Abstract
Macrophage activation participates pivotally in the pathophysiology of chronic inflammatory diseases, including atherosclerosis. Through the receptor EP4, prostaglandin E2 (PGE2) exerts an anti-inflammatory action in macrophages, suppressing stimulus-induced expression of certain proinflammatory genes, including chemokines. We recently identified a novel EP4 receptor-associated protein (EPRAP), whose function in PGE2-mediated anti-inflammation remains undefined. Here we demonstrate that PGE2 pretreatment selectively inhibits lipopolysaccharide (LPS)-induced nuclear factor κB1 (NF-κB1) p105 phosphorylation and degradation in mouse bone marrow-derived macrophages through EP4-dependent mechanisms. Similarly, directed EPRAP expression in RAW264.7 cells suppresses LPS-induced p105 phosphorylation and degradation, and subsequent activation of mitogen-activated protein kinase kinase 1/2. Forced expression of EPRAP also inhibits NF-κB activation induced by various proinflammatory stimuli in a concentration-dependent manner. In co-transfected cells, EPRAP, which contains multiple ankyrin repeat motifs, directly interacts with NF-κB1 p105/p50 and forms a complex with EP4. In EP4-overexpressing cells, PGE2 enhances the protective action of EPRAP against stimulus-induced p105 phosphorylation, whereas EPRAP silencing in RAW264.7 cells impairs the inhibitory effect of PGE2-EP4 signaling on LPS-induced p105 phosphorylation. Additionally, EPRAP knockdown as well as deficiency of NF-κB1 in macrophages attenuates the inhibitory effect of PGE2 on LPS-induced MIP-1β production. Thus, PGE2-EP4 signaling augments NF-κB1 p105 protein stability through EPRAP after proinflammatory stimulation, limiting macrophage activation.
Macrophages participate in the pathogenesis of many chronic inflammatory diseases, including atherosclerosis (1–3), the metabolic syndrome (4, 5), cancer (6, 7), and autoimmunity. Thus, the regulation of macrophage activation holds a key to understanding the pathophysiology and rational treatment of these conditions.
In response to various stimuli, the sequential actions of cyclooxygenase (COX)3 and PGE synthase produce PGE2 from arachidonic acid. PGE2 has four known G-protein-coupled receptors, designated EP1–EP4. Each EP receptor shows distinct tissue distribution and tightly regulated expression, suggesting the pivotal roles of PGE2-EP receptor signaling in maintaining local homeostasis under a variety of pathophysiological settings (8).
Macrophages express EP4 more abundantly than other PGE receptors, such as EP2. EP4 as well as EP2 mainly couple to Gs asaGα subunit and increase intracellular cAMP levels upon PGE2 ligation. In animals, EP4 signaling protects against experimental inflammatory bowel disease (9) and reduces inflammatory bone resorption (10). In addition, we previously reported that PGE2 markedly suppressed production of a number of chemokines in LPS-stimulated human primary macrophages (11). Notably, PGE2 pretreatment selectively diminished responses to various proinflammatory stimuli by macrophages but not by vascular endothelial cells or smooth muscle cells. Further data indicated involvement of EP4 signaling in this anti-inflammatory function of PGE2 (11). We then isolated a novel EP4 receptor-associated protein (EPRAP), using the yeast two-hybrid screening method with a human bone marrow cDNA library (12).
EPRAP contains eight ankyrin repeat motifs but has no predicted enzymatic or catalytic domain. The EPRAP counterpart in mice, Fem1a (13), has homology with Caenorhabditis elegans FEM-1, which participates in nematode sex determination (14), yet the biological functions of mammalian Fem1 families remain unknown.
Because ankyrin repeat commonly mediates protein-protein interactions between molecules that figure importantly in signal transduction, including Notch and inhibitor of NF-κB(IκB) family proteins (15, 16), we hypothesized that EPRAP interacts with proinflammatory signaling molecules and inhibits macrophage activation. Here we demonstrate that EPRAP associates directly with NF-κB1 p105/p50. Through EP4/EPRAP-dependent mechanisms, PGE2 attenuates stimulus-induced phosphorylation and degradation of p105, an important cytoplasmic inhibitor of NF-κB and MEK activation. PGE2 also augments IL-10 production in LPS-stimulated macrophages independently of EP4 and EPRAP.
These observations add new insights into the mechanisms that may mitigate unchecked macrophage activation at sites of inflammation and suggest EP4-EPRAP signaling as a novel target for the treatment of chronic inflammatory diseases.
EXPERIMENTAL PROCEDURES
Reagents, Antibodies, and cDNA Constructs—PGE2 and Butaprost were from Cayman Chemical (Ann Arbor, MI), LPS (Escherichia coli O55: B5) was from Calbiochem. Human recombinant TNFα and IL-1β were from Pierce. Antibodies against phospho-p105 (Ser933), phospho-IκBα (Ser32/36), phospho-MEK1/2 (Ser217/221), MEK1/2, p65, and β-actin were from Cell Signaling Technology (Beverly, MA); anti-mouse p105/p50 antibody was from Abcom (Cambridge, MA); anti-FLAG antibody was from Sigma; anti-V5 and anti-lamin B1 antibodies were from Invitrogen; and antibodies against p105/p50, Tpl2, α-tubulin, and GFP were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The expression vectors for C terminus FLAG-tagged human EP4, C terminus V5-tagged human EPRAP and deletion mutants, and C terminus GFP-tagged human NF-κB family proteins (p65, p50, p105, and p105-ΔN) were constructed by PCR and subcloned into p3XFLAG-CMV-14 (Sigma), pcDNA3.1/V5-His (Invitrogen), and pGFP2-N vector (PerkinElmer Life Sciences), respectively. Mouse EP4 as well as EPRAP expression constructs were also generated by PCR and subcloned into pIRES2-eGFP vector (Clontech). All cDNA constructs were verified by DNA sequencing.
Mice—Homozygotic EP4-deficient mice (EP4–/–) and wild type (WT) on the same genetic background (EP4+/+) were obtained by crossing mice heterozygous for ptger4 gene mutation (EP4+/–) and verified as previously described (17). Because EP4–/– mice only survive on a recombinant inbred strain, all EP4–/– and EP4+/+ mice used in this study were on the mixed background, composed of 129/Olac, C57BL/6, and DBA/2 (17). IL-10–/– mice (18), nfkb1–/– mice (19), and their WT controls (nfkb1+/+) were purchased from the Jackson Laboratory (Bar Harbor, ME). Mice were bred in a specific pathogen-free environment. All experiments were performed under protocols approved by the Animal Research Committee of Harvard Medical School and in accordance with institutional guidelines.
Cell Culture—Mouse bone marrow-derived macrophages (BMDM) were prepared as previously described (20) with minor modifications. Briefly, bone marrow cells were isolated from 8–10-week-old male mice and plated in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum and 10 ng/ml mouse recombinant macrophage colony-stimulating factor (Cell Sciences, Canton, MA). After 7 days of culture, adherent macrophages were harvested for further experiments. RAW264.7 cells (ATCC number TIB-71) and HEK293 cells (ATCC number CRL-1573) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 2 mm glutamine. Transient transfection in HEK293 cells was performed with FuGENE HD Reagent (Roche Applied Science), and transfection in RAW264.7 cells was performed using Nucleofector Kit (Amaxa, Germany) according to the manufacturer's instructions.
RNA Isolation and Quantitative Real Time PCR (qPCR)—Total RNA was isolated using an RNeasy Micro kit (Qiagen, Valencia, CA), and cDNA was synthesized with QuantiTect Reverse Transcription (Qiagen). The mRNA level for each target gene was quantified by SYBR Green-based qPCR using the iCycler iQ real time PCR detection system (Bio-Rad). Primer sequences used for this study are available as supplemental material (Table S1). Data were normalized using β-actin as a reference gene, and quantification of the results was performed by the standard curve method using a plasmid containing each target PCR fragment as a cDNA control (21). All reactions were performed in triplicate.
Protein Analyses—To detect the interaction between EPRAP and NF-κB1, cells were lysed with Buffer A (20 mm Hepes, 150 mm sodium chloride, 1 mm EDTA, 1 mm EGTA, 10% glycerol, 1% Brij 58, and protease inhibitor mixture (Sigma)). Protein extracts were precleared by nonimmune control IgG, followed by immunoprecipitation with antibody against either V5, GFP, or p105/p50, coupled with Protein A/G-agarose beads (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). To detect the binding of EP4 to EPRAP or NF-κB1, cells were lysed with Buffer B (20 mm Hepes, 150 mm sodium chloride, 2 mm magnesium chloride, 10% glycerol, 2% ASB-14 (Sigma)), followed by immunoprecipitation with anti-FLAG antibody. To examine the interaction of p105 and Tpl2 in RAW264.7 cells, cells were lysed with Buffer C (50 mm Hepes, 150 mm sodium chloride, 1 mm EDTA, 1 mm EGTA, 10% glycerol, 1% Nonidet P-40 (Sigma)), followed by immunoprecipitation with anti-mouse p105/p50 antibody. Immune complexes were eluted from the beads and subjected to immunoblotting.
