Abstract
Objectives
To develop mechanisms to formulate queries over the semantic representation of cancer-related data services available through the cancer Biomedical Informatics Grid (caBIG).
Design
The semCDI query formulation uses a view of caBIG semantic concepts, metadata, and data as an ontology, and defines a methodology to specify queries using the SPARQL query language, extended with Horn rules. semCDI enables the joining of data that represent different concepts through associations modeled as object properties, and the merging of data representing the same concept in different sources through Common Data Elements (CDE) modeled as datatype properties, using Horn rules to specify additional semantics indicating conditions for merging data.
Validation
In order to validate this formulation, a prototype has been constructed, and two queries have been executed against currently available caBIG data services.
Discussion
The semCDI query formulation uses the rich semantic metadata available in caBIG to build queries and integrate data from multiple sources. Its promise will be further enhanced as more data services are registered in caBIG, and as more linkages can be achieved between the knowledge contained within caBIG's NCI Thesaurus and the data contained in the Data Services.
Conclusion
semCDI provides a formulation for the creation of queries on the semantic representation of caBIG. This constitutes the foundation to build a semantic data integration system for more efficient and effective querying and exploratory searching of cancer-related data.
Introduction
There is a large, ever-growing, and increasingly complex body of bioinformatics and genetic data publicly available through the World Wide Web; this wealth of information is quite varied in nature and objectives, and provides immense opportunities to cancer biology researchers, while posing significant challenges in terms of housing, accessing, and analyzing these data sets. 1 This increased availability of data has catalyzed a systems view of biomedicine, where the integration of biology, medicine, computation, and technology is proposed to comprehend biological information processing. 2 Arguably, cancer is an almost ideal domain of expertise in which to apply the concept of systems biology, as it involves complex biological processes and staggering amounts of disparate experimental data that needs to be connected and integrated. 3
The essential problem in data integration is not in how to store it or retrieve it, but in how best to distill insights and associate these interpretations with the data. 4 For this, knowledge needs to be organized for higher-level reasoning. 5 Knowledge representation techniques that can explicitly describe the meaning, that is, the semantics of the data, as enabled by the Semantic Web developments, are needed to improve interoperability for biological data representation and management. 6 Formal representation of knowledge using such technologies allows for complex queries, and for automated reasoning that can uncover inconsistencies. 7 Semantic representation of the information stored in multiple data sources is essential for defining correspondence among entities belonging to different sources, resolving conflicts among sources, and ultimately automating the integration process. 8 Ontologies hold the promise of providing a unified semantic view of the data by providing a means of representing knowledge. 9
The National Cancer Institute (NCI) is at the forefront in the implementation of semantic technologies and collaborative environments. The national-scale cancer Biomedical Informatics Grid (caBIG) program aims to create a network of cancer clinical and research centers to better leverage their combined strength and expertise. Towards this objective, caBIG is developing standards, guidelines, data and analytical services, and open-source software tools to enable more effective sharing of data, all supported by an underlying service-oriented infrastructure called caGrid. 10
One important use case for caBIG is the ability to get information about some specific concept that is provided by a multitude of grid-enabled data services published on caGrid infrastructure. 11 As such, the idea is to enable users to define a query such as, for example, “find all tissue samples for expression profiles for genes related to the EGF signaling pathway,” or “identify genes that segregate with the increased prostate cancer rate observed in African Americans,” and process such a query across all data sources containing relevant information. caBIG provides programming methods to access such information in a standardized manner, and also provides linkages between conceptual representations and data.
In this paper, the Semantic caBIG Data Integration (semCDI) query formulation is introduced for the purpose of specifying and executing queries across multiple caBIG data services at a high level of semantic abstraction. This enables researchers to work with conceptual representations rather than with the sometimes arcane details of data storage formats. semCDI is specifically focused on queries over concepts modeled in several data sources by using the rich semantic metadata made explicit by the NCI caBIG Initiative. The query formulation views caBIG semantics and data as an ontology, using World Wide Web Consortium (W3C) standards such as OWL and SPARQL to represent this ontology and the queries posed against it. In addition, semCDI expands caBIG semantics through the use of Horn rules in order to establish additional conditions and constraints to guide the integration of data from multiple sources. The semCDI formulation is the technological foundation necessary to build a system that enables researchers to find data relevant for their endeavors within the caBIG data services through the examination of the semantic models and abstractions of this data.
Background
The caBIG project utilizes a four-layer approach for interoperability: interface integration at the syntactic level, and information models, semantic metadata, and ontology-based controlled terminologies at the semantic level. 12 The information models and semantic metadata are object-oriented constructions contained in the Cancer Data Standards Repository (caDSR), 13 providing a framework and protocols for specifying, maintaining and sharing metadata across diverse domains. The controlled terminology component of caBIG is maintained in the NCI Thesaurus, 13–15 a reference terminology published by the Enterprise Vocabulary Services (EVS), a partnership between the NCI Office of Communications and the NCI Center for Bioinformatics.
The caBIG program contains a rapidly expanding collection of tools and datasets relevant to cancer research. In order to achieve compatibility, data services in caBIG must map elements of its data sources into object models annotated to provide semantic meanings as described in caDSR and EVS. 10 Data services must also allow querying through an XML-based caGrid query language called CQL, 16 and must be capable of working within the underlying caGrid collaborative computing technology. Data services currently available through caBIG include caArray, a source of microarray data, 17 the Grid Enablement of the Protein Information Resource (gridPIR), 18 and annotations on microarrays through the caBIG Function Express Server (caFE); 19 the incorporation of several other sources of information is being undertaken.
