Abstract
Signaling mucins are cell adhesion molecules that activate RAS/RHO guanosine triphosphatases and their effector mitogen-activated protein kinase (MAPK) pathways. We found that the Saccharomyces cerevisiae mucin Msb2p, which functions at the head of the Cdc42p-dependent MAPK pathway that controls filamentous growth, is processed into secreted and cell-associated forms. Cleavage of the extracellular inhibitory domain of Msb2p by the aspartyl protease Yps1p generated the active form of the protein by a mechanism incorporating cellular nutritional status. Activated Msb2p functioned through the tetraspan protein Sho1p to induce MAPK activation as well as cell polarization, which involved the Cdc42p guanine nucleotide exchange factor Cdc24p. We postulate that cleavage-dependent activation is a general feature of signaling mucins, which brings to light a novel regulatory aspect of this class of signaling adhesion molecule.
Introduction
Signal transduction pathways control multiple aspects of cellular behavior, including regulating the connectivity between cells and their adhesion to external surfaces. Among the signaling proteins that regulate cell adhesion are members of the mucin family of proteins. Signaling mucins are cell-surface proteins composed of a heavily glycosylated extracellular domain that contains a Ser/Thr/Pro-rich mucin homology domain (MHD; Silverman et al., 2003), which is connected to a cytoplasmic domain that regulates RAS/RHO GTPases and their effector MAPK pathways (Carraway et al., 2007). Proteolytic processing separates the domains and results in the shedding of the extracellular glycodomain from the cell (Singh and Hollingsworth, 2006). Signaling mucins also contribute to the regulation of other pathways including the Wnt, nuclear factor-κβ, and estrogen receptor pathways (Huang et al., 2003; Wei et al., 2006).
In Saccharomyces cerevisiae, the signaling mucin Msb2p regulates MAPK activity by interaction with the RHO GTPase Cdc42p (Cullen et al., 2004), a global regulator of cell signaling and polarity (Johnson, 1999). Cdc42p and its effector kinase Ste20p regulate multiple MAPK pathways including the filamentous growth (Peter et al., 1996; Leberer et al., 1997), pheromone response (Simon et al., 1995), and high osmolarity glycerol response (HOG) pathways (Tatebayashi et al., 2006). Differential MAPK activation is regulated in part by pathway-specific MAPKs (Madhani et al., 1997) and recruitment of core signaling modules to pathway-dedicated receptor complexes by scaffolding proteins (Butty et al., 1998; Harris et al., 2001). Msb2p is required for activation of the filamentous growth pathway (Cullen et al., 2004) and may function in a minor way in the HOG pathway (O'Rourke and Herskowitz, 2002). Indeed, Msb2p associates with the tetraspan protein Sho1p (Cullen et al., 2004), which operates in both the filamentous growth and HOG pathways (Maeda et al., 1995; Cullen et al., 2004; Zarrinpar et al., 2004). Although it is unclear how Msb2p becomes activated, loss of the MHD induces the signaling function of the protein, which suggests an inhibitory role for this domain in MAPK signaling (Cullen et al., 2004).
In the present study, we show that Msb2p, like its mammalian counterparts, is processed into secreted and cell-associated forms. This discovery, coupled with the fact the secreted domain of Msb2p is inhibitory, suggests a mechanism where processing and release of the extracellular domain activates the protein. We confirmed this hypothesis and identified an aspartyl protease, Yps1p, that is required for Msb2p processing. Intriguingly, the gene encoding Yps1p is induced in response to nutrient limitation, which provides a straightforward mechanism for regulated processing of Msb2p. We speculate that cleavage-dependent activation may be a general feature of mucin receptors.
Results and discussion
Msb2p is processed into distinct polypeptides
The extracellular portion of the Msb2p protein was detected in supernatants by Western blotting, which indicates that it is secreted from the cell. To determine if secretion results from processing in the extracellular domain, a version of the protein was examined in which the extracellular (HA, at 500 residues) and cytoplasmic (GFP, at 1,306 residues) domains were differentially tagged. Conditioned media derived from cells expressing HA-Msb2p-GFP showed that the extracellular but not cytoplasmic domain was secreted (Fig. 1 A). This result confirms topological and subcellular fractionation data that indicate that Msb2p is a membrane-spanning protein (Cullen et al., 2004). Although the extracellular domain of Msb2p migrated as a single heavily glycosylated product at >250 kD, the cytoplasmic domain migrated as multiple intermediate forms (Fig. 1 A), which indicates that it is unstable. Consistent with this result, the extracellular portion of Msb2p tagged with GFP was found evenly distributed throughout the cell (Fig. 1 B), whereas the cytoplasmic domain tagged with GFP was predominately localized to vacuoles (Fig. 1 B). These results indicate that Msb2p is processed into multiple polypeptides with different stabilities and localization patterns.
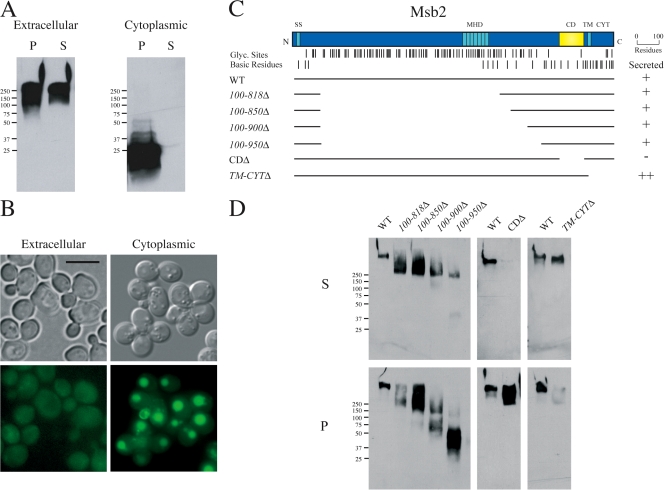
Figure 1.
