Abstract
Reversible acetylation of lysine residues within a protein is considered a biologically relevant modification that rivals phosphorylation (Kouzarides, T. (2000) EMBO J. 19,1176 -1179). The enzymes responsible for such protein modification are called histone acetyltransferases (HATs) and deacetylases (HDACs). A role of protein phosphorylation in regulating muscle contraction is well established (Solaro, R. J., Moir, A. J., and Perry, S. V. (1976) Nature 262 ,615 -617). Here we show that reversible protein acetylation carried out by HATs and HDACs also plays a role in regulating the myofilament contractile activity. We found that a Class II HDAC, HDAC4, and an HAT, PCAF, associate with cardiac myofilaments. Primary cultures of cardiomyocytes as well as mouse heart sections examined by immunohistochemical and electron microscopic analyses revealed that both HDAC4 and PCAF associate with the Z-disc and I- and A-bands of cardiac sacromeres. Increased acetylation of sarcomeric proteins by HDAC inhibition (using class I and II HDAC inhibitors or anti-HDAC4 antibody) enhanced the myofilament calcium sensitivity. We identified the Z-disc-associated protein, MLP, a sensor of cardiac mechanical stretch, as an acetylated target of PCAF and HDAC4. We also show that trichostatin-A, a class I and II HDAC inhibitor, increases myofilament calcium sensitivity of wild-type, but not of MLP knock-out mice, thus demonstrating a role of MLP in acetylation-dependent increased contractile activity of myofilaments. These studies provide the first evidence that HATs and HDACs play a role in regulation of muscle contraction.
Histone deacetylases (HDACs)2 and acetyltransferases (HATs) constitute two separate families of enzymes, which were originally characterized as nuclear enzymes modifying histones (3, 4). In mammals over a dozen HDAC family members have been identified, which can be classified into three different classes based on their structure, complex formation, and expression pattern (5, 6). All members of the HDAC family contain a highly homologous catalytic domain; however, sequences outside the catalytic domain are highly divergent, suggesting that these enzymes might have different biological functions and a broader substrate repertoire beyond histones (7). Recently, several non-histone proteins, including MyoD, YY1, Ku70, p53, and tubulin have been identified as substrates of HDACs (reviewed in Refs. 1 and 7).
HDAC4 is member of class II HDACs. Members of this group are highly expressed in the heart, brain, and skeletal muscle and possess a unique ability of shuttling in and out of nucleus. In myocytes phosphorylation of HDAC4 by calcium/calmodulin-dependent kinase-II generates binding sites for the 14-3-3 protein and promotes its export from the nucleus to the cytoplasm (8). In contrast MAPK/ERK1-dependent phosphorylation causes importation of HDAC4 into the nucleus (9). In the nucleus HDAC4 associates with MEF2 and serum response factor and represses the transcription of muscle genes harboring MEF2 and serum response factor binding sites (10, 8). The role of HDAC4 outside the nucleus is virtually unknown. In this report we present evidence for the first time showing that HDAC4 and an acetyltransferase, PCAF, associate with cardiac sarcomeres and play a role in regulation of cardiac muscle contraction.
MATERIALS AND METHODS
Plasmid Constructs—The constructs Myc-HDAC4 (11), pcGAL4-HDAC4 (11), GST-HDAC4-AA208-310 (12), GST-MLP full-length, GST-MLP-LIM1 or LIM2 (13), and FLAG-hMLP (14) have been described elsewhere. Truncations of human HDAC4 were generated by PCR from pcGAL4-HDAC4 and inserted into the appropriate restriction sites of pcDNA3-FlagA. The following primers were used: HDAC4-AA1-210, forward primer (5′-CGGGATCCCATGAGCTCCCAAAGCCATCCAGATGG-3′) and reverse (5′-CCGCTCGAGCTAGGAACTGTGCTGCGTTTTCCCGTACC-3′); HDAC4-AA304-620, forward primer (5′-CGGAATTCAACAACAGCTCCGGGAGCGTCAGC-3′) and reverse (5′-CCGCTCGAGCTAGGACACGGGGATGCCGGCGGCCTCC-3′); and for HDAC4-AA612-1084, forward primer (5′-CGGAATTCATGGAGGCCGCCGGCATCCCCGTGTCC-3′) and reverse (5′-CGGATATCCTACAGGGGCGGCTCCTCTTCCATGG-3′). Wild-type and mutant GST constructs for the MLP-LIM1 spanning from 62 to 119 amino acids were constructed by PCR from GST-MLP full-length plasmid using the following primers: WT forward, 5′-CCGATCTAGACTATGGGCGCAAGTATGGCCCCAAGGGG-3′ ; K69A forward, 5′-CCGATCTAGACCGCAAGTATGGCCCCGCGGGGATCGGG-3′; and a common reverse primer, 5′-CCGCTCGAGCTATTACTTCTCTGATTCTCCAAACTTCGC-3′. Purified PCR products were inserted into XbaI-XhoI sites of pGex-KG vector. All PCRs were carried out using proofreading Pfu Turbo Cx Hotstart DNA polymerase (Stratagene).
Antibodies Used—The following antibodies and conjugates were used in this study: rabbit anti-HDAC4 (ab12172, Abcam, and sc11418, Santa Cruz); mouse anti-HDAC4 (sc-46672, Santa Cruz); rabbit anti-HDAC1 (sc-7872, Santa Cruz); rabbit anti-histone H2A (2578, Cell Signaling); rabbit anti-cMyc (sc-789, Santa Cruz); chicken MLP (ab14013, Abcam); rabbit anti-PCAF (sc-8999, Santa Cruz); rabbit anti-acetyl lysine (06-933, Upstate); mouse anti-α sarcomeric actinin (A7811, Sigma); goat anti-actin (sc-1616, Santa Cruz); mouse anti-GST (sc-138, Santa Cruz); goat anti-integrin β1 (sc-6622, Santa Cruz); goat anti-RNA polymerase II (sc-5943, Santa Cruz); mouse anti-cMyc (9E10) agarose beads (sc-40 AC, Santa Cruz); anti-rabbit SIRT1 (ab28170, Abcam), anti-cardiac myosin (ab15, Abcam), anti-FLAG M2 affinity gel (A2220, Sigma), rhodamine-phalloidin (R415, Invitrogen); goat anti-rabbit IgG-HRP (sc-2054, Santa Cruz); donkey anti-mouse IgG-HRP (sc-2096, Santa Cruz); donkey anti-goat IgG-HRP (sc-2056, Santa Cruz) and bovine anti-chicken IgY-HRP (sc-2917, Santa Cruz); donkey anti-mouse IgG-fluorescein isothiocyanate (sc-2099, Santa Cruz); donkey anti-goat IgG-Alexa Fluor 594 (A11058, Invitrogen) and donkey anti-rabbit IgG-Alexa Fluor 594 (A21207, Invitrogen). HDAC4 peptide used for blocking of the antibody activity was from Abcam.