For immunoblot analyses, cells were lysed with radioimmune precipitation buffer (50 mm Tris, pH 8.0, 150 mm sodium chloride, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) containing protease and phosphatase inhibitor mixtures (Sigma). To quantify the level of the phosphorylated forms of p105, bolts were scanned and analyzed by densitometry with Image J software (National Institutes of Health, Bethesda, MD). The blot intensity of phosphorylated p105 was normalized to that of β-actin.
Enzyme-linked Immunosorbent Assay (ELISA)—Quantitative determinations of MIP-1β and IL-10 concentration in cell culture supernatants were performed by sandwich ELISA (R&D Systems, Minneapolis, MN). Measurement of each sample was performed in triplicate.
NF-κB Reporter Gene Assay—HEK293 cells were cultured in 24-well plates and transiently transfected with a secreted alkaline phosphatase reporter construct containing an NF-κB consensus enhancer element (-GGGAATTTCC-) (Clontech), a LacZ expression vector (Invitrogen), and increasing amounts of a human EPRAP cDNA expression construct as well as mock plasmid for adjusting the total amount of DNA to 0.3 μg/well. At 48 h after transfection, cells were stimulated with either TNFα (25 ng/ml) or IL-1β (10 ng/ml) for 16 h. Secreted alkaline phosphatase reporter activity was measured in supernatant from cell-conditioned media (Clontech). Transfection was performed in triplicate in each experiment, and transfection efficiency was normalized by β-galactosidase activity in cell extracts collected from each well (Clontech).
NF-κB Translocation and DNA Binding Assay—Nuclear and cytoplasmic extracts were obtained as previously described (22). To quantify the DNA binding activity of p50 or p65 to NF-κB consensus oligonucleotide, ELISA-based DNA binding assays were performed (Panomics, Redwood City, CA) according to the manufacturer's instructions. Briefly, nuclear extracts were incubated with the biotinylated oligonucleotides containing an NF-κB consensus binding site (NF-κB probe). The oligonucleotides with activated NF-κB molecules were then immobilized on a streptavidin-coated plate. An antibody directed against the NF-κB subunit, p50 or p65, detected each NF-κB molecule bound to the probe, followed by incubation with a horseradish peroxidase-conjugated secondary antibody. The amounts of p50 or p65 subunits bound to the NF-κB consensus probe were quantified by a colorimetric method. The specific binding was verified by competition studies using nonlabeled oligonucleotides corresponding with the probe. All reactions were performed in triplicate.
RNA Interference—Mouse EPRAP silencing was performed by transfection in RAW264.7 cells with a pool of EPRAP-specific siRNA duplexes (Dharmacon, Lafayette, CO) at 100 nm. As a control, RAW264.7 cells were transfected with nonspecific pooled siRNA duplexes (Dharmacon).
Statistical Analysis—Data are presented as mean ± S.E. Statistical comparisons between groups were made by Student's t test or one-way analysis of variance. p < 0.05 was considered statistically significant.
RESULTS
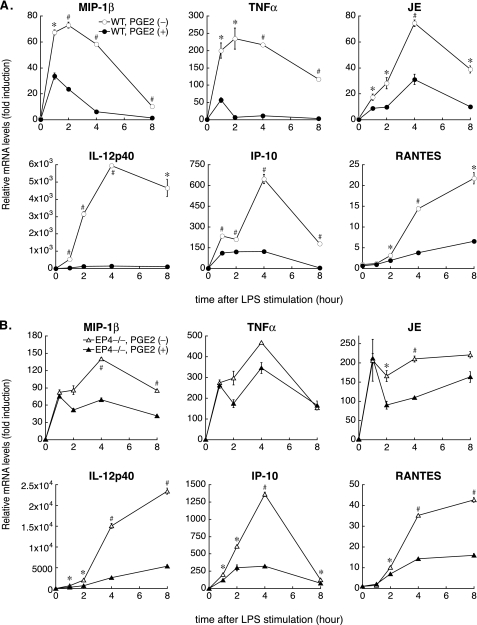
PGE2 Has a Prolonged Anti-inflammatory Effect on LPS-stimulated Mouse BMDM—To clarify the molecular mechanisms underlying the EP4-mediated anti-inflammatory properties of PGE2, we used mouse BMDM, with or without target gene mutations. First, demonstrating that PGE2 exerts an anti-inflammatory effect in WT BMDM (EP4+/+, on the same genetic background as EP4–/– BMDM) entailed performing kinetic analyses of LPS-induced gene expression with qPCR. In addition to inflammatory cytokines (e.g. TNFα and IL-12p40), the levels of expression of several chemokines increase in macrophages activated by proinflammatory stimuli, such as LPS. JE/mouse MCP-1/CCL2 is a well known chemoattractant for monocytes; MIP-1β/CCL4, IP-10/CXCL10, and RANTES/CCL5 function as chemoattractants and activators for T lymphocytes. Activated macrophages, especially classically activated macrophages, produce these chemokines (23, 24), enhancing local inflammatory responses. As in human primary macrophages (11), PGE2 pretreatment markedly suppressed LPS-induced proinflammatory gene expression (MIP-1β, TNFα, JE, IL-12p40, IP-10, and RANTES) in mouse BMDM (Fig. 1A). In each case, except for RANTES, PGE2 attenuated rapid gene induction at the onset of LPS stimulation and maintained inhibition for prolonged periods.
FIGURE 1.
Kinetic analyses of LPS-induced gene expression in WT or EP4–/– BMDM with or without PGE2 pretreatment. A, WT (EP4+/+, as described under “Experimental Procedures”) BMDM were pretreated with 50 nm PGE2 (PGE2 (+)) or vehicle (PGE2 (–)) for 1 h, followed by LPS stimulation (5 ng/ml) for various times. Total RNA was isolated and subjected to qPCR for indicated messages. mRNA levels of β-actin served as an internal control for normalization between samples. B, in EP4–/– macrophages, PGE2 does not inhibit rapid mRNA induction at the onset of LPS stimulation, especially for MIP-1β or TNFα. LPS-induced gene expression was measured by qPCR using EP4–/– BMDM with or without PGE2 pretreatment as described in A. In both studies (A and B), data represent at least three independent experiments on cells from different donors. Results are expressed as the mean -fold induction ± S.E. compared with unstimulated, vehicle-treated cells. *, p < 0.05; #, p < 0.01 for the indicated comparison (PGE2 (–) versus PGE2 (+) at each time point).
PGE2 Suppresses LPS-induced Phosphorylation and Degradation of NF-κB1 p105 through EP4-dependent Mechanisms—Clarifying the particular role of EP4 signaling in PGE2-mediated anti-inflammatory actions involved kinetic analyses of LPS-induced gene expression through qPCR using EP4–/– BMDM. The levels of mRNAs encoding the studied proinflammatory molecules after LPS stimulation in EP4–/– macrophages exceeded those in WT (EP4+/+) BMDM (Fig. 1, A and B). In addition, PGE2 pretreatment did not suppress rapid mRNA induction at the onset of LPS stimulation in EP4–/– macrophages, especially for MIP-1β, TNFα, and JE/MCP-1 (Fig. 1B). These data suggested that EP4 signaling interfered with proinflammatory signaling pathways promptly evoked by LPS.
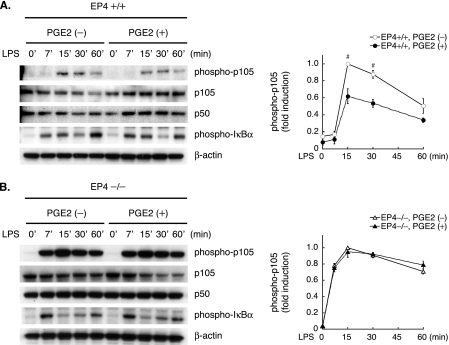
The transcription factor NF-κB participates centrally in gene expression induced by LPS and many other proinflammatory stimuli. Testing whether PGE2 modulates NF-κB activation entailed immunoblot analyses of extracts from LPS-stimulated BMDM with or without PGE2 pretreatment. Notably, PGE2-pretreated WT (EP4+/+) macrophages demonstrated considerably reduced accumulation of phosphorylated NF-κB1 p105 (Fig. 2A, left, representative blots; right, quantification by densitometry). Phosphorylation of the C-terminal domain leads to proteosomal degradation of p105 (25, 26); accordingly, PGE2 pretreatment attenuated the decline in p105 levels post-LPS (Fig. 2A). Upon LPS stimulation, the IκB kinase (IKK) complex predominantly mediates phosphorylation of the IκB proteins as well as that of p105 (27); however, PGE2 pretreatment did not reduce IκBα phosphorylation (Fig. 2A), suggesting that inhibition of IKK activity may not mediate the protective action of PGE2 against inducible p105 phosphorylation.
