Abstract
One of the major determinants of organic solvent tolerance is the increase in membrane phospholipids. Here we report for the first time that an increase in the synthesis of phosphatidic acid is responsible for enhanced phospholipid synthesis that confers tolerance to the organic solvent in Saccharomyces cerevisiae. This increase in phosphatidic acid formation is because of the induction of Ict1p, a soluble oleoyl-CoA:lysophosphatidic acid acyltransferase. YLR099C (ICT1) was reported to be maximally expressed during solvent tolerance (Miura, S., Zou, W., Ueda, M., and Tanaka, A. (2000) Appl. Environ. Microbiol. 66, 4883–4889); however, its physiological significance was not understood. In silico analysis revealed the absence of any transmembrane domain in Ict1p. Domain analysis showed that it has a hydrolase/acyltransferase domain with a distinct lipid-binding motif and a lysophospholipase domain. Analysis of ict1Δ strain showed a drastic reduction in phosphatidic acid suggesting the role of Ict1p in phosphatidic acid biosynthesis. Overexpression of Ict1p in S. cerevisiae showed an increase in phosphatidic acid and other phospholipids on organic solvent exposure. To understand the biochemical function of Ict1p, the gene was cloned and expressed in Escherichia coli. The purified recombinant enzyme was found to specifically acylate lysophosphatidic acid. Specific activity of Ict1p was found to be higher for oleoyl-CoA as compared with palmitoyl- and stearoyl-CoAs. This study provides a mechanism for organic solvent tolerance from the point of membrane dynamics in S. cerevisiae.
Tolerance to organic solvents was reported in several microorganisms, which includes Pseudomonas strains (1) and Saccharomyces cerevisiae (2). Steady accumulation of these solvents in plasma membrane resulted in the loss of structural integrity (3). Because the plasma membrane acts as a selectively permeable barrier of the cell, such a toxicity causes an impairment in the ionic and the metabolic balances, pH gradient, electrical potential, etc., leading to cell lysis.
Cell surface modifications were found to be one major reason for such a tolerance. Other important factors are metabolic degradation of organic solvents, pumps responsible for extrusion (2), changes in saturated fatty acid contents, and conversion from cis to trans fatty acids (4). For example in Pseudomonas putida DOT-T1, the conversion of cis-9,10-methylene hexadecanoic acid to unsaturated cis-9-hexadecenoic acid was observed on exposure to toluene. However, the mechanism for the conversion is unknown (5). Change in the phospholipid content was shown to be an important factor in providing tolerance against the organic solvents. Elegant work on mechanisms of solvent tolerance in P. putida strain Idaho showed that the strain was able to repair the damaged membranes through efficient turnover and increased phospholipid biosynthesis. A detailed analysis of phospholipid head group turnover revealed the presence of a large amount of phosphatidylglycerol followed by phosphatidylethanolamine (PE)3 and cardiolipin. When the P. putida strain was grown in the presence of o-xylene, the amount of PE was high, and it was proposed that the increase in phospholipids was necessary to stabilize the cell membrane structure (3). When Escherichia coli was exposed to 0.25% toluene, a dramatic effect in the ultrastructure of the cell was observed. On treatment with 0.25% of phenethyl alcohol, intracellular membrane formation was observed in E. coli (6).
An isooctane-tolerant strain of S. cerevisiae KK-12 was reported to have increased saturated fatty acid content (2). Among the various genes up-regulated on isooctane treatment, ICT1 (Increased Copper Tolerance 1) was found to have maximal expression (7). The gene product Ict1p belongs to the group of α/β-hydrolase family of proteins. Domain analysis suggests that it has a hydrolase/acyltransferase domain with a distinct lipid-binding and a lipase motif (8, 9). Based on the above observations, we investigated the relationship of ICT1 (YLR099C) and its role in phospholipid metabolism in S. cerevisiae with isooctane treatment.
We initially determined the differences in the phospholipid composition in the presence and in the absence of isooctane. We then studied the phospholipid profiles of ict1Δ and ICT1 overexpressed strains on isooctane treatment. Finally, we elucidated the enzymatic function of the purified recombinant Ict1p. Our results clearly show that on treatment with 4% isooctane, the overall phospholipid biosynthesis in the cell was markedly enhanced. Intriguingly, ict1Δ strain showed a reduced biosynthesis of major phospholipids both in the presence and in the absence of isooctane. However, the amount of phosphatidic acid (PA) was found to be drastically reduced in the ict1Δ strain. Based on the above observations, we report here that Ict1p is a soluble lysophosphatidic acid acyltransferase that is highly expressed during organic solvent stress, to sustain enhanced requirement of membrane phospholipid for the repair of damaged membrane.
EXPERIMENTAL PROCEDURES
Materials—S. cerevisiae (BY4741: MATa; his 3Δ1; leu2Δ0; met15Δ0; ura3Δ0) and deletion mutant strain of ICT1 (BY4741 background) were obtained from Invitrogen. [1-14C]Oleoyl-CoA (54 mCi/mmol), [3H]1-oleoyl-lysophosphatidic acid (47 Ci/mmol), and other radiochemicals were obtained from PerkinElmer Life Sciences. [32P]Orthophosphate (5000 Ci/mmol) was obtained from Board of Radiation and Isotope Technology, Bhabha Atomic Research Centre (Mumbai, India). Silica Gel 60F254 TLC plates were purchased from Merck. Synthetic complete medium and yeast nitrogen base media were obtained from Sigma. Yeast extract and peptone were obtained from Difco. The DNA purification kit and nickel-nitrilotriacetic acid (Ni2+-NTA) matrix were from Qiagen. All chemicals and solvents were purchased from Sigma unless specifically mentioned.
