Abstract
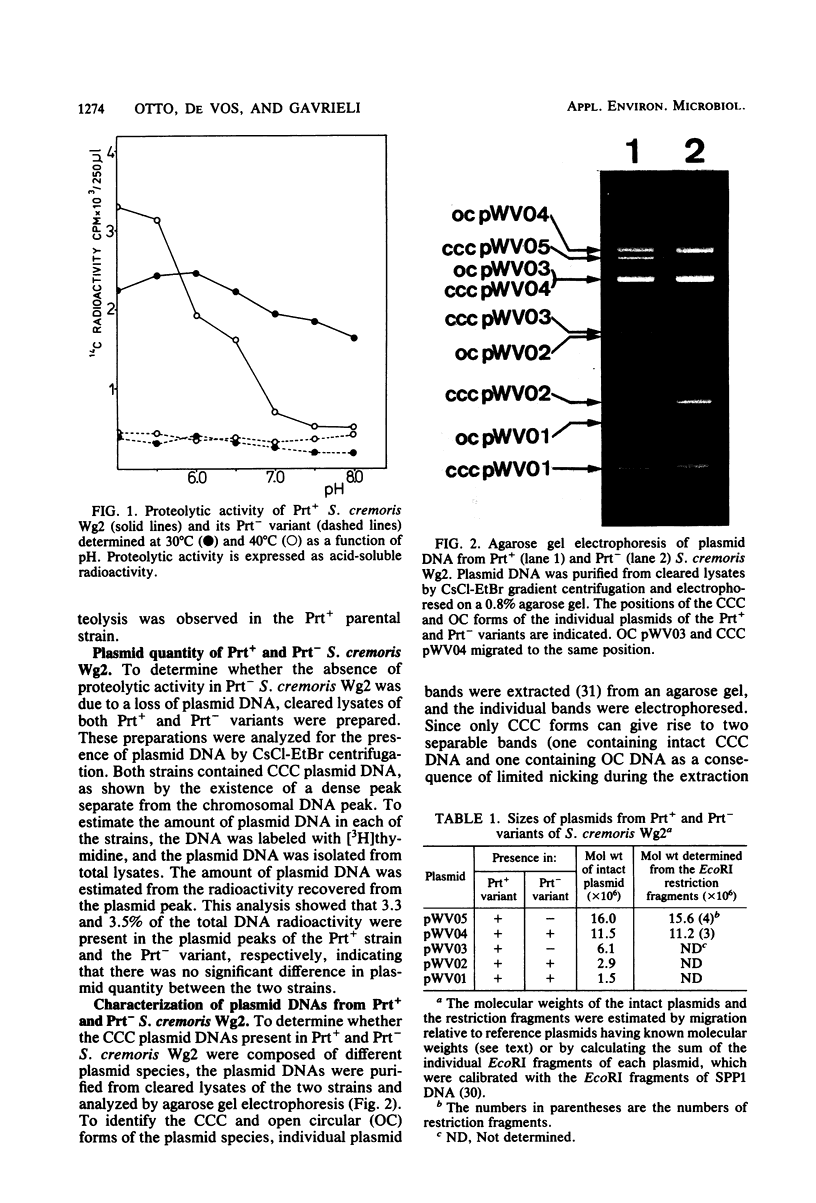
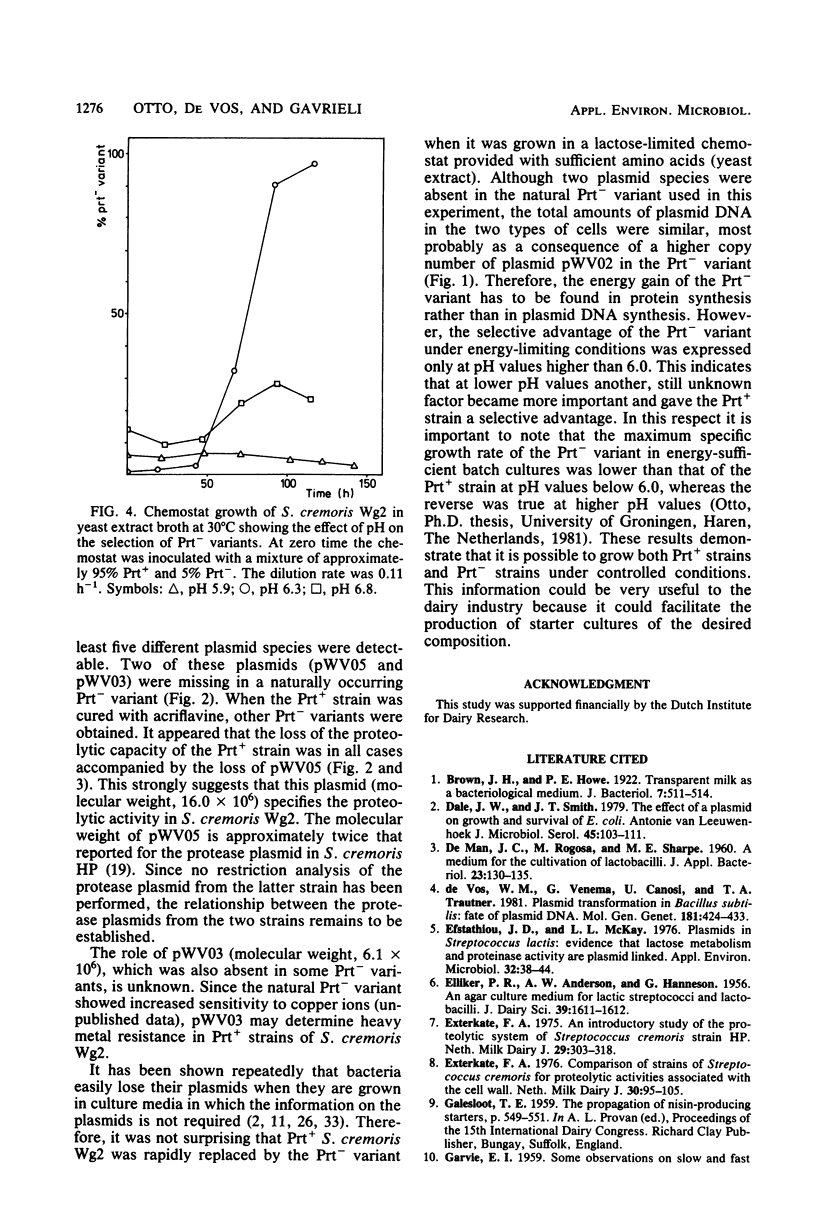
Cleared lysates of a proteolytic (Prt+) strain and a naturally occurring non-proteolytic (Prt−) variant of Streptococcus cremoris Wg2 contain equal amounts of covalently closed circular plasmid DNA. An analysis of this plasmid DNA by agarose gel electrophoresis revealed the presence of at least five different plasmid species in the Prt+ strain and only three plasmid species in the Prt− variant. Curing studies with acriflavine indicated that a 16-megadalton plasmid determined proteolytic activity in the Prt+ strain. In energy-limited chemostats inoculated with both strains it was observed that the Prt+ strain was replaced by the Prt− variant. This effect was most apparent when the pH of the culture was fixed at a value above 6.3. No selection for the Prt− variant was observed at pH 5.9. Since the two types of organisms contain equal amounts of plasmid DNA, it was concluded that the energy gain of the Prt− variants at pH values above 6.0 probably has to be found in protein synthesis rather than in plasmid DNA synthesis.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown J. H., Howe P. E. Transparent Milk as a Bacteriological Medium. J Bacteriol. 1922 Sep;7(5):511–514. doi: 10.1128/jb.7.5.511-514.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale J. W., Smith J. T. The effect of a plasmid on growth and survival of E. coli. Antonie Van Leeuwenhoek. 1979;45(1):103–111. doi: 10.1007/BF00400783. [DOI] [PubMed] [Google Scholar]
- Efstathiou J. D., McKay L. L. Plasmids in Streptococcus lactis: evidence that lactose metabolism and proteinase activity are plasmid linked. Appl Environ Microbiol. 1976 Jul;32(1):38–44. doi: 10.1128/aem.32.1.38-44.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin D., Slater J. H. The influence of the growth environment on the stability of a drug resistance plasmid in Escherichia coli K12. J Gen Microbiol. 1979 Mar;111(1):201–210. doi: 10.1099/00221287-111-1-201. [DOI] [PubMed] [Google Scholar]