Several approaches to data integration have been proposed in the last few years. Integration approaches used in existing systems can be broadly classified into two categories: data warehousing, which deals with the translation of data into a centralized repository, and data mediation or federation, which employs query translation to decompose a global query into local queries at each integrated data source; mediator-based systems are more suitable to integrate large amounts of information from different sources over which the user has little or no control. 20 A number of mediator-based systems have recently been detailed, including a data integration framework based on XML and grid technology, 21 and systems using ontologies for data representation. 22,23,24
Within caBIG, the Cancer Translational Research Informatics Platform (caTRIP) project focuses on an object-oriented approach to integrate data from a specific set of grid services (caTissue CORE, Clinical Annotation Engine, Tumor Registry, and caIntegrator SNP services). It uses a mediator-based, federated query engine and an extension to the caGrid query language called Distributed CQL (DCQL) to present a single interface where these services can be discovered and subsequently queried in a metadata-driven manner. In caTRIP, queries that involve merging of data from multiple sources must be specified by linking together data elements from different sources at the attribute level. 25
Formulation Process
caBIG Semantic Structure
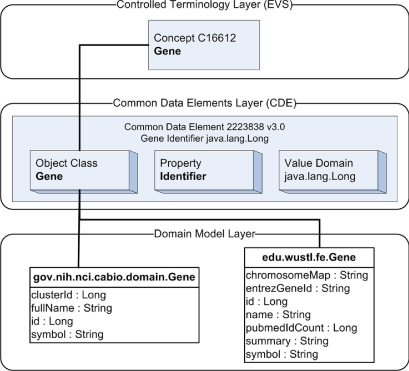
Semantic metadata is organized in caBIG in three layers of abstraction, as illustrated in ▶. At the top level, semantic concepts are organized through the NCI Thesaurus, and accessed through the Enterprise Vocabulary Services (EVS). These concepts are related to each other through associations, and to values through the use of Common Data Elements (CDEs), stored and accessed through the cancer Data Standards Repository (caDSR).
Figure 1.
Layers of semantic interoperability in caBIG.
Data sources compliant to caBIG are required to expose a domain information model, which contain the information necessary to translate the caBIG semantic abstractions into specific values within data sources. Domain information models are encoded as UML class diagrams, where each UML class is linked to a concept within the NCI Thesaurus, each relationship between UML classes is linked to an association, and each relationship between a UML class and an attribute value is linked to a CDE. Package names are used to identify the data source represented by a UML class.
In addition to the associations encoded in caDSR, the NCI Thesaurus contains relationships between concepts called roles. These roles are used to make logical assertions between concepts, 14 and result in fairly expressive graphs of semantic relationships among basic and clinical science concepts. 26 However, the use of this semantic structure for data integration is hampered by the absence of links between specific concepts in NCI Thesaurus and the data points within data sources which represent such concepts. Thus, the formulation presented in this paper does not consider the use of roles for data integration; the issues preventing their use and some potential solutions are presented in the Discussion further below.
OWL Representation of caBIG Semantics
The semantic information of caBIG contained in NCI Thesaurus concepts, caDSR associations, CDEs, domain information models, and data can be modeled as an ontology. The semCDI query formulation conceptualizes this ontology as a Web Ontology Language (OWL) graph; OWL is a vocabulary extension of the Resource Description Framework (RDF). 27 Following OWL standard nomenclature, then, NCI Thesaurus concepts and UML classes will be modeled as classes; an ontology class representing a UML class is linked to one representing an NCI Thesaurus concept through a subClassOf relationship, corresponding to the mapping between UML classes and their corresponding NCI Thesaurus concepts maintained in caDSR. UML classes are differentiated from each other through the use of XML namespaces, such as those listed in Table 1 (available as a JAMIA online supplement at www.jamia.org). The linkage between ontology classes and their corresponding NCI Thesaurus concepts and UML classes is maintained through annotation properties. OWL class declarations depicting both NCI Thesaurus concepts and UML classes are shown in Table 2 (available as a JAMIA online supplement at www.jamia.org). Note that in the definition of the class Gene corresponding to the NCI Thesaurus concept of the same name, the attributes evs:code and evs:definition provide an identification and human-readable description of the concept, respectively.
In caBIG, data is accessed through UML classes; each object so accessed can be considered an instance, or in OWL terms, an individual member of the UML class, and by extension, of the superclass NCI Thesaurus concept. 14 Membership in a specific UML class indicates data origin. In this sense, an individual member of class cafe:Gene is a member of the class Gene that has been retrieved from caFE.
Associations define the relationships between two objects, called object properties in OWL. Thus, for example, the Gene class has associations with Protein, Biochemical Pathway, and Organism. Table 3 (available as a JAMIA online supplement at www.jamia.org) shows the definition of object properties for the class Gene.
Objects also have relationships to value-based attributes, and these relationships, called datatype properties in OWL, are encoded in CDEs. Thus, for example, Gene is related to a value of type String by the CDEs Gene-Gene Symbol and Gene Name, among others, and to a value of type Long by the CDE Gene Pubmed Identifier Count. An example OWL representation of some datatype properties for class Gene can be seen in Table 4 (available as a JAMIA online supplement at www.jamia.org).
Semantic Querying through caBIG
In terms of the view of caBIG semantic metadata as an ontology, querying can be conceived as the search and retrieval of individuals members of one or more classes, from one or more data sources. We recognize two important types of queries: joins and merges.