FIGURE 2.
PGE2 suppresses LPS-induced phosphorylation and degradation of NF-κB1 p105 through EP4-dependent mechanisms. Results of immunoblot analyses using LPS-stimulated WT (EP4+/+)(A) or EP4–/– (B) BMDM with or without PGE2 pretreatment as described in the legend to Fig. 1 (left, representative blots; right, densitometric quantification of phosphorylated p105 in cell extracts). Data are expressed relative to the values at 15 min after LPS stimulation in the absence of PGE2 pretreatment (defined as 1.0) in mean ± S.E. (from three different experiments). #, p < 0.01 for the indicated comparison (PGE2 (–) versus PGE2 (+) at each time point).
On the contrary, PGE2 pretreatment did not suppress LPS-induced p105 phosphorylation and the subsequent decline in p105 levels in EP4–/– macrophages (Fig. 2B, left, representative blots; right, quantification of phosphorylated p105 by densitometry). Thus, PGE2 attenuates LPS-induced p105 phosphorylation and degradation primarily through EP4-dependent mechanisms.
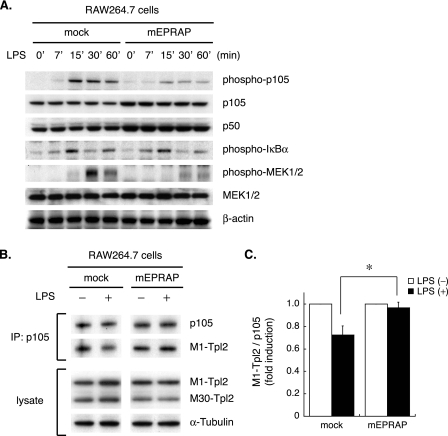
Forced EPRAP Expression Suppresses p105 Phosphorylation/Degradation and Subsequent MEK1/2 Activation in a Mouse Macrophage-like Cell Line—Clarifying the role of EPRAP in anti-inflammatory signaling in macrophages involved transiently enforced mouse EPRAP expression in RAW264.7 cells. Interestingly, similar to PGE2-pretreated macrophages, EPRAP-overexpressing cells had markedly impaired LPS-induced p105 phosphorylation and subsequent decline compared with the mock-transfected control (Fig. 3A). On the other hand, IKK-mediated IκBα phosphorylation did not differ significantly (Fig. 3A).
FIGURE 3.
Forced EPRAP expression inhibits stimulus-induced p105 phosphorylation, degradation, and subsequent MEK1/2 activation. A, RAW264.7 cells were transiently transfected with mouse EPRAP expression construct (mEPRAP) or mock plasmid and then stimulated with LPS (10 ng/ml) for the indicated durations, followed by immunoblotting (representative of three different experiments). B, enforced EPRAP expression impairs inducible release of Tpl2 from p105. RAW264.7 cells transiently expressing either mouse EPRAP or mock plasmid were stimulated with LPS (50 ng/ml) or vehicle for 15 min. The same amounts of extracts were immunoprecipitated (IP) with anti-p105/p50 antibody, followed by immunoblotting with anti-Tpl2 or anti-p105/p50 antibody. To assess the protein levels of Tpl2, immunoblot analyses were performed using whole lysates (representative of three different experiments). C, densitometric quantification of M1-Tpl2/p105 ratio. Results are expressed relative to the values of samples without LPS treatment (defined as 1.0, from three different experiments) in mean ± S.E. *, p < 0.05 for the indicated comparison.
p105 directly interacts with mitogen-activated protein kinase kinase kinase Tpl2/Cot and inhibits its activation (28–30). Upon stimulation, proteolytic degradation of p105 liberates Tpl2, which in turn phosphorylates and activates MEK1/2. Notably, in accordance with the attenuated decline in p105 levels, EPRAP-overexpressing RAW264.7 cells had markedly reduced MEK1/2 phosphorylation induced by LPS (Fig. 3A). To explore the interaction of Tpl2 with p105, cell extracts were immunoprecipitated with p105 and subjected to immunoblotting. As demonstrated in previous reports (31), alternative translation initiation results in expression of two isoforms of Tpl2, M1- and M30-Tpl2 (Fig. 3B, bottom), and the longer isoform, M1-Tpl2, dissociates from p105 upon LPS stimulation (30). Notably, forced EPRAP expression in RAW264.7 cells resulted in impaired release of M1-Tpl2 from p105 after LPS stimulation (Fig. 3, B (top), representative blots; C, quantification of M1-Tpl2/p105 ratio by densitometry), suggesting that EPRAP attenuates MEK1/2 activation by inhibiting LPS-induced degradation of p105, a Tpl2 scaffold.
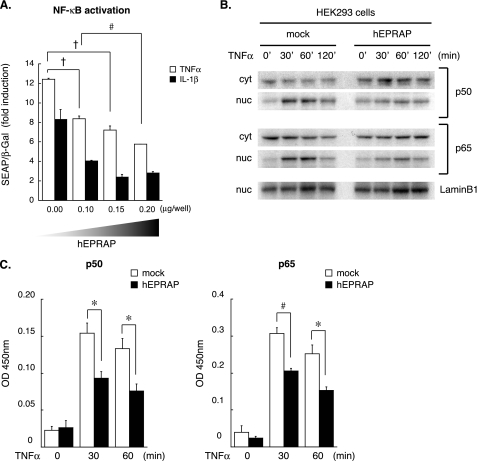
Enforced EPRAP Expression Attenuates NF-κB Activation Induced by Various Stimuli—In addition to limiting MEK activation, p105 functions as a cytoplasmic NF-κB inhibitor through its association with p50, p65/Rel-A, or c-Rel subunit (32, 33), which thereby remains sequestered in the cytoplasm. Examining the effect of EPRAP on NF-κB activation entailed analyzing the reporter gene in HEK293 cells co-transfected with various amounts of human EPRAP expression plasmid. Similar to RAW264.7 cells, EPRAP-overexpressing HEK293 cells showed substantial inhibition of TNFα-induced phosphorylation and degradation of p105 (see Fig. 6). Notably, forced EPRAP expression attenuated NF-κB activation evoked by TNFα and IL-1β in a concentration-dependent manner (Fig. 4A). In addition, EPRAP overexpression markedly suppressed TNFα-induced nuclear translocation of both p50 and p65 at each time point (Fig. 4B). Determining the DNA binding activity of each NF-κB subunit involved ELISA-based assays. In accordance with the inhibition of nuclear translocation, EPRAP-overexpressing cells demonstrated impaired TNFα-induced interactions of p50 and p65 with the NF-κB recognition site, in contrast to the mock-transfected control (Fig. 4C).
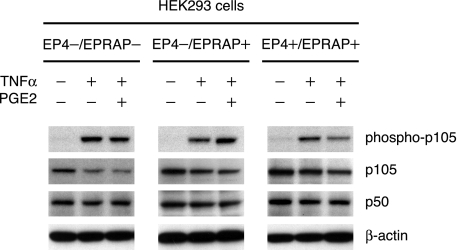
FIGURE 6.
PGE2 enhances the EPRAP-mediated antagonism of stimulus-induced p105 phosphorylation through EP4. HEK293 cells were co-transfected with expression plasmids for either human EP4, EPRAP, or mock control (EP4-/EPRAP-, EP4-/EPRAP+, or EP4+/EPRAP+). At 48 h after transfection, cells were incubated with PGE2 (100 nm) or vehicle for 1 h, followed by TNFα (25 ng/ml) for 15 min. Cell extracts were immunoblotted with the indicated antibodies (representative of three independent experiments). Note that only in EP4/EPRAP-overexpressing cells (EP4+/EPRAP+) did PGE2 demonstrate substantial inhibition of TNFα-induced p105 phosphorylation.
FIGURE 4.
EPRAP overexpression attenuates NF-κB activation and nuclear translocation induced by proinflammatory stimuli. A, EPRAP overexpression attenuates NF-κB activation induced by various stimuli in a dose-dependent manner. NF-κB reporter gene analyses were performed using HEK293 cells cotransfected with the secreted alkaline phosphatase reporter plasmid, LacZ expression plasmid, and increasing amounts of human EPRAP expression construct (hEPRAP) as described under “Experimental Procedures.” Transfection efficiency was normalized by β-galactosidase (β-Gal) activity. Data are representative of three independent experiments. Results are expressed relative to the values of unstimulated cells in mean ± S.E. #, p < 0.01; †, p < 0.005 for the indicated comparison. B, nuclear translocation of NF-κB subunits. HEK293 cells transfected with either human EPRAP expression plasmid or mock plasmid were stimulated by TNFα (25 ng/ml) for increasing durations. Nuclear (nuc) and cytoplasmic (cyt) extracts were analyzed by immunoblotting using antibodies against p50, p65, or lamin B1 (loading control) (representative of three different experiments). C, DNA binding activity of p50 and p65 in the nuclear extracts of TNFα-stimulated HEK293 cells was determined by an ELISA-based DNA-binding assay. Data are representative of three independent experiments. Results were expressed as the mean OD value (450 nm) ± S.E. *, p < 0.05; #, p < 0.01 for the indicated comparison.