Growth Conditions—Yeast cells were grown in YEPD (1% yeast extract, 2% peptone, and 2% dextrose) medium (pH 7.0) with aeration at 30 °C. For growth in organic solvent-containing medium, the yeast cells were grown in 5 ml of YEPD medium for 12 h. The cells were harvested and transferred to 25 ml of YEPD media overlaid with 4% (v/v) isooctane, so that the final A600 of 0.1 was achieved. Following this the cells were grown at 30 °C at 180 rpm. Cell growth was studied by measuring the A600 of the cultures at frequent intervals.
[32P]Orthophosphoric Acid Labeling—The cells were grown to saturation in 5 ml of YEPD cultures and then transferred to 25 ml of YEPD medium either in the presence or in the absence of isooctane such that the absorbance was 0.1. To this 100 μCi of [32P]orthophosphate (5000 Ci/mmol) was added. Cells (absorbance of 10) were grown for 12 h and harvested by centrifugation.
Phospholipid Extraction and Identification—Phospholipids from yeast cells were extracted after 12 h of growth (with and without 4% isooctane) according to Bligh and Dyer (10). Briefly, to the cell pellet 400 μl of methanol and 200 μl of chloroform were added followed by vortexing. To this, 400 μl of acidified water (2% phosphoric acid) was added and vigorously vortexed. Chloroform extracts containing total lipids were subjected to two-dimensional TLC with Silica Gel 60F254 TLC plates. The solvents for the first dimension were chloroform/methanol/ammonia (65:25:5, v/v); solvents for the second dimension were chloroform/methanol/acetone/acetic acid/water (50:10:20:15:5, v/v). Individual phospholipids were located by comparing the Rf values of the unknown with the Rf values of the standard. The radioactive spots were developed by autoradiography for 12 h. The spots were scraped from the TLC plate, and the incorporation was counted in liquid scintillation counter (PerkinElmer Life Sciences).
Purification of Recombinant Ict1p—For expression of recombinant Ict1p in E. coli, DNA fragments containing the coding sequence of ICT1 were generated by PCR amplification of the genomic DNA from S. cerevisiae using forward primers (5′-CCCCGGATCCATGTGGACAAACACTTTCAAATGGTGC-3′) and reverse primer (5′-CCCCGAATTCTTACTTTGACAGGAACGAGACTAAA-3′). The forward primer had a BamHI followed by the beginning of an open reading frame; the reverse primer had an EcoRI site. PCR (1 min of denaturation at 94 °C, 1 min of annealing at 55 °C, and 1 min of elongation at 72 °C) was performed using Pfu polymerase for 30 cycles with a 10 pmol concentration of each primer in a final volume of 50 μl. The purified PCR product was digested with BamHI and EcoRI and ligated directionally in predigested pRSET A vector. The construct was transformed into E. coli BL21 (DE3) cells, and the insert was sequenced. The transformed E. coli cells were induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h, and the cell lysate was resolved on a 12% SDS-PAGE. Recombinant Ict1p was purified by Ni2+-NTA affinity column chromatography. Briefly, the cell pellet was resuspended in lysis buffer containing 50 mm Tris-HCl (pH 8.0) and 300 mm NaCl (buffer A). Cells were disrupted by sonication. The supernatant (10,000 × g) was allowed to bind to the Ni2+-NTA matrix. The column was washed with buffer A containing 80 mm imidazole. The bound protein was eluted with 250 mm imidazole in buffer A. One-milliliter fractions were collected and checked for activity. The fractions were analyzed for the presence of protein in 12% SDS-PAGE followed by Coomassie Blue staining.
Antisera Production—Rabbits were immunized by subcutaneous injection of 250 μg of purified Ict1p, emulsified in Freund's complete adjuvant. Three booster doses of 125 μg of protein emulsified in Freund's incomplete adjuvant were administered at 3 weekly intervals. Ten days after the last injection, blood was collected, and serum was separated and stored at -20 °C. The antibody production, specificity, and the titer were analyzed by enzyme-linked immunosorbent assay (11).
Construction of Yeast Expression Plasmid—For the overexpression of ICT1 in S. cerevisiae, we utilized a plasmid pPS189 bearing a constitutive promoter, PTEF2 (12). The ICT1 open reading frame was subcloned from pRSET A vector to pPS189 at BamHI-EcoRI sites.
Expression of ICT1 and Preparation of Cell Extracts—BY4741 strain of S. cerevisiae was transformed with pPS189 cloned with and without ICT1 (13). The transformants were selected on the plates containing synthetic medium (SM) lacking uracil. For the preparation of cell extracts, the cells were grown in a 5-ml SM culture overnight. Cells (A = 5) were taken and suspended in 10 mm Tris-HCl (pH 7.5), 2% SDS. The cells were lysed using glass beads (0.45–0.6 μm). The proteins were separated by SDS-PAGE and transferred onto a nitrocellulose membrane for immunoblotting as described (14). The overexpressed Ict1p was detected using anti-Ict1p antibodies at a dilution of 1:2500 (v/v).
Growth Studies on Yeast Strains Overexpressing ICT1—Yeast cells were grown overnight in 5 ml of SM without uracil. The cells were then transferred to 25 ml of SM without uracil and overlaid with 4% (v/v) isooctane, so that the final A600 of 0.1 was achieved. Following this, the cells were grown at 30 °C at 180 rpm. Cell growth was monitored by measuring the A600 of the cultures at frequent intervals.
Labeling of Overexpressed ICT1 Strain—Strain bearing ICT1 overexpression plasmid and vector alone was grown to saturation in 5 ml of SM lacking uracil and then transferred to 25 ml of the same, either in the presence or in the absence of isooctane such that the absorbance is 0.1. To this, 100 μCi of [32P]orthophosphate was added. Cells were grown for 20 h and harvested (A = 10) by centrifugation.
