Abstract
Arabinogalactan proteins (AGPs) are proteoglycans of higher plants, which are implicated in growth and development. We recently have shown that two AGPs, NaAGP1 (from Nicotiana alata styles) and PcAGP1 (from Pyrus communis cell suspension culture), are modified by the addition of a glycosylphosphatidylinositol (GPI) anchor. However, paradoxically, both AGPs were buffer soluble rather than membrane associated. We now show that pear suspension cultured cells also contain membrane-bound GPI-anchored AGPs. This GPI anchor has the minimal core oligosaccharide structure, d-Manα(1–2)-d-Manα(1–6)-d-Manα(1–4)-d-GlcN-inositol, which is consistent with those found in animals, protozoa, and yeast, but with a partial β(1–4)-galactosyl substitution of the 6-linked Man residue, and has a phosphoceramide lipid composed primarily of phytosphingosine and tetracosanoic acid. The secreted form of PcAGP1 contains a truncated GPI lacking the phosphoceramide moiety, suggesting that it is released from the membrane by the action of a phospholipase D. The implications of these findings are discussed in relation to the potential mechanisms by which GPI-anchored AGPs may be involved in signal transduction pathways.
Arabinogalactan proteins (AGPs) are a family of proteoglycans found in higher plants, occurring in many different tissues: on the plasma membrane, in the cell wall and in the extracellular matrix (for reviews see refs. 1–4). They typically contain <10% protein, which is composed mainly of proline/hydroxyproline, alanine, serine, and threonine. The major part of AGPs (>90%) consists of polysaccharide, composed mainly of β-(1–3)-galactan chains with β-(1–6)-galactosyl side chains terminated primarily with arabinosyl residues.
AGPs are implicated in many aspects of plant growth and development, including cell fate, cell proliferation, and cell expansion (5). However, because of their complexity, the function of a single AGP is yet to be defined, which has led to a major effort to determine the structure and function of individual AGPs.
During the last five years several AGP genes have been cloned (6–9). The corresponding deduced protein backbone sequences were broadly classified as “classical” and “nonclassical” (2, 3). The classical AGPs have an N-terminal secretion signal, a central proline/hydroxyproline-, alanine-, serine-, and threonine-rich region of variable length and a short C-terminal hydrophobic region. The nonclassical AGPs do not all share a common domain structure, but all have an N-terminal secretion signal, a proline/hydroxyproline-, alanine-, serine-, and threonine-rich region, and a variable, but hydrophilic, C-terminal region.
We recently have shown that in two classical AGPs, NaAGP1 (from Nicotiana alata styles) and PcAGP1 [from Pyrus communis (pear) cell suspension culture], the C-terminal hydrophobic regions are replaced by a type of lipid membrane anchor known as a glycosylphosphatidylinositol (GPI). It is likely that all of the known classical AGPs (of which there are currently more than 20) are similarly modified (10, 11), suggesting that GPI anchors may be important for their function.
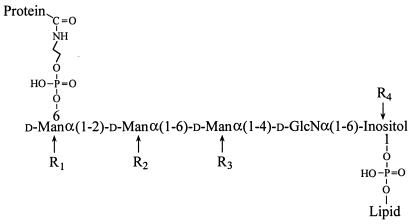
In the few GPI anchors that have been characterized to date (from animals, protozoa, and yeast; ref. 12), a conserved core oligosaccharide is linked at the nonreducing end, via ethanolamine phosphate, to the C terminus of the protein, with the reducing end linked to an inositol-phospholipid moiety (Fig. 1), which is embedded in the outer leaflet of the plasma membrane. The core oligosaccharide may be substituted at various positions by mono- or oligo-saccharides, and/or by up to two ethanolamine phosphate residues.
Figure 1.
Structure of the GPI anchors from animals, protozoa, and yeast. The GPI anchor core is conserved, but may carry additional substituents (R groups). Where present, R1 is usually α-Man, R2 is ethanolamine phosphate, R3 can be mono-, oligo-, poly-saccharides, or ethanolamine phosphate, and R4 is palmitate. The lipid can be a diacyl-, lysoacyl-, alkylacyl-, or lysoalkyl-glycerol or a ceramide (19).
In a recent report from Nothnagel and colleagues (13), the presence of a GPI anchor on AGPs from plasma membranes of Rosa cells in suspension culture was inferred from the presence of trace amounts of Man, GlcN, inositol, tetracosanoic acid, and phytosphingosine. The susceptibility of the AGPs to phospholipase C (PLC) digestion provided strong evidence that the AGPs in this cell line were indeed GPI anchored (13).
The function of GPI anchors is not fully understood other than as a means of attaching proteins to the plasma membrane. However, GPI anchors can lead to increased mobility in the plasma membrane, exclusion from clathrin-coated pits, targeting of proteins to the apical surface of polarized epithelial cells, and possible action in signal transduction pathways (ref. 12, and references therein).
Although the presence of GPI anchors on several AGPs has been shown, their detailed structures are unknown. Indeed, no plant GPI anchor structure has yet been determined. We now report the structure of the pear AGP GPI anchor, demonstrate the presence of membrane-bound GPI-anchored AGP on pear suspension cultured cells, and also show that the extracellular form of PcAGP1 contains a truncated GPI lacking the lipid moiety, suggesting that it has been released from the cell surface by the action of a phospholipase.