Cell Culture and Transfection—Primary cultures of 2-day-old neonatal rat heart myocytes were carried out using an established procedure described previously (10). Adult rabbit heart myocytes were isolated from New Zealand White rabbits (2.2-2.7 kg) by retrograde collagenase perfusion (15). Rabbits were sedated with intramuscular injection of ketamine (40 mg/kg) and xylazine (6 mg/kg), the heart was rapidly excised and perfused in a retrograde Langendorff mode for 35 min with type II collagenase (Worthington Biochemical Corp.) in calcium-free Tyrode solution. After perfusion the ventricles were gently dissociated in 2.5% bovine serum albumin (Sigma) in Tyrode solution and cells were filtered through 200-μm2 pore nylon mesh (Spectrum Labs). Calcium concentration was increased from 50 to 800 μm over a period of 1 h. Cells were suspended in M199 medium (Sigma) supplemented with penicillin (100 units/ml, Invitrogen), streptomycin (0.1 mg/ml, Invitrogen), 10 μm cytosine-1-β-d-arabinofuranoside (Sigma), and 5% fetal bovine serum. Cells (20,000-50,000) were seeded on laminin (1 μg/cm2, Sigma)-coated coverslips in 20-mm2 culture plates with 2 ml of growth medium. Cells were allowed to attach for 12 h and, thereafter, fresh supplemented medium was applied to cells. Twenty-four to 30 h after seeding cells were used for immunostaining as described below. COS-7 cells were grown in Dulbecco's modified Eagle's medium supplemented with penicillin-streptomycin and 10% fetal bovine serum (complete growth medium). Cells were transfected with appropriate plasmids using Superfect transfection reagent (Qiagen) according to the manufacturer's protocol. All animal protocols were approved by the University of Chicago's Institutional Animal Care and Use Committee.
Subcellular Fractionation, Immunoprecipitations, and Western Analyses—Subcellular protein fractions of mouse hearts were prepared using a Proteoextract subcellular Proteome Extraction Kit (Calbiochem) according to the manufacturer's protocol. For protein sample preparations from heart papillary muscles or skinned fibers, tissue was homogenized in an appropriate volume of 6× SDS-PAGE sample loading buffer without β-mercaptoethanol. Protein concentration was determined using the detergent-compatible BCA kit (Pierce). β-Mercaptoethanol was added before boiling the samples. The whole cell lysate was prepared by incubating cell pellets (2 × 107 cells) in 1 ml of RIPA buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.4, 1 mm EDTA, 1% Triton X-100, 0.1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, protease inhibitor mixture, Sigma), passed through 21-gauge needle to shear the DNA and incubated on ice for 40 min with intermittent vortexing every 10 min. Lysates were centrifuged at 10,000 × g for 10-15 min in the cold and clear supernatants were used as whole cell lysates. For immunoprecipitation, whole cell lysate, lysate prepared from skinned fibers, or nuclear extracts from mouse heart (500-700 μg) were pre-cleared with Protein A/G plus (Santa Cruz) for 30 min at 4 °C. Beads were pelleted at 1000 × g for 30 s and pre-cleared supernatants were incubated with 10-20 μg of primary antibody-agarose conjugates at 4 °C overnight on a rotator. When agarose or a gel conjugate was unavailable, lysates were incubated with primary antibody or an equivalent amount of control IgG for 2 h at 4 °C and then overnight along with Protein A/G plus beads to collect the immune complexes. Beads were collected by centrifugation, washed several times with RIPA buffer, one wash with PBS, and resuspended in SDS-PAGE sample loading buffer. Immune complexes and 75-100 μg of input proteins were resolved by SDS-PAGE. Western blot analyses were performed using appropriate antibodies as mentioned in figure legends.
GST Pull-down Assay—GST, GST-MLP full-length, GST-LIM1, GST-LIM2, and GST-HDAC4-AA208-311 were purified as described before (16). For pull-down assays, 50 μg of GST or GST fusion proteins purified using glutathione-Sepharose 4B beads (GE Healthcare) were incubated for 2 h at 4 °C on a rotator with the in vitro translated [35S]methionine-labeled HDAC4-Myc or hMLP-FLAG proteins as appropriate. Beads were washed initially with a buffer containing 200 mm NaCl, 50 mm Tris-HCl (pH 7.5), 0.5% Nonidet P-40, 1 mm dithiothreitol, protease inhibitor mixture (Sigma) and 1% bovine serum albumin. Bound proteins were sequentially washed again with the same buffer containing 350 and 600 mm NaCl, three times with each buffer followed by a rinse in PBS. Bound complexes were resolved by SDS-PAGE and detected by autoradiography.
In Vitro Acetylation Assay of MLP—GST-MLP full-length, GST.LIM1, GST.LIM2, GSTΔLIM1 (aa 62-119), or GSTΔLIM1.K69A were used as a substrate for in vitro acetylation. Briefly, 6 μg of substrate protein bound to glutathione-Sepharose beads (GE Healthcare) were resuspended in 1× HAT buffer (Upstate Biotechnology). A typical acetylation reaction was comprised of 1 μg of active PCAF enzyme (Upstate Biotechnology), 3 μg of active p300-HAT enzyme, 250 nCi of [1-14C]acetyl-CoA (Moravek Biochemicals), and 10 mm sodium butyrate in 1× HAT buffer. Reactions were incubated at 30 °C for 30-60 min on a rotator. Reactions were terminated by adding SDS sample buffer and resolved on 12% SDS-PAGE gel. Proteins were transferred to Hybond-P membrane (GE Healthcare) and detected by autoradiography. The same membrane was also used for Western analysis with anti-GST antibody to demonstrate that equal amounts of GST proteins were used for the assay.
In Vitro Acetylation-Deacetylation Assay of MLP—For this assay, unlabeled FLAG-MLP was translated in vitro using the TnT coupled Transcription/Translation rabbit reticulocyte lysate kit (Promega) according to the manufacturer's instructions. FLAG-MLP was immunoprecipitated from the reaction using anti-FLAG M2 affinity beads. Beads with bound proteins were washed 4-5 times with RIPA buffer followed by a PBS wash. Final wash was given in 1× HAT buffer (Upstate Biotechnology). The acetylation reaction was carried out as described above except only active PCAF enzyme was used that was found to be sufficient to carry out acetylation. For the deacetylation assay, acetylated FLAG-MLP bound to beads was washed as above and resuspended in 1× HDAC buffer (Upstate Biotechnology). This acetylated FLAG-MLP substrate was either incubated just in 1× HDAC buffer (control) or with 10 μg of HeLa cell lysates or with equal amounts of Myc-HDAC4 or Myc tag protein immobilized onto Myc (9E10, Santa Cruz)-agarose beads in 1× HDAC buffer. To obtain Myc tag proteins, COS7 cells were transfected with the respective plasmids and 24 h post-transfection proteins were immunoprecipitated using the Myc-agarose beads. These beads were washed with 1× RIPA buffer 4-5 times, once with PBS, and once with a final wash of 1× HDAC buffer and resuspended in 20 μl of the same buffer. Five and fifteen μl of these resuspended beads were used for the deacetylation reaction. Reaction mixtures were incubated for 2 h at 37 °C on a rotator. Proteins were resolved by 12% SDS-PAGE and subjected to autoradiography after transferring to a Hybond-P membrane.
Immunostaining—Neonatal rat cardiomyocytes (10,000-20,000) plated on 1% laminin-coated coverslips were used for immunostaining after 2 and 7 days of culturing. Cells were washed briefly in 1× PBS and fixed for 15 min in 3.7% formaldehyde at room temperature. After 3 washes of 1× PBS, cells were permeabilized with 0.1% Triton X-100 for 5 min and washed again 3 times with 1× PBS. Cells were incubated at room temperature for 1 h in blocking solution (5% donkey serum, 1% bovine serum albumin in 1× PBS) and then in a primary antibody overnight at 4 °C in a humid chamber. The next day, after 3 washes of 1× PBS containing 0.02% Tween 20 and 1 wash of 1× PBS (10 min each wash), cells were incubated for 1 h with appropriate secondary antibody conjugated with either fluorescein isothiocyanate (green) or Alexa Fluor 594 (red). Cells were washed as before and mounted in Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (Vector Laboratories). Adult rabbit cardiomyocytes were similarly processed for immunostaining. For F-actin visualization, rhodamine-phalloidin (Invitrogen) staining of cells was done according to the manufacturer's instructions. For immunofluorescent staining of adult mouse heart, 7-μm thick cryosections of ventricular tissue were fixed in chilled acetone for 5 min at room temperature, rinsed in 1× PBS, and then processed as above. All microscopy and imaging analyses were done in the digital light microscopy core facility at the University of Chicago.