Sequence Analysis and Construction of Phylogenetic Tree—Domain analysis was done based on the conserved domain data base of NCBI. Hydrophobicity plot was analyzed by TMPRED. Multiple sequence alignment was constructed using “bootstrap resampling.” This bootstrap replicate alignment was then utilized to construct phylogenetic trees by the neighbor-joining method (15). Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 2.1 software. The phylogenetic tree was visualized using MEGA. The result was analyzed using the bootstrap method (1000 replicates) to provide confidence levels for the tree topology (16). The tree topology was generated by neighbor-joining, with bootstrap support at critical nodes indicated as percentage.
LPA Acyltransferase Activity—The assay mixtures consisted of all of the components of lysis buffer containing 20 μm [1-14C]oleoyl-CoA (54 mCi/mmol), 1 μg of enzyme, and 100 μm LPA (1-oleoyl) in a total volume of 100 μl (17). The incubation was carried out at 30 °C for 15 min and stopped by extracting lipids as described above. In addition, acyltransferase activity was also assayed using 50 μm labeled acyl acceptor [1-oleoyl-9,10-3H]LPA (47 Ci/mmol) along with 20 μm oleoyl-CoA. Lipids were separated on either a two-dimensional TLC plate using chloroform/methanol/ammonia/water (65:25:0.9:3, v/v) as the solvent of the first dimension, and chloroform/methanol/acetic acid/water (40:20:5:0.5, v/v) as the solvent of the second dimension (18) or on a one-dimensional silica TLC plate. The solvent system that was used for one-dimensional TLC was composed of chloroform/methanol/acetone/acetic acid/water (50:10:20:15:5, v/v). Individual phospholipids were located by comparing the Rf values of the unknown with the Rf values of the standard. The plates were developed overnight for autoradiography to localize 14C-labeled lipids. [14C]PA spots were then scraped from the TLC plate and counted with 5 ml of toluene-based scintillation mixture.
RESULTS
Enhanced Phospholipid Accumulation on Isooctane Exposure—To investigate the mechanism of organic solvent tolerance, we first determined the growth kinetics of S. cerevisiae in the presence and in the absence of isooctane (data not shown). As it is known that one of the important mechanisms of organic tolerance is rapid turnover of membrane phospholipids (3), we wanted to ascertain whether changes in total membrane phospholipid were responsible for the growth advantage of S. cerevisiae in the presence of 4% isooctane. To analyze the membrane phospholipid profile, S. cerevisiae cells were grown in the presence of isooctane along with [32P]orthophosphate for 12 h. It was found that on exposure to isooctane there was a 1.8-fold increase in the incorporation of label into phospholipids.
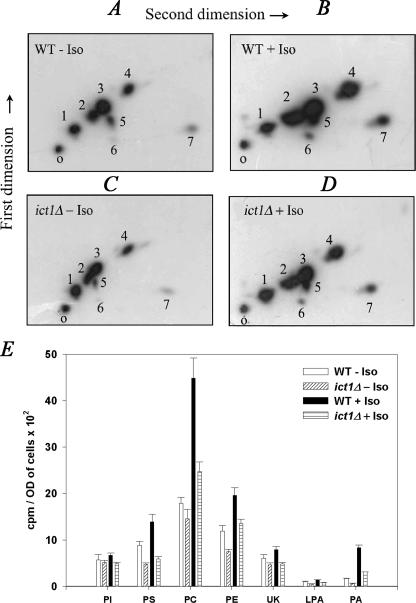
When the total phospholipid of S. cerevisiae was separated on a two-dimensional TLC, six components were easily distinguished (Fig. 1, A and B). They were identified as phosphatidylinositol (PI), LPA, phosphatidylserine (PS), phosphatidylcholine (PC), PE, and PA, and one of the spots remained unknown. The identification was based on the comparison of their Rf values to that of the known standards. PC and PE were detected as the major phospholipids followed by PS and PI. On exposure to isooctane there was nearly a 2-fold increase in the incorporation of label into PC, PE, and PS. However, PA showed a 4.9-fold increase with isooctane treatment (Fig. 1E). These results suggested a correlation between the cell survival and the increased phospholipid formation.
FIGURE 1.
ICT1 knock-out shows decreased phospholipid accumulation in the presence of isooctane. A–D, both wild type (WT) and ict1Δ cells were grown overnight and transferred to 25 ml of YEPD medium containing 100 μCi of [32P]orthophosphate to a final absorbance of 0.1 and then grown for 12 h in the presence and in the absence of 4% isooctane (Iso). Lipids were extracted and resolved on two-dimensional silica TLC by chloroform/methanol/ammonia (65:25:5, v/v) as first dimension and chloroform/methanol/acetone/acetic acid/water (50:10:20:15:5, v/v) as second dimension solvent systems. Lipids are identified by numbers ranging from 1 to 7. 1, PI; 2, PS; 3, PC; 4, PE; 5, unknown; 6, LPA; and 7, PA. O represents the origin. E, amount of [32P]orthophosphate incorporated into various phospholipids. The amount of [32P]orthophosphate incorporated into lipids is presented as counts/min per A600 of cells per 12 h of labeling. Values are the mean of three separate experiments, each performed in duplicate.
Phospholipid Profile of ict1Δ Strain Shows a Deficient PA Formation—Initially, the growth characteristic of ict1Δ in the presence of isooctane was determined. To our surprise, the null mutant behaved in a similar way to that of the wild type (data not shown). To analyze the phospholipid profile in the presence of isooctane, we performed comparative labeling studies with ict1Δ and BY4741 strains in the presence or in the absence of isooctane. ict1Δ and the wild type were grown in the presence of [32P]orthophosphate to examine the total phospholipid synthesis. It was interesting to observe that there was nearly a 3-fold decrease in the PA formation in ict1Δ (Fig. 1, C–E, PA bars) compared with the wild type (Fig. 1, A, B, and E, PA bars) both in the absence or in the presence of isooctane. The overall phospholipid formation in ict1Δ was reduced, suggesting that reduced PA formation probably resulted in the decrease of all the major phospholipids. These data suggest that ict1Δ is partially defective in PA formation.
