Abstract
Human galectins have functionally divergent roles, although most of the members of the galectin family bind weakly to the simple disaccharide lactose (Galβ1-4Glc). To assess the specificity of galectin-glycan interactions in more detail, we explored the binding of several important galectins (Gal-1, Gal-2, and Gal-3) using a dose-response approach toward a glycan microarray containing hundreds of structurally diverse glycans, and we compared these results to binding determinants on cells. All three galectins exhibited differences in glycan binding characteristics. On both the microarray and on cells, Gal-2 and Gal-3 exhibited higher binding than Gal-1 to fucose-containing A and B blood group antigens. Gal-2 exhibited significantly reduced binding to all sialylated glycans, whereas Gal-1 bound α2-3- but not α2-6-sialylated glycans, and Gal-3 bound to some glycans terminating in either α2-3- or α2-6-sialic acid. The effects of sialylation on Gal-1, Gal-2, and Gal-3 binding to cells also reflected differences in cellular sensitivity to Gal-1-, Gal-2-, and Gal-3-induced phosphatidylserine exposure. Each galectin exhibited higher binding for glycans with poly-N-acetyllactosamine (poly(LacNAc)) sequences (Galβ1-4GlcNAc)n when compared with N-acetyllactosamine (LacNAc) glycans (Galβ1-4GlcNAc). However, only Gal-3 bound internal LacNAc within poly(LacNAc). These results demonstrate that each of these galectins mechanistically differ in their binding to glycans on the microarrays and that these differences are reflected in the determinants required for cell binding and signaling. The specific glycan recognition by each galectin underscores the basis for differences in their biological activities.
The galectin family of β-galactoside-binding proteins has over a dozen human members, and each galectin may have different biological roles and recognize different glycan receptors (1-4). These conclusions are supported by recent studies on the first three vertebrate galectins identified, termed galectin (Gal)2-1, Gal-2, and Gal-3. Gal-1 inhibits mast cell degranulation (5), whereas Gal-3 induces degranulation in mast cells independently of IgE-mediated antigen stimulation (6). Gal-1 blocks leukocyte chemotaxis (7), whereas Gal-3 has the opposite effect, inducing leukocyte chemotaxis (8) and the release of pre-formed interleukin-8 from neutrophils (9), which further augments chemotaxis of leukocytes (10). In addition, whereas Gal-1 inhibits acute inflammatory responses through various mechanisms, including suppression of phospholipase A2-induced edema (11) and inhibition of neutrophil extravasation (7), Gal-3 enhances the extravasation of neutrophils, and Gal-3 null mice also exhibit attenuated leukocyte infiltration following challenge (12). Interestingly, patients with reduced Gal-2 expression were found to have reduced risk for myocardial infarction, suggesting that Gal-2 may also have pro-inflammatory roles (13). Furthermore, Gal-1, Gal-2, and Gal-3 have all been reported to signal T cells through different receptors (14-16). These types of studies suggest that Gal-1, Gal-2, and Gal-3 recognize distinct receptors on leukocytes.
There is compelling evidence, however, that different galectins may also recognize related receptors. For example, Gal-3 attenuates Gal-1 inhibition of growth in neuroblastoma cells at the receptor level, and both Gal-1 and Gal-3 induce superoxide production in human neutrophils (17-19). Gal-1 and Gal-2 both induce surface exposure of phosphatidylserine (PS) in activated human neutrophils in the absence of apoptosis through a Ca2+-dependent pathway (20). Therefore, although Gal-1, Gal-2, and Gal-3 may recognize discrete glycoconjugates, they may also recognize some common receptors. In this way, galectins likely exhibit unique versatility in a wide range of biological functions (3, 4).
Although some differences have been reported in glycan recognition by these galectins (21-28), there are many questions remaining about their glycan recognition and subsequent effects on galectin binding. It has been suggested that differences in the biological effects of Gal-1 and Gal-3 result from differences in tertiary structure, rather than ligand binding properties (29, 30), because Gal-3 was thought to behave primarily as a monomer (31). Tertiary structure differences may contribute to differences in cellular responses to different galectins (30, 32); however, Gal-3 can form homo-oligomeric structures (32), which supports the likelihood that the major differences in biological functions by these lectins are because of differences in glycan recognition.