Materials and Methods
Plant Material.
P. communis (pear) cell suspension culture was initiated from fruit and maintained as described (8, 14).
Purification of AGPs from the Growth Medium of Suspension-Cultured Pear Cells.
Secreted AGPs were purified as described (10), except that the Yariv precipitation was replaced by anion-exchange chromatography and RP-HPLC (see below).
Purification of Membrane AGPs from Suspension-Cultured Pear Cells.
Cells from 2 liters of suspension culture (packed cell volume ≈30%) were washed once with 100 mM Tris⋅HCl, pH 7.6/10 mM EDTA/0.1% β-mercaptoethanol, then suspended in the same buffer containing 1% SDS at 90°C and stirred for 15 min. The cells then were disrupted by sonication, stirred for an additional 15 min at 90°C, then cooled and centrifuged. The pellet was washed once with the SDS-containing buffer, and cold acetone (2 vol) was added to the combined supernatants. After 30 min at 4°C the insoluble material was recovered by centrifugation, suspended in 20 mM Tris buffer containing 1% Triton X-100 and stirred for 30 min. The solubilized material was freeze-dried, washed twice with methanol, and dissolved in water. Pectin was precipitated by the addition of calcium chloride (10 mM). The supernatant was freeze-dried, then dissolved in water and precipitated with chloroform/methanol 1:1 (vol/vol; 20 vol). The precipitate was resuspended and subjected to two more precipitations with chloroform/methanol/water (10:10:1 vol/vol/vol). It then was dissolved in water, and the AGPs were precipitated by the addition of β-glucosyl Yariv reagent (15). The hydrophobic lipid-containing AGPs were separated from the hydrophilic AGPs and remaining proteins by RP-HPLC.
Quantitation of AGP.
AGP was determined by β-glucosyl Yariv reagent binding in a radial gel diffusion assay (16).
Liquid Chromatography.
Anion exchange chromatography of ethanol-precipitated material from suspension culture medium was performed on DEAE-Sepharose CL-6B (2 × 42 cm; Amersham Pharmacia) at 1 ml/min. The sample was dissolved in 20 mM imidazole⋅HCl, pH 7.0 and applied to the column, which was equilibrated in the same buffer. Bound material was eluted with a linear gradient to 2 M imidazole⋅HCl over 16 hr, collecting 10-ml fractions.
All other liquid chromatography was performed on a System Gold HPLC (Beckman) (10). Solvent A was 0.1% (vol/vol) aqueous trifluoroacetic acid (TFA) and solvent B was 0.08% (vol/vol) TFA in 80% aqueous acetonitrile.
Preparative RP-HPLC of hydrophilic AGPs was carried out on a SB-300 C8 column (9.4 × 250 mm; Zorbax, Activon, Thornleigh, Australia) and hydrophobic AGPs on a Macro-Prep methyl HIC column (5 × 50 mm; Bio-Rad). Analytical RP-HPLC was carried out on a RP-300 C8 column (2.1 × 30 mm; Brownlee Lab). In all cases, samples were applied to the column that previously was equilibrated in solvent A. AGPs were eluted with linear gradients: 0–20% solvent B in 40 min at 4 ml/min for the Zorbax column, 0–100% solvent B in 60 min at 1 ml/min for the HIC column, and 0–100% solvent B in 10 min at 0.4 ml/min for the Brownlee column.
Oligosaccharides were fractionated by gel filtration chromatography on a BioGel P2 column (10 × 300 mm; Bio-Rad) eluted with 25% solvent B at a flow rate of 0.5 ml/min, and by RP-HPLC on a Hypercarb column (3 × 100 mm; Hypersil, Cheshire, U.K.) eluted with a gradient of 30%–50% solvent B in 30 min at a flow rate 0.4 ml/min.
In all cases chromatography was monitored by absorption at 215 and 280 nm. Oligosaccharides labeled with 6-aminoquinolyl-carbamyl (AQC) (see below) were monitored by fluorescence with excitation at 260 nm and emission at 470 nm. Individual peaks were collected manually except for the fractionation of the hydrophilic AGPs where 10-ml fractions were collected and gel filtration of unlabeled oligosaccharides where 0.5-ml fractions were collected.
Deglycosylation of AGPs by Anhydrous Hydrogen Fluoride (HF).
AGPs were deglycosylated by anhydrous HF based on the method of Mort and Lamport (17) as described by Du et al. (6).
Dephosphorylation of AGPs.
AGPs (10 mg secreted or 100 μg membrane) were dephosphorylated by treatment with 40% aqueous HF at 0°C for 60 hr (18). The reagents were removed under a stream of nitrogen at 0°C and the products were fractionated on BioGel P2.
Hydrazinolysis of AGP.
Secreted AGP (2 mg) was dissolved in anhydrous hydrazine and heated at 100°C under nitrogen for 16 hr. Hydrazine was removed under reduced pressure and the products were fractionated on BioGel P2.
Nitrous Acid Deamination.
The GlcN-inositol linkage in the core GPI oligosaccharide of AGPs (200 μg) was cleaved by treatment with nitrous acid (18) and the inositol-phospholipid was extracted into dichloromethane.