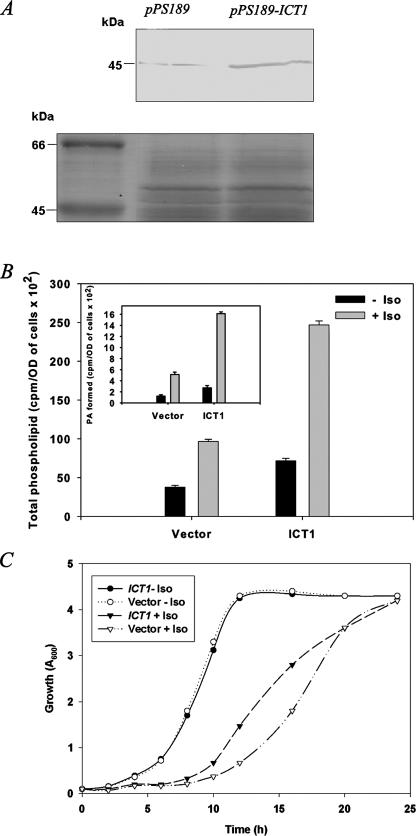
ICT1 Overexpression Enhances the Levels of Membrane Phospholipids and Reduces Lag Phase on Isooctane Exposure—To study the role of ICT1 in modulating the membrane phospholipid profile on isooctane exposure, S. cerevisiae was transformed with pPS189-ICT1. Western blotting with anti-Ict1p antibodies confirmed the expression and the proper molecular mass of the protein (Fig. 2A). To study the membrane phospholipid profile of the ICT1 overexpressing strain, the cells were grown in SM without uracil, overlaid with or without 4% isooctane. Overexpression of ICT1 (without isooctane) increased the total cellular phospholipid levels by 1.9-fold. Interestingly, in the presence of isooctane, the total cellular phospholipid enhanced by 2.5-fold as compared with the vector-transformed cells. This increase was nearly 6.5-fold when compared with vector-transformed cells in the absence of isooctane (Fig. 2B). The increase of PA was nearly 3.1-fold as compared with the control, on isooctane treatment (Fig. 2B, inset). Thus, the overexpression of ICT1 along with isooctane exposure led to a much higher activity than without isooctane.
FIGURE 2.
Overexpression of ICT1. A, yeast cells overexpressing ICT1 and the vector control were grown overnight, and cells (A = 5) were lysed using glass beads. The proteins were separated on a 12% SDS-PAGE, and immunoblotting was performed using anti-Ict1p antibodies at the dilution of 1:2500 (v/v), confirming the enhanced expression in the ICT1-transformed cells. A segment of the Coomassie Blue-stained gel is given as a loading control for the immunoblot. B, yeast cells overexpressing ICT1 and the vector control were grown for 20 h in the presence of 100 μCi of [32P]orthophosphate, and cells (A = 10) were used for lipid extraction and resolved on two-dimensional silica TLC by chloroform/methanol/ammonia (65:25:5, v/v) as first dimension and chloroform/methanol/acetone/acetic acid/water (50:10:20:15:5, v/v) as second dimension solvent systems. The amounts of [32P]orthophosphate incorporated into phospholipids were scraped and counted. Inset, represents the amount of [32P]orthophosphate incorporated into PA. The amount of [32P]orthophosphate incorporated into lipids is presented as counts/min per A600 of cells per 20 h of labeling. Each data point represents the mean of three independent experiments ± S.D. C, exponentially growing cultures of ICT1-overexpressing strain and vector control, grown in synthetic medium lacking uracil, were added to a fresh medium (with or without 4% isooctane (Iso)) to a final absorbance of 0.1 and grown at 30 °C. Absorbance was measured at regular intervals.
To ascertain that the overexpression of ICT1 could rescue the growth delay of the organism on isooctane treatment, the ICT1-overexpressing strain was grown both in the presence and in the absence of 4% isooctane in SM without uracil. The strain overexpressing ICT1 was found to have an improved doubling time as compared with the vector control (Fig. 2C). The partial rescue of the doubling time suggests that enhanced expression of ICT1 could possibly have a role in maintaining the membrane homeostasis on exposure to organic solvent.
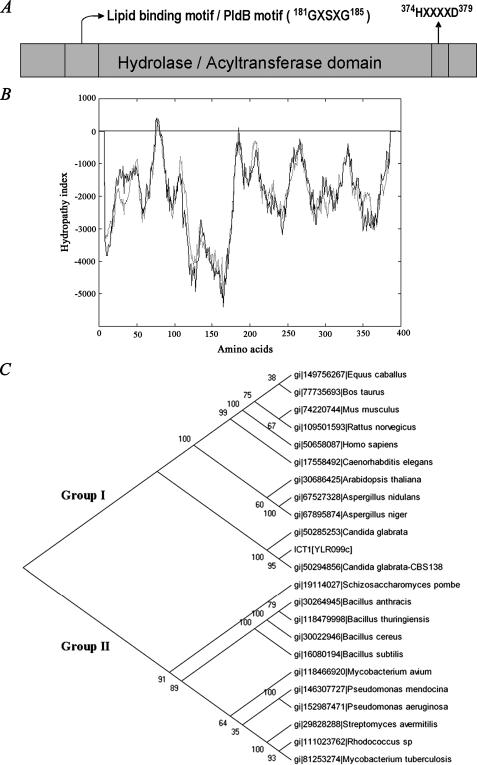
Ict1p Belongs to the Family of α/β-Hydrolases—Ict1p is a 394-amino acid protein with a pI of 9.87. Ict1p is a member of hydrolase family of proteins, which has derived its name from a highly conserved tertiary fold consisting of alternating α-helices and β-sheets (8, 9). The rodent and human genomes contain at least 15 orthologous sets of predicted α/β-hydrolases, none of which are functionally annotated in terms of catalytic activity or substrate selectivity (19). In addition to the hydrolase domain, Ict1p possesses an esterase (pfam 00756), hydrolase/acyltransferase domain (COG 0596), and lysophospholipase domain (COG 2267) (Fig. 3A). The first structural motif of Ict1p, 181GXSXG185 (Fig. 3A) forms a highly conserved stretch of amino acids in majority of the known lipases, phospholipases, lysophospholipases, esterases, and serine proteases (20). A distinct structural motif 374HXXXXD379 is present in Ict1p (Fig. 3A) at the C-terminal region. It is well documented that glycerolipid acyltransferases from the bacteria, plant, and animal kingdoms share a highly conserved domain containing the invariant histidine and aspartic acid residues separated by four less conserved residues (21–23). Hydropathy plot of Ict1p predicted the absence of any transmembrane domain (Fig. 3B). To determine the relationship of Ict1p with its close homologues among prokaryotes, higher eukaryotes, and mammals, a phylogenetic tree was made (Fig. 3C). The cladogram was mainly divided in two groups. Group I includes higher eukaryotes like plants and mammals, and group II includes mainly prokaryotes. Ict1p was more closely related to group I. Ict1p was found to be closely associated to Candida homologues, but it was found to be diverged from Schizosaccharomyces pombe. Ict1p homologue in S. pombe was more closely related to group II.