Msb2p is processed in the cleavage domain and the extracellular portion of the protein is secreted. (A) Western blot of supernatant (S) and pellet (P) fractions derived from cells containing HA-Msb2p-GFP. (left) Anti-HA antibodies. (right) Anti-GFP antibodies. The band at 25 kD migrates at the predicted size of GFP. (B) Localization of Msb2p containing GFP inserted into the extracellular and cytoplasmic domains. (top) Differential interference contrast. (bottom) FITC. Bar, 5 μm. (C) Deletion analysis of MSB2. Top lines, potential glycosylation sites; bottom lines, basic residues. (D) Western blot of S and P fractions derived from cells expressing HA-Msb2p deletions. Molecular mass standards (kD) are indicated on the left of the gel blots.
Deletion analysis of the MSB2 ORF was performed to identify the region of the protein required for secretion (Fig. 1 C). Versions of Msb2p containing deletions in the extracellular domain (100-818Δ, 100–850Δ, and 100-900Δ) were secreted (Fig. 1, C and D), as was the version 100-950Δ, although to a lesser extent (Fig. 1, C and D). Additional deletion analysis was performed using HA-Msb2p as the template because epitope fusions in the cytoplasmic domain were unstable and difficult to detect. An extracellular region of the protein adjacent to the transmembrane (TM) domain was identified that was required for secretion (cleavage domain [CD] 1,045–1,145 residues; Fig. 1, C and D). The CD overlaps with the predicted cleavage site by Western blotting (the 55-kD product in Fig. 1 A) and resides between the glycosylation-rich extracellular domain and basic C terminus (Fig. 1 C). A version of Msb2p lacking the TM and cytoplasmic domains was secreted at higher levels (TM-CYTΔ; Fig. 1, C and D), which is consistent with the idea that the TM domain anchors Msb2p to the plasma membrane.
The aspartyl protease Yps1p processes Msb2p
To identify the protease required for Msb2p processing, protein processing mutants from the ordered deletion collection (Giaever et al., 2002) were examined for defects in secretion of HA-Msb2p. One mutant stood out as partially defective (yps1Δ; Table S3, available at http://www.jcb.org/cgi/content/full/jcb.200704079/DC1). Yps1p is a member of a family of glycosylphosphatidylinositol (GPI)-anchored aspartyl proteases called yapsins (Yps1p, Mkc7/Yps2p, Yps3p, Yps6p, and Yps7p; Krysan et al., 2005). HA-Msb2p was poorly secreted in the yps1Δ mutant, more so than other yps single mutants (Fig. 2 A). Combinations of yps mutants showed combinatorial defects with yps1Δ including yps1Δ mkc7Δ, yps1Δ yps3Δ, and the quintuple mutant 5ypsΔ (Fig. 2 A), which indicates a minor role for other Yps proteins in Msb2p processing. Introduction of YPS1 on a centromere-based plasmid rescued the majority of the secretion defect of HA-Msb2p in the 5ypsΔ strain (Fig. S1 A), confirming that Yps1p is the primary Yps protease required for Msb2p processing.
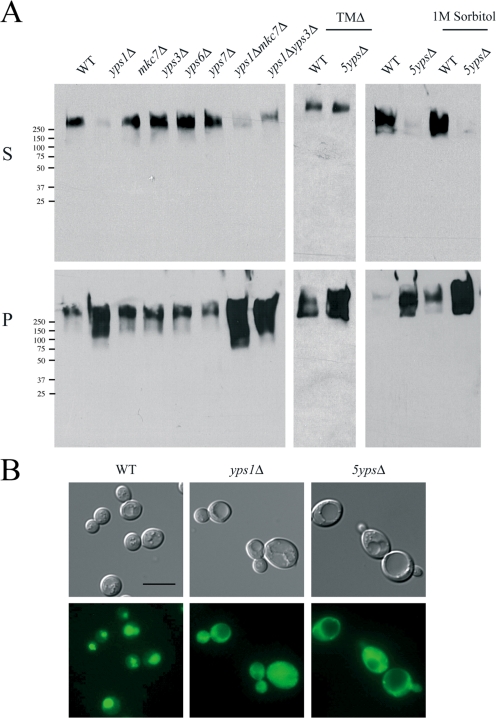
Figure 2.
The role of yapsin proteases in Msb2p processing. (A) Western blot analysis of the secretion defect of HA-Msb2p in yps mutants. Molecular mass standards (kD) are indicated on the left of the gel blots. P, pellet; S, supernatant. (B) Msb2p-GFP localization in WT, yps1Δ, and 5ypsΔ strains. Bar, 10 μm.