Joins
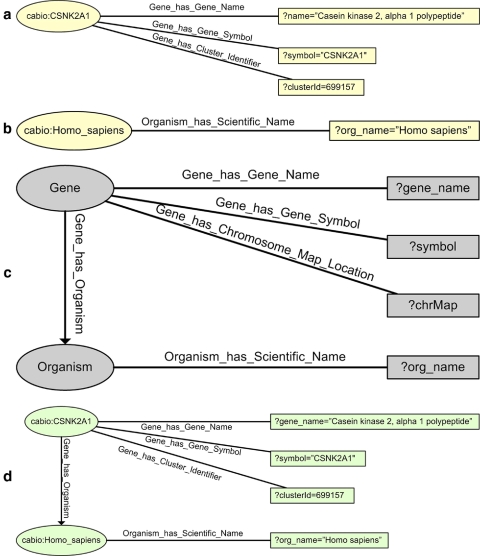
Queries must be capable of retrieving joins, that is, individual members of different classes semantically related to each other. Consider for example an individual member of class cabio:Gene, as illustrated in ▶(a), and an individual of class cabio:Taxon as illustrated in ▶(b); cabio:Taxon is a subclass of the NCI Thesaurus class Organism. A query such as the one illustrated in ▶ (c), which seeks to retrieve information about genes, including the name of the organism to which the genes belong, then returns the join on the object property Gene_has_Organism of the individuals in ▶ (a) and (b), as shown in ▶ (d). The power of caBIG semantics in facilitating data integration for these cases is apparent, as the join conditions are directly specified by the object properties within the OWL representation of caBIG data sources.
Figure 2.
Example of joining two individuals from different classes: (a) individual from Gene class; (b) individual from Organism class; (c) graphical representation of query; (d) joining of 2 individuals.
Merges
Consider the question of finding information about some concept in several data sources; for example, consider querying for information about genes in caFE Server and caBIO. A simple query for this purpose could be graphically represented as in Figure 3(a) (available as a JAMIA online supplement at www.jamia.org). The execution of this query would return every individual member of cafe:Gene and cabio:Gene, with its corresponding attributes. Two such results, one from caFE and one from caBIO, are illustrated in ▶ (b) and Figure 3(b) (available as a JAMIA online supplement at www.jamia.org). Note that these two results, while referring to the same gene symbol—and thus, presumably, to the same gene—are actually two separate results.
When classes exist across multiple sources, however, it is more likely that the objective of a query be to discover whether individuals members of the same NCI Thesaurus concept class can be in fact considered a single individual; we call this type of join a merge. For this, it is necessary to determine one or more datatype properties of Gene—that is, CDEs which have the class Gene as its subject—that are shared by both subclasses. In this case, both subclasses share CDEs Gene-Gene Symbol and Gene Name, and so it is natural to use one or both of these CDEs as a merge condition. This results in a set of individuals members of class Gene which contain attributes from both caBIO and caFE, such as the example in Figure 3(c) (available as a JAMIA online supplement at www.jamia.org).
Model Description
The creation and execution of queries on the OWL representation of caBIG semantics is done using the SPARQL Protocol and RDF Query Language (SPARQL). SPARQL, a W3C Recommendation, defines a robust, standardized query language over RDF datasets. 28 In this section, we present the methodology by which queries are formulated.
Query Representation and Processing in SPARQL
A SPARQL query consists of three main parts: an algebra expression, a dataset, and a query form; the algebra expression, in turn, contains a graph pattern expression and solution modifiers. 28 The SPARQL representation of the query graphically depicted in Figure 3(a) (available as a JAMIA online supplement at www.jamia.org) is shown in Table 5 (available as a JAMIA online supplement at www.jamia.org) where the three parts of the query can be identified as follows:
• The graph pattern expression under the WHERE clause defines the criteria for choosing individuals from the data sources and binding variables to values. The OPTIONAL clauses in the graph pattern of the query indicate that it is not required that an object contain a certain attribute; if these OPTIONAL clauses were not present, an individual would be required to have values for all properties. As this query does not contain solution modifiers, the graph pattern expression by itself is the algebra expression of the query.
• The dataset specification in the FROM clause indicates the data sources—that is, the source OWL graphs—from which the query is to be made. For this query, and for all the examples depicted in this paper, the default graph, which is always included in the query, is the collection of classes and properties from NCI Thesaurus and the caDSR.
• The query form SELECT clause indicates a projection of the results into a table of variable bindings.
Joins of individuals of classes related by semantic associations are straightforwardly represented in SPARQL, by querying on the relationships represented by object properties. Consider the query graphically depicted in ▶(c), and for simplicity suppose that data is to be retrieved only from caBIO; the SPARQL formulation of this query is shown in Table 5 (available as a JAMIA online supplement at www.jamia.org).
Merges, on the other hand, are not easily represented in SPARQL; the query shown in Table 5 (available as a JAMIA online supplement at www.jamia.org) still results in a set of disjoint Gene objects coming from the different data sources used, such as the example in Figure 3(b) (available as a JAMIA online supplement at www.jamia.org). It is necessary, then, to have a mechanism to encode merge conditions as additional semantics. The semCDI query formulation presented here proposes to use definite Horn rules for this encoding.
Rules
Definite Horn rules are clauses of the form conclusion ← condition, where condition is a conjunction of atomic Boolean-valued formulas without negation, 29 and conclusion is a fact determined to be true if the condition is true; axioms are defined as conclusions with empty (or true) condition. These Horn rules define (possibly) conditional statements that are not asserted in the ontology defined by caBIG. By design, they are defined outside of a query; in this way, rules can be used by multiple queries independently.
The World Wide Web Consortium has established a Working Group charged with the development of a Rules Interchange Format (RIF), which has published a first working draft of its Basic Language Dialect (RIF-BLD), 29 and of its compatibility with RDF and OWL. 30 In this paper, we use the presentation syntax derived by the RIF working group to denote these rules.