Electron Microscopy—Freshly isolated mouse heart ventricular tissue was fixed in 0.2% gluteraldehyde and embedded in L.R. white resin. Post-embedding staining of sections was done using a primary antibody rabbit anti-HDAC4 (ab12172, Abcam), rabbit anti-PCAF (sc-8999, Santa Cruz), or rabbit antihistone H2A (2578, Cell Signaling). Primary antibody staining was detected using goat anti-rabbit IgG conjugated to 10-nm gold beads (Ted Pella Inc.). Transmission electron microscopy and imaging were carried out at the electron microscopy core facility at the University of Chicago.
Skinned Cardiac Papillary Muscle Fibers—Mice were anesthetized using isoflurane followed by cervical dislocation. The heart was rapidly removed and placed in ice-cold PBS. The heart was then transferred to relaxing solution containing: 40 mm BES (pH 7.0), 20 mm KCl, 1 mm free Mg2+, 5 mm MgATP, 10 mm creatine phosphate, 20 mm EGTA, 1 mm dithiothreitol, 0.01 mm leupeptin, 0.1 mm phenylmethylsulfonyl fluoride (180 mm ionic strength). Papillary muscles were dissected out and cut into strips and skinned for 4 h in relaxing solution with 1% Triton X-100 at 4 °C. Skinned fibers were then treated overnight with HDAC inhibitor, acetyl-CoA, or vehicle at 4 °C.
Steady-state Force-pCa Relationship—Skinned muscle bundles were mounted in a 750-μl bath and were attached to a length controller (model 322B, Aurora Scientific) at one end and a force transducer (model 403A, Aurora Scientific) at the other end using aluminum T-clips (KEM-MIL-CO). Average bundle dimensions were ∼1-mm long by 180-μm wide. Sarcomere length was then set using laser diffraction methods (10 milliwatt HeNe laser, Spectra-Physics, Mountain View, CA) to 1.9 μm. The muscle bundle was then fully activated twice prior to generation of force-pCa curves.
Statistical Analysis—Data are expressed as mean ± S.E. Data from the normalized force-pCa curves were fitted using a nonlinear regression (variable slope) equation of the graphical software GraphPad (San Diego, CA) to obtain pCa50 and Hill coefficient values. Statistical differences among groups were determined using either Student's t test (for two groups) or one-way analysis of variance (for more than two groups) followed by Fisher's least significant difference test for post hoc pairwise comparisons if necessary.
RESULTS
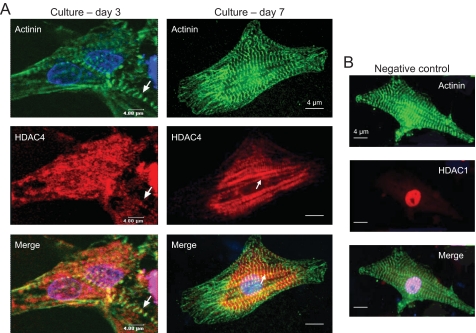
To examine the expression pattern of HDAC4 in cardiac myocytes, we double-stained neonatal rat cardiomyocytes with HDAC4 and α-actinin-specific antibodies and localized the protein expression by using confocal microscopy. As expected, staining for α-actinin, a Z-disc-associated protein, uncovered striations of sarcomeres, the contractile units of cardiomyocytes (Fig. 1). When myocytes were examined for HDAC4 localization, we found that the deacetylase was present both in the cytoplasm and in the nucleus of the cell. Much to our surprise we noticed that the myocytes that were grown for a relatively longer period (7 days) and had well developed sarcomeres showed a striated pattern of HDAC4 staining (Fig. 1). This striated pattern of HDAC4 expression was, however, difficult to see in younger myocytes cultured for 3 days or less, not having well defined sarcomeres (Fig. 1). To validate these results further, we performed a similar experiment with cardiomyocytes isolated from adult rabbit hearts (Fig. 2). Again, as with neonatal rat myocytes, we found HDAC4 localization to sarcomeres of cardiomyocytes. These results were intriguing because, so far, no HDAC isoform of any class has been shown to be associated with cardiac myofilaments.
FIGURE 1.
HDAC4 is localized to sarcomeres of neonatal rat heart myocytes. A, cardiomyocytes grown for 3 or 7 days were stained with α-actinin and HDAC4-specific antibodies. Nuclei were localized by 4′,6-diamidino-2-phenylindole staining. The white arrow indicates the striated pattern of HDAC4 staining in myocytes. B, a negative control in which neonatal rat heart myocytes were stained with α-actinin and HDAC1-specific antibodies. Note the exclusive nuclear localization of HDAC1 in the cardiomyocyte.
FIGURE 2.
Localization of HDAC4 on sarcomeres of adult rabbit heart myocytes. Adult rabbit heart myocytes were stained with rhodamine-phalloidin to visualize F-actin or stained with HDAC4 primary antibody and fluorescein isothiocyanate-conjugated secondary antibody. Note that HDAC4 was localized both in nuclei and to sarcomeres of the cell.
To confirm these results, we analyzed the adult mouse heart sections stained with HDAC4 and α-actininspecific antibodies. Both α-actinin and HDAC4 revealed striated patterns of expression that overlapped neatly (Fig. 3A), indicating that HADC4 is likely to be a Z-disc-associated deacetylase. When these heart sections were analyzed at higher magnification, we found that HDAC4 was localized not only to the Z-disc, but also between Z-discs where no α-actinin staining (no yellow in merge) could be seen (Fig. 3B). As a negative control, we stained cardiomyocytes with four other antibodies against histones and HDACs (e.g. HDAC1, SIRT1, H2A, and HDAC6), and none of them showed a striated pattern of staining (Figs. 1B and 3D, HDAC1 staining). We also included another negative control in which heart sections were stained with HDAC4 antibody that was blocked with the peptide, and again found no striated pattern of staining (Fig. 3C). These results strongly indicated that the observed HDAC4 staining of sarcomeres was specific.
FIGURE 3.
HDAC4 is localized to Z-disc and I- and A-bands of the sarcomere. A, adult mouse heart sections stained with α-actinin and HDAC4-specific antibodies. Nuclei were localized by 4′,6-diamidino-2-phenylindole (DAPI) staining. The white arrows indicate that HDAC4 was localized to the Z-discs of the sarcomere. B, analysis of the mouse heart sections at higher magnification. Arrows indicate localization of HDAC4 not only to the Z-discs, but also between these structures (see merge picture, white arrow). C, a negative control in which HDAC4 antibody activity was blocked with the peptide. D, a negative control in which the heart section was stained with HDAC1 specific antibody. E-G, representative electron micrographs of mouse heart sections stained with HDAC4 or H2A antibodies. Numbers at the bottom indicate magnification. N and MYO in panel E represent the position of the nucleus and myofilaments, respectively. High density black dots (arrows) indicate localization of HDAC4. In panel G, stained with H2A antibody, different regions of the sarcomere (Z-disc, I- and A-bands, and M line) are shown. Note that HDAC4 is present at Z disc and on I- and A-bands, but not at the M line of the sarcomere (panel F).
To obtain firm evidence for HDAC4 localization to sarcomeres, we next performed immunoelectron microscopy. Immunogold particles, as reflected by high-density black dots, depicting HDAC4, were found to be localized in the nucleus as well as to the Z-discs of sarcomeres (Fig. 3E). At higher magnification, HDAC4 was found mostly at the Z-disc, but a small portion of the deacetylase was also present at the “I” and “A” bands of sarcomeres (Fig. 3F). However, no HDAC4 was found at the M-line of sarcomeres. These results were substantiated by use of three different sources of HDAC4 antibody as well as by use of different negative controls, such as no primary antibody, nonspecific IgG, or H2A antibody (Fig. 3G). Taken together, these results demonstrated that HDAC4 associates with cardiac sarcomeres.