Yps1p has been shown to process heterologously expressed glycoprotein substrates by cleavage in the extracellular domain at monobasic residues (Bourbonnais et al., 2000). By these criteria, Msb2p is a likely substrate of Yps1p and the first physiological target to be identified. Several lines of evidence support this possibility. First, yps mutants are not generally defective in protein secretion, and several proteins have been examined that are secreted in yps mutants to wild type (WT) or higher levels (unpublished data). The Msb2p protein was delivered to the cell surface in yps mutants based on the fact that HA-Msb2p lacking its TM domain was secreted in the 5ypsΔ mutant (Fig. 2 A). Intriguingly, Msb2p-GFP was absent from vacuoles in yps mutants (Fig. 2 B). Several other cell-surface proteins showed a similar localization pattern (unpublished data), which suggests a general requirement for Yps proteins in delivery of a subset of cell-surface proteins to vacuoles. Second, although yps mutants have defects in cell integrity and cell wall biosynthesis (Krysan et al., 2005), secretion of HA-Msb2p was not restored by conditional rescue of the cell integrity defect of the 5ypsΔ mutant grown in 1 M sorbitol (Fig. 2 A). Likewise, cell-wall mutant combinations defective for β-1,3 and β-1,6-glucan synthesis that mimic yps phenotypes showed elevated secretion of HA-Msb2p (Fig. S1 B). Third, the yps1Δ mutant was defective in processing Msb2p-TAP, resulting in accumulation of the full-length protein and loss of a processing intermediate (Fig. S1 C).
Cleavage of Msb2p is required for MAPK activation and is stimulated by nutrient limitation–dependent induction of YPS1 expression
Msb2p lacking the MHD hyperactivates the filamentous growth pathway (Cullen et al., 2004). Versions of Msb2p lacking larger portions of the extracellular domain were also hyperactive, more so than Msb2pΔMHD (Fig. 3 A), which confirms that the extracellular domain (from 100 to 950 residues) functions in an inhibitory capacity. Cleavage of the inhibitory domain may provide an explanation for how the protein becomes activated. In support of this possibility, the noncleavable version of Msb2p, Msb2pΔCD, was defective for MAPK activity (Fig. 3 A) and filamentous growth (not depicted). Versions of Msb2p lacking the TM and CYT domains were similarly defective for signaling (Fig. 3 A).
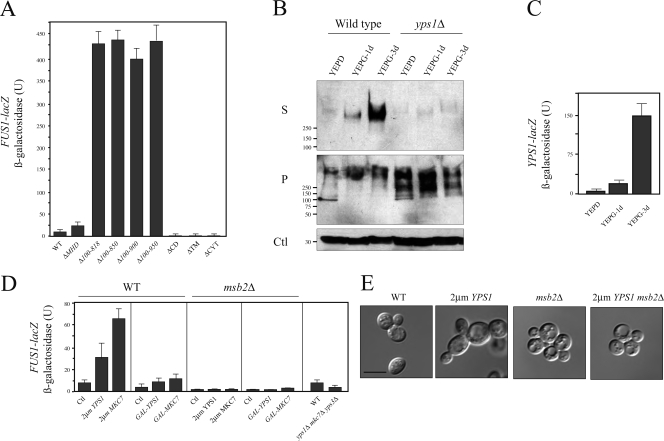
Figure 3.
Cleavage of Msb2p induces MAPK signaling and is regulated at the level of YPS1 expression. (A) FUS1-lacZ expression in strains containing Msb2p deletions. (B) Western blot of HA-Msb2p in WT and yps1Δ strains grown in mid-log phase yeast extract peptone dextrose (YEPD) or YEP-GAL (YEPG) medium for 1 and 3 d. Dpm1p was used as a loading control (Ctl). Molecular mass standards (kD) are indicated on the left of the gel blots. (C) YPS1-lacZ expression in cells incubated in mid-log phase in SCD (Glu) or SC-Gal (Gal) medium for 1and 3 d. (D) FUS1-lacZ expression in indicated strains overexpressing YPS1 and MKC7. Error bars represent the standard deviation of the values derived from at least two separate experiments. (E) Cell morphology in strains containing high copy (2 μm) YPS1. Bar, 5 μm.
To determine if Msb2p processing is regulated, secretion of Msb2p was examined in cells incubated under different conditions, particularly nutrient limitation, a known inducer of filamentous growth (Cullen and Sprague, 2000). Secretion of HA-Msb2p was induced >10-fold by nutrient limitation, and the enhanced secretion was Yps1p dependent (Fig. 3 B). Unprocessed Msb2p accumulated in the yps1Δ mutant under this condition (Fig. 3 B). To determine how Yps1p regulates Msb2p processing in a nutrient-dependent manner, we turned to the expression of the YPS1 gene, which is highly regulated (Krysan et al., 2005). Expression of YPS1 was induced under nutrient-limiting conditions that promote filamentous growth (Fig. 3 C). Overexpression of YPS1 or MKC7/YPS2 from a high-copy plasmid or inducible promoter stimulated Msb2p-dependent MAPK activity (Fig. 3 D) and polarized growth (Fig. 3 E and not depicted). Cells lacking Yps1p were partially defective for MAPK activity, and this effect was exacerbated in cells lacking Mkc7p and Yps3p (Fig. 3 D). Therefore, Msb2p processing is regulated by starvation-dependent induction of its cognate protease. Cleavage by Yps1p may trigger Msb2p activation or be a prerequisite for activation.