Release of Ceramides.
Ceramides were released from membrane AGPs (200 μg) by treatment with 1 M sodium hydroxide at 37°C for 16 hr (19) and then extracted into dichloromethane.
Permanganate/Periodate Oxidation.
Alkene groups in isolated ceramides were oxidized with potassium permanganate and sodium periodate (20). The resulting fatty acids were extracted into dichloromethane, converted into methyl esters (see below), and analyzed by GC-MS.
Fluorescent Labeling of the Core GPI Oligosaccharides.
GPI oligosaccharides liberated by aqueous HF dephosphorylation were AQC-labeled with 6-aminoquinolyl-N-hydroxysuccinimidylcarbamate (21), then desalted on BioGel P2 and separated by RP-HPLC.
Glycosidase Digestions.
Glycosidases were assayed before use for contaminating activities by using nitrophenyl glycosides as substrates: none were detected (not shown). AQC-labeled oligosaccharides were digested overnight at 37°C with: α-mannosidase (Sigma; from Jack bean, 10 units/ml in 50 mM sodium acetate, pH 5.0), β-galactosidase (Sigma, from Aspergillus niger, 1 unit/ml in 50 mM sodium acetate, pH 4.0; Oxford Glycosciences, Rosedale, NY, from Streptococcus pneumoniae 1 unit/ml in 100 mM sodium acetate, pH 6.0; Calbiochem, from bovine testes, 5 units/ml in 100 mM sodium acetate pH 4.3), or α-galactosidase (Boehringer Mannheim, from coffee beans, in 50 mM ammonium acetate pH 6.0). The products were fractionated by RP-HPLC and analyzed by electrospray ionization (ESI)-MS.
Compositional Analyses.
Monosaccharides, fatty acids, and sphingoid bases were released by solvolysis in 1M methanolic HCl at 80°C for 16 hr. For fatty acid analysis, the products were extracted into dichloromethane and analyzed directly by GC-MS. For monosaccharide and sphingoid base analyses, the products were converted to trimethylsilyl derivatives by reaction with Tri-Sil (Pierce) at 70°C for 10 min and analyzed by GC-MS. Linkage analysis of oligosaccharides was performed as described (22). The d-configuration of GlcN was determined by GC-MS of the but-2-yl glycoside acetates (23).
GC-MS.
GC-MS was performed on a CP-SIL5 column (Chrompack, Walnut Creek, CA) or a BPX70 column (SGE, Victoria, Australia) fitted to a HP6890/5973 GC-MS (Hewlett–Packard). Data were acquired in full scan mode from 40–450 atomic mass units or selected ion mode monitoring ions at m/z 116, 132, 204, 297, 299, 312, 338, 340, and 342 for sphingoid bases.
ESI-MS.
ESI-MS was performed as described (10). Lipid samples were introduced by infusion in dichloromethane/methanol (1:1 vol/vol) and analyzed in both positive and negative ion modes.
Results
Purification of Secreted PcAGP1.
AGPs recovered from pear cell suspension culture filtrate by ethanol precipitation and partially purified by anion-exchange chromatography and ammonium sulfate precipitation were fractionated by RP-HPLC. The AGP content of each collected fraction was determined by Yariv diffusion gel assay and the protein content by absorbance at 215 nm in analytical RP-HPLC (Fig. 2). Two distinct populations of AGPs, differing in their relative protein content, were apparent. AGPs in fractions 2 and 3 had a low protein content (≈1%) whereas AGPs in fractions 6–10 had a high protein content (≈20%). AGPs in fractions 4 and 5 had intermediate values, suggesting that the two types of AGPs coeluted in these fractions. Because earlier work (ref. 8 and unpublished results) had indicated that PcAGP1 eluted early in RP-HPLC and had a low protein content, only fractions 2 and 3 were studied further. These fractions together comprised 40% of the total AGP, but only 11% of the total protein. Fractions 2 and 3 were combined and a portion deglycosylated with anhydrous HF and analyzed by ESI-MS. The spectrum (Fig. 3) showed the presence of a protein with a Mr of 9,354, which matches the calculated value for PcAGP1 with 28 of the 30 Pro residues hydroxylated, N-terminal pyroglutamate and ethanolamine on the C-terminal amino acid (Ser-97) as previously shown (10).
Figure 2.
RP-HPLC of pear-secreted AGP. Secreted AGP, which had been partially purified by anion-exchange chromatography, was fractionated by RP-HPLC and 10-ml fractions were collected. (Upper) The protein content of each fraction, determined by absorbance at 215 nm in analytical RP-HPLC, is plotted. (Lower) The AGP content of each fraction, determined by Yariv radial diffusion gel assay, is plotted. The shaded regions indicate the fractions that were pooled and used for further study.
Figure 3.
ESI-MS of deglycosylated PcAGP1. A portion of the pooled RP-HPLC fractions 2 and 3 of pear-secreted AGP (see Fig. 2) was deglycosylated with anhydrous HF and analyzed by ESI-MS. Numbers above each signal refer to the charge state (z) and the m/z value of the most intense ion. Each signal contains several ions separated by 16/z, because of heterogeneity of proline hydroxylation. The molecular weight can be calculated from each ion as (m/z)z-z.