Previous studies on glycan recognition by galectins and most other glycan-binding proteins (GBPs) have been limited because of the availability and diversity of glycans tested (22). This limitation arises from the difficulty of synthesizing a large diverse library of glycan structures (33). In addition, the methods of analysis may have also hindered the identification of differential specificity. For example, we recently found that the specificity of Gal-1 for glycans depends not only on the structure of glycans but also the mode of their presentation (34), either in solid phase or in solution. In equilibrium gel filtration assays, similar to other solution-based assays (21, 22), Gal-1 binds glycans with a single N-acetyllactosamine (LacNAc) unit (Galβ1-4GlcNAc) equivalently to those with poly-N-acetyllactosamine (poly(LacNAc)) sequences (Galβ1-4GlcNAc)n; however, the dimeric form of Gal-1 showed a significant preference for the poly(LacNAc)-containing glycans in solid phase assays (34, 35). Significantly, Gal-1 failed to recognize internal Lac-NAc units within poly(LacNAc) (34, 35), suggesting that this preference likely reflects favorable poly(LacNAc) conformational constraints of the terminal LacNAc unit that are enhanced by immobilization (34). Gal-1 also recognized poly-(LacNAc)-containing glycans on leukocyte surfaces with a similar affinity as observed for immobilized poly(LacNAc) glycans (34), corroborating the reliability of the solid phase binding studies. Similarly, analyses of Gal-1, Gal-2, and Gal-3 utilizing frontal affinity chromatography or isothermal calorimetry, in which the glycans are free in solution, also do not reveal profound differences in carbohydrate recognition (21, 22, 36), further suggesting that galectin-glycan interactions may be most usefully tested in the context of immobilized glycan presentation. However, whether Gal-2 and Gal-3 behave like Gal-1 in showing a different glycan preference when glycans are immobilized has not been studied, nor has the in vitro binding data been correlated to binding determinants on cell surfaces.
These issues prompted us to evaluate Gal-1, Gal-2, and Gal-3 interactions using immobilized glycans in a glycan microarray format that includes several hundred structurally diverse glycans (33, 34, 37), along with parallel studies of binding to a variety of human cells. To more accurately determine the binding specificity of Gal-1, Gal-2, and Gal-3 using the glycan microarray format, we evaluated binding of each galectin over a broad concentration range. This allowed us to extrapolate a binding isotherm for each galectin toward each respective glycan. To determine whether the specificity obtained using this method reflected similar binding patterns toward cells, we tested the binding toward promyelocytic HL60 cells, which respond to signals by these galectins resulting in exposure of surface PS (38, 39), and tested binding to human erythrocytes. Our results provide novel insights into differential glycan recognition by each of these galectins and provide support for using glycan microarrays in conjunction with cell binding studies to explore glycan recognition by glycan-binding proteins (33).
EXPERIMENTAL PROCEDURES
Preparation of Human Gal-1, Gal-2, and Gal-3—Human galectins were prepared as outlined previously (35, 40). Each recombinant galectin was purified by affinity chromatography on lactosyl-Sepharose, and bound lectin was eluted with 100 mm lactose in PBS, 14 mm βME. Prior to derivatization, βME was removed from galectin samples by utilizing a PD-10 gel filtration column (GE Healthcare), followed by the addition of lactose (100 mm final concentration) to help maintain the stability of each galectin and reduce the likelihood of adduct formation at or near the carbohydrate recognition domain. Alexa Fluor 488-labeled forms of galectins were prepared using either Alexa Fluor 488 C5-maleimide or Alexa Fluor 488 carboxylic acid, succinimidyl ester, dilithium salt-reactive dyes (Molecular Probes) as described (35). We found that derivatization of Gal-2 with Alexa dyes substantially decreased the stability of the protein; thus, all studies assessing Gal-2 glycan recognition utilized biotinylated Gal-2. Galectins were biotinylated by incubating 3-5 mg/ml of each galectin with 2 mm EZ-link™ Sulfo-NHS-LC-Biotin (sulfosuccinimidyl-6-(biotinamido)hexanoate) (Pierce) for 2 h at 4 °C. Unconjugated EZ-link™ Sulfo-NHS-LC-Biotin and free lactose were separated from galectin using a PD-10 gel filtration column. Galectins were re-chromatographed over lactosyl-Sepharose to remove any inactive material following labeling. Bound galectin was eluted with 100 mm lactose, then applied to a PD-10 gel filtration column to remove lactose, and stored at 4 °C in 14 mm βME in PBS until further use. Control lectins, peanut agglutinin (Arachis hypogaea) (200 μg/ml), Ricinus communis agglutinin I (RCA-I) (2 μg/ml), Sambucus nigra agglutinin (200 μg/ml), and Lycopersicon esculentum agglutinin (LEA) (200 μg/ml) were purchased from Vector Laboratories and utilized under the same assay conditions as Gal-1, Gal-2, and Gal-3. Cholera toxin subunit B (Molecular Probes) was also assayed under the same conditions at 200 μg/ml.