Activated Msb2p functions through Sho1p
Msb2p interacts with the tetraspan protein Sho1p (Cullen et al., 2004). The C-terminal domain of Msb2p was sufficient to recapitulate the Msb2p–Sho1p interaction (Fig. 4 A). To better understand the role of Sho1p in Msb2p-dependent signaling, alleles of SHO1 were isolated that hyperactivate the filamentous growth pathway. A dominant gain-of-function allele was identified by random mutagenesis that encodes a P120L change in the short DGPKK linker of the second extracellular loop of the protein (Fig. 4 B). The mutation at P120L did not affect Sho1p protein levels (Fig. 4 B) but stimulated the filamentous growth pathway based on hyperpolarized growth of haploid and diploid cells (Fig. 4 C), agar invasion phenotypes (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200704079/DC1), and induction of filamentous growth pathway reporters (Fig. S2 B). Sho1pP120L functioned normally in the HOG pathway (Fig. S2, C–F). Other alleles of SHO1 that differentially influence its function between pathways map to the cytoplasmic SH3 domain and affect the Sho1p–Pbs2p interaction (Marles et al., 2004; Zarrinpar et al., 2004; Tatebayashi et al., 2006). The P120L mutation maps to the tetraspan domain that is thought to be dispensable for Sho1p function in the HOG pathway (Raitt et al., 2000) and may identify a region of the protein that specifically regulates Sho1p function in the filamentous growth pathway.
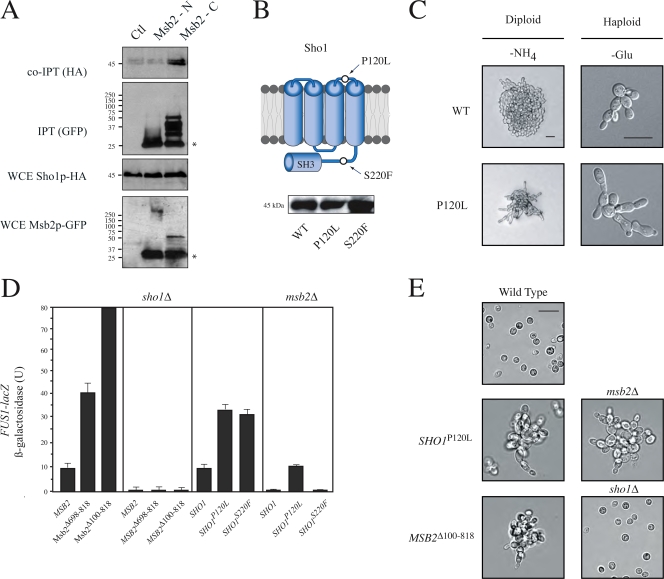
Figure 4.
Msb2p induces MAPK activation through Sho1p. (A) Co-IPT analysis of Sho1p-HA with N- and C-terminally tagged Msb2p-GFP. IPT, immunoprecipitation; WCE, whole cell extract. Proteins were tagged and precipitated with the indicated epitopes. The asterisks indicate the bands that correspond to GFP. Molecular mass standards (kD) are indicated on the left of the gel blots. (B) Alleles of SHO1 (P120L and S220F) that hyperactivate the filamentous growth pathway. (bottom) Western blot of Sho1p-HA protein levels. (C) Sho1pP120L induces hyperpolarized growth in haploid (in SC-Glu) and diploid cells (in SLAHD). (D) FUS1-lacZ expression in strains containing alleles of SHO1 and MSB2. Error bars represent the standard deviation of the values derived from at least two separate experiments. (E) Cell morphology induced by Sho1pP120L and Msb2p-HAΔ100-818 in the indicated strains grown in SCD medium. Bars: (C) 5 μm; (E) 20 μm.
Gain-of-function alleles of MSB2 and SHO1 were used to explore the functional relationships between the cell-surface proteins. Activated versions of Msb2p were dependent on Sho1p for MAPK signaling (Fig. 4 D) and hyperpolarized growth (Fig. 4 E). In contrast, Sho1pP120L partially induced MAPK signaling (Fig. 4 D) and hyperpolarized growth (Fig. 4 E) in the absence of Msb2p. Sho1pP120L also restored MAPK activity to the Msb2pΔCD strain and the yps1Δ mkc2Δ yps3Δ mutant (not depicted). This property reflects a change in the activation state of Sho1p, as another gain-of-function allele that was present at higher levels (S220F; Fig. 4 B), or overexpression of SHO1, failed to bypass msb2Δ (Fig. 4 D, shown for S220F). These results indicate that MAPK signaling is initiated by Msb2p and propagated through Sho1p. This conclusion may oversimplify the relationship between the proteins as they show some degree of interdependence (Fig. 4 D, SHO1P120L in WT compared with msb2Δ) but is nevertheless consistent with the idea that cleavage of Msb2p is an early or initial event in MAPK activation.