Single Merge Conditions
The simplest merging of individuals from multiple sources consists in defining a shared datatype property as a single merge key. In order to indicate such a merge, a rule is defined so that the chosen property is asserted to be inverse-functional, meaning that if two individuals share the same value in such a property, then they must be the same individual. An example rule, with empty condition, establishing Gene_has_Gene_Symbol as an inverse-functional property, is shown in Table 7 (available as a JAMIA online supplement at www.jamia.org), where the query from Table (available as a JAMIA online supplement at www.jamia.org) is redefined. Note that the graph pattern for Gene_has_Gene_Symbol is not within an OPTIONAL clause anymore, since the symbol must exist in order to use it as a merge condition.
One important point to make regards the definition of datatype properties as inverse-functional. As is noted in the OWL specifications, this places the ontology model used here within the OWL Full flavor. While this is required in order to accurately represent the characteristics of the data sets, this also means that the ontology becomes undecidable in terms of reasoning. However, Horn rules are themselves undecidable, and the inverse-functional datatype properties are only being used in the context of these rules; therefore, there is no added complexity in this choice of representation.
Multiple Merge Conditions
The merging of two individuals that are instances of some base class such as Gene may require that merge keys be established at multiple values, otherwise some of the data obtained may not be semantically valid. In other words, the use of a single CDE as a key to determine equivalence between individuals may not be sufficient. Consider again the query defined in Table 7 (available as a JAMIA online supplement at www.jamia.org); it produces some undesirable results, since genes from different organisms may actually share the same symbol, but may have otherwise different characteristics, such as a different chromosome map location. Table 8 (available as a JAMIA online supplement at www.jamia.org) shows a more complete query, which includes joins between Gene and Organism, and between Gene and Biochemical Pathway; this second join is included in order to restrict the number of results to those related to the EGF signaling pathway, as explained in the section on Validation by Example. This query includes rules to define a multiple merge condition: the first rule simply asserts that organisms are identified by their scientific name, and the second rule defines that two genes are equal if they have the same organism and if they have the same gene symbol. semCDI utilizes Horn logic because multiple conditions with existential qualification such as the one shown here cannot be expressed in RDF or OWL axioms.
Merge Conditions over Dissimilar CDEs
Ideally, two UML classes should use the same CDE to model the same datatype property. Due to the complexity of caBIG, however, it is expected that this may not be so in all cases. Consider for example the question of relating a set of proteins from caBIO with information from GeneConnect containing identifiers from alternate sources such as GenBank and Ensembl. The UML class cabio:Protein uses the CDE Protein Primary Accession Number, while genec:Protein uses Protein UniprotKB Primary Accession Number Genomic Identifier to refer to the same value.
In order to merge Protein individuals from these two data sources, it is necessary to establish that the properties Protein_has_Primary_Accession_Number and Protein_has_UniprotKB_Primary_Accession_Number_Genomic_Identifier are equivalent and can therefore be combined. As shown in Table 9 (available as a JAMIA online supplement at www.jamia.org), this can be encoded in a rule as an equivalent property axiom, and then a query merging cabio:Protein and genec:Protein individuals can be generated. Figure 4(b) (available as a JAMIA online supplement at www.jamia.org) illustrates the result of merging the two individuals shown in Figure 4(a) (available as a JAMIA online supplement at www.jamia.org).
Derived Merge Conditions
In some cases, two data sources model the same data in somewhat different manners, using different CDEs. For example, the Pathways Interaction Database uses the CDE Biochemical Pathway Short Name for class pid:Pathway, while caBIO uses the CDE Biochemical Pathway Name for class cabio:Pathway. Both CDEs model in essence the same data, although since caBIO models pathways for organisms other than humans, the pathway name contains the string “h_” in front of every human pathway.
In order to merge the Biochemical Pathway individuals in caBIO with those in PID, then, and knowing that all pathways in PID refer to the organism homo sapiens, it is necessary to concatenate the string “h_” with the value of Biochemical_Pathway_has_Short_Name for class pid:Pathway, and equate this resulting string with the value of Biochemical_Pathway_has_Name for class cabio:Pathway. This is illustrated in Table 10 (available as a JAMIA online supplement at www.jamia.org): note that in the definition of the derived equality, we borrow the function fn:concat from the XQuery/XPath functions and operators; 31 in general, these functions are used to specify more complex rules for derived merges.