- Iordănescu S. Recombinant plasmid obtained from two different, compatible staphylococcal plasmids. J Bacteriol. 1975 Nov;124(2):597–601. doi: 10.1128/jb.124.2.597-601.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P., Sklan D., Gordin S. Effect of diluent on bacterial counts in milk and milk products. J Dairy Sci. 1974 Jan;57(1):127–128. doi: 10.3168/jds.S0022-0302(74)84842-3. [DOI] [PubMed] [Google Scholar]
- Klaenhammer T. R., McKay L. L., Baldwin K. A. Improved lysis of group N streptococci for isolation and rapid characterization of plasmid deoxyribonucleic acid. Appl Environ Microbiol. 1978 Mar;35(3):592–600. doi: 10.1128/aem.35.3.592-600.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl S. A., Larsen L. D., McKay L. L. Plasmid Profiles of Lactose-Negative and Proteinase-Deficient Mutants of Streptococcus lactis C10, ML(3), and M18. Appl Environ Microbiol. 1979 Jun;37(6):1193–1195. doi: 10.1128/aem.37.6.1193-1195.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey R. W., Chopra I. Genetic studies of a multi-resistant strain of Staphylococcus aureus. J Med Microbiol. 1974 May;7(2):285–297. doi: 10.1099/00222615-7-2-285. [DOI] [PubMed] [Google Scholar]
- Larsen L. D., McKay L. L. Isolation and characterization of plasmid DNA in Streptococcus cremoris. Appl Environ Microbiol. 1978 Dec;36(6):944–952. doi: 10.1128/aem.36.6.944-952.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald I. J. Occurence of lactose-negative mutants in chemostat cultures of lactic streptococci. Can J Microbiol. 1975 Mar;21(3):245–251. doi: 10.1139/m75-035. [DOI] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Efstathiou J. D. Transductional evidence for plasmid linkage of lactose metabolism in streptococcus lactis C2. Appl Environ Microbiol. 1976 Jul;32(1):45–52. doi: 10.1128/aem.32.1.45-52.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A. Plasmid distribution and evidence for a proteinase plasmid in Streptococcus lactis C2-1. Appl Microbiol. 1975 Apr;29(4):546–548. doi: 10.1128/am.29.4.546-548.