Sho1p contributes to guanine nucleotide exchange factor (GEF) polarization by multiple mechanisms
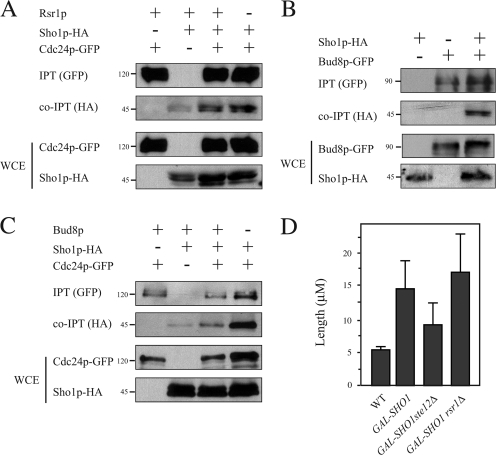
Msb2p interacts with the GTPase Cdc42p (Cullen et al., 2004) and has genetic interactions with the GEF Cdc24p (Bender and Pringle, 1992). CDC24 showed genetic interactions with SHO1 and other filamentous growth pathway components (Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200704079/DC1). The Cdc24p protein also associated with Sho1p (Fig. 5 A); an interaction with Msb2p was not detected (not depicted). Given the complex associations between receptors and GEFs in other contexts (Nern and Arkowitz, 1999), it is possible that the Sho1p–Cdc24p interaction is mediated by other proteins. We also tested for an interaction between Sho1p and the positional marker Bud8p (Harkins et al., 2001), which is required for unipolar budding during filamentous growth (Taheri et al., 2000; Cullen and Sprague, 2002) and for GEF recruitment during vegetative growth by the bud-site selection GTPase Rsr1p (Shimada et al., 2004). Sho1p and Bud8p were found to associate (Fig. 5 B). These results are consistent with the idea that Sho1p contributes to Bud8p-dependent recruitment of Cdc24p by a mechanism that requires MAPK activation. Sho1p and Cdc24p might interact by affiliation with bud-site selection proteins; however, the Sho1p–Cdc24p interaction was independent of Rsr1p (Fig. 5 A) and Bud8p (Fig. 5 C). Hyperactive alleles of SHO1, or its overexpression, induced hyperpolarized growth in a manner that was partially independent of bud-site selection cues and the MAPK pathway (Fig. 5 D). These results indicate that Sho1p may also have a function in stabilization of Cdc24p at polarized sites.
Figure 5.
Sho1 interacts with Cdc24p and Bud8p and contributes to Cdc24p polarization by multiple mechanisms. (A) Sho1p and Cdc24p interact by co-IPT analysis. Proteins were tagged and precipitated with the indicated epitopes. (B) Sho1p and Bud8p interact by co-IPT analysis. (C) The interaction between Sho1p and Cdc24p is independent of Bud8p. Molecular mass standards (kD) are indicated on the left of the gel blots. (D) The length of cells overexpressing SHO1 in WT, ste12Δ, and rsr1Δ strains compared with a WT control. More than 50 cells were examined for each strain. Error bars represent the standard deviation of the values derived from at least two separate experiments.
Summary
We have identified a novel MAPK activation mechanism in yeast by processing and release of the inhibitory domain of Msb2p. Cleavage-dependent activation has been found for Notch (Schweisguth, 2004) and PAR receptors (Barry et al., 2006) and may extend to other members of the signaling mucin family. We have also identified a glycosylphosphatidylinositol (GPI)-anchored aspartyl protease that regulates Msb2p function. In mammalian cells, secreted aspartyl proteases are coregulated with signaling mucins (Liaudet-Coopman et al., 2006), which suggests the possibility that aspartyl proteases may be general regulators of signaling mucin receptors.
Materials and methods
Strains, plasmids, and microbiological techniques
Yeast strains are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200704079/DC1) and plasmids in Table S2. β-galactosidase assays were performed as described and represent at least two independent trials (Cullen et al., 2000). FUS1-HIS3 expression was used to confirm FUS1-lacZ reporter data and was measured by spotting equal amounts of cells onto synthetic medium lacking histidine and containing 4-amino-1,2,4-triazole. Unless otherwise indicated, the locations of the epitope tags are at 500 residues for Msb2p and at 367 residues for Sho1p. HA-Msb2p-GFP contains a GFP epitope at position 1,306 residues. All fusion proteins were functional with respect to filamentation phenotypes and MAPK reporter activity. Other functional C-terminal Msb2p fusions show similar processing intermediates as Msb2p-GFP.
Yeast and bacterial strains were manipulated by standard methods (Sambrook et al., 1989; Rose et al., 1990). The budding pattern was based on established methodology (Chant and Pringle, 1995) and by visual inspection as described previously (Cullen and Sprague, 2002) using the single cell invasive growth assay (Cullen and Sprague, 2000). The plate-washing assay (Roberts and Fink, 1994) and pseudohyphal growth assay (Gimeno et al., 1992) were performed as described previously.
The plasmid containing the MSB2 gene (pMSB2; PC1287) was identified in a genetic screen for high-copy suppressors of the signaling defect of an msb2Δ mutant (PC948) using a galactose-inducible plasmid library (Ramer et al., 1992). The MSB2 gene was expressed in the opposite orientation with respect to the GAL promoter and was GAL independent. Plasmid pHA-MSB2 (PC1456) was created by GAP repair using homologous recombination of plasmid pMSB2 in strain PC999. pMSB2-GFP was constructed by PCR amplification and insertion of the GFP cassette into pMSB2 at position 400 residues and was constructed by A. Mirando (University of Rochester Medical Center, Rochester, NY). Plasmids containing filamentous growth pathway targets fused to lacZ and plasmid V84 were provided by C. Boone (University of Toronto, Ontario, Canada; Roberts et al., 2000). Plasmid pIL30-URA3 containing FgTy-lacZ was provided by B. Errede (University of North Carolina, Chapel Hill, NC; Laloux et al., 1994), and pFRE-lacZ was provided by H. Madhani (University of California, San Francisco, San Francisco, CA; Madhani et al., 1997). Other plasmids containing versions of Sho1p were provided by H. Saito (University of Tokyo, Tokyo, Japan) and A. Davidson (University of Toronto). The HA fusions at the MSB2 locus were made at 500 residues or other regions of the Msb2p protein as indicated (Schneider et al., 1995). Gene disruptions and GAL1 promoter fusions were made by PCR-based methods (Baudin et al., 1993; Longtine et al., 1998), including the use of antibiotic resistant markers (Goldstein and McCusker, 1999). Integrations were confirmed by PCR analysis and phenotype.