Validation by Example
Experimental Setup
To validate the query formulation described in this paper, we implemented a prototype query processor in the Java programming language, capable of querying data services through caGrid. The input to this prototype is the location of each data service to be queried, the name of the NCI Thesaurus concepts and CDEs to be queried, and of the join and merge conditions specified by the queries and rules. For the purposes of these experiments, all caGrid data services were queried through their respective URIs as specified in the caGrid Portal, 32 except for the Pathways Interaction Database (PID), which does not yet have an operating data service. Instead, a PID service was created locally using the caCORE Software Development Kit (SDK) and the PID domain information model, populating the data with information downloaded from the PID website. 26 Further, the locations of the caBIG data services were provided manually, although a final implementation would use the caGrid Index Service to automatically discover these locations, as detailed in online caGrid documentation. 11
Translation of Queries
The functioning of the prototype query processor is illustrated in Figure 5 (available as a JAMIA online supplement at www.jamia.org). It parses the queries and rules in order to divide the incoming query into an execution plan consisting of a sequence of source-specific SPARQL queries that incorporate additional terms required to resolve the rules. Each of these source-specific SPARQL queries is then converted into an equivalent query expressed in CQL (CaGrid Query Language), and then submitted to the appropriate data service. CQL, which must be supported by all caGrid data services, is an XML based language that uses Query-by-Example (QBE) syntax, where the user provides an example conforming to the objects and associations within the information model exposed by the data service being queried. Details on CQL can be found in online caGrid documentation. 34
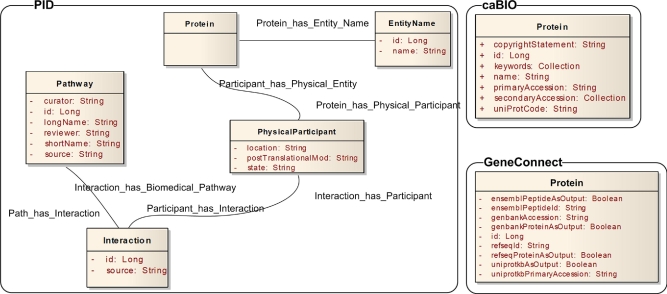
Consider for example the query in Table 12 (available as a JAMIA online supplement at www.jamia.org), which seeks to find all human proteins related to the EGF signaling pathway present in caBIO, GeneConnect, and PID; the relevant portions of the domain information models of these three sources is illustrated in ▶. This query is similar to the one outlined in Table 9 (available as a JAMIA online supplement at www.jamia.org), but with the addition of a derived merge. This query generates an execution plan consisting of a query against PID, as shown in Table 13 (available as a JAMIA online supplement at www.jamia.org), and a set of queries against caBIO and GeneConnect using the results from the query against PID, of which one example is shown in Table 14 (available as a JAMIA online supplement at www.jamia.org). Each of these queries is then translated into a CQL query based on the correspondence between ontology classes and properties and the UML domain information model for a data service, as encoded in the annotation properties of the ontology elements. The translation of the query in Table 14 (available as a JAMIA online supplement at www.jamia.org) is shown in Table 15 (available as a JAMIA online supplement at www.jamia.org).
Figure 6.
Relevant portions of domain information models for PID, GeneConnect, and caBIO.
Queries and Results
Two queries were run on this setup, seeking information into genes and proteins that interact with the EGF signaling pathway. This pathway was chosen since we expect to use the querying methodology outlined here to study, in future work, the reported correlation between EGFR expression and brain tumor progression; 35 for now, the restriction into genes and proteins interacting with this pathway serves as a useful way to reduce the size of the result data for presentation purposes.
The first query run through our prototype is the query shown in Table 8 (available as a JAMIA online supplement at www.jamia.org); it seeks to find all human genes related to the EGF signaling pathway from caBIO and the caFE Server. The results obtained for this query are shown in ▶. Note that for each gene symbol there are two rows, each containing a different literal bound to the variable ?gene_name. The literal with the first letter capitalized is retrieved from caBIO, while the other literal, with the first letter in lowercase, comes from caFE Server. This is further illustrated in Figure 7 (available as a JAMIA online supplement at www.jamia.org), which shows that the same gene object, with symbol CSNK2A1, has two distinct values for the property Gene_has_Gene_Name. When such an object is projected into a table of variable bindings, as specified by the SPARQL SELECT clause, it generates two separate rows. SPARQL does not contain mechanisms to select only one binding under these circumstances; the ability to do so within the semCDI query formulation is a matter of ongoing work.
Table 11.
Table 11 Query Results for EGF Signaling Pathway Genes
| Symbol | Gene_name | Org_name | ChrMap | ClusterId |
|---|---|---|---|---|
| CSNK2A1 | Casein kinase 2, alpha 1 polypeptide | Homo sapiens | 20p13 | 699157 |
| CSNK2A1 | casein kinase 2, alpha 1 polypeptide | Homo sapiens | 20p13 | 699157 |
| CUTL1 | Cut-like 1, CCAAT displacement protein (Drosophila) | Homo sapiens | 7q22.1 | 654389 |
| CUTL1 | cut-like 1, CCAAT displacement protein (Drosophila) | Homo sapiens | 7q22.1 | 654389 |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | Homo sapiens | 11p15.5 | 37003 |
| HRAS | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Homo sapiens | 11p15.5 | 37003 |
| PLP2 | Proteolipid protein 2 (colonic epithelium-enriched) | Homo sapiens | Xp11.23 | 77422 |
| PLP2 | proteolipid protein 2 (colonic epithelium-enriched) | Homo sapiens | Xp11.23 | 77422 |
| PRKCA | Protein kinase C, alpha | Homo sapiens | 17q22-q23.2 | 680598 |
| PRKCA | protein kinase C, alpha | Homo sapiens | 17q22-q23.2 | 680598 |
| STAT5A | Signal transducer and activator of transcription 5A | Homo sapiens | 17q11.2 | 437058 |
| STAT5A | signal transducer and activator of transcription 5A | Homo sapiens | 17q11.2 | 437058 |
The second query is the one shown in Table 12 (available as a JAMIA online supplement at www.jamia.org). The query incorporates three rules. The first one establishes the equality between two different CDEs modeling protein primary accession numbers, as has already been discussed. The second rule establishes the Protein_has_Primary_Accession_Number property as inverse-functional. The third rule establishes protein equality on the UniProt code through a derived condition, where the string “_HUMAN” must be appended to the protein entity name, which is retrieved from PID. In addition, this query includes a FILTER condition to ensure that only proteins related to the EGF signaling pathway are retrieved; this condition is obtained by navigating through a concatenation of property relationships, which in this case are exclusively used by PID. The results of running this query are shown in ▶.
Table 16.