Binding of Galectin to Aminoalkyl Glycosides Immobilized on Activated (N-Hydroxysuccinimidyl) Glass Surface—Glycan microarrays were prepared as described previously (33, 37) and obtained from the National Institutes of Health/NIGMS-funded Consortium for Functional Glycomics. For galectin recognition of glycans on the printed glycan microarray, a solution of between 0.1 and 10 μm galectin in PBS containing 0.005% Tween 20 and 14 mm βME was incubated for 1 h at 25 °C. The slide was then immersed in PBS containing 0.005% Tween 20, drained, and then overlaid with FITC-streptavidin. After 1 h at room temperature in a dark humid chamber, the slide was washed by successive immersion in PBS, 0.01% Tween 20 (three times) and water, 0.1% Tween 20 (twice). The slide was briefly rinsed with distilled water and dried under microfiltered air. An image of bound fluorescence was obtained using a microarray scanner (Scan Array Express, PerkinElmer Life Sciences). The integrated spot intensities were determined using Metamorph software (Universal Imaging, Downingtown, PA).
Measurement of Galectin Binding Affinity Using Surface Plasmon Resonance (SPR)—All surface plasmon resonance (SPR) experiments were performed at 25 °C on a Biacore 3000 instrument (Biacore AB (part of GE Healthcare)) largely as outlined previously (33, 37, 41, 42). Biotinylated glycosides were captured on research grade streptavidin-coated sensor chips (Sensor Chip SA, Biacore Inc.) that were pretreated according to the manufacturer's instructions. A solution of each biotinylated glycoside (10 fmol/ml) was injected at 2 ml/min in PBS, pH 7, containing 0.005% Tween 20 (running buffer) for varying lengths of time (3-7 min) until an optimal amount of glycan was captured on each independent surface. Three related glycosides were studied using one streptavidin sensor chip. A control (nonbinding) glycan, arabinose, was also captured on the same sensor chip, and the specific binding of nonderivatized recombinant Gal-1, Gal-2, or Gal-3 for the test glycans was measured using the in-line reference subtraction feature of the Biacore 3000 instrument. Increasing concentrations of Gal-1, Gal-2, or Gal-3 (0.1-100 μm) were injected at a flow rate of 60 ml/min over all four surfaces of the sensor chip. Bound Gal-1, Gal-2, or Gal-3 were eluted with the running buffer after the injection was complete. The equilibrium binding data of Gal-1, Gal-2, or Gal-3 were analyzed by nonlinear curve fitting using the BIAevaluation software (Biacore Inc.).
Cell Culture—Promyelocytic leukemia HL60 cells were obtained from ATCC and maintained at 37 °C and 5% CO2 in complete RPMI medium (RPMI 1640 supplemented with 10% FBS, 2 mm glutamine, 100 milliunits/ml penicillin, 100 μg/ml streptomycin). Chinese hamster ovary (CHO) cells were maintained in complete Dulbecco's modified Eagle's medium (Dulbecco's modified Eagle's medium supplemented with 10% FBS, 2 mm glutamine, 100 milliunits/ml penicillin, 100 μg/ml streptomycin) at 37 °C and 5% CO2.
Enzymatic Deglycosylation—Prior to enzymatic deglycosylation, HL60 cells were fixed by washing three times in PBS at 4 °C, followed by resuspension in 2% paraformaldehyde buffered in PBS, pH 7.4, at 4 °C. Cells were allowed to fix overnight on a shaker at 4 °C. Following fixation, cells were washed three times in PBS and then two times in the appropriate buffer as recommended by the manufacturer. For enzymatic digestion of cell surface glycans, fixed cells were washed in the following buffers. Cells were washed in 50 mm sodium citrate, pH 6.0, and incubated with 250 milliunits of Salmonella typhimurium α2-3-neuraminidase (New England Biolabs) at 107 cells/ml for 12 h at 37 °C. Cells were washed in 50 mm sodium citrate with 100 mm sodium chloride, pH 6.0, and incubated with 250 milliunits of Clostridium perfringens α2-3-α2-6-neuraminidase (New England Biolabs) at 107 cells/ml for 12 h at 37 °C. Cells were washed in 50 mm sodium acetate, pH 5.8, and incubated with 200 milliunits of Escherichia freundii endo-β-galactosidase (Seikagaku Kogyo) at 107 cells/ml for 24 h at 37 °C. Cells were washed with 50 mm sodium phosphate, pH 5.8, and incubated with 200 milliunits of Bacteroides fragilis endo-β-galactosidase (Calbiochem; QA Labs) at 107 cells/ml for 24 h at 37 °C. Cells were washed in 50 mm sodium phosphate, pH 5.0, and incubated with 100 milliunits jack bean β-galactosidase (Glyko) at 107 cells/ml for 12 h. Buffer control treatments lacking enzymes were used for each individual condition.