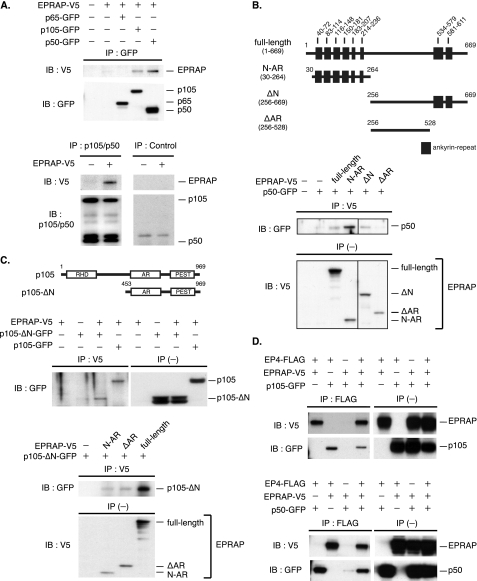
EPRAP Associates Directly with NF-κB1 p105/p50 and Forms a Complex with EP4—EPRAP contains multiple ankyrin repeat motifs, which mediate many protein-protein interactions, at both the N and C terminus (Fig. 5B, top, schematic representation of EPRAP domain structure). The inhibition of NF-κB1 documented above generated our hypothesis that EPRAP directly associates with NF-κB proteins. Indeed, in co-transfected HEK293 cells, V5-tagged EPRAP was co-immunoprecipitated with GFP-tagged p105 and p50 (Fig. 5A, top). In addition, an antibody against endogenous p105/p50 immunoprecipitated transiently expressed V5-tagged EPRAP (Fig. 5A, bottom). On the other hand, EPRAP did not bind to p65 (Fig. 5A, top), indicating that EPRAP selectively interacts with NF-κB1 p105/p50. Moreover, co-immunoprecipitation studies using a series of deletion mutants revealed that ankyrin repeat motifs in EPRAP are critical to optimal interaction with p50 (Fig. 5B).
FIGURE 5.
EPRAP directly interacts with NF-κB1 p105/p50 and forms a complex with EP4. A, EPRAP binds to NF-κB1 p105/p50. HEK293 cells were co-transfected with expression plasmids for V5-tagged EPRAP (EPRAP-V5) and GFP-tagged NF-κB proteins (p65-, p105-, and p50-GFP). Cell lysates were immunoprecipitated (IP) with rabbit anti-GFP antibody, followed by immunoblotting (IB) with anti-V5 or anti-GFP mAb (top). Cell extracts isolated from HEK293 cells with or without transient expression of EPRAP-V5 were immunoprecipitated with rabbit anti-p105/p50 antibody or control rabbit IgG, followed by immunoblotting with anti-V5 or anti-p105/p50 mAb (bottom). B, schematic representations for human EPRAP and deletion mutants. The positions of ankyrin repeat motif are indicated (top). HEK293 cells were co-transfected with expression plasmids for p50-GFP and V5-tagged EPRAP mutants. Cell lysates were immunoprecipitated with anti-V5 mAb, followed by immunoblotting with rabbit anti-GFP antibody (bottom). C, schematic representations for human NF-κB1 and deletion mutant. RHD, Rel homology domain; AR, ankyrin repeat; PEST, Pro-Glu-Ser-Thr-rich domain (top). HEK293 cells were co-transfected with expression plasmids for EPRAP-V5 and GFP-tagged p105 mutants (fulllength, p105-ΔN-GFP) (middle) or those for p105-ΔN-GFP and V5-tagged EPRAP mutants (N-AR, ΔAR, or fulllength) (bottom). Cell lysates were immunoprecipitated with anti-V5 mAb, followed by immunoblotting with anti-GFP antibody. D, EP4 forms a complex with EPRAP and NF-κB1 p105/p50. HEK293 cells were co-transfected with expression plasmids for FLAG-tagged EP4 (EP4-FLAG), EPRAP-V5, and p105-GFP (top) or p50-GFP (bottom). Cell lysates were immunoprecipitated with rabbit anti-FLAG antibody, followed by immunoblotting with anti-V5 or anti-GFP mAb. In these studies (B–D), expression of each recombinant protein was verified by immunoblotting using whole cell extracts (IP (–)).
Strikingly, EPRAP co-immunoprecipitated not only fulllength p105 but also p105 C terminus, p105-ΔN (amino acid residues 453–969) (Fig. 5C, top, schematic representation of p105 domain structure; middle, representative blots). These findings indicate that EPRAP associates with both the N terminus p50 subunit and the C terminus of p105. The interaction of EPRAP with p105 C terminus, which contains conserved target sequences for the IKK complex Pro-Glu-Ser-Thr-rich domain, probably contributes crucially to EPRAP-mediated protection against p105 phosphorylation and degradation. The C terminus of p105 also contains multiple ankyrin repeat motifs, which interact with NF-κB subunits p50 and p65 (Fig. 5C, top). Interestingly, full-length EPRAP and the N terminus with ankyrin repeat motifs (N-AR; amino acid residues 30–264 of EPRAP) along with the deletion mutant lacking ankyrin repeat (ΔAR; amino acid residues 256–528) interacted with the p105 C terminus (Fig. 5C, bottom).
Initially isolating EPRAP as an EP4-associated protein enabled EPRAP and NF-κB1 to form a complex with EP4. Indeed, co-immunoprecipitation experiments demonstrated that EP4 interacted with not only EPRAP but also p105 and p50 (Fig. 5D). The direct interaction and complex formation among EP4, EPRAP, and NF-κB1 may participate in the anti-inflammatory function mediated by PGE2.
PGE2-EP4 Signaling Enhances the Protective Action of EPRAP against Stimulus-induced p105 Phosphorylation—Clarifying the role of PGE2-EP4 signaling in the inhibitory effect of EPRAP on inducible p105 phosphorylation involved immunoblot analyses of extracts from TNFα-stimulated HEK293 cells transiently expressing either EP4 or EPRAP (EP4–/EPRAP–, EP4–/EPRAP+, EP4+/EPRAP+). EPRAP-overexpressing cells (EP4–/EPRAP+, EP4+/EPRAP+) had reduced p105 phosphorylation induced by TNFα regardless of enforced EP4 expression (Fig. 6). Notably, in EP4/EPRAP-overexpressing cells (EP4+/EPRAP+), PGE2 pretreatment markedly impaired TNFα-induced p105 phosphorylation, whereas the cells without enforced EP4 expression (EP4–/EPRAP–, EP4–/EPRAP+) demonstrated no significant attenuation of p105 phosphorylation by PGE2 (Fig. 6). Thus, PGE2-EP4 signaling augments the inhibitory effect of EPRAP against stimulus-induced p105 phosphorylation.
At this time point (15 min after TNFα stimulation), PGE2 pretreatment did not yet manifest a significant inhibitory effect on p105 degradation in each group; however, EP4/EPRAP-overexpressing cells (EP4+/EPRAP+) had a markedly reduced p105 decline induced by TNFα in contrast to the cells without enforced EP4 expression (EP4–/EPRAP–, EP4–/EPRAP+) (Fig. 6), indicating the pivotal role of EP4-EPRAP signaling in p105 protein stability under proinflammatory settings.
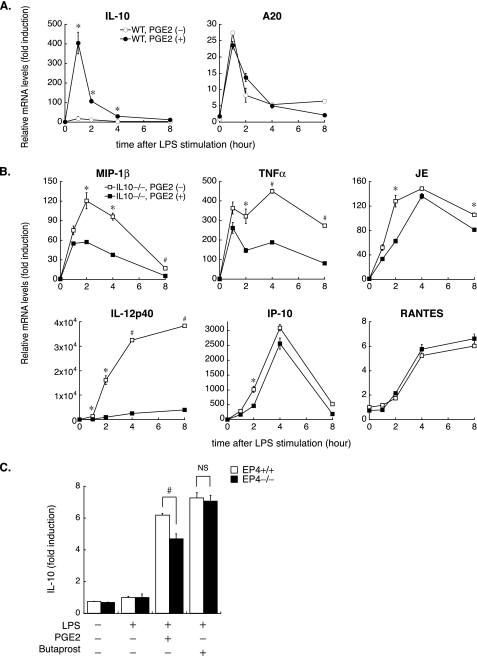
IL-10 Participates in the Anti-inflammatory Function of PGE2 Independently of EP4—Testing the possibility that PGE2 increased expression of other anti-inflammatory molecules entailed qPCR analyses using WT BMDM, which revealed that PGE2 synergistically augmented IL-10 expression with LPS at both the mRNA and protein levels (Fig. 7, A and D). As previously demonstrated (34), LPS also induced the expression of A20/TNFAIP3 (TNFα-induced protein 3), an inducible inhibitor of NF-κB activation, but no noteworthy difference emerged among samples with or without PGE2 pretreatment at each time point (Fig. 7A).