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A. Simultaneous loss of proteinase- and lactose-utilizing enzyme activities in Streptococcus lactis and reversal of loss by transduction. Appl Microbiol. 1974 Sep;28(3):342–346. doi: 10.1128/am.28.3.342-346.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Zottola E. A. Loss of lactose metabolism in lactic streptococci. Appl Microbiol. 1972 Jun;23(6):1090–1096. doi: 10.1128/am.23.6.1090-1096.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Edelman I., Schwesinger M. D., Gruss A. D., Swanson E. C., Pattee P. A. Genetic translocation in Staphylococcus aureus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):400–404. doi: 10.1073/pnas.76.1.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff S. W., Luh J., Ganesan A. T., Behrens B., Thompson R., Montenegro M. A., Morelli G., Trautner T. A. The genome of Bacillus subtilis phage SPP1: the arrangement of restriction endonuclease generated fragments. Mol Gen Genet. 1979 Jan 10;168(2):165–172. doi: 10.1007/BF00431442. [DOI] [PubMed] [Google Scholar]
- Thuring R. W., Sanders J. P., Borst P. A freeze-squeeze method for recovering long DNA from agarose gels. Anal Biochem. 1975 May 26;66(1):213–220. doi: 10.1016/0003-2697(75)90739-3. [DOI] [PubMed] [Google Scholar]
- Wouters J. T., van Andel J. G. R-plasmid persistence in Escherichia coli grown in chemostat cultures [proceedings]. Antonie Van Leeuwenhoek. 1979;45(2):317–318. doi: 10.1007/BF00418597. [DOI] [PubMed] [Google Scholar]
- de Vos W. M., Venema G., Canosi U., Trautner T. A. Plasmid transformation in Bacillus subtilis: fate of plasmid DNA. Mol Gen Genet. 1981;181(4):424–433. doi: 10.1007/BF00428731. [DOI] [PubMed] [Google Scholar]
- van Embden J. D., Veltkamp E., Stuitje T., Andreoli P. M., Nijkamp H. J. Integration of a transposable DNA sequence which mediates ampicillin resistance into Clo DF13 plasmid DNA: determination of the site and orientation of TnA insertions. Plasmid. 1978 Feb;1(2):204–217. doi: 10.1016/0147-619x(78)90039-2. [DOI] [PubMed] [Google Scholar]