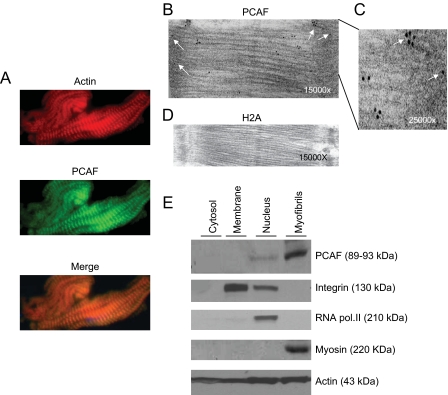
The effect of HDACs is counteracted by HATs (17). We, therefore, wanted to investigate whether a member of the HAT family was also localized to sarcomeres. Mouse heart sections were immunostained with antibodies against PCAF and p300, two commonly known HATs. We found that PACF antibody exhibited an obvious striated pattern of expression, which over-lapped with actin staining, indicating that PCAF is likely to be a sarcomere-associated acetyltransferase (Fig. 4A). However, there was no staining of myofilaments with the p300 antibody (data not shown). We next performed an immunoelectron microscopic analysis to confirm these results. High-density immunogold particles (black dots), representing PCAF, were localized to the Z-disc and the I- and A-bands of sarcomeres (Fig. 4, B and C). These are the same sarcomere structures with which HDAC4 was found to be associated. Because, PCAF immunostaining was less intense than the HDAC4, which could be related to the reactivity of the antibody, we decided to confirm PCAF localization to myofilaments by using an alternative method. For this purpose, we prepared four subcellular fractions of the adult mouse myocardium: (i) enriched in cytoskeleton and myofibrils (myofibrils), (ii) nuclear fraction (nucleus), (iii) containing intracellular proteins associated with other subcellular organelles (membrane), and (iv) cytosolic fraction (cytosol). These subcellular fractions were characterized by using antibodies against fraction-specific proteins. As shown in Fig. 4E, RNA polymerase II was present only in the nuclear fraction, and myosin was seen only in the myofibrillar fraction, whereas integrin was present in both the nuclear and the membrane-bound fractions. Actin was present in all four fractions, and as expected, it was enriched in myofibrillar and nuclear fractions. When these fractions were analyzed for PCAF expression, we found that the acetyltransferase was present only in the nuclear and myofibrillar fractions, and it was, surprisingly more enriched in the myofibrillar fraction (Fig. 4E). This higher level of PCAF expression in the myofibrillar fraction may be related to the multitude of sarcomere units present in a cell as opposed to one or two nuclei of cardiomyocytes.
FIGURE 4.
Localization of PCAF to cardiac sarcomeres. A, representative pictures of adult mouse heart sections stained with rhodamine-phalloidin for F-actin. PCAF was localized by immunostaining using PCAF-specific primary antibody and fluorescein isothiocyanate-conjugated secondary antibody. B, an electron micrograph of the heart section stained with PCAF antibody. High density black dots (white arrows) indicate the localization of PCAF. C, enlarged section of the Z-disc and I-band region of the electron micrograph in panel B. D, heart section stained with H2A antibody, a negative control. E, different subcellular fractions of a mouse heart were prepared as described under “Materials and Methods.” Fractions were characterized by Western blot analyses using antibodies against fraction-specific proteins as given at the right side of each panel. Note that PCAF was present both in the nuclear and the myofibrillar fractions, but not in the membrane or the cytosolic fractions of the heart.
Knowing that HDAC4 and PCAF associate with cardiac sarcomeres, we next examined the functional significance of these enzymes. Skinned fibers from mouse heart papillary muscles were used for this purpose. Skinning of myofibers with Triton X-100 (1%) removes all membranous structures including the nucleus, sarcolemma, sarcoplasmic reticulum, mitochondria, and other subcellular organelles, leaving only intact myofilaments (18). After skinning, myofilaments were treated overnight either with vehicle (Me2SO) or 200 nm trichostatin A (TSA), a class I and II HDAC inhibitor. The next morning fibers were mounted in a bath, and force-pCa data were obtained at a constant sarcomere length (1.9 μm). TSA treatment significantly increased the calcium sensitivity of myofilaments, as quantified by pCa50 (Fig. 5, A and B; vehicle, 5.72 ± 0.02; TSA, 5.84 ± 0.02; p < 0.05), without any significant changes in the maximally activated force, Fmax (Fig. 5B; vehicle: 53.0 ± 3.3; TSA, 51.0 ± 6.1 milli-newton mm2-) or the Hill coefficient (vehicle, 3.55 ± 0.40; TSA, 4.22 ± 0.70). To test whether this effect was related to the chemical structure of TSA, we studied the effect of two other structurally dissimilar HDAC inhibitors, MS-275 and Scriptaid (19). Results indicated that all three HDAC inhibitors caused a similar increase in the myofilament calcium sensitivity (Fig. 5B), suggesting that HDAC inhibition, and not any particular structure of the inhibitors, was responsible for this observation. To test whether HDAC4 inhibition was indeed involved in altered myofilament contractile activity, we performed the same experiment with myofibers treated with anti-HDAC4 antibody. Fibers treated with nonspecific IgG were used as negative controls. As shown in Fig. 5, A-C, treatment of myofibers with HDAC4 antibody significantly increased calcium sensitivity of myofilaments without changing the maximally activated force. However, IgG treatment had no effect on mechanical parameters of myofibers. Thus, these data indicated that HDAC4 does in fact participate in regulating the myofilament contractile activity.
FIGURE 5.
Class I and II HDAC inhibitors increase Ca2+ sensitivity of myofilaments. A, force-pCa curves generated from skinned fibers that were treated with vehicle (Me2SO or IgG at 50 μg/ml, n = 5), the HDAC inhibitor trichostatin A (200 nm, n = 6), or anti-HDAC4 antibody (50 μg/ml, n = 6). Note, there is a significant increase (Student's t test, p < 0.05) in Ca2+ sensitivity as denoted by a leftward shift in the force-pCa curve generated from TSA and HDAC4 antibody-treated fibers, compared with vehicle-treated fibers. Values are mean ± S.E. B, Ca2+ sensitivity of skinned fibers treated with other HDAC inhibitors, MS-275 (5 μm) and Scriptaid (10 μm), and acetyl-CoA (AcCoA, 1 mm). There is a significant increase in Ca2+ sensitivity in all skinned fibers treated with HDAC inhibitors and acetyl-CoA (one-way analysis of variance, p < 0.05). Values are mean ± S.E. (n = 4-7). C, maximal activated force generated from different treatments. D and E, after measuring mechanical activity, skinned fibers were analyzed by Western blot analysis. Papillary (Pap.) muscle samples not subjected to skinning were utilized as controls. Note the presence of HDAC4 and PCAF in both papillary muscle and skinned fiber samples. However, other nuclear proteins such as SIRT1 and H2A were not detected in skinned fibers. *, signifies statistically significant compared with control (vehicle).
HDAC inhibition is expected to increase the acetylation of the target proteins. Therefore, to complement HDAC inhibition experiments, we also obtained the force-pCa data from skinned fibers treated with 1 mm acetyl-coenzyme A. Similar to the effects of TSA, an increase in calcium sensitivity was observed following acetyl-CoA treatment (Fig. 5B; acetyl-CoA, 5.84 ± 0.04; p < 0.05 versus vehicle). Thus, the HDAC inhibition and acetyl-CoA results suggested that the reversible acetylation of myofilament proteins can regulate the calcium sensitivity of myofilaments.