Table 16 Results of Query for Proteins related to the EGF Signaling Pathway
| accession_num | Protname | Protcode |
|---|---|---|
| P00533 | Epidermal growth factor receptor precursor | EGFR_HUMAN |
| P01133 | Pro-epidermal growth factor precursor | EGF_HUMAN |
| P23458 | Tyrosine-protein kinase JAK1 | JAK1_HUMAN |
| P20936 | Ras GTPase-activating protein 1 | RASA1_HUMAN |
| P42224 | Signal transducer and activator of transcription 1-alpha/beta | STAT1_HUMAN |
Discussion
Significance
The query formulation presented in this paper enables the use of the rich semantic metadata available through caBIG in order to construct queries over multiple caGrid-enabled data sources at a high level of semantic abstraction. This will help accomplish the goal of allowing researchers to more easily find data relevant to their questions and investigations, and to do so more efficiently, minimizing the need for scientists to comprehend and analyze the arcane structures of data stores. We believe that this also will expand the capability of researchers to perform explorative investigation of data, searching for relationships between concepts that may be corroborated—or contradicted, for the matter—by existing data, and pointing to avenues for improving their understanding of the processes that surround cancer genesis and progression.
Limitations and Future Work
While there has been substantial progress within caBIG, and in particular in the implementation of caGrid, this is still a work in progress, and some inconsistencies and limitations can be found in the availability of data and in its semantic modeling. We are currently working towards addressing these limitations and implementing the semCDI query formulation within an application for the semantic integration of caBIG data services. This software application is currently a work in progress and thus not yet publicly available.
Availability of Data and Scalability
The number of data services available through caGrid is relatively small, especially compared to the number of domain information models available: currently, there are over 50 domain information models available through the caBIG UML Model Browser, 36 while only 15 distinct data services are operational through the caGrid portal. 32 Additionally, through testing, we have determined that some of these existing services have problems handling large amounts of data, returning error messages when the size of the result set is expected to be large. Both the number of available services and their ability to handle larger amounts of data are expected to improve rapidly as the caBIG infrastructure matures and as more data providers make their services compatible with it.
In addition to the limitations on the scalability of existing caGrid data services, it is also important to note that the scalability of the query formulation process presented here has not been established. The size of the ontology representations of caBIG data services, as measured by their number of classes and properties, must be studied in terms of the human-computer interaction mechanisms for construction of queries, while the number of available data services and the size of the result sets will influence the query processing and execution mechanisms.
Availability and Semantics of Data Elements
The number of CDEs useful for semantic merging of data is relatively limited. At the time of writing of this paper, there were 10,471 CDEs that were registered in caBIG with status of “RELEASED”. Of these, we found that 7,850 are not linked with any data source; most of these CDEs are used as building blocks to construct forms for clinical trials. Another set of 2,034 CDEs are linked with a single classification scheme. This means that only 587 CDEs are available for data merging, and of these, over half connect only two data sources.
Not all shared CDEs are appropriate to be used as merge keys. For example, classes cafe:Gene and cabio:Gene have another CDE in common, Gene Identifier. A merge on this CDE results in individuals such as the example in Figure 8 (available as a JAMIA online supplement at www.jamia.org); note that this individual has two different values for gene symbol, and in reality refers to two distinct genes. The issue in this case is that the CDE Gene Identifier refers to an identifier local to each data source. Therefore, while the meaning of the concept “Gene Identifier” is the same for all data sources using this CDE, the meaning of a specific value for this attribute is not the same across these sources. In terms of the semCDI query formulation, there does not exist a mechanism to preclude the use of inappropriate merge keys, as Horn rules do not permit negation. We are currently investigating ways in which such statements can be characterized, such as the use of disjointness or complement axioms. It should be noted that the specific issue of local identifiers is being addressed by an initiative to create global caBIG identifiers for every piece of data. 13
Different data sources may contain similar data not intended to be used as merge keys, such as names and descriptions. Such repeated data causes multiple result sets for each individual when projections into variable bindings are made, as shown in ▶. We are currently working to resolve this issue; potential solutions being analyzed include the ability to specify uniqueness in results through restrictions to preferred data sources, and the ability to post-process the binding tables. We are also considering options to specify restrictions in merge keys through OWL axioms or Horn rules.
Knowledge and Inferencing
Another important issue for semCDI regards the use of the knowledge contained within the NCI Thesaurus. While the NCI Thesaurus was initially conceived as a terminology system, it has evolved into an expressive graph of semantic relationships among molecular, biological, genomic, phenotypical, and pharmacological concepts. 26 These relationships and properties hold substantial promise for a variety of research uses, including the possibilities of querying over complex relationships in multiple data sources. The NCI Thesaurus is only linked to the caDSR through the equivalence of concepts to object classes in CDEs; this linkage, although crucial, is not sufficient: it is also necessary to link data instances to subclasses in NCI Thesaurus that model more specific concepts. For example, the thesaurus contains a class called EGFR Gene as a subclass of Gene, which has roles that associate it to diseases, molecular processes, and abnormalities. On the other hand, our semCDI query formulation can extract an individual member of class Gene with symbol “EGFR”. A linkage between this individual and the NCI Thesaurus concept for EGFR Gene does not currently exist in the semantic metadata in caBIG. This kind of richer, tighter relationship between concepts in the NCI Thesaurus and individuals retrieved from data services would permit a system to use the semantics from the thesaurus itself and provide more information to a researcher, more effectively, and would further enable the use of ontology reasoning mechanisms, in order to infer additional information on these individuals not explicitly stated either in the thesaurus or in the data sources. While such inferred information could be processed on each data source, it has been noted that it may be more efficient to perform reasoning during query processing. 37
We are currently exploring ways to automatically detect potential linkages between concepts in NCI Thesaurus and individuals from data services, through mechanisms that we have developed for the alignment of ontologies, 38 and we are devising mechanisms to incorporate reasoning into the processing of queries.