Protein analysis
Secreted proteins were analyzed from conditioned media that was passed through a 2-μm filter to remove cells. Western blots were performed as described previously (Cullen et al., 2004). Monoclonal mouse anti-HA antibodies (12CA5.16.4), monoclonal goat anti-GFP antibodies (Rockland Scientific International), and monoclonal antibodies to Dpm1p were used.
Coimmunoprecipitation (co-IPT) analysis
To examine the interaction between Sho1p and Cdc24p, co-IPT analysis was performed on extracts derived from PC1652. To measure the interaction in a strain lacking Rsr1p, co-IPT analysis was performed from extracts derived from PC2281. For the Sho1p–Bud8 interaction, PC2249 was transformed with pBUD8-GFP (PC1883) and a control plasmid.
Co-IPT analysis was based on previous methodology (Kemp and Sprague, 2003). 100 ml of exponentially grown cells were pelleted at room temperature for 5 min at 3,000 g. Cells were resuspended in 4 ml of DTT buffer (50 mM Tris-HCl, pH 7.5, and 10 mM DTT) and incubated at room temperature for 15 min. The cells were pelleted at room temperature for 5 min at 15,800 g, and pellets were resuspended in 3.75 ml of spheroplast buffer (1.2 M sorbitol, 50 mM potassium phosphate, pH 7.4, 1 mM MgCl, and 250 μg/ml of zymolyase) and incubated in 30°C on an end-to-end rotator for 1 h. Spheroplasts were centrifuged for 2 min at 2,300 g. Cell pellets were washed in 1 ml of 1.2 M sorbitol and centrifuged for 4 min at low speed as before. Spheroplasts were lysed in 1 ml of ice-cold IPT buffer (50 mM Tris-HCl, pH 8, 1 mM EDTA, 50 mM NaCl, and 1.5% Igepal CA-630) supplemented with 1× protease inhibitor cocktail, EDTA free, and 1 mM PMSF before use. Lysates were centrifuged at 4°C for 20 min at 15,800 g. Supernatants were precleared by incubating with 75 μl of immobilized Protein G plus (Thermo Fisher Scientific) on an end-to-end rotator for 30 min. Beads were pelleted at 4°C for 10 min at 15,800 g. The protein concentration of precleared supernatants was determined by the Bradford Coomassie blue assay. 5–7 mg of proteins were immunoprecipitated with a polyclonal antibody in 1:75 dilution for 1 h, and complexes were pulled down by adding 75 μl of protein G and incubating for another 1 h. IPT complexes were pelleted at 15,800 g for 1 min. Beads were washed in IPT buffer three times for 1 min each time. 50 μl of Thorner buffer (8 M urea, 5% SDS, 40 mM Tris-HCl, pH 6.8, 0.1 M EDTA, 0.4 mg/ml Bromophenol blue, and 1% β-mercaptoethanol) was added to beads, and IPT complexes were released from beads by boiling for 10 min. Beads were removed by centrifuging for 1 min at 15,800 g, after which the pellet was discarded. Supernatants (protein complexes) were either immediately processed by SDS-PAGE or frozen at −80°C.
Microscopy
Differential interference contrast and fluorescence microscopy of the GFP protein using FITC filter sets were performed using an fluorescent microscope (Axioplan 2; Carl Zeiss, Inc.) with a Plan-Apochromat 100×/1.4 (oil) objective (NA 0.17; Carl Zeiss, Inc.). For most experiments, cells were visualized by resuspending in water at 25°C. Digital images were obtained with a camera (Axiocam MRm; Carl Zeiss, Inc.). Axiovision 4.4 software (Carl Zeiss, Inc.) was used for image acquisition and analysis. Images were further analyzed in Photoshop (Adobe), where adjustments of brightness and contrast were made.
SHO1 mutagenesis
Mutagenesis of the SHO1 gene was performed by plasmid mutagenesis based on a protocol for direct transformation into yeast (Rose and Fink, 1987). Efficiency was 5% as determined by the frequency of sho1 null mutations. An allele of SHO1 was used (S171C and NF300GSA; Marles et al., 2004) that fully complemented the sho1Δ mutant. Hyperactive alleles were selected on media lacking histidine and containing 4-amino-1,2,4-triazole using the FUS1-HIS3 reporter (PC1531) integrated into ste4Δ Σ1278b strains (Cullen et al., 2004). 283,680 URA3+ colonies were screened and ∼100 ATA resistant isolates were identified, of which six were plasmid dependent; five of the six resulted in the P120L change and were identified in separate pools. Plasmid-dependent mutations were analyzed by sequence analysis at the Roswell Park sequencing facility.