After measuring mechanical activity, we homogenized the skinned fibers in Laemmli buffer and subjected them to Western analysis to determine the localization of HDAC4 and PCAF with myofilaments. Protein bands corresponding to HDAC4 and PCAF were detected from skinned fibers as well as from intact papillary muscle samples (Fig. 5, D and E). However, no evidence for the presence of other nuclear proteins, such as SIRT1 or H2A, was found on the skinned fibers. On the contrary, these molecules were present in the papillary muscle samples. Thus, these data again confirmed that HDAC4 and PCAF associate with myofilament structures.
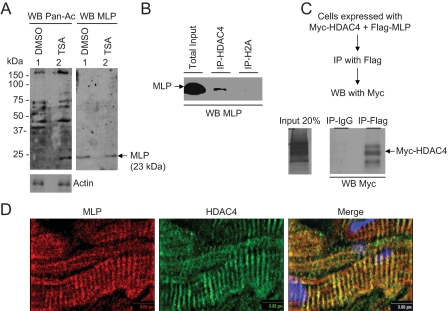
We next asked whether TSA treatment did indeed alter the acetylation of sarcomeric proteins. Skinned myofibers, incubated overnight with TSA or vehicle, were subjected to Western analysis with an acetyl-lysine antibody. We found a notable increase in the acetylation of several myofilament proteins following TSA treatment. Among them, a protein running below 25 kDa was consistently recognized by the acetyl-lysine antibody from TSA-treated samples (Fig. 6A). To identify this protein, we probed the same membrane with antibodies against different sarcomeric proteins with molecular masses of less than 25 kDa: for example, troponin-I, troponin-C, myosin light chain-1, myosin light chain-2, and MLP (20). We found that the acetylated protein band running below the 25-kDa marker was identical to the band recognized by the MLP antibody, suggesting that MLP of sarcomeres might be a substrate of acetylation (Fig. 6A). These experiments also gave us a clue that MLP could be a sarcomeric partner protein binding to HDAC4. To explore this possibility, we first performed co-immunoprecipitation experiments using a cardiac cell lysate. As shown in Fig. 6B, MLP was successfully pulled down by the HDAC4 antibody, but not by the H2A antibody or by IgG alone, which served as negative controls. We also performed an inverse experiment in which HDAC4 was sought to be pulled down by MLP. For this experiments we co-transfected COS7 cells with plasmids encoding FLAG-MLP and Myc-HDAC4. Cell extract was prepared and subjected to immunoprecipitation with an anti-FLAG antibody. The resulting beads were analyzed by the Western blot analysis using anti-Myc antibody. As shown in Fig. 6C, Myc-HDAC4 was co-immunoprecipitated with FLAG-MLP and not with IgG alone. To obtain further evidence for interaction of HDAC4 with MLP, we examined the co-localization of both proteins on sarcomeres. Mouse heart sections were immunostained with HDAC4 and MLP antibodies, and the expression pattern of proteins was examined by use of color-tagged secondary antibodies. As before, HDAC4 revealed a striated pattern of expression and that overlapped with MLP staining of sarcomeres (Fig. 6D), suggesting that both are Z-disc-associated proteins. These data indicated that both native and recombinant MLP and HDAC4 bind to each other in vivo, and that they associate with the Z-discs of cardiac sarcomeres.
FIGURE 6.
MLP of sarcomeres is acetylated and recognized by HDAC4. A, skinned fibers treated overnight with 200 nm TSA or vehicle (Me2SO) were analyzed by Western blot (WB) using an acetyl-lysine antibody (Pan-Ac). The same membrane was probed with an anti-MLP antibody (WB-MLP). Note the 23-kDa band representing MLP was highly acetylated following TSA treatment. B, co-immunoprecipitation (Co-IP) of native MLP with HDAC4. Mouse heart lysate was subjected to co-IP using either anti-HDAC4 or anti-H2A antibodies. The resultant beads were analyzed by Western analysis using anti-MLP antibody (WB-MLP). C, co-IP of recombinant HDAC4 with MLP. COS7 cells were co-transfected with plasmids expressing Myc-HDAC4 and FLAG-MLP. Cell lysate was subjected to IP with anti-FLAG antibody and the resultant beads analyzed by Western blot using an anti-Myc antibody. D, co-localization of MLP and HDAC4 on the Z-discs of cardiac sarcomeres. A typical adult mouse heart section stained with MLP (red) and HDAC4 (green) antibodies is shown.
Because proteins in vivo could associate by either direct or indirect means, we performed GST pull-down assays to demonstrate a direct physical interaction between these two proteins. Full-length HDAC4 and its different truncated versions were in vitro synthesized as 35S-labeled proteins. Synthesis of proteins was verified by SDS-PAGE and samples having >90% of the expected molecular weight of proteins utilized for further analysis. As shown in Fig. 7C, the full-length HDAC4 was pulled down with GST-MLP, but not the truncated version of HDAC4 having 1-210, 304-620, or 612-1084 aa fragments, indicating that the MLP-binding region of HDAC4 must lie between aa 210 and 304. To test the binding potential of this region we obtained a GST-HDAC4 containing 208-311-aa fragment of the deacetylase. This fusion protein was examined for its ability to pull down in vitro synthesized full-length MLP. As shown in Fig. 7E, 35S-labeled MLP was successfully pulled down by GST-HDAC4-(208-311), but not by GST alone. In this series of experiments we also examined the binding ability of two truncated versions of MLP, one containing the LIM1 domain and the other having the LIM2 domain of MLP. We found that HDAC4 binds to the LIM1 domain, and not to the LIM2 domain of the MLP protein (Fig. 7D). From these results we conclude that the N-terminal fragment of HDAC4 spanning the 208-304-aa region interacts with the LIM1 domain of MLP.
FIGURE 7.
GST pull-down assay to localize protein-binding domains. A, Coomassie-stained GST fusion proteins used in the assay. B, input (20%) of in vitro-synthesized radiolabeled HDAC4 peptides of different length. C, radiolabeled HDAC4 peptides were incubated with GST or GST-MLP on beads, as indicated at the bottom of the picture. Beads were repeatedly washed and proteins bound to beads were analyzed by SDS-PAGE followed by autoradiography. D, radiolabeled HDAC4 was subjected to a pull-down assay with GST-MLP or GST having the LIM1 or LIM2 domains of MLP. Note HDAC4 binds to the LIM1 domain of MLP. E, in vitro synthesized radiolabeled MLP was incubated with GST or GST-HDAC4 containing a 208-311-aa fragment of the deacetylase. Note that this truncated peptide of HDAC4 was sufficient to bind to MLP. F, summary of MLP binding ability to different segments of HDAC4.
Results described above demonstrated that MLP can be acetylated when it is present on sarcomeres. To provide additional evidence to this observation, we examined acetylation of MLP in in vitro assay conditions. Recombinant GST-MLP was incubated with [1-14C]acetyl-CoA in an acetylation reaction buffer containing GST-PCAF and GST-p300HAT as provided with the acetylation kit. The protein acetylation was detected by SDS-PAGE followed by autoradiography. We found that GST-MLP was highly acetylated, but not the GST alone (Fig. 8A). In this assay, we also analyzed the acetylation ability of two truncated fragments of MLP containing either LIM1 or the LIM2 domain. We found that the LIM1 domain of MLP was acetylated to the same extent as the full-length MLP; however, no acetylation of the LIM2 domain was detected (Fig. 8A). By analyzing different point mutations within the LIM1 domain we identified conserved lysine 69, situated within the motif YGP-KGIG, as a target amino acid acetylated by HATs (Fig. 8C). In these blots, we could also detect two slow migrating bands of autoacetylated GST-PCAF and GST-p300HAT, which served as positive controls.