FIGURE 7.
PGE2 augments IL-10 expression in LPS-stimulated macrophages but not through EP4-dependent mechanisms. A and B, LPS-induced gene expression was measured by qPCR using WT (A) or IL-10–/– (B) BMDM with or without PGE2 pretreatment as described in the legend to Fig. 1. Note that PGE2 augments the expression of IL-10, which participates in the anti-inflammatory function of PGE2, especially for IP-10 and RANTES. Data represent at least three independent experiments on cells from different donors. Results are expressed as the mean -fold induction ± S.E. compared with unstimulated, vehicle-treated cells. *, p < 0.05; #, p < 0.01 for the indicated comparison (PGE2 (–) versus PGE2 (+) at each time point). C, increased IL-10 production by PGE2 is not mediated through EP4-specific signaling. WT (EP4+/+) or EP4–/– BMDM were pretreated with either vehicle Butaprost (10 μm) or PGE2 (50 nm) for 1 h, followed by LPS (24 h). IL-10 protein levels in cell culture supernatants were measured by ELISA. Data are expressed relative to the values of samples from LPS-stimulated, vehicle-pretreated cells (defined as 1.0, from three different experiments) in mean ± S.E. #, p < 0.01 for the indicated comparison.
Clarification of the role of IL-10 in PGE2-mediated anti-inflammation involved kinetic analyses of LPS-induced gene expression with qPCR using IL-10–/– BMDM. Despite the impaired inhibitory effect of PGE2, especially for IP-10 and RANTES gene induction, PGE2 pretreatment still suppressed expression of other genes, including MIP-1β, TNFα, and IL-12p40, in IL-10–/– macrophages (Fig. 7B).
Previous studies demonstrated that PGE2 induces IL-10 expression in macrophages (35, 36). In mouse BMDM, EP4 constitutes the most abundant receptor for PGE2, whereas reverse transcription-PCR analysis also detects EP2 and EP3 (expression levels: EP4 > EP2 ≫ EP3; data not shown). Notably, pretreatment with the selective EP2 agonist Butaprost, instead of PGE2, exerted a similar effect on IL-10 protein expression in LPS-stimulated wild type (EP4+/+) macrophages (PGE2 + LPS versus LPS alone, 6.19 ± 0.11; Butaprost + LPS versus LPS alone, 7.26 ± 0.33, p = 0.069) (Fig. 7C). EP4-deficient macrophages had lower IL-10 levels after PGE2 stimulation than WT EP4+/+ cells (PGE2 + LPS versus LPS alone: EP4+/+, 6.19 ± 0.11; EP4–/–, 4.70 ± 0.30, p < 0.01) (Fig. 7C), but they showed a similar response to Butaprost (Butaprost + LPS versus LPS alone: EP4+/+, 7.26 ± 0.33; EP4–/–, 7.06 ± 0.37, p = 0.687) (Fig. 7C), suggesting not only EP4 but also EP2 figures importantly in increased IL-10 expression by PGE2. These data demonstrated that IL-10 participates in the anti-inflammatory effect of PGE2, but augmented IL-10 expression by PGE2 does not occur mainly through EP4-dependent pathways.
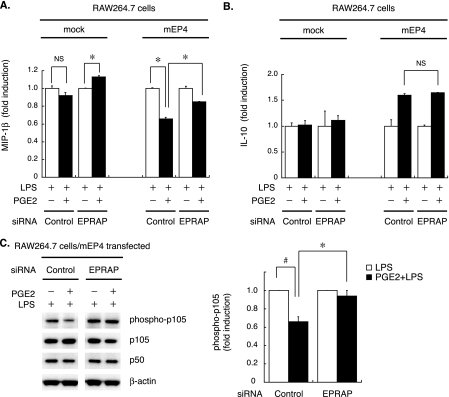
EPRAP Silencing in Mouse Macrophages Impairs the Anti-inflammatory Effect of PGE2—As previously reported (37), the RAW264.7 cell line constitutively expresses mRNAs encoding prostaglandin E receptors EP2, EP3, and EP4. However, unstimulated RAW264.7 cells have much lower levels of EP4 and EP2 mRNA than do BMDM (≫10%, evaluated by qPCR; data not shown); thus, PGE2 does not produce a significant anti-inflammatory effect in naive or mock-transfected RAW264.7 cells (Fig. 8A, left). Transient transfection with mouse EP4 allowed RAW264.7 cells to respond to PGE2 in terms of the inhibition of LPS-induced MIP-1β production (Fig. 8A, far right), but EPRAP knockdown by a specific siRNA significantly impaired the inhibitory effect of PGE2 (percentage inhibition by PGE2: control siRNA, 34.1 ± 1.64%; EPRAP siRNA, 14.9 ± 0.14%, p < 0.05) (Fig. 8A, right two columns). In these experiments, EPRAP mRNA levels fell to 33 ± 0.4% with EPRAP-specific siRNA (versus control siRNA, determined by qPCR; data not shown). Interestingly, in mock-transfected RAW264.7 cells, PGE2 augmented MIP-1β production by EPRAP silencing (LPS + PGE2 versus LPS alone; 1.13 ± 0.02, p < 0.05) (Fig. 8A), suggesting pivotal anti-inflammatory roles for EPRAP in PGE2 signaling.
FIGURE 8.
EPRAP knockdown in macrophages impairs the anti-inflammatory action of PGE2. A and B, RAW264.7 cells were transfected with mouse EP4 expression construct (mEP4) or mock plasmid, together with either EPRAP-specific or control siRNA. At 48 h after transfection, cells were incubated with or without PGE2 (100 nm, 1 h), followed by LPS (10 ng/ml, 24 h). MIP-1β (A) or IL-10 (B) production was measured in cell culture supernatants by ELISA. Data represent three independent experiments. Results are expressed relative to the values of samples without PGE2 pretreatment (defined as 1.0) in mean ± S.E. *, p < 0.05 for the indicated comparison. Note that in EP4-overexpressing cells, PGE2 suppressed MIP-1β production, but ERPAP knock-down impaired this effect. EPRAP silencing did not affect IL-10 production. C, EPRAP knockdown impairs the protective effect of PGE2-EP4 signaling on LPS-induced p105 phosphorylation. EP4-transfected RAW264.7 cells with or without EPRAP silencing were incubated with PGE2 (100 nm, 1 h), followed by LPS (50 ng/ml, 15 min). Protein extracts were subjected to immunoblotting (left) and quantified by densitometry (right). Data are expressed relative to the values of samples without PGE2 pretreatment (defined as 1.0, from three different experiments) in mean ± S.E. *, p < 0.05 for the indicated comparison.
In EP4-transfected RAW264.7 cells, PGE2 augmented IL-10 expression with LPS; however, EPRAP silencing did not affect IL-10 production by PGE2 (LPS + PGE2 versus LPS alone; control siRNA, 1.60 ± 0.03; EPRAP siRNA, 1.64 ± 0.01, p = 0.225) (Fig. 8B, right two columns). These data indicate that the anti-inflammatory action of the EP4-EPRAP pathway does not depend on IL-10 expression.
Immunoblot analyses using cell extracts from EP4-transfected RAW264.7 cells with or without EPRAP silencing verified the role of EPRAP in PGE2/EP4-mediated anti-inflammation. PGE2 attenuated LPS-induced p105 phosphorylation and degradation in control siRNA-transfected cells, whereas EPRAP knock-down significantly impaired this inhibitory effect of PGE2 (Fig. 8C, left, representative blots; right, quantification of phosphorylated p105 by densitometry). Taken together, these data indicate that EPRAP contributes crucially to the anti-inflammatory function of PGE2-EP4 signaling, which selectively inhibits stimulus-induced p105 phosphorylation and degradation.
