated in medium 199 (Gibco-BRL) supplemented with 5 mmol/liter creatine, 2 mmol/liter l-carnitine, 5 mmol/liter taurine, and 0.2% mmol/liter BSA at a field density of 10,000 cells per squared centimeter on 35-mm culture dishes precoated with laminin (Sigma). After 1 h, the media were changed to remove the nonadherent cells, and then the cells were infected with adenovirus shuttle vectors of the WT-SNTA1 and A390V-SNTA1 mutant.
Immunoprecipitation.
For immunoprecipitations, mouse heart or brain homogenates (2 mg of protein) and preimmune serum were used. Immunoprecipitations were carried out by using 5 μg of anti-Nav1.5 antibody. The immune complexes were analyzed by SDS/PAGE (4–15% gradient gels; Bio-Rad) and transfected to PVDF membranes (Invitrogen) for Western blot with antibodies to Nav1.5, nNOS, PMCA, and SNTA at a 1:250 dilution.
GST Pull-Down Assay.
To generate bait for pull-down experiments, Rosetta gami 2 bacteria (Novagen) expressing the GST-labeled C terminus of SCN5A were collected and sonicated in PBS with 1% Triton X-100 and then purified and fixed on MagneGST particles (Promega) according to the manufacturer's instructions. The purity and concentration of the fusion proteins were determined by SDS/PAGE followed by Coomassie blue. GST bait was incubated in protein-binding buffer overnight at 4°C with the translation products that were obtained by the transient transfection of nNOS, PMCA4b, and WT-SNTA1 or A390V-SNTA1. The samples were then washed four times in binding buffer. The bound proteins were liberated by boiling in Laemmli sample buffer with 50 mM DTT and were analyzed by SDS/PAGE and immunoblotting with anti-nNOS, anti-PMCA, and anti-SNTA1 antibodies.
Electrophysiology.
INa was recorded using the whole-cell patch clamp in HEK293 cells transfected with SCN5A, nNOS, PMCA4b, and either wild type SNTA1 or A390V-SNTA1. Cells were continuously perfused with bath (extracellular) solution containing 140 mM NaCl, 4 mM KCl, 1.8 mM CaCl2, 0.75 mM MgCl2, and 5 mM HEPES (pH 7.4 set with NaOH). The pipette (intracellular) solution contained 120 mM CsF, 20 mM CsCl2, 5 mM EGTA, and 5 mM HEPES (pH 7.4 set with CsOH) at room temperature. L-NMMA was added to the culturing medium to block NO prior to the experiment. Electrodes were pulled using borosilicate (P-87, Sutter Instrument Co.) and heat-polished with a microforge (MF-83, Narishige) with electrode resistance ranged from 1.5 to 2.0 MΩ. INA was measured and analyzed as previously described (28) using standard voltage-clamp protocols with an Axopatch 200B amplifier and pClamp 9.2 software.
S-Nitrosylation Biotin Switch Assay.
The samples that were prepared in HENS buffer (250 mM HEPES/NaOH, pH 7.7/1 mM EDTA/0.1 mM neocuproine/1% SDS) first were incubated with NO donors (GSNO and NOR3) at a final concentration of 100 μM for GSNO and 30 μM for NOR3 in darkness for 20 min at room temperature. The samples were then passed through a spin column (Bio-Rad) once to remove the NO donors and then blocked with methyl methanethiosulfonate (Pierce) at a final concentration of 4 mM at 50°C. After 20 min, methyl methanethiosulfonate was removed by passing the samples through a spin column three times. The samples then were incubated with 10 mM ascorbic acid (Sigma–Aldrich) to release the NO from the sulfhydryl group and subsequently biotinylated by incubating with 0.4 mM biotin-HPDP (Pierce). The biotinylated proteins were precipitated by incubating the samples with 50 μl of neutravidin-agarose (Pierce). The neutravidin–agarose then was pelleted and washed five times with HENS buffer. The biotinylated proteins were eluted by SDS/PAGE sample buffer to be detected by anticardiac sodium channel antibody.
Statistical Analysis.
All data points are shown as the mean value, and the bars represent the SEM. Determinations of statistical significance were performed with a Student t test for comparisons of two means or with ANOVA for comparisons of multiple means. A P value of <0.05 was considered statistically significant.
Acknowledgments.