FIGURE 8.
MLP is acetylated on lysine 69 residue. A, GST-MLP and its derivatives were subjected to acetylation by incubating with [1-14C]acetyl-CoA in a buffer containing GST-p300HAT and GST-PCAF. Reactions carried out with GST alone or no enzyme served as negative control. B, the same membrane was subjected to Western (WB) analysis with an anti-GST antibody. C, lysine 69 of the LIM1 domain of MLP is acetylated. GST, GST-MLP, GSTΔLIM1 (62-119-aa fragment of LIM1 domain), or GSTΔLIM1K69A mutant were subjected to in vitro acetylation. Arrows with the open head indicate position of autoacetylated HATs and with the closed head represent the position of GST-MLP and its derivatives. D, deacetylation of MLP by HDAC4. In a deacetylation reaction buffer, acetylated FLAG-MLP was incubated with different additions, as indicated. In lanes 2 and 3 and 4 and 5, equal increments of Myc beads (5 and 15 μl) containing no HDAC4 (lanes 2 and 3) or wild-type HDAC4 (lanes 4 and 5) were added, respectively. In lane 6, HeLa extract having deacetylase activity was used as a positive control. Lower panel, Western blot with FLAG antibody (WB-Flag) was used as a loading control. E, quantification (mean of 3 experiments) of acetylated (Ac) FLAG-MLP.
We next asked whether MLP could be deacetylated by HDAC4. For this experiment, in vitro synthesized full-length FLAG-MLP was immunoprecipitated with the FLAG antibody and then subjected to in vitro acetylation with PCAF. We noticed that PCAF alone was sufficient to acetylate FLAG-MLP and inclusion of p300 in the reaction mixture had no additional effect. To test deacetylase activity of HDAC4, acetylated FLAG-MLP was incubated in a deacetylation reaction buffer with in vitro synthesized full-length HDAC4. We found, however, no detectable deacetylation of FLAG-MLP by this experiment (results not shown). We reasoned that this negative result could be either due to lack of catalytic activity of in vitro synthesized HDAC4, or the enzyme needs other accessory factors for its deacetylase activity. To test this possibility, we then examined the deacetylase activity of in vivo synthesized HDAC4. Cells were overexpressed with plasmids synthesizing Myc-HDAC4 or Myc tag alone. Proteins were immunoprecipitated using Myc-specific antibody and the resulting beads were incubated with acetylated FLAG-MLP. As shown in Fig. 8, D and E, we found that beads containing Myc-HDAC4 had the ability to deacetylate FLAG-MLP in a concentration-dependent manner (lanes 4 and 5), but not the Myc only protein (lanes 2 and 3). In this assay, HeLa cell extract having the deacetylase activity was used as a positive control (lane 6). These results thus demonstrated that HDAC4 from the in vivo source has the ability to deacetylate MLP.
Finally, to test whether MLP is required for an acetylation-mediated change in myofilament calcium sensitivity we examined the effect of TSA on skinned papillary muscle fibers obtained from MLP knock out (-/-) mice (21). For the control of the MLP-/- mice, we utilized skinned fibers obtained from the same background strain, C57BL6 mice. As shown in Fig. 9A, there was a significant increase in calcium sensitivity of the skinned cardiac fibers from the wild-type mice (C57BL6 strain) treated with 200 nm TSA (Fig. 9A). However, no effect of TSA treatment was observed in skinned cardiac fibers obtained from MLPKO mice (Fig. 9B), whereas HDAC4 and PCAF both were present on these fibers, as detected by the Western analysis (Fig. 9C). These results demonstrated that MLP is involved in the acetylation-mediated change in myofilament calcium sensitivity.
FIGURE 9.
MLP is required for acetylation-mediated change in calcium sensitivity of myofilaments. A and B, force-pCa curves generated in skinned fibers from C57BL6 wild-type (WT, n = 6) or MLP knock-out (MLPKO, n = 6) mice treated with 200 nm TSA or vehicle (Me2SO). MLP knock-out mice utilized in this study were young males (4 weeks old) with no apparent signs of cardiac abnormality. In skinned fibers from WT mice, TSA treatment resulted in a significant leftward shift (as shown in the inset) of the force-pCa curve denoting an increase in Ca2+ sensitivity (p < 0.05). In contrast, in the skinned fibers from the MLP knock-out mice no such change in Ca2+ sensitivity was observed. In both cases, TSA treatment did not significantly alter maximally activated force (WT, vehicle, 38.0 ± 2.1; WT, TSA, 44.2 ± 6.0; MLPKO, vehicle, 42.1 ± 6.1; MLPKO, TSA, 43.4 ± 3.6 milli-newton mm2-) and Hill coefficient (WT, vehicle, 2.33 ± 0.06; WT, TSA, 2.35 ± 0.19; MLPKO, vehicle, 1.70 ± 0.21; MLPKO, TSA, 1.44 ± 0.06). Values are mean ± S.E. C, following measurement of mechanical parameters, skinned fibers were homogenized and analyzed by the Western analysis. Papillary (Pap.) muscle samples not subjected to skinning were utilized as controls. Note the presence of both HDAC4 and PCAF in the wild-type (WT) and MLPKO mice. H2A was utilized as negative control of myofilaments and actin as a loading control.
DISCUSSION
In this paper we report two novel findings. First, reversible acetylation of sarcomeric proteins, like phosphorylation, plays a role in regulation of muscle contraction. By using various biochemical and biophysical techniques, we found that HDAC4 and PCAF bind to cardiac sarcomeres and inhibition of HDACs by several inhibitors increases calcium sensitivity of cardiac myofilaments. Second, we identified MLP, a Z-disc-associated protein, as a target of PCAF and HDAC4. MLP was acetylated in vitro as well as in vivo when associated with myofilaments in its native state. We delineated lysine 69 of the LIM1 domain as an acetylated residue of MLP. In contrast to the data from wild-type mice, myofilaments from MLP-/- mice were resistant to the TSA-mediated change in calcium sensitivity, thus suggesting a role of MLP in the acetylation-dependent increase in myofilament contractile activity. These studies underscore the importance of reversible acetylation in regulation of cardiac muscle contraction.
HDAC4 belongs to the class II family of histone deacetylases. Although most members of this class are localized mainly in the cytoplasm and nucleus, one member, HDAC7, has been also shown to be present in mitochondria (8, 9, 22). Under normal growth conditions HDAC4 is localized in both the cytoplasm and nucleus, and the proportion of HDAC4 between the two compartments varies greatly depending upon cell-type and the stage of cell development. In proliferating C2C12 myoblasts and NIH 3T3 fibroblasts, HDAC4 is mostly expressed in the cytoplasm and it is imported into the nucleus following activation of the Ras-MAPK pathway or DNA damage (9, 23). In contrast, HDAC4 expression in cardiomyocytes is nearly 50:50 between the nucleus and cytoplasm (8). HDAC4 is phosphorylated by calcium/calmodulin-dependent kinase-II during hypertrophy of myocytes that promotes exportation of HDAC4 from the nucleus to the cytoplasm (8). Inside the nucleus HDAC4 represses transcriptional activity of MEF2, leading to repression of muscle gene expression. A recent study has shown that the repression of MEF2 transcription activity by HDAC4 is independent of its deacetylase domain (24). Similarly, others have reported that the catalytic domain of HDAC4 has minimal or no deacetylase activity of its own; and whatever enzymatic activity seen with in vivo precipitated HDAC4 is from factors associating with the HDAC4 complex. In our study, we found that the TSA-induced increase in myofilament calcium sensitivity can be reproduced by incubating fibers with an anti-HDAC4 antibody as well as by treating fibers with acetyl-CoA. These observations suggest that: (a) acetylation of fibers indeed increases the calcium sensitivity of myofilaments, and (b) HDAC4 inhibition does in fact contribute to this effect.