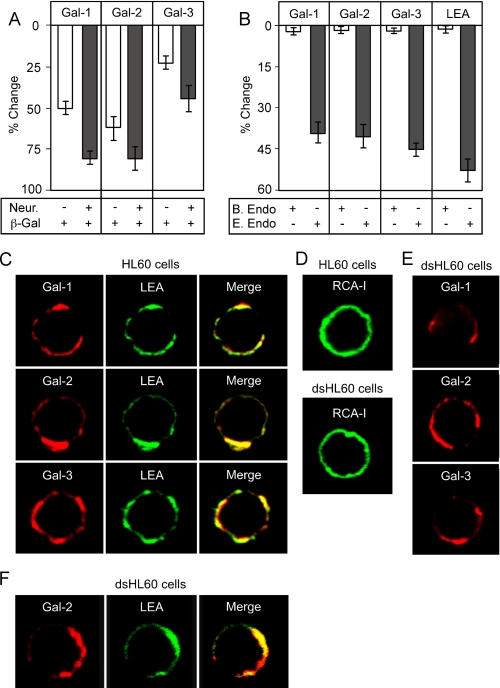
Galectin Binding to Cells—For lectin binding, cells were washed twice in PBS at 4 °C and incubated with biotinylated Gal-1, Gal-2, Gal-3, or the indicated plant lectins (LEA, RCA-I, and Maackia amurensis lectin II; Vector Laboratories) at a concentration of between 5 and 10 μg/ml at 4 °C for 1 h. As controls, cells were incubated with 50 mm lactose along with the galectins. Cells were washed three times and then incubated with Alexa Fluor 488 streptavidin or Alexa Fluor 633 streptavidin (Molecular Probes) at 4 °C for 1 h. Cells were then washed twice, followed by resuspension in 400 μl of PBS for analysis by flow cytometry using a FACSCalibur flow cytometer (BD Biosciences). The bars in each graph represent the % change in binding when compared with the binding of control buffer treated cells from each enzymatic pair. Error bars in each graph represent standard deviation of duplicate analysis.
Measuring Galectin-induced PS Exposure—For annexin-V staining, cells were treated for 1 h with 100 milliunits of Arthrobacter ureafaciens neuraminidase or buffer control (RPMI 1640 media or Hanks' balanced salt solution). Cells were washed following treatment in complete RPMI, followed by resuspension in complete RPMI at 106 cells/ml. Cells were treated with 20 μm Gal-1, Gal-2, or Gal-3 or at the concentrations indicated at 37 °C and 5% CO2 for 4 h followed by disengagement with 50 mm lactose and staining with annexin-V (CalTag) as outlined previously (38). Galectin binding toward cells treated with A. ureafaciens neuraminidase was accomplished as outlined above.
Confocal Microscopy—Cells were incubated with FITC-LEA (Vector Laboratories) and either Gal-1-biotin, Gal-2-biotin, or Gal-3-biotin at 4 °C for 1 h. Cells were washed and incubated with Alexa Fluor 633 streptavidin (Molecular Probes) for an additional 1 h at 4 °C and washed and placed on glass coverslips pretreated with poly-l-lysine at 4 °C. Cells were allowed to settle on the coverslip, followed by the addition of 2% paraformaldehyde buffered in PBS, pH 7.4, and incubation at 4 °C. Alternatively, cells were treated with 100 milliunits of A. ureafaciens neuraminidase as outlined above, followed by staining with RCA-FITC, LEA-FITC, Gal-1-biotin, Gal-2-biotin, or Gal-3-biotin as indicated. Cells were analyzed using a Leica confocal microscope (Department of Pathology confocal core facility, Emory University).
Hemagglutination—HL60 cells were cultured and treated with neuraminidase as outlined above. Erythrocytes of different blood group antigen specificity were obtained from Immucor, Inc. Cells were plated in round bottom 96-well plates, mixed with serial dilutions of each galectin, and allowed to agglutinate. The last concentration at which agglutination occurred was defined as the agglutination end point.
RESULTS
Gal-1, Gal-2, and Gal-3 Differentially Recognize O- and N-Glycans—For analyses on the glycan microarrays, we characterized the binding of Gal-1, Gal-2, and Gal-3 over a wide range of concentrations (∼8 to ∼0.2 μm). This allowed us to extrapolate a binding isotherm in an effort to more accurately estimate the glycan preference and specificity of each galectin. Such dose dependence is important to define. Historically, studies using the glycan array display are done at saturating binding conditions and high lectin concentrations (43), making it difficult to distinguish subtle differences in specificity that may occur.