FIGURE 3.
Domain structure, hydropathy plot, and phylogenetic tree of Ict1p. A, domain structure of Ict1p. The domains are retrieved from conserved domain data base at NCBI. α/β-Hydrolase fold (pfam 0059; COG 0596); PldB, lysophospholipase (COG 2267). 374HXXXXD379 is a signature motif for all the glycerolipid acyltransferases. B, hydropathy plot of Ict1p. Hydrophobicity plot was analyzed using TMPRED. C, phylogenetic tree of Ict1p with other related species are based on sequence alignments calculated using the neighbor-joining method to show the evolutionary relationship of members. The length of the branches is proportional to the degree of divergence and thus corresponds to the statistical significance of the phylogeny between the protein sequences. The numbers at the nodes of branches represents bootstrap values.
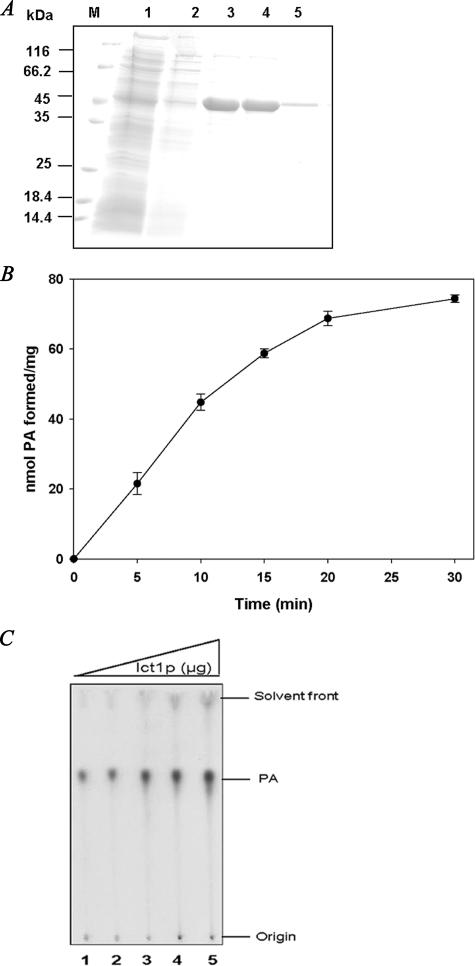
ICT1 Encodes an LPA Acyltransferase—A drastic decrease in PA in ict1Δ either in the absence (Fig. 1, A and C) or in the presence of isooctane (Fig. 1, B and D), suggested a role of ICT1 in PA formation. PA can be produced de novo from acylation of LPA. Alternatively, PA can be synthesized either by phospholipase D-mediated hydrolysis of phospholipids or through phosphorylation of diacylglycerol. Toward this, ICT1 was cloned in pRSET A, expressed in BL21 (DE3), and purified using nickel-nitrilotriacetic acid column (Fig. 4A). Immunoblot analysis of the transformed cells with anti-His tag was found to be highly specific for Ict1p (data not shown). Based on the sequence information of Ict1p, it was predicted that this protein could synthesize PA either via phospholipase D-like activity or via direct acyl-CoA-dependent acylation of LPA. Recombinant Ict1p was then analyzed for lipase and phospholipase activities using various substrates. It was found that there was no lipase ([3H]triacylglycerol) and phospholipase ([14C]PC and [14C]PE) activities in the recombinant Ict1p (data not shown). The purified recombinant enzyme was therefore analyzed for LPA acyltransferase activity. Ict1p showed a time (Fig. 4B) and enzyme concentration (Fig. 4C)-dependent increase in the incorporation of [1-14C]oleoyl-CoA into LPA to form PA. The enzymatic product was further analyzed by two-dimensional TLC, and a single discrete spot of PA was observed (data not shown). These data suggested that Ict1p indeed has a lysophosphatidic acid acyltransferase activity.
FIGURE 4.
Purification of recombinant Ict1p. A, purified Ict1p from E. coli BL21 (DE3) using nickel-nitrilotriacetic acid affinity purification column. Visualization of the induced Ict1p in Coomassie Blue-stained SDS-polyacrylamide gel. M denotes the mobility of molecular weight marker. Lane 1, unbound; lane 2, wash; lanes 3–5, purified Ict1p. B, time-dependent formation of PA. Assays were performed using 100 μm LPA and 20 μm [14C]oleoyl-CoA and 5 μg of enzyme in a final volume of 0.5 ml. Aliquots of 100 μl were withdrawn at different time intervals, and the reaction was terminated by adding methanol/chloroform (2:1, v/v) mixture. The activity was assessed as described under “Materials and Methods.” Values are the mean of three separate experiments, each performed in triplicate. C, LPA acyltransferase activity was monitored under the standard assay conditions with increasing amounts of enzyme. Lanes 1–5 represent assays performed using 0.125, 0.25, 0.5, 0.75, and 1 μg of the purified recombinant Ict1p.