Experiments carried out to delineate sarcomeric partner of HDAC4 led to the identification of MLP, a Z-disc protein, as a target of the deacetylase. Our attempts to demonstrate deacetylation of MLP by in vitro synthesized HDAC4 were unsuccessful, which is consistent with other reports (24). However, when HDAC4 was precipitated from an in vivo source the resulting beads were able to deacetylate MLP successfully. These results suggest that either a cofactor associating with HDAC4 or a protein modification may enhance the deacetylase activity of HDAC4. Recently, Fischle et al. (25) have reported that the active complex of HDAC4 contains HDAC3 and corepressors SMRT and N-CoR, and the deacetylase activity of the complex was due to the presence of HDAC3. HDAC3 is primarily a nuclear protein (26); however, one group has shown a notable amount of HDAC3 expression also in the cytoplasm (27). We tried to demonstrate co-localization of HDAC3 and HDAC4 on cardiac sarcomeres; but remained unsuccessful. We also tested the possibility of whether HDAC6, an enzymatically active member of class II HDAC that is present in the cytoplasm, could associate with sarcomeres; however, these results were again negative (28).
Other possibilities could be that HDAC4 is modified by a post-translational mechanism and/or it binds to an unknown protein of sarcomeres that potentiates its deacetylase activity. Recently, HDAC4 has been shown to be sumoylated, a post-translation modification that alters its ability to repress gene transcription as well as reduces its histone deacetylase activity (29). It is note-worthy that most of the studies carried out to date for deacetylase activity of HDAC4 have utilized a canonical acetyl-lysine residue of histones as a substrate. A recent report has shown that HDAC4 is nearly a thousand-fold more active on a non-canonical synthetic substrate having trifluoroacetyl-lysine, which is proposed to destabilize the amide bond of lysine (24). Based on this report, it is reasonable to think that there could be other natural substrates, perhaps outside the nucleus, with less stable amide bonds that are targeted by HDAC4. Future studies directed toward the goal of finding out the mechanism of deacetylation of the sarcomeric target proteins by HDAC4 should be able to distinguish between these possibilities.
A recent study has reported that chronic HDAC inhibition can attenuate the pathologic aspects of the cardiac hypertrophy secondary to pressure overload (30). Specifically, TSA-induced HDAC inhibition for 3 weeks in mice subjected to pressure overload resulted in attenuation of hypertrophy and increased left ventricular systolic function and contractile state as compared with vehicle-treated mice with pressure overload. The increased left ventricular contractile performance in TSA-treated mice was partially attributed to an attenuation of the myosin heavy chain isoform switch (from α-myosin heavy chain to β-myosin heavy chain) that is associated with pressure-overload cardiac hypertrophy. Our data regarding enhanced myofilament contractile activity (as indexed by increased calcium sensitivity) following TSA treatment are qualitatively consistent with these in vivo results. However, because our observations were made with relatively acute HDAC inhibition (overnight incubation of muscle fibers with TSA versus 3-week TSA treatment in the in vivo study), they are likely due to a direct effect of increased myofilament protein acetylation and not due to any myosin heavy chain isoform switch. It is possible that this direct effect of acetylation was also present in the in vivo study. If so, the increased acetylation of myofilament proteins may be another adaptive process that preserves myofilament contractile activity in pressure overload cardiac hypertrophy.
A role of reversible phosphorylation of sarcomeric proteins in regulation of myofilament contractile activity is well established (reviewed in Refs. 2 and 31). For example, it is well known that protein kinase A-dependent phosphorylation of cardiac troponin I at the serine residue leads to decreased calcium sensitivity of myofilaments. In contrast, phosphorylation of cardiac troponin I at the threonine residue by p21-activated kinase-3 is associated with increased myofilament calcium sensitivity. Whereas the protein kinase C-dependent phosphorylation of myofilaments reduces the maximal calcium-activated force, its effects on myofilament calcium sensitivity is known to be isoform-specific. In the present study we report a acetylation-induced increase in myofilament calcium sensitivity of about 0.12 pCa50 units. This magnitude of change is comparable with that due to protein kinase A-mediated phosphorylation (ΔpCa50 ∼ 0.15) (32), although the two responses are in the opposite directions. Thus, it appears that reversible acetylation of sarcomeric proteins may be another significant pathway for post-translational regulation of myofilament contractile activity.
Our data showing reversible acetylation of MLP and lack of effect of HDAC inhibitors on MLP-null myofibers clearly indicate that MLP is involved in a acetylation-dependent change in myofilament contractile activity. However, a basic question remains, how does MLP acetylation enhances myofilament contractile activity? Based on current knowledge of the role of MLP and other Z-disc proteins in the regulation of myofilament contraction, at least two possibilities could be envisioned. First, a growing body of evidence supports the notion of a functionally active Z-disc, i.e. Z-disc-associated proteins are involved in regulating intracellular signaling pathways and myofilament activation. For example, CapZ, the cardiac actin-capping protein, has been shown to play an important role in regulating the protein kinase C signaling to the myofilaments (33, 34). Cardiac muscles from a transgenic mouse with reduced levels of CapZ protein has been shown to exhibit increased myofilament calcium sensitivity. This effect is mediated by the reduction in the amount of myofilament-bound β2 isoform of protein kinase C and consequently decreased phosphorylation of myofilament proteins (troponin I, troponin T, and myosin binding protein C) (34). It is likely that the MLP acetylation status plays a similar role in regulating protein kinase signaling to the myofilaments. The second possible mechanism is based on the observations that MLP interacts with a number of other Z-disc proteins (reviewed in Ref. 35), including the titin filament capping protein T-cap/telethonin (21). Titin, a sarcomeric giant elastic protein, acts as a molecular spring and is responsible for the passive force generation in the sarcomere (36). Because titin connects thick and thin filaments in an oblique manner, titin-based passive force has a radial component that can modulate interfilament lattice spacing, which, in turn, can alter myofilament calcium sensitivity: higher passive force pulls filaments together reducing lattice spacing and consequently, increasing myofilament calcium sensitivity (37). It is possible that MLP acetylation affects the interaction between MLP and T-cap/telethonin, which in turn, can change titin-based passive tension and myofilament calcium sensitivity. It is also important to consider that, although we found a large proportion of HDAC4 present on the Z-disc, a notable amount of HDAC4 was also detected at the I- and A-bands of the sarcomeres. Thus, a possibility remains that there could be other sarcomeric target proteins, in addition to MLP, that are post-translationally modified by reversible acetylation. Studies are underway in our laboratories to identify new targets of HDAC4 and to find out the mechanism of acetylation mediated regulation of cardiac muscle contraction. Whatever may be the ultimate mechanism(s), the data presented in this study clearly demonstrate that protein acetylation is another layer of regulation controlling muscle contraction. These data provide impetus to understand the role of HDACs outside the nucleus and suggest that these enzymes might have a much broader role in regulating the cell function than previously realized.
Acknowledgments
We thank the following investigators for their generous gift of plasmids used in this study: Drs. T. Kouzarides, E. N. Olson, S. F. Konieczny, and N. E. Bowles. MLP knock-out mice were kindly provided by Dr. M. Hoshijima and C57BL6 strain wild-type mice were obtained from Dr. C. P. O'Donnell. We also thank Dr. Francesca Davis for technical assistance in the beginning part of the study.
This work was supported in part by National Institutes of Health Grants RO1 HL-68083, HL-77788, and HL83423 and McGinnis Endowed Chair funds. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HDAC, histone deacetylase; HAT, histone acetyl-transferase; TSA, trichostatin A; MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated kinase; MEF, muscle enhancer factor; GST, glutathione S-transferase; HRP, horseradish peroxidase; PBS, phosphate-buffered saline; aa, amino acid; BES, 2-[bis(2-hydroxyethyl)amino]ethanesulfonic acid; MLP, muscle LIM protein.