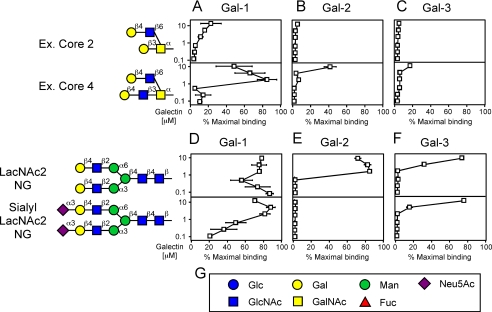
We first compared Gal-1, Gal-2, and Gal-3 in parallel to evaluate recognition of O- and N-glycans. Following biotinylation, each galectin was re-chromatographed over lactosyl-Sepharose to ensure that homogeneous preparations of active proteins were examined on the array. Gal-1 did not bind to the various core O-glycan structures (data not shown), although it did bind to some core structures that were extended to contain a terminal LacNAc unit, as demonstrated for binding to extended core 2 and core 4 (Fig. 1A). Although Gal-2 showed binding toward extended core 4, Gal-1 showed significantly more binding than Gal-2 (Fig. 1, A and B). In striking contrast, Gal-3 displayed very little binding toward all O-glycans tested (Fig. 1C).
FIGURE 1.
Gal-1, Gal-2, and Gal-3 recognition of O-glycans and N-glycans. Trivial names followed by the structures of each glycan tested are shown. Recognition of each representative glycan is displayed as the percent bound when compared with the highest bound ligand at each concentration tested by each respective galectin tested in this study. Glycan recognition of O-glycans is shown for Gal-1 (A), Gal-2 (B), and Gal-3 (C). Glycan recognition of N-glycans is shown for Gal-1 (D), Gal-2 (E), and Gal-3 (F). G, legend of symbols for monosaccharides used in this study.
Gal-1 exhibited the strongest binding toward the biantennary N-glycan (LacNAc2 NG) (Fig. 1D). In our studies the term “strong binding” refers to those glycans still recognized by the galectin at very low concentrations (≤μm). Gal-2 also exhibited strong binding toward LacNAc2 NG (Fig. 1E). By comparison, Gal-3 bound weakly to LacNAc2 NG (Fig. 1F). Interestingly, the presence of α2-3-sialic acid on the terminal LacNAc unit of LacNAc2 NG completely blocked LacNAc2 NG recognition by Gal-2 (Fig. 1E) and reduced binding by Gal-1 (Fig. 1D), whereas it had no significant effect on recognition of LacNAc2 NG by Gal-3 (Fig. 1F). The presence of α2-6-sialic acid on LacNAc2 NG blocked glycan recognition by all three galectins (data not shown). These results demonstrate that Gal-1, Gal-2, and Gal-3 differ significantly in their recognition of N- and O-glycans. Interestingly, RCA-I, a plant lectin that recognizes terminal Gal residues (44, 45), bound strongly to all N- and O-glycans terminating in LacNAc, demonstrating the presence and accessibility of these glycans (data not shown). Peanut agglutinin bound to the core 1 structure (data not shown), consistent with previous results (46, 47). Thiodigalactoside blocked all galectin-glycan recognition, although sucrose had no effect on galectin binding (data not shown), further demonstrating the carbohydrate dependence of these interactions.
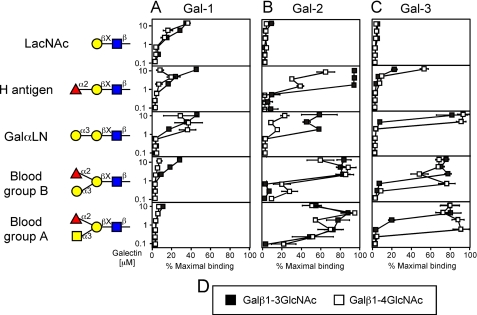
Gal-1, Gal-2, and Gal-3 Exhibit Differential Recognition of LacNAc Derivatives—To examine galectin interactions with LacNAc in more detail, we specifically evaluated galectin interactions with LacNAc-containing glycans. Unexpectedly, Gal-2 and Gal-3 showed very weak binding to LacNAc (Fig. 2, B and C), although both bound more strongly to some derivatives of LacNAc (Fig. 2, B and C). By contrast, Gal-1 bound LacNAc (Fig. 2A), although binding was weak relative to LacNAc2 NG (Fig. 1A). Gal-1, Gal-2, and Gal-3 failed to recognize LacDiNAc (GalNAcβ1-4GlcNAc) or Galα3Gal (data not shown).
FIGURE 2.
Gal-1, Gal-2, and Gal-3 recognition of LacNAc and LacNAc-derivative glycans. Trivial names followed by the structures of each glycan tested are shown. Recognition of each representative glycan is displayed as the percent bound when compared with the highest bound ligand by each respective galectin tested in this study. Glycan recognition is shown for Gal-1 (A), Gal-2 (B), and Gal-3 (C). D, legend for type 1 and type 2 structures. Black squares = type 1 LacNAc, white squares = type 2 LacNAc.