GPI-Core Oligosaccharide of PcAGP1.
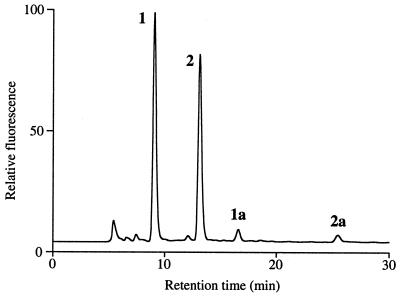
The core oligosaccharide of the GPI of PcAGP1 was selectively released by treatment with aqueous HF, which preferentially cleaves phosphoester bonds, leaving glycosidic and peptide bonds intact. The products were fractionated by gel filtration and collected fractions, which eluted after the large AGP peak at the void volume, were screened by ESI-MS for the presence of possible oligosaccharides. Two components, 1 and 2, were detected (Table 1) that had molecular weights consistent with oligosaccharides having the compositions hexose (Hex)4-hexosamine (HexN)-inositol and Hex3HexN-inositol, respectively. Fractions containing 1 and 2 were pooled, and the amino groups of the glucosamine residues were converted to the fluorescent AQC derivatives. RP-HPLC of the products (Fig. 4) revealed two major fluorescent components (54 mol% and 46 mol%), which were shown by ESI-MS (Table 1) to be the AQC derivatives of oligosaccharides 1 and 2, respectively. Two minor fluorescent oligosaccharides (≈3%), 1a and 2a, also were detected, which had molecular weights 82 atomic mass units greater than the AQC oligosaccharides, 1 and 2, respectively (Table 1). This difference corresponds to a fluorophosphate residue, which is the likely intermediate formed during aqueous HF cleavage of the phosphodiesters. This conclusion was supported by MS-MS experiments, which showed that the 82-atomic mass unit substituent was present on terminal Hex residues of the oligosaccharides (data not shown).
Table 1.
ESI-MS data and deduced compositions of the PcAGP1 GPI-anchor oligosaccharides and their derivatives
| Oligosaccharide | Molecular ion [M + H]+
|
Deduced composition | ||
|---|---|---|---|---|
| Native | AQC derivative | Per-O-methylated AQC derivative | ||
| 1 | 990.5 | 1,160.2 | 1,468.3 | Hex4HexN-inositol |
| 2 | 828.4 | 998.2 | 1,264.3 | Hex3HexN-inositol |
| 1a | ND | 1,242.2 | — | FP-Hex4HexN-inositol |
| 2a | — | 1,080.3 | — | FP-Hex3HexN-inositol |
| 1b | 1,113.6 | — | — | EtNOP-Hex4HexN-inositol |
| 2b | 951.7 | — | — | EtNOP-Hex3HexN-inositol |
ND, not determined; FP, fluorophosphate; EtNOP, ethanolamine phosphate.
Figure 4.
RP-HPLC of the AQC-labeled oligosaccharides released from pear-secreted PcAGP1 by aqueous HF treatment. Peaks 1, 2, 1a, and 1b contained oligosaccharides and were collected for further analysis.
Only mannose and myo-inositol were detected in compositional analyses of oligosaccharide 2 (AQC-GlcN is not detectable by this method). Linkage analysis (Table 2) indicated that it was a linear tetraglycosyl-inositol composed of terminal, 2- and 6-linked mannosyl residues, a 4-linked glucosaminyl residue, and a mono-substituted inositol. ESI-MS of the product of α-mannosidase treatment of oligosaccharide 2 (Table 3) showed that all three mannosyl residues had been removed, and furthermore, that they had the d-configuration. The d-configuration was established for the GlcN residue by GC-MS of the but-2-yl glycoside acetates. Thus, two structures, differing in the sequence of the two internal Man residues, are possible for oligosaccharide 2. However, analytical results obtained for oligosaccharide 1 (see below) indicate that the correct structure is d-Manα(1–2)-d-Manα(1–6)-d-Manα(1–4)-d-GlcN-inositol.
Table 2.
Linkage analyses of AQC derivatives of PcAGP1 GPI-anchor oligosaccharides
| Linkage | Molar proportion
|
|
|---|---|---|
| Oligosaccharide 1 | Oligosaccharide 2 | |
| t-inositol | 0.3* | 0.4* |
| t-Manp | 1.2 | 1.2 |
| t-Galp | 1.1 | — |
| 2-Manp | 1.0 | 1.0 |
| 6-Manp | NP | 1.0 |
| 4,6-Manp | 0.8 | — |
| 4-GlcpN | + | + |
NP, not present. +, The presence of 4-GlcN was determined in a separate methylation of N-acetylated mixed oligosaccharides 1 and 2 because AQC-GlcN did not give an observable product in GC-MS.
The low recovery of the inositol derivative is caused by the stability of the GlcN-inositol linkage.
Table 3.