Western blots and protein analysis
Proteins were separated by SDS-PAGE on 10% precast gels (Bio-Rad Laboratories) and transferred to nitrocellulose membranes (protran BA85, VWR International, LLC). Membranes were incubated in 10 ml of blocking buffer (5% nonfat dry milk, 10 mM Tris-HCl, pH 8, 150 mM NaCl, and 0.05% Tween 20) either for 1 h at 25°C or 16 h at 4°C. Preblocked membranes were incubated in blocking buffer containing primary antibodies for 1 h at 25°C. Blots were washed three times for 5 min each in TBST (10 mM Tris-HCl, pH 8, 150 mM NaCl, and 0.05% Tween 20). Blots were incubated in HRP-conjugated secondary antibodies for 1 h at room temperature. ECL Plus Western blotting reagent (GE Healthcare) was used to detect proteins. Predicted N- and O-glycosylation sites were determined by O-GLYCBASE version 2.0 (Center for Biological Sequence Analysis, Technical University of Denmark; Hansen et al., 1997). The secondary structure of Msb2p domains was determined by PROF and NORS algorithms (Rost et al., 2004).
Online supplemental material
Fig. S1 shows the secretion defect of Msb2p in yps and cell wall mutant combinations. Fig. S2 shows analysis of the SHO1P120L allele. Fig. S3 shows genetic data between filamentous growth pathway components and Cdc24p. Table S1 lists yeast strains. Table S2 lists plasmids used in this study. Table S3 shows analysis of secretion of the Msb2p-HA protein in protease mutants. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200704079/DC1.
Supplementary Material
Acknowledgments
We thank B. Errede, H. Saito, A. Davidson, C. Boone, and H. Madhani for reagents. We also give thanks to B. Andrews, E. Bi, J. Berry, J. Thorner, J. Pringle, P. Pryciak, D. Lew, and R. Fuller for helpful discussions.
P.J. Cullen is supported by grants from the National Institutes of Health (1R03DE018425-01), American Cancer Society (TBE-114083), and the American Heart Association (GM 0535393T). D.J. Krysan is supported by a National Institutes of Health grant (K08AI062978).
Abbreviations used in this paper: CD, cleavage domain; co-IPT, coimmunoprecipitation; GEF, guanine nucleotide exchange factor; HOG, high osmolarity glycerol response; MHD, mucin homology domain; TM, transmembrane; WT, wild type.
References
- Barry, G.D., G.T. Le, and D.P. Fairlie. 2006. Agonists and antagonists of protease activated receptors (PARs). Curr. Med. Chem. 13:243–265. [DOI] [PubMed] [Google Scholar]
- Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, A., and J.R. Pringle. 1992. A Ser/Thr-rich multicopy suppressor of a cdc24 bud emergence defect. Yeast. 8:315–323. [DOI] [PubMed] [Google Scholar]
- Bourbonnais, Y., C. Larouche, and G.M. Tremblay. 2000. Production of full-length human pre-elafin, an elastase specific inhibitor, from yeast requires the absence of a functional yapsin 1 (Yps1p) endoprotease. Protein Expr. Purif. 20:485–491. [DOI] [PubMed] [Google Scholar]
- Butty, A.C., P.M. Pryciak, L.S. Huang, I. Herskowitz, and M. Peter. 1998. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science. 282:1511–1516. [DOI] [PubMed] [Google Scholar]
- Carraway, K.L. III, M. Funes, H.C. Workman, and C. Sweeney. 2007. Contribution of membrane mucins to tumor progression through modulation of cellular growth signaling pathways. Curr. Top. Dev. Biol. 78:1–22. [DOI] [PubMed] [Google Scholar]
- Chant, J., and J.R. Pringle. 1995. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129:751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, P.J., and G.F. Sprague Jr. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA. 97:13619–13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, P.J., and G.F. Sprague Jr. 2002. The roles of bud-site-selection proteins during haploid invasive growth in yeast. Mol. Biol. Cell. 13:2990–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, P.J., J. Schultz, J. Horecka, B.J. Stevenson, Y. Jigami, and G.F. Sprague Jr. 2000. Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics. 155:1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, P.J., W. Sabbagh Jr., E. Graham, M.M. Irick, E.K. van Olden, C. Neal, J. Delrow, L. Bardwell, and G.F. Sprague Jr. 2004. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 18:1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever, G., A.M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 418:387–391. [DOI] [PubMed] [Google Scholar]
- Gimeno, C.J., P.O. Ljungdahl, C.A. Styles, and G.R. Fink. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 68:1077–1090. [DOI] [PubMed] [Google Scholar]
- Goldstein, A.L., and J.H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 15:1541–1553. [DOI] [PubMed] [Google Scholar]
- Hansen, J.E., O. Lund, K. Rapacki, and S. Brunak. 1997. O-GLYCBASE version 2.0: a revised database of O-glycosylated proteins. Nucleic Acids Res. 25:278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkins, H.A., N. Page, L.R. Schenkman, C. De Virgilio, S. Shaw, H. Bussey, and J.R. Pringle. 2001. Bud8p and Bud9p, proteins that may mark the sites for bipolar budding in yeast. Mol. Biol. Cell. 12:2497–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, K., R.E. Lamson, B. Nelson, T.R. Hughes, M.J. Marton, C.J. Roberts, C. Boone, and P.M. Pryciak. 2001. Role of scaffolds in MAP kinase pathway specificity revealed by custom design of pathway-dedicated signaling proteins. Curr. Biol. 11:1815–1824. [PubMed] [Google Scholar]
- Huang, L., J. Ren, D. Chen, Y. Li, S. Kharbanda, and D. Kufe. 2003. MUC1 cytoplasmic domain coactivates Wnt target gene transcription and confers transformation. Cancer Biol. Ther. 2:702–706. [PubMed] [Google Scholar]