Modifications of LacNAc, as commonly occurs in vivo (48), resulted in significant differences in glycan recognition by these three galectins. For example, α1-2-fucosylation of LacNAc did not alter Gal-1 recognition (Fig. 2A), although the same modification significantly increased recognition by Gal-2 and Gal-3 (Fig. 2, B and C). Similarly, the addition of a Galα1-3 terminal sequence to LacNAc also increased Gal-2 and Gal-3 binding (Fig. 2, B and C), although it had no effect on Gal-1 binding (Fig. 2A). Interestingly, the addition of both Fucα1,2 and Galα1-3 to LacNAc (representing the blood group B antigen), significantly improved binding by both Gal-2 and Gal-3 (Fig. 2, B and C), while significantly reducing recognition by Gal-1 (Fig. 2A). Similar results were obtained with the blood group A antigen (Fig. 2, A-C). Although Gal-1 and Gal-2 exhibited no preference for type 1 or type 2 LacNAc (Galβ1-3GlcNAc versus Galβ1-4GlcNAc, respectively) either alone or in the context of modification (Fig. 2, A and B), Gal-3 displayed higher binding toward type 2 LacNAc, compared with type 1 LacNAc, following modification (Fig. 2C). These results demonstrate that common modifications of LacNAc cause significant changes in glycan recognition by Gal-1, Gal-2, and Gal-3.
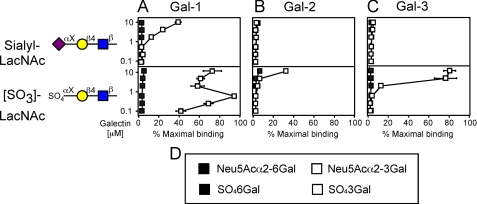
To further define the effect of substitutions of LacNAc on galectin binding, we examined the effects of both sulfation and sialylation of glycans. Gal-1 recognized α2-3-sialylated LacNAc and nonsialylated LacNAc equally (Fig. 2A and Fig. 3A), whereas α2-6-sialylation eliminated recognition (Fig. 3A). Interestingly, although α2-3-sialylation of LacNAc blocked recognition by RCA-I, α2-6-sialylation had no effect on RCA-I recognition (data not shown). S. nigra agglutinin, previously demonstrated to prefer α2-6-sialylated LacNAc (49), also exhibited recognition of α2-6-sialylated LacNAc glycans (data not shown). These results show that the failure of Gal-1, Gal-2, and Gal-3 to recognize these glycans was not a reflection of their lack of accessibility on the microarray. Sialylation of LacNAc-containing glycans by either α2-3 or α2-6 linkage completely blocked recognition by both Gal-2 and Gal-3 (Fig. 3, B and C). Furthermore, all three galectins failed to recognize GM1. This ganglioside was previously identified as a potential ligand for Gal-1 (data not shown) (50, 51). As a control for this finding, we found that cholera toxin subunit B, previously demonstrated to recognize GM1 (52), bound tightly to GM1 in this solid phase format (data not shown). Because Gal-2 failed to recognize any sialylated compounds, we sought to partly test whether this simply reflected a lack of tolerance by Gal-2 for charge modification at the 3-OH of galactose. Interestingly, 3-O-sulfation of Gal residues in LacNAc significantly increased glycan recognition by Gal-2 (Fig. 3B), whereas 4-O- or 6-O-sulfation of Gal blocked binding (Fig. 3B and data not shown). Gal-3 also showed an increase in LacNAc recognition following specific sulfation of Gal (Fig. 3C). Gal-1 exhibited the most significant increase in LacNAc recognition following sulfation (Fig. 3A), consistent with previous results (53). These results demonstrate that all three galectins demonstrate preference for sulfated versus unsulfated glycans. This contrasts strongly in regard to sialylated glycans, because only Gal-1 bound to sialylated LacNAc.
FIGURE 3.
Gal-1, Gal-2, and Gal-3 recognition of sulfated LacNAc and sialylated LacNAc. Trivial names followed by the structures of each glycan tested are shown. Recognition of each representative glycan is displayed as the percent bound when compared with the highest bound ligand by each respective galectin tested in this study. Glycan recognition is shown for Gal-1 (A), Gal-2 (B), and Gal-3 (C). D, legend describing linkages of sulfate and sialic acid. Black squares represent binding toward the glycan with attachment of sialic acid or sulfate to the 6-OH of galactose. White squares represent binding toward the glycan with attachment of sialic acid or sulfate to the 3-OH of galactose.