ESI-MS and deduced compositions of PcAGPI AQC oligosaccharides 1 and 2 and their glycosidase digestion products
| AQC oligosaccharide | Glycosidase | [M + H]+ | Deduced composition |
|---|---|---|---|
| 1 | Untreated | 1,160.2 | Gal1Man3GlcN-inositol |
| α-Gal | 1,160.2 | Gal1Man3GlcN-inositol | |
| β-Gal | 998.2 | Man3GlcN-inositol | |
| α-Man | 836.2 | Gal1Man1GlcN-inositol | |
| α-Man/β-Gal | 674.1 | Man1GlcN-inositol | |
| β-Gal/α-Man | 512.2 | GlcN-inositol | |
| 2 | Untreated | 998.2 | Man3GlcN-inositol |
| α-Man | 512.2 | GlcN-inositol |
α-Gal, α-galactosidase; β-Gal, β-galactosidase; α-Man, α-mannosidase.
Mannose, galactose, and myo-inositol were detected in compositional analyses of oligosaccharide 1 and linkage analysis (Table 2) indicated that it was related to oligosaccharide 2, but with an additional galactosyl residue linked to O-4 of the 6-linked mannosyl residue. The galactosyl residue was resistant to hydrolysis by α-galactosidase but was removed by β-galactosidase, giving a product that had the same molecular weight as oligosaccharide 2 (Table 3), coeluted with oligosaccharide 2 on RP-HPLC, and could be completely demannosylated by α-mannosidase (Table 3). Two mannosyl residues could be removed from oligosaccharide 1 by α-mannosidase (Table 3), confirming the site of galactosylation as O-4 of the 6-linked mannosyl residue. Thus, oligosaccharide 1 has the structure d-Manα(1–2)-d-Manα(1–6)-[D-Galβ(1–4)]-d-Manα(1–4)-d-GlcN-inositol.
Because the aqueous HF treatment may have cleaved substituents such as phosphate and ethanolamine phosphate from the GPI oligosaccharides, an alternative cleavage procedure utilizing anhydrous hydrazine also was used. This reagent cleaves peptide bonds but not glycosidic or phosphate bonds. The products were fractionated by gel filtration, and fractions were screened by ESI-MS for the presence of oligosaccharides. Two oligosaccharides, 1b and 2b, were detected (Table 1) that had molecular weights 123 atomic mass units higher than oligosaccharides 1 and 2, respectively. This mass difference corresponds to a single ethanolamine phosphate residue, which would be expected because this is the linker between the GPI oligosaccharides and the C-terminal amino acid (see Fig. 1). Thus, no other ethanolamine phosphate or phosphate substituents are present on the GPI anchor oligosaccharides.
GPI Anchor of Membrane AGP.
Membrane AGPs were obtained from detergent extracts of whole cells and were separated from secreted AGPs by RP-HPLC, by virtue of their greater hydrophobicity. We refer to these as membrane AGPs, although their membrane origin cannot be unequivocally stated because they were isolated from whole cells. A portion of the membrane AGPs was dephosphorylated with aqueous HF and the GPI oligosaccharides converted to AQC derivatives. Two peaks were detected in RP-HPLC, which coeluted with AQC oligosaccharides 1 and 2, and these were present in the same proportions as in the secreted PcAGP1. Thus, we conclude that the core GPI-oligosaccharide structures of the membrane and secreted AGPs are identical. Furthermore, no oligosaccharides corresponding to those reported on “non-GPI” glycolipids (24) were detected, suggesting that these were not present in the purified AGP preparations.
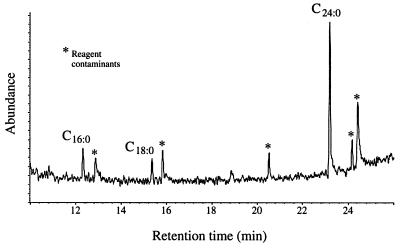
To determine the structure of the GPI-lipid moiety, we first determined the fatty acid composition of the membrane AGPs. Only one major fatty acid, tetracosanoic acid, was detected by GC-MS of the methyl esters (Fig. 5). Minor amounts of hexadecanoic and octadecanoic acids also were detected, but these were also present in similar amounts in a control sample that did not contain AGP. These are very common environmental contaminants and are invariably detected in low-level analyses of fatty acids.
Figure 5.
A GC-MS reconstituted ion chromatogram of the fatty acid methyl esters from pear GPI-anchored AGP. Peaks marked * are reagent contaminants. The peaks corresponding to hexadecanoic acid (C16:0), octadecanoic acid (C18:0), and tetracosanoic acid (C24:0) were assigned on the basis of their retention times and mass spectra by comparison with authentic standards.
The presence of tetracosanoic acid in the membrane AGP suggested that the lipid might be a ceramide. Thus, a portion of the membrane AGP was examined for the presence of sphingoid bases. Because of the difficulty of analyzing these bases at low levels (pmol) and the coelution of some sugar derivatives on GC-MS, it was not possible to identify any bases directly. However, by using selected ion monitoring for ions that are characteristic of the most common bases (sphingosine, dihydrosphingosine, and phytosphingosine) the presence of phytosphingosine was confirmed.