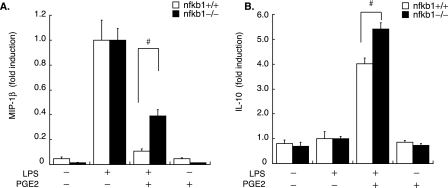
NF-κB1 Knockdown in Macrophages Impairs the Anti-inflammatory Effect of PGE2—Finally, verifying the contribution of NF-κB1 p105 to PGE2-mediated anti-inflammation entailed measuring LPS-induced MIP-1β release in cell-conditioned media of BMDM isolated from NF-κB1 knock-out mice (nfkb1–/–) or WT (nfkb1+/+). Levels of EP4 and EPRAP expression did not differ significantly between nfkb1–/– and nfkb1+/+ BMDM (by reverse transcription-PCR analyses; data not shown). Compared with WT, nfkb1–/– macrophages had significantly impaired inhibition by PGE2 of LPS-induced MIP-1β production (percentage inhibition by PGE2: nfkb1+/+, 89.4 ± 2.08%; nfkb1–/–, 61.1 ± 5.21%, p < 0.01) (Fig. 9A). Strikingly, instead of a reduced anti-inflammatory effect, PGE2 augmented IL-10 production post-LPS more prominently in nfkb1–/– macrophages than in WT (LPS + PGE2 versus LPS alone; nfkb1+/+, 4.01 ± 0.24; nfkb1–/–, 5.41 ± 0.25, p < 0.01) (Fig. 9B), although PGE2 alone did not induce substantial IL-10 expression in either group. These data indicate that NF-κB1 participates importantly in PGE2-mediated anti-inflammation but not through the augmentation of IL-10 expression.
FIGURE 9.
Deficiency of NF-κB1 in macrophages attenuates the anti-inflammatory effect of PGE2. BMDM isolated from nfkb1–/– or WT (nfkb1+/+) mice were pretreated with or without PGE2 (50 nm, 1 h), followed by LPS stimulation (5 ng/ml, 24 h). MIP-1β (A) or IL-10 (B) production was measured in cell culture supernatants by ELISA. Data are representative of three independent experiments. Results are presented as the mean -fold induction ± S.E. compared with the values of LPS treatment alone (defined as 1.0). #, p < 0.01 for the indicated comparison.
DISCUSSION
Macrophages exposed to proinflammatory stimuli, such as LPS, substantially increase the expression of both COX-2 and microsomal PGE synthase-1, resulting in markedly increased PGE2 synthesis (38–40). In addition to those inducible enzymes, COX-1 and cytosolic PGES, constitutively expressed synthases for PGE2, produce PGE2 in response to various stimuli in cultured macrophages (40, 41). Because we did not pretreat cells with a COX inhibitor (e.g. indomethacin) or a PGES inhibitor before LPS challenge, endogenous PGE2 levels rose even in cells without exogenous PGE2 pretreatment. Therefore, in wild type (EP4+/+) macrophages, but not in EP4-deficient cells, endogenous PGE2 may provoke an EP4-dependent reduction in proinflammatory gene expression. The enhanced and prolonged levels of mRNAs that encode proinflammatory genes in LPS-stimulated EP4–/– macrophages compared with wild type (EP4+/+) cells indicate the importance of the inhibitory effect of endogenous PGE2-EP4 signaling on the expression of those genes. For instance, qPCR analyses demonstrated that the maximal mRNA levels of MIP-1β rose up to 1.93 ± 0.04-fold greater in LPS-stimulated EP4-deficient macrophages than in wild type cells, 2.00 ± 0.03-fold for TNFα, and 2.97 ± 0.20-fold for JE/MCP-1 (Fig. 1, A and B).
The nfkb1 gene encodes the p105 protein, the precursor of the p50 subunit. The similar levels of p105 and p50 in most cell types may result from co-translational processing of p105 producing p50. On the other hand, exposing cells to proinflammatory stimuli triggers phosphorylation at the C terminus and the complete degradation of p105 (42), a cytoplasmic inhibitor of NF-κB and a scaffold for Tpl2. In addition to Tpl2–/– macrophages (43), RAW264.7 cells overexpressing a p105 mutant, which resists IKK-mediated phosphorylation, had markedly impaired MEK1/2 activation induced by LPS (44), indicating the essential role of p105 in inhibition of MEK signaling activation.
Consistent with our observations, PGE2/EP4-mediated inhibition of stimulus-induced MEK activation occurs in other cell types (45–47). We furnish here novel evidence supporting the involvement of EP4-EPRAP signaling in the suppression of MEK activation through inhibiting inducible p105 phosphorylation and degradation. Although the expression of EPRAP in other cell types still needs corroboration, EP4-EPRAP signaling may provide endogenous inhibitory machinery against the axis of p105 degradation, Tpl2 and MEK activation triggered by proinflammatory stimuli in many cell types, including macrophages.
Co-immunoprecipitation studies indicated that EPRAP interacted directly with the N terminus p50 subunit as well as the C terminus of p105, which may participate pivotally in the protective action of EPRAP against stimulus-induced p105 phosphorylation, yet the role of EPRAP-p50 association in PGE2-mediated anti-inflammation remains incompletely understood. Forced EPRAP expression restrained p50 and p65 nuclear translocation, although co-immunoprecipitation studies did not detect direct interaction of EPRAP with p65. The C terminus of p105, rescued by EPRAP overexpression, might associate with the NF-κB subunits and retain them in the cytoplasm; however, genetic studies in mice lacking the C terminus of p105 suggested that endogenous p105 only figures essentially in the cytoplasmic retention of p50 (48). Previous studies indicated that MEK signaling participates pivotally in persistent NF-κB activation (49) and enhances the transcriptional activity of p65 (50). Therefore, inhibition of MEK signaling may explain part of the suppression of NF-κB activation by EPRAP, or through the interaction with an unknown NF-κB-associating protein, EPRAP might inhibit the nuclear translocation of p65 and suppress NF-κB activation.
Determining the breadth of EPRAP involvement in counter-inflammatory responses will require further investigation, but the current data establish a role for EPRAP-mediated inhibition of NF-κB activation to the attenuation of inflammatory responses attributed to PGE2/EP4 signaling. Although the effect of PGE2 or EP4 signaling on NF-κB activation probably varies by cell type (51), previous studies demonstrated that PGE2 and EP4 signaling suppressed NF-κB DNA binding activities in various inflammatory cells (46, 52, 53).
In investigating the underlying mechanisms of PGE2-mediated anti-inflammation, we also found that PGE2 synergistically augmented IL-10 production with LPS, yet the enhanced IL-10 expression did not depend on EP4, EPRAP, or NF-κB1. In addition, a previous study using human monocytes indicated that IL-10 induced NF-κB1 p105/p50 expression and increased p50/p50 homodimers in the nucleus, an important element in the anti-inflammatory action of IL-10, while not affecting p105 protein stability after TNFα stimulation (54). These data suggest that IL-10 does not participate pivotally in the anti-inflammatory effect of EP4-EPRAP signaling, although IL-10 does appear to contribute to the anti-inflammatory action of PGE2, particularly in the late stage of LPS challenge. Previous studies have shown that long term IL-10 pretreatment (e.g. for 24 h) inhibited activation of LPS-induced phosphatidylinositol 3-kinase (55), early growth response-1, and mitogen-activated protein kinase (56). In addition, PGE2 enhanced IL-10 signaling and functions in human monocytes (57).
Thus far, yeast two-hybrid screenings have identified several other molecules as p105-interacting proteins that may inhibit its proteolysis (58–60). Among them, EPRAP is the only one that acquires an ability to govern p105 protein stability by an agonist (PGE2)-receptor (EP4) interaction. As we showed in EP4/EPRAP-overexpressing cells, ligation of EP4 by PGE2 enhanced the protective action of EPRAP, which directly associates with the EP4 C terminus (12), against TNFα-induced p105 phosphorylation. Although neither PGE2 nor TNFα treatment influenced mutual binding among EP4, EPRAP, and NF-κB1 in co-immunoprecipitation studies (data not shown), these findings strongly suggested that PGE2-EP4 signaling led to either activation or modulation of the EP4-EPRAP interaction. Perhaps noteworthy in this context, multiple serine and threonine residues in both the ankyrin repeat motifs and other domains in EPRAP provide potential phosphorylation sites, as do those in the cytoplasmic tail of EP4, according to computational predictions. Therefore, agonist-induced activation of protein kinases or phosphatases might regulate EP4-EPRAP signaling through the modulation of EP4/EPRAP/NF-κB1 interaction, although clarifying the possible modification or structural change in these molecules will require further experiments.