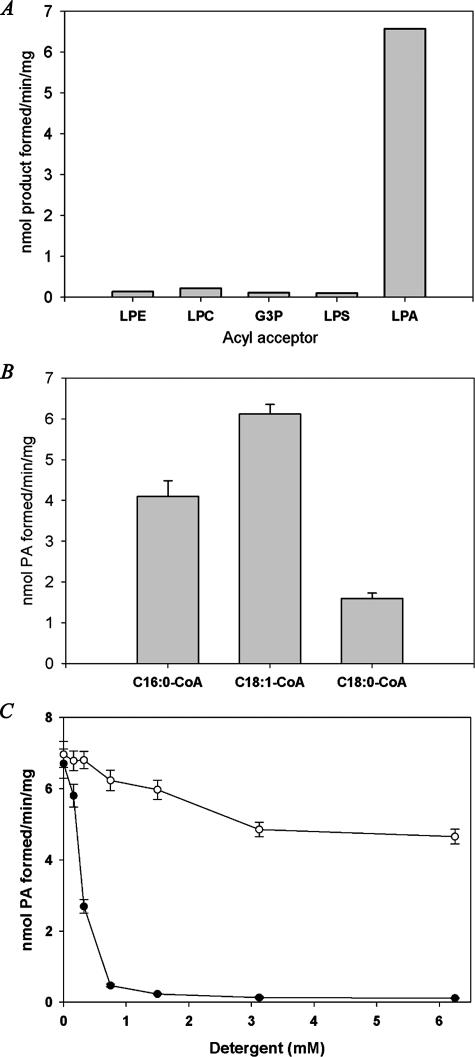
Substrate Preference of the Recombinant Ict1p—To study the substrate specificity of Ict1p for lysophospholipids, acyltransferase activity was performed using [14C]oleoyl-CoA and various unlabeled lysophospholipids. It was found that Ict1p was highly selective to LPA, showing nearly no activity with other lysophospholipids such as lysophosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE), and lysophosphatidylserine (LPS) (Fig. 5A). LPA acyltransferase activity was also measured using [3H]LPA and unlabeled acyl-CoAs. The highest activity was observed with oleoyl-CoA (6.12 nmol of PA formed per min/mg protein) as compared with palmitoyl-CoA. When stearoyl-CoA was used as an acyl donor, only 26% of the activity was found under the standard assay conditions (Fig. 5B).
FIGURE 5.
Characterization of purified Ict1p. A, purified Ict1p was incubated with 100 μm of various acyl acceptors such as Glc-3-P (G3P), LPA, LPC, LPE, and LPS, and the reaction products were resolved on silica-TLC using chloroform/methanol/acetone/acetic acid/water (50:10:20:15:5, v/v) as the solvent system, and corresponding acylated phospholipid products were measured. Values are the average of two separate experiments, and each was performed in triplicate. B, LPA acyltransferase activity was monitored using 50 μm of [3H]LPA and 50 μm of palmitoyl-CoA or oleoyl-CoA or stearoyl-CoA. C, effect of Triton X-100 and CHAPS on LPA acyltransferase activity. Values are the mean of three separate experiments, and each was performed in triplicate.
LPA acyltransferase activity was found to be sensitive to Triton X-100, but it was fairly stable over wide range of CHAPS concentrations. The activity was reduced to 7% at 0.75 mm of Triton X-100; however, 92% of the activity was recovered at the same concentration of CHAPS. At 3.125 mm of Triton X-100, the activity was reduced to 2%; however, only 30% decrease in the activity was observed with CHAPS under the same concentration (Fig. 5C).
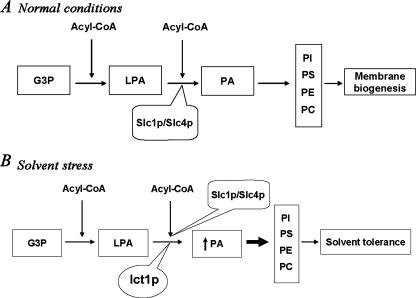
Together these findings indicated that Ict1p is a soluble LPA acyltransferase that may play an important role in increasing PA biosynthesis on organic solvent exposure, in addition to the membrane-bound Slc1p and Slc4p/Lpt1p. Increase in the PA formation could eventually lead to an increase in PC, PE, and PS (the major phospholipids in S. cerevisiae), thereby facilitating tolerance to the solvent stress (Fig. 6, A and B).
FIGURE 6.
Biosynthesis and utilization of phosphatidic acid in S. cerevisiae on isooctane stress. Metabolites used are as follows: Glc-3-P (G3P), LPA, PA, PI, PS, PE, and PC. Enzymes used are as follows: Slc1/Slc4, 1-acylglycerol-3-phosphate acyltransferase; ICT1, increased copper tolerance 1. A, Glc-3-P (G3P) is esterified to LPA, which in turn is acylated by membrane-bound Slc1p/Slc4p to form PA. PA is the major intermediate for all other phospholipids that constitute the plasma membrane of S. cerevisiae. B, on organic solvent exposure, Ict1p is overexpressed, resulting in enhanced synthesis of PA. Increased synthesis of PA leads to increased synthesis of PC, PE, PI, and PS that are necessary for the repair of damaged membranes.