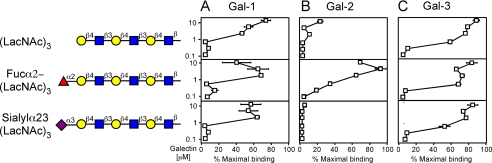
Gal-1, Gal-2, and Gal-3 Recognize Poly(LacNAc) Structures—Previous studies implicated poly(LacNAc) as a common determinant for galectin binding (34, 54-56). We examined the binding of Gal-1, Gal-2, and Gal-3 toward poly(LacNAc)-containing glycans in the microarray format. Interestingly, all three galectins preferred longer poly(LacNAc) structures compared with the single LacNAc unit (Fig. 4, A-C, Fig. 2, A-C); Gal-1 and Gal-3 exhibited the most significant preference in binding (Fig. 4, A and C). Importantly, tomato lectin (LEA), which primarily recognizes internal LacNAc within poly(LacNAc) (57), also showed higher binding toward poly(LacNAc) glycans in the microarray (data not shown). Furthermore, modification of poly(LacNAc) with terminal sialic acid or substitutions of the terminal Gal residues with a Fucα1,2 residue had no effect on Gal-1 or Gal-3 recognition of poly(LacNAc) (Fig. 4, A and C). By contrast, α1,2-fucosylation significantly increased Gal-2 recognition of poly(LacNAc), whereas α2-3-sialylation of poly(Lac-NAc) completely eliminated recognition by Gal-2 (Fig. 4B).
FIGURE 4.
Gal-1, Gal-2, and Gal-3 recognition of poly(LacNAc). Trivial names followed by the structures of each glycan tested are shown. Recognition of each representative glycan is displayed as the percent bound when compared with the highest bound ligand by each respective galectin tested in this study. Glycan recognition is shown for Gal-1 (A), Gal-2 (B), and Gal-3 (C).
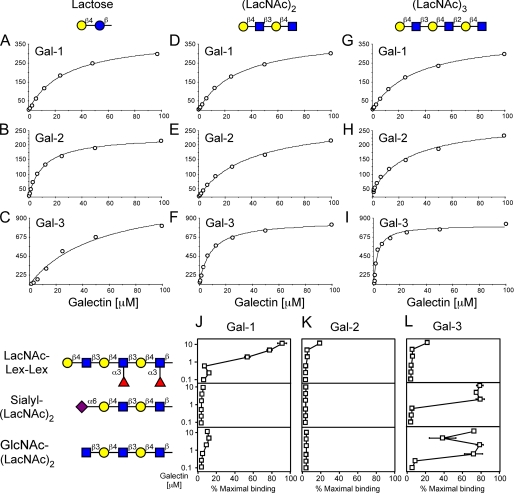
Gal-1, Gal-2, and Gal-3 Display Differential Recognition of Poly-(LacNAc) Glycans and Chimera Poly(LacNAc) Glycans—Previous studies demonstrated that Gal-1 prefers poly(Lac-NAc) over LacNAc when they are immobilized on a solid phase compared with solution binding, and that Gal-1 primarily recognizes the terminal LacNAc unit in poly(LacNAc)-containing glycans (34, 35). To further explore whether Gal-2 or Gal-3 might recognize terminal or internal LacNAc motifs within poly(LacNAc), we analyzed galectin interactions with lactose, (LacNAc)2, and (LacNAc)3 using SPR to measure binding in a solution-based equilibrium format. Gal-1, Gal-2, and Gal-3 exhibited rapid on and off rates for each immobilized glycan tested (data not shown), demonstrating that each lectin bound each glycan, that recognition was dose-dependent, and that these interactions were readily reversible. Importantly, Gal-3 bound to lactose using this solution-based assay, although it failed to recognize lactose or LacNAc when immobilized in the glycan array (Fig. 3C). This demonstrates that the context of glycan presentation can significantly influence glycan recognition by Gal-3, similar to the earlier findings with Gal-1 (34). Binding isotherms generated from the SPR data provided Kd values for each galectin-glycan interaction (Table 1). Gal-1 bound equivalently to lactose, (LacNAc)2, and (LacNAc)3 (Fig. 5, A, D, and G, and Table 1), corroborating our earlier findings and further demonstrating that Gal-1 principally recognizes the terminal LacNAc unit (34, 35). Conversely, Gal-2 showed lower binding toward (LacNAc)2 and (LacNAc)3 in solution (Fig. 5, B, E, and H, and Table 1), which demonstrates that Gal-2 does not recognize internal LacNAc motifs. By contrast, Gal-3 exhibited significant increases in affinity with each LacNAc extension (Fig. 5, C, F, and I, and Table 1).
TABLE 1.