We thank research subjects from across the United States and North America for participating and Jing Wang (University of Wisconsin) for technical assistance in the myocyte experiments. This work was supported by the University of Wisconsin Cellular and Molecular Arrhythmia Research Program (J.C.M.), the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program (M.J.A.), the Established Investigator Award from the American Heart Association (to M.J.A.), and National Institutes of Health Grants HD42569 (to M.J.A.), HL71092 (to J.C.M.), and DK17238 (to G.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Lehnart SE, et al. Inherited arrhythmias: A National Heart, Lung, and Blood Institute and Office of Rare Diseases workshop consensus report about the diagnosis, phenotyping, molecular mechanisms, and therapeutic approaches for primary cardiomyopathies of gene mutations affecting ion channel function. Circulation. 2007;116:2325–2345. doi: 10.1161/CIRCULATIONAHA.107.711689. [DOI] [PubMed] [Google Scholar]
- 2.Abriel H, Kass RS. Regulation of the voltage-gated cardiac sodium channel Nav1.5 by interacting proteins. Trends Cardiovasc Med. 2005;15:35–40. doi: 10.1016/j.tcm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Vatta M, et al. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation. 2006;114:2104–2112. doi: 10.1161/CIRCULATIONAHA.106.635268. [DOI] [PubMed] [Google Scholar]
- 4.Medeiros-Domingo A, et al. SCN4B-encoded sodium channel β4 subunit in congenital long-QT syndrome. Circulation. 2007;116:134–142. doi: 10.1161/CIRCULATIONAHA.106.659086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L, et al. Mutation of an A-kinase-anchoring protein causes long-QT syndrome. Proc Natl Acad Sci USA. 2007;104:20990–20995. doi: 10.1073/pnas.0710527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nathan C, Xie QW. Nitric oxide synthases: Roles, tolls, and controls. Cell. 1994;78:915–918. doi: 10.1016/0092-8674(94)90266-6. [DOI] [PubMed] [Google Scholar]
- 7.Barouch LA, et al. Nitric oxide regulates the heart by spatial confinement of nitric oxide synthase isoforms. Nature. 2002;416:337–339. doi: 10.1038/416337a. [DOI] [PubMed] [Google Scholar]
- 8.Ahern GP, Hsu SF, Klyachko VA, Jackson MB. Induction of persistent sodium current by exogenous and endogenous nitric oxide. J Biol Chem. 2000;275:28810–28815. doi: 10.1074/jbc.M003090200. [DOI] [PubMed] [Google Scholar]
- 9.Arking DE, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 10.Aarnoudse AJ, et al. Common NOS1AP variants are associated with a prolonged QTc interval in the Rotterdam Study. Circulation. 2007;116:10–16. doi: 10.1161/CIRCULATIONAHA.106.676783. [DOI] [PubMed] [Google Scholar]
- 11.Williams JC, et al. The sarcolemmal calcium pump, α-1 syntrophin, and neuronal nitric-oxide synthase are parts of a macromolecular protein complex. J Biol Chem. 2006;281:23341–23348. doi: 10.1074/jbc.M513341200. [DOI] [PubMed] [Google Scholar]
- 12.Oceandy D, et al. Neuronal nitric oxide synthase signaling in the heart is regulated by the sarcolemmal calcium pump 4b. Circulation. 2007;115:483–492. doi: 10.1161/CIRCULATIONAHA.106.643791. [DOI] [PubMed] [Google Scholar]
- 13.Gavillet B, et al. Cardiac sodium channel Nav1.5 is regulated by a multiprotein complex composed of syntrophins and dystrophin. Circ Res. 2006;99:407–414. doi: 10.1161/01.RES.0000237466.13252.5e. [DOI] [PubMed] [Google Scholar]
- 14.Jaffrey SR, et al. Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 15.Attwell D, et al. The steady state TTX-sensitive (“window”) sodium current in cardiac Purkinje fibres. Pflügers Arch. 1979;379:137–142. doi: 10.1007/BF00586939. [DOI] [PubMed] [Google Scholar]
- 16.Bennett PB, Yazawa K, Makita N, George AL., Jr Molecular mechanism for an inherited cardiac arrhythmia. Nature. 1995;376:683–685. doi: 10.1038/376683a0. [DOI] [PubMed] [Google Scholar]
- 17.Cronk LB, et al. Novel mechanism for sudden infant death syndrome: Persistent late sodium current secondary to mutations in caveolin-3. Heart Rhythm. 2007;4:161–166. doi: 10.1016/j.hrthm.2006.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahern GP, Klyachko VA, Jackson MB. cGMP and S-nitrosylation: Two routes for modulation of neuronal excitability by NO. Trends Neurosci. 2002;25:510–517. doi: 10.1016/s0166-2236(02)02254-3. [DOI] [PubMed] [Google Scholar]
- 19.Murad F. Shattuck Lecture. Nitric oxide and cyclic GMP in cell signaling and drug development. N Engl J Med. 2006;355:2003–2011. doi: 10.1056/NEJMsa063904. [DOI] [PubMed] [Google Scholar]
- 20.Hess DT, et al. Protein S-nitrosylation: Purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 21.Nunez L, et al. Nitric oxide blocks hKv1.5 channels by S-nitrosylation and by a cyclic GMP-dependent mechanism. Cardiovasc Res. 2006;72:80–89. doi: 10.1016/j.cardiores.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 22.Evans JR, Bielefeldt K. Regulation of sodium currents through oxidation and reduction of thiol residues. Neuroscience. 2000;101:229–236. doi: 10.1016/s0306-4522(00)00367-5. [DOI] [PubMed] [Google Scholar]
- 23.Ahmmed GU, et al. Nitric oxide modulates cardiac Na+ channel via protein kinase A and protein kinase G. Circ Res. 2001;89:1005–1013. doi: 10.1161/hh2301.100801. [DOI] [PubMed] [Google Scholar]
- 24.Tester DJ, Will ML, Haglund CM, Ackerman MJ. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2005;2:507–517. doi: 10.1016/j.hrthm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 25.Sherman J, Tester DJ, Ackerman MJ. Targeted mutational analysis of ankyrin-B in 541 consecutive, unrelated patients referred for long QT syndrome genetic testing and 200 healthy subjects. Heart Rhythm. 2005;2:1218–1223. doi: 10.1016/j.hrthm.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz PJ, Moss AJ, Vincent GM, Crampton RS. Diagnostic criteria for the long QT syndrome. An update. Circulation. 1993;88:782–784. doi: 10.1161/01.cir.88.2.782. [DOI] [PubMed] [Google Scholar]
- 27.Tester DJ, Will ML, Ackerman MJ. Mutation detection in congenital long QT syndrome: Cardiac channel gene screen using PCR, dHPLC, and direct DNA sequencing. Methods Mol Med. 2006;128:181–207. doi: 10.1385/1-59745-159-2:181. [DOI] [PubMed] [Google Scholar]
- 28.Makielski JC, et al. A ubiquitous splice variant and a common polymorphism affect heterologous expression of recombinant human SCN5A heart sodium channels. Circ Res. 2003;93:821–828. doi: 10.1161/01.RES.0000096652.14509.96. [DOI] [PubMed] [Google Scholar]