To provide direct evidence of a phytosphingosine-based ceramide, the membrane AGP was deaminated with nitrous acid, which cleaves the linkage between the glucosamine and inositol on the GPI oligosaccharide, releasing the inositol-phospholipid. ESI-MS of this lipid gave a molecular ion ([M-H]−) at m/z 908.6, which matches the calculated value for an inositol-phosphoceramide containing phytosphingosine and tetracosanoic acid. Other types of inositol-phospholipid (e.g., diacyl-, alkylacyl- or lysoalkyl-glycerol) were not detected, nor was there any evidence of inositol palmitoylation, but, because of the low signal/noise ratio of the spectrum, we could not rule out their presence.
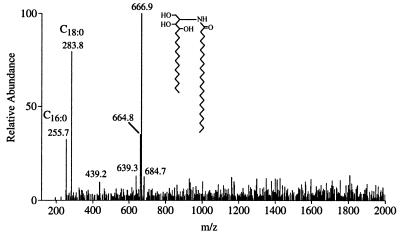
Membrane AGP also was subjected to alkaline hydrolysis under conditions that selectively cleaves the ceramide-phosphate linkage while leaving intact the amide linkage between the fatty acid and the sphingoid base (19). ESI-MS of the released ceramide (Fig. 6) showed, in addition to ions caused by free hexadecanoic and octadecanoic acid contaminants, a strong molecular ion ([M-H]−) at m/z 666.9, which matches the calculated value for a ceramide composed of phytosphingosine and tetracosanoic acid. A minor signal (≈5%) at m/z 639.3 suggests the presence of a docosanoic acid homologue. The alkaline hydrolysis also should release glycerolipids, and although ester-linked fatty acids would be lost, alkyl ethers would be stable. However, no alkylglycerol was detected, indicating the absence of lysoalkyl- and alkylacyl-glycerolipids.
Figure 6.
Negative-ion ESI-MS of the ceramide released from pear GPI-anchored AGP by base treatment. The ions at m/z 255.7 and 283.8 are the result of the presence of the contaminants hexadecanoic (C16:0) and octadecanoic (C18:0) acids, respectively.
An ion at m/z 664.9 (≈30%) was consistently observed in ESI-MS of the ceramide, suggesting the presence of an unsaturated variant. Because no unsaturated fatty acids had been detected, the alkene was probably in the sphingoid base. To determine the location of the alkene, the ceramide was oxidized with permanganate-periodate, which cleaves alkenes (and vicinal diols) generating carboxyl groups at the oxidized carbons. Decanoic acid was the major product, which would be expected from oxidation of 4-hydroxy-8-sphingenine, a common unsaturated sphingoid base in plant ceramides (20). Pentadecanoic acid, which is the expected oxidation product of phytosphingosine, also was detected although in much lower amounts than expected (≈10%). Thus, the structure of the pear AGP GPI anchor is as shown in Fig. 7.
Figure 7.
Structure of the GPI anchor from membrane pear AGP. The secreted PcAGP1 has an identical structure, but lacks the phosphoceramide substituent on the inositol residue.
Discussion
We describe here the structure of the GPI anchor of both membrane and secreted forms of AGPs from pear suspension cultured cells. The core oligosaccharide structure of the GPI anchor is heterogeneous, comprising two oligosaccharides of similar abundance. The structure of the smaller oligosaccharide [d-Manα(1–2)-d-Manα(1–6)-d-Manα(1–4)-d-GlcN-inositol] is consistent with the minimal core oligosaccharide of the animal, protozoan, and yeast GPI anchors. The larger oligosaccharide has the same core, but has a β-galactosyl substituent on O-4 of the 6-linked mannosyl residue. This is a new structure, although other glycosyl substituents have been described in other systems, e.g., a β-N-acetylgalactosaminyl substitution at the same position is present in rat Thy-1 (25) and Torpedo acetylcholinesterase (26), and an α-galactosyl substitution at O-3 of the same mannosyl residue occurs in Trypanosoma brucei variant surface glycoprotein (27). An unusual feature of the AGP GPI anchor is in the lipid moiety, which is a ceramide rather than glycero-lipid. Ceramide lipids previously have been identified in GPI anchors from Dictyostelium discoideum and Saccharomyces cerevisiae (28–30). The pear AGP ceramide is composed primarily of phytosphingosine and tetracosanoic acid, although there is evidence for the presence of 4-hydroxy-8-sphingenine (≈30%) and docosanoic acid (<5%). Recently, GPI-anchored AGPs from rose suspension cultured cells also were shown to contain tetracosanoic acid, most likely in a ceramide lipid with phytosphingosine as the sphingoid base (13).
Pear suspension cultured cells produce several secreted AGPs (8, 9), but PcAGP1 is the only one known to have a GPI anchor. None of the cDNAs encoding other pear AGPs have the C-terminal GPI- anchor signal sequence. However, as there are numerous genes for putative GPI-anchored AGPs in Arabidopsis (11), and at least two GPI-anchored AGPs are produced in the styles of Nicotiana alata (ref. 10 and unpublished results), it is possible that more than one GPI-anchored AGP is produced by pear cells. Because the GPI- oligosaccharide structures of the total GPI-anchored membrane AGPs and the purified extracellular PcAGP1 are identical, if there are several GPI-anchored AGPs produced by the pear cells, they all must have the same GPI structure.