Our study provides new information regarding the function of EPRAP, a novel multiple ankyrin repeat-containing protein. The ankyrin repeat, a versatile protein-protein interaction module, figures importantly in a diverse set of cellular functions, including signal transduction, cell cycle regulation, and various transport phenomena, despite the absence of detected enzymatic activity for any ankyrin repeat domain (15, 16). Consequently, research has linked defects in ankyrin repeat-containing proteins to several human diseases. Indeed, a recent study reported that the EPRAP/FEM1A gene participates in smooth muscle cell differentiation and is reduced in human rhabdomyosarcoma (61). Another human genetic study indicated the potential of EPRAP as a candidate gene for polycystic ovary syndrome, the most common endocrine disorder among women of reproductive age (62). Furthermore, mice with a target gene mutation for Fem1b, a homolog of C. elegans FEM-1 with 51% amino acid similarity to mouse EPRAP/Fem1a (13), displayed abnormal glucose tolerance due to defective glucose-stimulated insulin secretion in β-cells (63). In addition to our study, these data support important roles of EPRAP in a variety of human diseases.
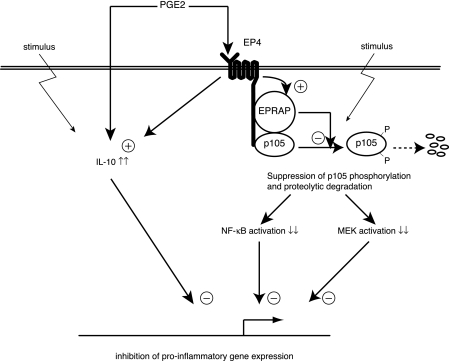
Fig. 10 depicts the anti-inflammatory mechanisms in macrophages attributed to PGE2-EP4 signaling on the basis of our study. PGE2-EP4 signaling attenuates NF-κB and MEK activation by inhibiting stimulus-induced p105 phosphorylation and degradation through EPRAP, which directly interacts with EP4 and p105. PGE2 also augments IL-10 production independently of EP4/EPRAP. Through these concerted yet distinct pathways, PGE2 can inhibit proinflammatory gene expression, thus muting unopposed macrophage activation.
FIGURE 10.
Schematic diagram depicting dual pathways for anti-inflammatory action of PGE2 in macrophages. Based on the data presented here, we propose that PGE2-EP4 signaling attenuates NF-κB as well as MEK activation under proinflammatory settings by inhibiting p105 phosphorylation and degradation through EPRAP, which directly interacts and forms a complex with NF-κB1 p105. PGE2 also increases production of anti-inflammatory cytokine IL-10 but through EP4- or EPRAP-independent mechanisms.
Regarding the importance of the anti-inflammatory function of PGE2 in human diseases, recent clinical trials have indicated that COX-2-selective inhibitors increase the risk of acute myocardial infarction and stroke even in patients without considerable cardiovascular risk factors (64, 65). Because PGE2 is one of the major COX-2-derived prostanoids, disruption of PGE2-mediated anti-inflammation in atheromata might favor the local activation of macrophages, promoting inflammation and, hence, complication of atherosclerotic plaques. Together, PGE2- and EP4/EPRAP-mediated anti-inflammatory signaling provide promising options for a novel therapeutic target in regulating excess macrophage activation associated with chronic inflammatory diseases.
Supplementary Material
Acknowledgments
We thank Dr. Beverly H. Koller (University of North Carolina, Chapter Hill, NC) for providing mice with the ptger4 gene mutation. We thank Dr. Pallavi R. Devchand for critical comments, Elissa Simon-Morrissey and Gihan Suliman for skillful technical assistance, and Joan Perry for excellent editorial assistance.
This work was supported by grants from the Donald W. Reynolds Foundation and NHLBI, National Institutes of Health, Grant HL-34636 (to P. L.). The authors have no conflicting financial interests to disclose. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
Footnotes
The abbreviations used are: COX, cyclooxygenase; EPRAP, prostaglandin E receptor type 4-associated protein; BMDM, bone marrow-derived macrophage(s); qPCR, quantitative real time PCR; PGE, prostaglandin E; LPS, lipopolysaccharide; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; GFP, green fluorescent protein; WT, wild type; ELISA, enzyme-linked immunosorbent assay; RANTES, regulated on activation, normal T cell expressed and secreted; IKK, IκB kinase; IL, interleukin; TNFα, tumor necrosis factor α; mAb, monoclonal antibody; siRNA, small interfering RNA.
References
- 1.Libby, P. (2002) Nature 420 868–874 [DOI] [PubMed] [Google Scholar]
- 2.Aikawa, M., and Libby, P. (2004) Cardiovasc. Pathol. 13 125–138 [DOI] [PubMed] [Google Scholar]
- 3.Hansson, G. K., and Libby, P. (2006) Nat. Rev. Immunol. 6 508–519 [DOI] [PubMed] [Google Scholar]
- 4.Libby, P., and Plutzky, J. (2007) Am. J. Cardiol. 99 27–40 [DOI] [PubMed] [Google Scholar]
- 5.Liang, C. P., Han, S., Senokuchi, T., and Tall, A. R. (2007) Circ. Res. 100 1546–1555 [DOI] [PubMed] [Google Scholar]
- 6.Colombo, M. P., and Mantovani, A. (2005) Cancer Res. 65 9113–9116 [DOI] [PubMed] [Google Scholar]
- 7.Sica, A., and Bronte, V. (2007) J. Clin. Invest. 117 1155–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugimoto, Y., and Narumiya, S. (2007) J. Biol. Chem. 282 11613–11617 [DOI] [PubMed] [Google Scholar]
- 9.Kabashima, K., Saji, T., Murata, T., Nagamachi, M., Matsuoka, T., Segi, E., Tsuboi, K., Sugimoto, Y., Kobayashi, T., Miyachi, Y., Ichikawa, A., and Narumiya, S. (2002) J. Clin. Invest. 109 883–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshida, K., Oida, H., Kobayashi, T., Maruyama, T., Tanaka, M., Katayama, T., Yamaguchi, K., Segi, E., Tsuboyama, T., Matsushita, M., Ito, K., Ito, Y., Sugimoto, Y., Ushikubi, F., Ohuchida, S., Kondo, K., Nakamura, T., and Narumiya, S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 4580–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takayama, K., Garcia-Cardena, G., Sukhova, G. K., Comander, J., Gimbrone, M. A., Jr., and Libby, P. (2002) J. Biol. Chem. 277 44147–44154 [DOI] [PubMed] [Google Scholar]
- 12.Takayama, K., Sukhova, G. K., Chin, M. T., and Libby, P. (2006) Circ. Res. 98 499–504 [DOI] [PubMed] [Google Scholar]
- 13.Ventura-Holman, T., Seldin, M. F., Li, W., and Maher, J. F. (1998) Genomics 54 221–230 [DOI] [PubMed] [Google Scholar]
- 14.Spence, A. M., Coulson, A., and Hodgkin, J. (1990) Cell 60 981–990 [DOI] [PubMed] [Google Scholar]
- 15.Li, J., Mahajan, A., and Tsai, M. D. (2006) Biochemistry 45 15168–15178 [DOI] [PubMed] [Google Scholar]
- 16.Mosavi, L. K., Cammett, T. J., Desrosiers, D. C., and Peng, Z. Y. (2004) Protein Sci. 13 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen, M., Camenisch, T., Snouwaert, J. N., Hicks, E., Coffman, T. M., Anderson, P. A., Malouf, N. N., and Koller, B. H. (1997) Nature 390 78–81 [DOI] [PubMed] [Google Scholar]
- 18.Kuhn, R., Lohler, J., Rennick, D., Rajewsky, K., and Muller, W. (1993) Cell 75 263–274 [DOI] [PubMed] [Google Scholar]
- 19.Sha, W. C., Liou, H. C., Tuomanen, E. I., and Baltimore, D. (1995) Cell 80 321–330 [DOI] [PubMed] [Google Scholar]
- 20.Warren, M. K., and Vogel, S. N. (1985) J. Immunol. 134 982–989 [PubMed] [Google Scholar]
- 21.Giulietti, A., Overbergh, L., Valckx, D., Decallonne, B., Bouillon, R., and Mathieu, C. (2001) Methods 25 386–401 [DOI] [PubMed] [Google Scholar]