- Johnson, D.I. 1999. Cdc42: An essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63:54–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp, H.A., and G.F. Sprague Jr. 2003. Far3 and five interacting proteins prevent premature recovery from pheromone arrest in the budding yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 23:1750–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, D.J., E.L. Ting, C. Abeijon, L. Kroos, and R.S. Fuller. 2005. Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryot. Cell. 4:1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloux, I., E. Jacobs, and E. Dubois. 1994. Involvement of SRE element of Ty1 transposon in TEC1-dependent transcriptional activation. Nucleic Acids Res. 22:999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer, E., C. Wu, T. Leeuw, A. Fourest-Lieuvin, J.E. Segall, and D.Y. Thomas. 1997. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 16:83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaudet-Coopman, E., M. Beaujouin, D. Derocq, M. Garcia, M. Glondu-Lassis, V. Laurent-Matha, C. Prebois, H. Rochefort, and F. Vignon. 2006. Cathepsin D: newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett. 237:167–179. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie III, D.J. Demarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Madhani, H.D., C.A. Styles, and G.R. Fink. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 91:673–684. [DOI] [PubMed] [Google Scholar]
- Maeda, T., M. Takekawa, and H. Saito. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 269:554–558. [DOI] [PubMed] [Google Scholar]
- Marles, J.A., S. Dahesh, J. Haynes, B.J. Andrews, and A.R. Davidson. 2004. Protein-protein interaction affinity plays a crucial role in controlling the Sho1p-mediated signal transduction pathway in yeast. Mol. Cell. 14:813–823. [DOI] [PubMed] [Google Scholar]
- Nern, A., and R.A. Arkowitz. 1999. A Cdc24p-Far1p-Gβγ protein complex required for yeast orientation during mating. J. Cell Biol. 144:1187–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke, S.M., and I. Herskowitz. 2002. A third osmosensing branch in Saccharomyces cerevisiae requires the Msb2 protein and functions in parallel with the Sho1 branch. Mol. Cell. Biol. 22:4739–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, M., A.M. Neiman, H.O. Park, M. van Lohuizen, and I. Herskowitz. 1996. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- Raitt, D.C., F. Posas, and H. Saito. 2000. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 19:4623–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramer, S.W., S.J. Elledge, and R.W. Davis. 1992. Dominant genetics using a yeast genomic library under the control of a strong inducible promoter. Proc. Natl. Acad. Sci. USA. 89:11589–11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, C.J., B. Nelson, M.J. Marton, R. Stoughton, M.R. Meyer, H.A. Bennett, Y.D. He, H. Dai, W.L. Walker, T.R. Hughes, et al. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 287:873–880. [DOI] [PubMed] [Google Scholar]
- Roberts, R.L., and G.R. Fink. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974–2985. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., and G.R. Fink. 1987. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 48:1047–1060. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., F. Winston, and P. Hieter. 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 205 pp.
- Rost, B., G. Yachdav, and J. Liu. 2004. The PredictProtein server. Nucleic Acids Res. 32:W321–W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning. A Laboratory Manual. Third Edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schneider, B.L., W. Seufert, B. Steiner, Q.H. Yang, and A.B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 11:1265–1274. [DOI] [PubMed] [Google Scholar]
- Schweisguth, F. 2004. Notch signaling activity. Curr. Biol. 14:R129–R138. [PubMed] [Google Scholar]
- Shimada, Y., P. Wiget, M.P. Gulli, E. Bi, and M. Peter. 2004. The nucleotide exchange factor Cdc24p may be regulated by auto-inhibition. EMBO J. 23:1051–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman, H.S., M. Sutton-Smith, K. McDermott, P. Heal, S.H. Leir, H.R. Morris, M.A. Hollingsworth, A. Dell, and A. Harris. 2003. The contribution of tandem repeat number to the O-glycosylation of mucins. Glycobiology. 13:265–277. [DOI] [PubMed] [Google Scholar]
- Simon, M.N., C. De Virgilio, B. Souza, J.R. Pringle, A. Abo, and S.I. Reed. 1995. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature. 376:702–705. [DOI] [PubMed] [Google Scholar]
- Singh, P.K., and M.A. Hollingsworth. 2006. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 16:467–476. [DOI] [PubMed] [Google Scholar]
- Taheri, N., T. Kohler, G.H. Braus, and H.U. Mosch. 2000. Asymmetrically localized Bud8p and Bud9p proteins control yeast cell polarity and development. EMBO J. 19:6686–6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebayashi, K., K. Yamamoto, K. Tanaka, T. Tomida, T. Maruoka, E. Kasukawa, and H. Saito. 2006. Adaptor functions of Cdc42, Ste50, and Sho1 in the yeast osmoregulatory HOG MAPK pathway. EMBO J. 25:3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, X., H. Xu, and D. Kufe. 2006. MUC1 oncoprotein stabilizes and activates estrogen receptor alpha. Mol. Cell. 21:295–305. [DOI] [PubMed] [Google Scholar]
- Zarrinpar, A., R.P. Bhattacharyya, M.P. Nittler, and W.A. Lim. 2004. Sho1 and Pbs2 act as coscaffolds linking components in the yeast high osmolarity MAP kinase pathway. Mol. Cell. 14:825–832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.