PcAGP1 isolated from the culture filtrate of pear cell suspension cultures contains the ethanolamine phosphate-linked GPI oligosaccharide attached to the C-terminal amino acid, but has no lipid, suggesting that the AGP has been released from the plasma membrane by the action of a phospholipase. The isolation from PcAGP1, by hydrazinolysis, of GPI oligosaccharides lacking phosphate on the inositol residue implies that this enzyme is a phospholipase D (PLD), although the successive actions of a PLC and a phosphatase would give the same net result. PLDs are known in many plant species (31), although they are not GPI-specific. A soluble GPI-specific PLD is found in the serum of several mammalian species, but it does not release GPI-anchored proteins from mammalian cells when exogenously added (32). GPI-anchored proteins are released, however, from cells that express the enzyme (32). The function of this enzyme is unknown. There is also evidence that endogenous (G)PI-specific PLCs release GPI-anchored proteins in several mammalian and protozoan species (12), but this has not been demonstrated in plants, although a plant PI-PLC (from peanuts) is known (33). Additionally, Nothnagel and colleagues (13) observed “facile inherent release of AGPs from plasma membrane vesicles in vitro,” suggesting that an endogenous phospholipase is associated with plasma membranes of rose cells in suspension culture.
The secreted form of PcAGP1 is highly abundant in the culture filtrate, suggesting that it is rapidly synthesized. However, the membrane GPI-AGP is present in much smaller amounts (≈1/500 of the secreted AGP isolated from the culture filtrate), which implies that PcAGP1 resides on the plasma membrane for only a short time before being released. The rapid turnover of plasma membrane AGPs is consistent with a possible function of AGPs as carriers of specific carbohydrate structures, on the hydroxyproline-linked O-glycans, which are markers of cellular identity. Such markers would be expected to be present on the plasma membrane and there would be some mechanism to rapidly update them as required. Release of the AGP from the plasma membrane by the phospholipase, in response to intercellular or intracellular signals, could control the cell surface expression of these markers. Alternatively, the markers could be continuously updated through constant turnover of GPI-AGP without the need for external control of the phospholipase. In either case, there presumably would be a secondary function for the released AGP, perhaps as an intercellular signal, or as a structural component of the extracellular matrix.
It is also possible that the other product of the phospholipase cleavage of GPI-AGP, phospholipid, could itself act as an intracellular messenger. It is known that phospholipase cleavage of protein-free GPI (i.e., membrane GPI that is not attached to protein) can generate intracellular messengers such as phosphatidylinositol and inositol-phosphoglycan (34). Another mechanism of signaling could involve the interaction of GPI-anchored AGPs with other proteins (generally transmembrane proteins) to activate signal transduction pathways. Such a mechanism has been proposed for cell-cell communication in rat neuronal tissues (35), and plasma membrane AGPs can interact with other plasma membrane proteins as well as cell wall/extracellular matrix components (ref. 4 and references therein).
Acknowledgments
We thank Professor Adrienne E. Clarke for her support and ongoing input into these studies and for her guidance in the preparation of this manuscript. We thank Ms. Susan Mau for maintaining the pear cell culture and Dr. David McManus and Ms. Judith Webster of the Cooperative Research Centre for Industrial Plant Biopolymers for the pear suspension cell culture filtrate and the initial purification of the AGP from the culture filtrate by anion-exchange chromatography, respectively. This work was supported by funds from a Special Research Centre grant from the Australian Research Council.
Abbreviations
- AGP
arabinogalactan protein
- AQC
6-aminoquinolyl-carbamyl
- ESI
electrospray ionization
- GPI
glycosylphosphatidylinositol
- Hex
hexose
- HexN
hexosamine
- HF
hydrogen fluoride
- PLC
phospholipase C
References
- 1.Clarke A E, Anderson R C, Stone B A. Phytochemistry. 1979;18:521–540. [Google Scholar]
- 2.Bacic A, Du H, Stone B A, Clarke A E. In: Essays in Biochemistry. Apps D K, editor. Vol. 31. London: Portland Press; 1996. pp. 91–101. [PubMed] [Google Scholar]
- 3.Du H, Clarke A E, Bacic A. Plant J. 1996;9:313–323. doi: 10.1046/j.1365-313x.1996.09030313.x. [DOI] [PubMed] [Google Scholar]