- 22.Feinberg, M. W., Shimizu, K., Lebedeva, M., Haspel, R., Takayama, K., Chen, Z., Frederick, J. P., Wang, X. F., Simon, D. I., Libby, P., Mitchell, R. N., and Jain, M. K. (2004) Circ. Res. 94 601–608 [DOI] [PubMed] [Google Scholar]
- 23.Mantovani, A., Sica, A., Sozzani, S., Allavena, P., Vecchi, A., and Locati, M. (2004) Trends Immunol. 25 677–686 [DOI] [PubMed] [Google Scholar]
- 24.Martinez, F. O., Sica, A., Mantovani, A., and Locati, M. (2008) Front. Biosci. 13 453–461 [DOI] [PubMed] [Google Scholar]
- 25.Lang, V., Janzen, J., Fischer, G. Z., Soneji, Y., Beinke, S., Salmeron, A., Allen, H., Hay, R. T., Ben-Neriah, Y., and Ley, S. C. (2003) Mol. Cell. Biol. 23 402–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Viatour, P., Merville, M. P., Bours, V., and Chariot, A. (2005) Trends Biochem. Sci. 30 43–52 [DOI] [PubMed] [Google Scholar]
- 27.Salmeron, A., Janzen, J., Soneji, Y., Bump, N., Kamens, J., Allen, H., and Ley, S. C. (2001) J. Biol. Chem. 276 22215–22222 [DOI] [PubMed] [Google Scholar]
- 28.Belich, M. P., Salmeron, A., Johnston, L. H., and Ley, S. C. (1999) Nature 397 363–368 [DOI] [PubMed] [Google Scholar]
- 29.Beinke, S., Deka, J., Lang, V., Belich, M. P., Walker, P. A., Howell, S., Smerdon, S. J., Gamblin, S. J., and Ley, S. C. (2003) Mol. Cell. Biol. 23 4739–4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waterfield, M. R., Zhang, M., Norman, L. P., and Sun, S. C. (2003) Mol. Cell 11 685–694 [DOI] [PubMed] [Google Scholar]
- 31.Aoki, M., Hamada, F., Sugimoto, T., Sumida, S., Akiyama, T., and Toyoshima, K. (1993) J. Biol. Chem. 268 22723–22732 [PubMed] [Google Scholar]
- 32.Rice, N. R., MacKichan, M. L., and Israel, A. (1992) Cell 71 243–253 [DOI] [PubMed] [Google Scholar]
- 33.Mercurio, F., DiDonato, J. A., Rosette, C., and Karin, M. (1993) Genes Dev. 7 705–718 [DOI] [PubMed] [Google Scholar]
- 34.Heyninck, K., De Valck, D., Vanden Berghe, W., Van Criekinge, W., Contreras, R., Fiers, W., Haegeman, G., and Beyaert, R. (1999) J. Cell Biol. 145 1471–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strassmann, G., Patil-Koota, V., Finkelman, F., Fong, M., and Kambayashi, T. (1994) J. Exp. Med. 180 2365–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shinomiya, S., Naraba, H., Ueno, A., Utsunomiya, I., Maruyama, T., Ohuchida, S., Ushikubi, F., Yuki, K., Narumiya, S., Sugimoto, Y., Ichikawa, A., and Oh-ishi, S. (2001) Biochem. Pharmacol. 61 1153–1160 [DOI] [PubMed] [Google Scholar]
- 37.Hubbard, N. E., Lee, S., Lim, D., and Erickson, K. L. (2001) Prostaglandins Leukot. Essent. Fatty Acids 65 287–294 [DOI] [PubMed] [Google Scholar]
- 38.Uematsu, S., Matsumoto, M., Takeda, K., and Akira, S. (2002) J. Immunol. 168 5811–5816 [DOI] [PubMed] [Google Scholar]
- 39.Kang, Y. J., Wingerd, B. A., Arakawa, T., and Smith, W. L. (2006) J. Immunol. 177 8111–8122 [DOI] [PubMed] [Google Scholar]
- 40.Samuelsson, B., Morgenstern, R., and Jakobsson, P. J. (2007) Pharmacol. Rev. 59 207–224 [DOI] [PubMed] [Google Scholar]
- 41.Murakami, M., and Kudo, I. (2006) Curr. Pharm. Des. 12 943–954 [DOI] [PubMed] [Google Scholar]
- 42.Beinke, S., and Ley, S. C. (2004) Biochem. J. 382 393–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dumitru, C. D., Ceci, J. D., Tsatsanis, C., Kontoyiannis, D., Stamatakis, K., Lin, J. H., Patriotis, C., Jenkins, N. A., Copeland, N. G., Kollias, G., and Tsichlis, P. N. (2000) Cell 103 1071–1083 [DOI] [PubMed] [Google Scholar]
- 44.Beinke, S., Robinson, M. J., Hugunin, M., and Ley, S. C. (2004) Mol. Cell. Biol. 24 9658–9667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pillinger, M. H., Rosenthal, P. B., Tolani, S. N., Apsel, B., Dinsell, V., Greenberg, J., Chan, E. S., Gomez, P. F., and Abramson, S. B. (2003) J. Immunol. 171 6080–6089 [DOI] [PubMed] [Google Scholar]
- 46.Gomez, P. F., Pillinger, M. H., Attur, M., Marjanovic, N., Dave, M., Park, J., Bingham, C. O., III, Al-Mussawir, H., and Abramson, S. B. (2005) J. Immunol. 175 6924–6930 [DOI] [PubMed] [Google Scholar]
- 47.Fushimi, K., Nakashima, S., You, F., Takigawa, M., and Shimizu, K. (2007) J. Cell. Biochem. 100 783–793 [DOI] [PubMed] [Google Scholar]
- 48.Ishikawa, H., Claudio, E., Dambach, D., Raventos-Suarez, C., Ryan, C., and Bravo, R. (1998) J. Exp. Med. 187 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang, B., Brecher, P., and Cohen, R. A. (2001) Arterioscler. Thromb. Vasc. Biol. 21 1915–1920 [DOI] [PubMed] [Google Scholar]
- 50.Vermeulen, L., De Wilde, G., Van Damme, P., Vanden Berghe, W., and Haegeman, G. (2003) EMBO J. 22 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poligone, B., and Baldwin, A. S. (2001) J. Biol. Chem. 276 38658–38664 [DOI] [PubMed] [Google Scholar]
- 52.D'Acquisto, F., Sautebin, L., Iuvone, T., Di Rosa, M., and Carnuccio, R. (1998) FEBS Lett. 440 76–80 [DOI] [PubMed] [Google Scholar]
- 53.Largo, R., Diez-Ortego, I., Sanchez-Pernaute, O., Lopez-Armada, M. J., Alvarez-Soria, M. A., Egido, J., and Herrero-Beaumont, G. (2004) Ann. Rheum. Dis. 63 1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Driessler, F., Venstrom, K., Sabat, R., Asadullah, K., and Schottelius, A. J. (2004) Clin. Exp. Immunol. 135 64–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhattacharyya, S., Sen, P., Wallet, M., Long, B., Baldwin, A. S., Jr., and Tisch, R. (2004) Blood 104 1100–1109 [DOI] [PubMed] [Google Scholar]
- 56.Kamimura, M., Viedt, C., Dalpke, A., Rosenfeld, M. E., Mackman, N., Cohen, D. M., Blessing, E., Preusch, M., Weber, C. M., Kreuzer, J., Katus, H. A., and Bea, F. (2005) Circ. Res. 97 305–313 [DOI] [PubMed] [Google Scholar]
- 57.Cheon, H., Rho, Y. H., Choi, S. J., Lee, Y. H., Song, G. G., Sohn, J., Won, N. H., and Ji, J. D. (2006) J. Immunol. 177 1092–1100 [DOI] [PubMed] [Google Scholar]
- 58.Ferrier, R., Nougarede, R., Doucet, S., Kahn-Perles, B., Imbert, J., and Mathieu-Mahul, D. (1999) Oncogene 18 995–1005 [DOI] [PubMed] [Google Scholar]
- 59.Li, Z., Zhang, J., Chen, D., and Shu, H. B. (2003) Biochem. Biophys. Res. Commun. 309 980–985 [DOI] [PubMed] [Google Scholar]
- 60.Parameswaran, N., Pao, C. S., Leonhard, K. S., Kang, D. S., Kratz, M., Ley, S. C., and Benovic, J. L. (2006) J. Biol. Chem. 281 34159–34170 [DOI] [PubMed] [Google Scholar]
- 61.Ventura-Holman, T., Hahn, H., Subauste, J. S., and Maher, J. F. (2005) Tumour Biol. 26 294–299 [DOI] [PubMed] [Google Scholar]
- 62.Maher, J. F., Hines, R. S., Futterweit, W., Crawford, S., Lu, D., Shen, P., Oefner, P., Kazi, M., Wilson, J. G., Subauste, J. S., and Cowan, B. D. (2005) Gynecol. Endocrinol. 21 330–335 [DOI] [PubMed] [Google Scholar]
- 63.Lu, D., Ventura-Holman, T., Li, J., McMurray, R. W., Subauste, J. S., and Maher, J. F. (2005) Mol. Cell. Biol. 25 6570–6577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andersohn, F., Suissa, S., and Garbe, E. (2006) Circulation 113 1950–1957 [DOI] [PubMed] [Google Scholar]
- 65.Solomon, S. D., Pfeffer, M. A., McMurray, J. J., Fowler, R., Finn, P., Levin, B., Eagle, C., Hawk, E., Lechuga, M., Zauber, A. G., Bertagnolli, M. M., Arber, N., and Wittes, J. (2006) Circulation 114 1028–1035 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.