DISCUSSION
Organic solvent tolerance mechanism is known to be associated with membrane remodeling in microorganisms (3, 24). To understand this phenomenon, we chose S. cerevisiae as the model system. Enhanced phospholipid biosynthesis was observed on treatment with 4% isooctane, suggesting a correlation between accumulation of membrane phospholipids and tolerance to organic solvent. These data are consistent with tolerance mechanisms exhibited by P. putida Idaho strain (3, 5), where the basal level of phospholipid biosynthesis was high on exposure to solvent. Increased phospholipid biosynthesis is a common phenomenon exhibited by organisms to various environmental stresses. Vibrio costicola was found to have a high phospholipid biosynthesis under salt stress (25). It was shown that MUQ1 involved in PE biosynthesis was substantially expressed on isooctane stress, validating an enhanced phospholipid accumulation as observed in our system (2). One of the main initiatives of the study was to identify the role of ICT1 in such a stress. PA was substantially decreased in ict1Δ strain indicating a partial loss of PA biosynthesis.
Ict1p is annotated as a soluble protein with α/β-hydrolase domain containing hydrolase/acyltransferase motif, esterase, and lipase/phospholipase motif (8, 9, 26). Interestingly, the region consisting of amino acids 69–87 showed a highly hydrophobic region that corresponds to the lipid-binding motif of the protein. The presence of lysophospholipid binding domain suggested that the enzyme can utilize lysophospholipids. However, we could not find any lysophospholipase activity. Lipases were found to have acyltransferase activity (27, 28). The presence of a Ser residue in the active site forming the catalytic triad (Ser-His-Asp) of these enzymes (29) and HX4D motif (22, 23) suggests the possible acyltransferase activity of the protein. The utilization of His as a general base was proposed to be a common feature of acyltransferases. Histidine is well suited for the abstraction of a proton from the hydroxyl group of the acyl acceptor to facilitate nucleophilic attack on the thioester of the acyl donor (21).
The overexpression studies with ICT1 demonstrate that the induction of this gene enhanced the overall phospholipid biosynthesis. This effect was magnified in the presence of isooctane, which could be due to the existence of other pathways that could possibly be activated on isooctane exposure. This suggests that ICT1 probably is a crucial gene in organic solvent tolerance; however, such a complex phenomenon could be supported by a battery of other gene products that are necessary for the process.
Ict1p was found to be acyl-CoA:LPA-dependent acyltransferase, and the reaction was time- and protein-dependent. Ict1p had a specific preference for oleoyl-CoA suggesting its intrinsic capability to incorporate unsaturated fatty acids. Incorporation of unsaturated fatty acids during solvent stress leads to enhanced mobility of the membranes (30). Our results provide direct evidence for the presence of soluble acyl-CoA-dependent LPA acyltransferase. PA being an essential metabolite in glycerolipid and triacylglycerol biosynthesis enjoys special privilege in cellular metabolism. Therefore, the role of PA in conditions like organic solvent stress is undisputable. PA formation being the rate-limiting step in membrane lipid biosynthesis (31) could actually activate the biosynthesis of all the major phospholipids, especially PC and PE (Fig. 6). Such a phenomenon could be advantageous to the organism for two reasons. (i) Because PC is the major phospholipid in yeasts (32), the enhanced supply of PC could help repair the damaged membrane quickly. (ii) An increase in the proportion of high melting point PE could help counter membrane fluidity in isooctane-exposed cells (33).
In light of our observation, it is tempting to speculate a soluble lipid biosynthetic pathway that may function under abnormal cellular stress. Isooctane exposure could lead to the induction of ICT1 and other pathways involved in phospholipid biosynthesis. Such an induction could lead to enhanced phospholipid biosynthesis eventually resulting in membrane repair.
Until now, the only known LPA acyltransferase that was extensively studied in yeast at the molecular level was SLC1 (34), a homologue of E. coli plsC (35). Recently, three groups simultaneously showed the presence of another lysophospholipid acyltransferase, encoded by SLC4/LPT1, a member of membrane-bound O-acyltransferase family of protein that could acylate LPA as well, adding to the partial redundancy along with Slc1p. The enzyme accepts, however, LPS, lysophosphatidylinositol, LPC, and LPE, thereby adding to the heterogeneity observed among the yeast phospholipids (36–38). All these enzymes also have distinct transmembrane domains and form a major member of membrane-bound biosynthetic machinery of phospholipid biosynthesis.
We believe that SLC1 and SLC4 could independently provide the necessary PA for the cells to replenish their membrane requirement. However, ICT1 on isooctane exposure could provide the additional requirements for PA at a time of emergency. Therefore, PA could be channeled either for membrane lipid biosynthesis or for signaling events downstream as necessary under such situations.
The solvent tolerance mechanism of S. cerevisiae is dependent on activation of many different genes and pathways. Here we show that the increased membrane phospholipid is one of the major determinants for organic solvent tolerance. This study is the first one of its kind to provide a mechanistic basis of solvent tolerance.
Acknowledgments
We thank Dr. D. K. Venkata Rao for helpful comments on the manuscript. We also thank Neha Agarwal for technical assistance.