Human-Computer Interactions
In order to achieve the goal of permitting researchers to use conceptual abstractions to formulate queries and explore data, we are working to design and develop simple, clean, and easily understandable human-computer interaction paradigms that allow scientists to create queries effectively and efficiently. These user interfaces are designed to contain the following functionality:
• Exploration of the ontology representation of caBIG created through automated procedures that generate a view of the semantics in NCI Thesaurus and caDSR as an ontology. This interface provides the ability to perform lexical searches on ontology concepts and to navigate through the properties within the ontology view, in order to select the concepts on which queries will be formulated.
• Graphical assembly of queries and rules, such that the query terms selected through exploration are concatenated into queries and rules, together with variables defined by the user. Both rules and queries are designed to be reusable. Rules are associated with queries through an assisted process where a user either defines a new rule, or selects a rule from a library of rules relevant to a query; this relevance is determined, and verified, by ensuring that some classes and/or properties in the rule are also referenced in the query.
• Presentation of results in tabular and graphical formats.
Query Processing
We are working towards the enhancement of our software components used for querying caGrid data services. Our current prototype requires the identification of data services to be queried through manually-fed URIs: it is necessary to integrate the caGrid Index Service in order to automatically discover the location of the desired data services.
The current application also requires some manual processing of queries in order to separate them into queries on different sources. We have defined elsewhere a query algebra for SPARQL meant to be used for query manipulation; the combination of this algebra and the semCDI query formulation is being developed to perform automated SPARQL query processing.
Conclusions
In this paper, the Semantic caGrid Data Integration (semCDI) query formulation has been detailed, as a methodology designed to create queries at a conceptual level against the rich semantic information contained in caBIG. We have developed a view of these caBIG semantics as an ontology represented in OWL, and have presented ways in which queries can be built using the SPARQL query language complemented with Horn rules. Two examples of queries against caGrid Data Services have been detailed to validate the formulation. We have also discussed the limitations to semCDI due to the quantity and structure of CDEs in caDSR, the need for mechanisms to specify unique variable bindings, and the lack of links between data instances and NCI Thesaurus specific concepts. The semCDI query formulation, in conclusion, enables the creation of queries on the semantic representation of caBIG, thus constituting the foundation needed to build a caBIG semantic data integration system.
Footnotes
This work is supported by NIH grant 1R43CA132293. The authors also wish to acknowledge the contribution of Mr. Thomas Taylor and Mr. Michael Ryan of INFOTECHSoft, Inc., and the insights given by Drs. Thomas Deisboeck at Massachusetts General Hospital and Drs. Robert Clark and Stephen Byers at Georgetown University. The intellectual property rights for the semCDI query formulation presented in this paper are held by INFOTECHSoft, Inc.
References
- 1.Collins FS, Green ED, Guttmacher AE, Guyer MS. A vision for the future of genomics research Nature 2003;422(6934):835-847. [DOI] [PubMed] [Google Scholar]
- 2.Kitano H. Systems biology: a brief overview Science 2002;295(5560):1662-1664. [DOI] [PubMed] [Google Scholar]
- 3.Deisboeck TS, Zhang L, Martin S. Advancing cancer systems biology: introducing the Center for the Development of a Virtual Tumor, CViT Cancer Informatics 2007;2:1-8. [PMC free article] [PubMed] [Google Scholar]
- 4.Neumann E. A life science Semantic Web: are we there yet? Sci STKE 2005;2005(283):pe22. [DOI] [PubMed] [Google Scholar]
- 5.Neumann E, Quan D. Biodash: A Semantic Web dashboard for drug development Pac Symp On Biocomp 2006;11:176-187. [PubMed] [Google Scholar]
- 6.Wang X, Gorlitsky R, Almeida JS. From XML to RDF: how semantic web technologies will change the design of ‘omic’ standards Nat Biotechnol 2005;23(9):1099-1103. [DOI] [PubMed] [Google Scholar]
- 7.Schroeder M, Neumann E. Editorial: Semantic web for the life sciences J Web Sem 2006(4):3.
- 8.Rodriguez MA, Egenhofer MJ. Determining Semantic Similarity among Entity Classes from Different Ontologies IEEE Trans on Knowledge and Data Eng 2003;15(2):442-456. [Google Scholar]
- 9.Gruber TR. Toward principles for the design of ontologies used for knowledge sharing. Technical report KSL 93-04, Knowledge Systems Laboratory, Stanford University. ftp://ftp.ksl.stanford.edu/pub/KSL_Reports/KSL-93-04.ps.gz 2003. accessed November 2006.
- 10.Saltz J, Oster S, Hastings S, Langella S, Kurc T, Sanchez W, Kher M, Manisundaram A, Shanbhag K, Covitz P. caGrid: design and implementation of the core architecture of the cancer biomedical informatics grid Bioinform 2006;22(15):1910Schroeder M and Neumann E 6. [DOI] [PubMed] [Google Scholar]
- 11.http://www.cagrid.org/mwiki/index.php?title=CaGrid:FAQ#What_is_the_relationship_of_the_caDSR.2C_EVS.2C_and_caGrid_metadata_and_how_can_I_use_them.3F 2006. Accessed on: June 9, 2008.
- 12.Tobias J, Chilukuri R, Komatsoulis GA, Mohanty S, Sioutos N, Warzel DB, Wright LW, Crowley RS. The CAP cancer protocols–a case study of caCORE based data standards implementation to integrate with the Cancer Biomedical Informatics Grid BMC Med Inform Decis Mak 2006;6:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Cancer Institute Center for Bioinformatics, caCORE 3.2 Technical Guide, Revised 12/22/2006.