- 4.Nothnagel E A. Int Rev Cytol. 1997;174:195–291. doi: 10.1016/s0074-7696(08)62118-x. [DOI] [PubMed] [Google Scholar]
- 5.Knox J P. FASEB J. 1995;9:1004–1012. doi: 10.1096/fasebj.9.11.7544308. [DOI] [PubMed] [Google Scholar]
- 6.Du H, Simpson R J, Moritz R L, Clarke A E, Bacic A. Plant Cell. 1994;6:1643–1653. doi: 10.1105/tpc.6.11.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du H, Simpson R J, Clarke A E, Bacic A. Trends Cell Biol. 1996;6:413–416. doi: 10.1016/s0962-8924(96)20036-4. [DOI] [PubMed] [Google Scholar]
- 8.Chen C-G, Pu Z-Y, Moritz R L, Simpson R J, Bacic A, Clarke A E, Mau S-L. Proc Natl Acad Sci USA. 1994;91:10305–10309. doi: 10.1073/pnas.91.22.10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mau S-L, Chen C-G, Pu Z-Y, Moritz R L, Simpson R J, Bacic A, Clarke A E. Plant J. 1995;8:269–281. doi: 10.1046/j.1365-313x.1995.08020269.x. [DOI] [PubMed] [Google Scholar]
- 10.Youl J J, Bacic A, Oxley D. Proc Natl Acad Sci USA. 1998;95:7921–7926. doi: 10.1073/pnas.95.14.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schultz C, Gilson P, Oxley D, Youl J J, Bacic A. Trends Plant Sci. 1998;3:426–431. [Google Scholar]
- 12.McConville M J, Ferguson M A J. Biochem J. 1993;294:305–324. doi: 10.1042/bj2940305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svetek J, Yadav M P, Nothnagel E A. J Biol Chem. 1999;274:14724–14733. doi: 10.1074/jbc.274.21.14724. [DOI] [PubMed] [Google Scholar]
- 14.Pech J-C, Latché A, Fallot J. Physiol Plant. 1979;46:260–264. [Google Scholar]
- 15.Gane A M, Craik D, Munro S L A, Howlett G J, Clarke A E, Bacic A. Carbohydr Res. 1995;277:67–85. doi: 10.1016/0008-6215(95)00197-2. [DOI] [PubMed] [Google Scholar]
- 16.Van Holst G-J, Clarke A E. Anal Biochem. 1985;148:446–450. doi: 10.1016/0003-2697(85)90251-9. [DOI] [PubMed] [Google Scholar]
- 17.Mort A J, Lamport D T A. Anal Biochem. 1977;82:289–309. doi: 10.1016/0003-2697(77)90165-8. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson M A J. In: Lipid Modification of Proteins. Hooper N M, Turner A J, editors. Oxford: IRL; 1992. pp. 191–230. [Google Scholar]
- 19.Smith S W, Lester R L. J Biol Chem. 1974;249:3395–3405. [PubMed] [Google Scholar]
- 20.Kaul K, Lester R L. Plant Physiol. 1975;55:120–129. doi: 10.1104/pp.55.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen S A, Michaud D P. Anal Biochem. 1993;211:279–287. doi: 10.1006/abio.1993.1270. [DOI] [PubMed] [Google Scholar]
- 22.Oxley D, Bacic A. Glycobiology. 1995;5:517–523. doi: 10.1093/glycob/5.5.517. [DOI] [PubMed] [Google Scholar]
- 23.Oxley D, Wilkinson S G. Carbohydr Res. 1991;212:213–217. doi: 10.1016/0008-6215(91)84058-m. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh T C-Y, Kaul K, Laine R A, Lester R L. Biochemistry. 1978;17:3575–3581. doi: 10.1021/bi00610a024. [DOI] [PubMed] [Google Scholar]
- 25.Homans S W, Ferguson M A J, Dwek R A, Rademacher T W, Anand R, Williams A F. Nature (London) 1988;333:269–272. doi: 10.1038/333269a0. [DOI] [PubMed] [Google Scholar]
- 26.Mehlert A, Silman I, Homans S W, Ferguson M A J. Biochem Soc Trans. 1992;21:43S. doi: 10.1042/bst021043s. [DOI] [PubMed] [Google Scholar]
- 27.Ferguson M A J, Homans S W, Dwek R A, Rademacher T W. Science. 1988;239:753–759. doi: 10.1126/science.3340856. [DOI] [PubMed] [Google Scholar]
- 28.Haynes P A, Ferguson M A J, Gooley A A, Redmond J W, Williams K L. Eur J Biochem. 1993;216:729–737. doi: 10.1111/j.1432-1033.1993.tb18192.x. [DOI] [PubMed] [Google Scholar]
- 29.Conzelman A, Puoti A, Lester R L, Desponds C. EMBO J. 1992;11:457–466. doi: 10.1002/j.1460-2075.1992.tb05075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fankhauser C, Homans S W, Thomas-Oates J E, McConville M J, Desponds C, Conzelman A, Ferguson M A J. J Biol Chem. 1993;268:26365–26374. [PubMed] [Google Scholar]
- 31.Wang X. In: Lipid Metabolism in Plants. Moore T S, editor. Boca Raton, FL: CRC; 1993. pp. 505–526. [Google Scholar]
- 32.Scallon B J, Fung W-J C, Tsang T C, Li S, Kado-Fong H, Huang K-S, Kochan J P. Science. 1991;252:446–448. doi: 10.1126/science.2017684. [DOI] [PubMed] [Google Scholar]
- 33.Butikofer P, Boschung M, Brodbeck U, Menon A K. J Biol Chem. 1996;271:15533–15541. doi: 10.1074/jbc.271.26.15533. [DOI] [PubMed] [Google Scholar]
- 34.Peles E, Nativ M, Lustig M, Grumet M, Schilling J, Martinez R, Plowman G D, Schlessinger J. EMBO J. 1997;16:978–988. doi: 10.1093/emboj/16.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munnik T, Irvine R F, Musgrave A. Biochim Biophys Acta. 1998;1389:222–272. doi: 10.1016/s0005-2760(97)00158-6. [DOI] [PubMed] [Google Scholar]