References
- 1.Kouzarides, T. (2000) EMBO J. 191176 -1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solaro, R. J., Moir, A. J., and Perry, S. V. (1976) Nature 262615 -617 [DOI] [PubMed] [Google Scholar]
- 3.Cheung, W. L., Briggs, S. D., and Allis, C. D. (2000) Curr. Opin. Cell Biol. 12 326-333 [DOI] [PubMed] [Google Scholar]
- 4.Struhl, K. (1998) Genes Dev. 12 599-606 [DOI] [PubMed] [Google Scholar]
- 5.Cress, W. D., and Seto, E. (2000) J. Cell. Physiol. 1841 -16 [DOI] [PubMed] [Google Scholar]
- 6.Khochbin, S., Verdel, A., Lemercier, C., and Seigneurin-Berny, D. (2001) Curr. Opin. Genet. Dev. 11 162-166 [DOI] [PubMed] [Google Scholar]
- 7.Glozak, M. A., Sengupta, N., Zhang, X., and Seto, E. (2005) Gene (Amst.) 363 15-23 [DOI] [PubMed] [Google Scholar]
- 8.Little, G. H., Bai, Y., Williams, T., and Poizat, C. (2007) J. Biol. Chem. 2827219 -7231 [DOI] [PubMed] [Google Scholar]
- 9.Zhou, X., Richon, V. M., Wang, A. H., Yang, X. J., Rifkind, R. A., and Marks, P. A. (2000) Proc. Natl. Acad. Sci. U. S. A. 9714329 -14333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, F. J., Gupta, M., Camoretti-Mercado, B., Schwartz, R. J., and Gupta, M. P. (2003) J. Biol. Chem. 27820047 -20058 [DOI] [PubMed] [Google Scholar]
- 11.Miska, E. A., Karlsson, C., Langley, E., Nielsen, S. J., Pines, J., and Kouzarides, T. (1999) EMBO J. 185099 -5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, C. L., McKinsey, T. A., Chang, S., Antos, C. L., Hill, J. A., and Olson, E. N. (2002) Cell 110479 -488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohapatra, B., Jimenez, S., Lin, J. H., Bowles, K. R., Coveler, K. J., Marx, J. G., Chrisco, M. A., Murphy, R. T., Lurie, P. R., Schwartz, R. J., Elliott, P. M., Vatta, M., McKenna, W., Towbin, J. A., and Bowles, N. E. (2003) Mol. Genet. Metab. 80 207-215 [DOI] [PubMed] [Google Scholar]
- 14.Flick, M. J., and Konieczny, S. F. (2000) J. Cell Sci. 113,1553 -1564 [DOI] [PubMed] [Google Scholar]
- 15.Mitcheson, J. S., Hancox, J. C., and Levi, A. J. (1998) Cardiovasc. Res. 39 280-300 [DOI] [PubMed] [Google Scholar]
- 16.Gupta, M. P., Amin, C. S., Gupta, M., Hay, N., and Zak, R. (1997) Mol. Cell. Biol. 173924 -3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischle, W., Kiermer, V., Dequiedt, F., and Verdin, E. (2001) Biochem. Cell Biol. 79 337-348 [PubMed] [Google Scholar]
- 18.Harrison, S. M., Lamont, C., and Miller, D. J. (1988) J. Physiol. 401115 -143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marks, P. A., Richon, V. M., Breslow, R., and Rifkind, R. A. (2001) Curr. Opin. Oncol. 13 477-483 [DOI] [PubMed] [Google Scholar]
- 20.Clark, K. A., McElhinny, A. S., Beckerle, M. C., and Gregorio, C. C. (2002) Annu. Rev. Cell Dev. Biol. 18 637-706 [DOI] [PubMed] [Google Scholar]
- 21.Knoll, R., Hoshijima, M., Hoffman, H. M., Person, V., Lorenzen-Schmidt, I., Bang, M. L., Hayashi, T., Shiga, N., Yasukawa, H., Schaper, W., McKenna, W., Yokoyama, M., Schork, N. J., Omens, J. H., McCulloch, A. D., Kimura, A., Gregorio, C. C., Poller, W., Schaper, J., Schultheiss, H. P., and Chien, K. R. (2002) Cell 111943 -955 [DOI] [PubMed] [Google Scholar]
- 22.Bakin, R. E., and Jung, M. O. (2004) J. Biol. Chem. 27951218 -51225 [DOI] [PubMed] [Google Scholar]
- 23.Basile, V., Mantovani, R., and Imbriano, C. (2006) J. Biol. Chem. 2812347 -2357 [DOI] [PubMed] [Google Scholar]
- 24.Lahm, A., Paolini, C., Pallaoro, M., Nardi, M. C., Jones, P., Neddermann, P., Sambucini, S., Bottomley, M. J., Lo Surdo, P., Carfi, A., Koch, U., De Francesco, R., Steinkuhler, C., and Gallinari, P. (2007) Proc. Natl. Acad. Sci. U. S. A. 10417335 -17340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischle, W., Dequiedt, F., Hendzel, M. J., Guenther, M. G., Lazar, M. A., Voelter, W., and Verdin, E. (2002) Mol. Cell 945 -57 [DOI] [PubMed] [Google Scholar]
- 26.Emiliani, S., Fischle, W., Van Lint, C., Al-Abed, Y., and Verdin, E. (1998) Proc. Natl. Acad. Sci. U. S. A. 952795 -2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang, W. M., Tsai, S. C., Wen, Y. D., Fejer, G., and Seto, E. (2002) J. Biol. Chem. 2779447 -9454 [DOI] [PubMed] [Google Scholar]
- 28.Bertos, N. R., Gilquin, B., Chan, G. K., Yen, T. J., Khochbin, S., and Yang, X. J. (2004) J. Biol. Chem. 27948246 -48254 [DOI] [PubMed] [Google Scholar]
- 29.Kirsh, O., Seeler, J. S., Pichler, A., Gast, A., Muller, S., Miska, E., Mathieu, M., Harel-Bellan, A., Kouzarides, T., Melchior, F., and Dejean, A. (2002) EMBO J. 212682 -2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong, Y., Tannous, P., Lu, G., Berenji, K., Rothermel, B. A., Olson, E. N., and Hill, J. A. (2006) Circulation 1132579 -2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Layland, J., Solaro, R. J., and Shah, A. M. (2005) Cardiovasc. Res. 6612 -21 [DOI] [PubMed] [Google Scholar]
- 32.Kajiwara, H., Morimoto, S., Fukuda, N., Ohtsuki, I., and Kurihara, S. (2000) Biochem. Biophys. Res. Commun. 272104 -110 [DOI] [PubMed] [Google Scholar]
- 33.Pyle, W. G., Hart, M. C., Cooper, J. A., Sumandea, M. P., de Tombe, P. P., and Solaro, R. J. (2002) Circ. Res. 901299 -1306 [DOI] [PubMed] [Google Scholar]
- 34.Pyle, W. G., La Rotta, G., de Tombe, P. P., Sumandea, M. P., and Solaro, R. J. (2006) J. Mol. Cell. Cardiol. 41537 -543 [DOI] [PubMed] [Google Scholar]
- 35.Hoshijima, M. (2006) Am. J. Physiol. 290H1313 -H1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Granzier, H., and Labeit, S. (2007) Muscle Nerve 36740 -755 [DOI] [PubMed] [Google Scholar]
- 37.Fukuda, N., Wu, Y., Farman, G., Irving, T. C., and Granzier, H. (2005) Pflugers Arch 449449 -457 [DOI] [PubMed] [Google Scholar]