This work was supported by a grant from the Department of Biotechnology (New Delhi, India) under the program support for nonconventional yeasts. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PE, phosphatidylethanolamine; LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; LPS, lysophosphatidylserine; PA, phosphatidic acid; PC, phosphatidylcholine; PI, phosphatidylinositol; PS, phosphatidylserine; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
References
- 1.Inoue, A., and Horikoshi, K. (1989) Nature 338 264-265 [Google Scholar]
- 2.Miura, S., Zou, W., Ueda, M., and Tanaka, A. (2000) Appl. Environ. Microbiol. 66 4883-4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinkart, H. C., and White, D. C. (1997) J. Bacteriol. 179 4219-4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heipieper, H. J., and De Bont, J. A. M. (1994) Appl. Environ. Microbiol. 60 4440-4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramos, J. L., Duque, E., Rodriguez-Herva, J. J., Godoy, P., Haidour, A., Reyes, F., and Fernandez-Barrero, A. (1997) J. Biol. Chem. 272 3887-3890 [DOI] [PubMed] [Google Scholar]
- 6.Woldringh, C. L. (1973) J. Bacteriol. 114 1359-1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsui, K., Hirayama, T., Kuroda, K., Shirahige, K., Ashikari, T., and Ueda, M. (2006) Appl. Microbiol. Biotechnol. 71 75-79 [DOI] [PubMed] [Google Scholar]
- 8.Ollis, D. L., Cheah, E., Cygler, M., Dijkstra, B., Frolow, F., Franken, S. M., Harel, M., Remington, S. J., Silman, I., and Schrag, J. (1992) Protein Eng. 5 197-211 [DOI] [PubMed] [Google Scholar]
- 9.Akiyama, M., Sawamura, D., Nomura, Y., Sugawara, M., and Shimuzu, H. (2003) J. Investig. Dermatol. 121 1029-1034 [DOI] [PubMed] [Google Scholar]
- 10.Bligh, E. G., and Dyer, W. J. (1959) Can. J. Biochem. Physiol. 37 911-917 [DOI] [PubMed] [Google Scholar]
- 11.Engvall, E. (1980) Methods Enzymol. 70 419-439 [DOI] [PubMed] [Google Scholar]
- 12.Singh, S. R., Rekha, N., Pillai, B., Singh, V., Naorem, A., Sampath, V., Srinivasan, N., and Sadhale, P. P. (2004) Nucleic Acids Res. 32 201-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schiestl, R. H., and Gietz, R. D. (1989) Curr. Genet. 16 339-346 [DOI] [PubMed] [Google Scholar]
- 14.Towbin, H., Staehelin, T., and Gordon, J. (1979) Proc. Natl. Acad. Sci. U. S. A. 76 4350-4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saitou, N., and Nei, M. (1987) Mol. Biol. Evol. 4 406-425 [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein, J. (1996) Methods Enzymol. 266 418-426 [DOI] [PubMed] [Google Scholar]
- 17.Gangar, A., Karande, A. A., and Rajasekharan, R. (2001) J. Biol. Chem. 276 10290-10298 [DOI] [PubMed] [Google Scholar]
- 18.Saulnier-Blache, J. S., Girard, A., Simon, M., Lafonton, M., and Valet, P. (2000) J. Lipid Res. 41 1947-1951 [PMC free article] [PubMed] [Google Scholar]
- 19.Simon, G. M., and Cravatt, B. F. (2006) J. Biol. Chem. 281 26465-26472 [DOI] [PubMed] [Google Scholar]
- 20.Wei, Y., Contreras, J. A., Sheffield, P., Osterlund, T., Derewenda, U., Kneusel, R. E., Matern, U., Holm, C., and Zygmunt, S. (1999) Nat. Struct. Biol. 6 340-345 [DOI] [PubMed] [Google Scholar]
- 21.Heath, R. J., and Rock, C. O. (1998) J. Bacteriol. 180 1425-1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okazaki, K., Sato, N., Tsuji, N., Tsuzuki, M., and Nishida, I. (2006) Plant Physiol. 141 546-556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saha, S., Enugutti, B., Rajakumari, S., and Rajasekharan, R. (2006) Plant Physiol. 141 1533-1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramos, J. L., Duque, E., Gallegos, M. T., Godoy, P., Ramos-Gonzalez, M. I., Rojas, A., Teran, W., and Segura, A. (2002) Annu. Rev. Microbiol. 56 743-768 [DOI] [PubMed] [Google Scholar]
- 25.Russell, N. J., Kogut, M., and Kates, M. (1985) J. Gen. Microbiol. 131 781-789 [Google Scholar]
- 26.Nardini, M., and Dijkstra, B. W. (1999) Curr. Opin. Struct. Biol. 9 732-737 [DOI] [PubMed] [Google Scholar]
- 27.Jonas, A. (2000) Biochim. Biophys. Acta 1529 245-256 [DOI] [PubMed] [Google Scholar]
- 28.Buckley, J. T. (1983) Biochemistry 22 5490-5493 [Google Scholar]
- 29.Hilton, S., and Buckley, J. T. (1991) J. Biol. Chem. 266 997-1000 [PubMed] [Google Scholar]
- 30.Ingram, L. O. (1986) Trends Biotechnol. 4 40-44 [Google Scholar]
- 31.Athenstaedt, K., and Daum, G. (1999) Eur. J. Biochem. 266 1-16 [DOI] [PubMed] [Google Scholar]
- 32.Reikhof, W. R., and Voelker, D. R. (2006) J. Biol. Chem. 281 36588-36596 [DOI] [PubMed] [Google Scholar]
- 33.Melchior, D. L. (1982) Curr. Top. Membr. Transp. 17 263-316 [Google Scholar]
- 34.Nagiec, M. M., Wells, G. B., Lester, R. L., and Dickson, R. C. (1993) J. Biol. Chem. 268 22156-22163 [PubMed] [Google Scholar]
- 35.Coleman, J. (1990) J. Biol. Chem. 265 17215-17221 [PubMed] [Google Scholar]
- 36.Benghezal, M., Roubaty, C., Veepuri, V., Knudsen, J., and Conzelmann, A. (2007) J. Biol. Chem. 282 30845-30855 [DOI] [PubMed] [Google Scholar]
- 37.Jain, S., Stanford, N., Bhagwat, N., Seiler, B., Costanzo, M., Boone, C., and Oelkers, P. (2007) J. Biol. Chem. 282 30562-30569 [DOI] [PubMed] [Google Scholar]
- 38.Tamaki, H., Shimada, A., Ito, Y., Ohya, M., Takase, J., Miyashita, M., Miyagawa, H., Nozaki, H., Nakayama, R., and Kumagai, H. (2007) J. Biol. Chem. 282 34288-34298 [DOI] [PubMed] [Google Scholar]