- 14.Hartel F, Coronado S, Dionne R, Fragoso G, Golbeck J. Modeling a Description Logic Vocabulary for Cancer Research J Biomed Inform 2005;38:114Schroeder M and Neumann E 29. [DOI] [PubMed] [Google Scholar]
- 15.Golbeck J, Fragoso G, Hartel F, Hendler J, Oberthaler J, Parsia B. The National Cancer Institutes Thesaurus and Ontology, Web Semantics Science, Services and Agents on the World Wide Web 2003;1:75-80. [Google Scholar]
- 16.http://www.cagrid.org/mwiki/index.php?title=Data_Services:CQL 2003. Accessed on: June 9, 2008.
- 17.National Cancer Institute caArray 1.4 Technical Guidehttp://gforge.nci.nih.gov/frs/download.php/1091/caArray_1_4_Technical_Guide.pdf 2003. Accessed on: June 9, 2008.
- 18.Wu CH, Nebert DW. Update on human genome completion and annotations: Protein Information Resource Hum Gen 2004;1:229-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Cancer Institute caBIG Function Express Maintenance and Extension Guide Version 1.0http://cabigcvs.nci.nih.gov/viewcvs/viewcvs.cgi/functionexpress/Deliverables/Maintenance and Extension Guide/ 2004. Accessed on: May 23, 2008.
- 20.Louie B, Mork P, Martin-Sanchez F, Halevy A, Tarczy-Hornoch P. Data integration and genomic medicine J Biomed Inform 2007;40(1):5-16Epub 2006 Mar 9. [DOI] [PubMed] [Google Scholar]
- 21.Kurc T, Janies DA, Johnson AD, Langella S, Oster S, Hastings S, Habib F, Camerlengo T, Ervin D, Catalyurek UV, Saltz JH. An XML-based system for synthesis of data from disparate databases J Am Med Inform Assoc 2006;13(3):289-301May-Jun;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohler J, Philippi S, Lange M. SEMEDA: ontology based semantic integration of biological databases Bioinform 2003;19(18):2420-2427Dec 12. [DOI] [PubMed] [Google Scholar]
- 23.Wang K, Tarczy-Hornoch P, Shaker R, Mork P, Brinkley JF. BioMediator data integration: beyond genomics to neuroscience data AMIA Annu Symp Proc 2005:779-783. [PMC free article] [PubMed]
- 24.Alonso-Calvo R, Maojo V, Billhardt H, Martin-Sanchez F, Garcia-Remesal M, Perez-Rey D. An agent and ontology-based system for integrating public gene, protein, and disease databases J Biomed Inform 2007;40(1):17-29Epub 2006 Mar 20. [DOI] [PubMed] [Google Scholar]
- 25.Duke Comprehensive Center caTRIP User Manual Version 1.0http://gforge.nci.nih.gov/docman/view.php/131/6482/caTRIP_End_User_manual.doc 2007. Accessed on: June 9, 2008.
- 26.Sioutos N, de Coronado S, Haber MW, Hartel FW, Shaiu WL, Wright LW. NCI Thesaurus: a semantic model integrating cancer-related clinical and molecular information J Biomed Inform 2007;40(1):30-43. [DOI] [PubMed] [Google Scholar]
- 27.Smith MK, Welty C, McGuinness DL. OWL web ontology language guide. W3C Recommendation, 10 February 2004. http://www.w3.org/TR/owl-guide/ 2007. Accessed on: June 9, 2008.
- 28.Prud'hommeaux E, Seaborne A. SPARQL Query Language for RDFhttp://www.w3.org/TR/rdf-sparql-query/ 2007. Accessed on: June 9, 2008.
- 29.Boley H, Kifer M. RIF Basic Logic Dialect. W3C Working Draft 30 October 2007. http://www.w3.org/TR/rif-bld/ 2007. Accessed on: Jan 18, 2008.
- 30.De Bruijn J. RIF RDF and OWL Compatibility. W3C Working Draft 30 October 2007. http://www.w3.org/TR/rif-rdf-owl/ 2007. Accessed on: Jan 18, 2008.
- 31.Malhotra A, Melton J, Walsh N. XQuery 1.0 and XPath 2.0 Functions and Operators. W3C Recommendation 23 January 2007. http://www.w3.org/TR/xpath-functions/ 2007. Accessed on: June 9, 2008.
- 32.http://cagrid-portal.nci.nih.gov 2007. Accessed on: June 9, 2008.
- 33.http://pid.nci.nih.gov/ 2007. Accessed on: June 9, 2008.
- 34.National Cancer Institute Center for Bioinformatics caGrid 1.1 Programmers Guidehttp://gforge.nci.nih.gov/frs/download.php/2385/caGrid-1-1_Programmers_Guide.pdf 2007. Accessed on: June 9, 2008.
- 35.Athale CA, Deisboeck TS. The effects of EGF-receptor density on multiscale tumor growth patterns J Theor Biol 2006;238(4):771-779Feb 21. [DOI] [PubMed] [Google Scholar]
- 36.http://umlmodelbrowser.nci.nih.gov/umlmodelbrowser/ 2006. Accessed on: June 9, 2008.
- 37.Anyanwu K, Sheth A. ρ-Queries: enabling querying for semantic associations on the semantic web. Proc 12th Intl Conf World Wide Web, Budapest, Hungary 2003:690-699.
- 38.Jean-Mary YR, Kabuka M. ASMOV: Ontology Alignment with Semantic Validation. Joint SWDB-ODBIS Workshop on Semantics, Ontologies, Databases, 2007. Accepted for publication.