Analysis of Gal-1, Gal-2, and Gal-3 interactions with glycans utilizing SPR
| Lactose (Kd) | LacNAc-LacNAc (Kd) | LacNAc-LacNAc-LacNAc (Kd) | |
|---|---|---|---|
| μm | μm | μm | |
| Gal-1 S1a | 13.7 | 15.1 | 16.7 |
| Gal-1 S2 | 14.4 | 16.9 | 19.2 |
| Gal-1 S3 | 9.0 | 11.0 | 12.1 |
| Gal-2 S1 | 24.9 | 60.5 | 62.6 |
| Gal-2 S2 | 28.3 | 77.6 | 66.7 |
| Gal-2 S3 | 32.3 | 58.4 | 54.5 |
| Gal-3 S1 | 53.5 | 8.4 | 2.7 |
| Gal-3 S2 | 60.7 | 9.5 | 3.2 |
| Gal-3 S3 | 51.4 | 9.5 | 3.9 |
S1, S2 and S3 represent separate analyses of each respective galectin under identical conditions.
FIGURE 5.
Binding isotherms representing Gal-1, Gal-2, and Gal-3 recognition of lactose, (LacNAc)2, and (LacNAc)3 glycans using SPR. The binding isotherms and Kd values are shown for lactose with Gal-1 (A), Gal-2 (B), and Gal-3 (C); for (LacNAc)2 with Gal-1 (D), Gal-2 (E), and Gal-3 (F); and for (LacNAc)3 with Gal-1 (G), Gal-2 (H), and Gal-3 (I). J-L, trivial names followed by the structures of each glycan tested are shown. Recognition of each representative glycan is displayed as the percent bound when compared with the highest bound ligand by each respective galectin tested in this study. Glycan recognition is shown for Gal-1 (J), Gal-2 (K), and Gal-3 (L).
To determine whether similar recognition patterns dictate poly(LacNAc) recognition following immobilization, we examined the binding of Gal-1, Gal-2, and Gal-3 toward poly(Lac-NAc) and modified poly(LacNAc) glycans, as was previously partly done for Gal-1 (35). Gal-1, Gal-2, and Gal-3 all recognized extended poly(LacNAc) (LacNAc)3, although all three galectins did not bind to glycans terminating in Lex (Lex-Lex-Lex) (data not shown). Similarly, RCA-I and LEA did not bind Lex-Lex-Lex (data not shown), consistent with previous results (34). Importantly, Gal-1 showed significant binding toward poly(LacNAc) with internally fucosylated LacNAc units (Lac-NAc-Lex-Lex) (Fig. 5J). This result demonstrates that Gal-1 recognizes the terminal LacNAc unit in poly(LacNAc) chains, similar to the results obtained in solution-based assays. Furthermore, Gal-3 did not bind LacNAc-Lex-Lex (Fig. 5L), which also shows that it primarily recognizes internal LacNAc units within poly(LacNAc). Importantly, RCA-I, like Gal-1, also recognized LacNAc-Lex-Lex, whereas LEA failed to demonstrate poly(LacNAc) recognition following internal LacNAc modification (data not shown). Furthermore, only Gal-3 recognized α2-6-sialylated poly(LacNAc) (Fig. 5L).
To complete our studies on the potential requirements of terminal LacNAc recognition within poly(LacNAc) on the solid phase array, we examined Gal-1, Gal-2, and Gal-3 recognition of poly(LacNAc) lacking a terminal galactose residue. The absence of the terminal galactose on poly(LacNAc) chains significantly reduced binding by Gal-1 and Gal-2 (Fig. 5, J and K), although this did not affect recognition by Gal-3 (Fig. 5L). Control experiments corroborated these results, because we found that RCA did not bind poly(LacNAc) lacking a terminal galactose residue, whereas LEA binding was unaltered and robust (data not shown). These results demonstrate that Gal-3 prefers glycans containing poly(LacNAc) because of recognition of internal LacNAc sequences, whereas the preference of poly(LacNAc) for Gal-1 and Gal-2 likely represents conformational constraints on the glycan that are enhanced when the glycans are immobilized on a surface. Taken together, these results demonstrate that Gal-1, Gal-2, and Gal-3 all bind poly(LacNAc) but that this recognition arises from fundamentally different mechanisms of interaction.
Gal-1, Gal-2, and Gal-3 Differentially Recognize Sialylated Cell Surface Glycans—We next sought to determine whether the differences seen in glycan specificity for each galectin reflected their binding to cell surface glycans. To test this we examined the binding of Gal-1, Gal-2, and Gal-3 toward promyelocytic leukemia HL60 cells prior to and following enzymatic digestion of specific glycan structures. Leukocytes exhibit signaling responses following exposure to galectins (38, 39). We used this response to ascertain whether the differential specificity observed on the array also occurs in the context of cell surface glycans, and to give information about the nature and composition of glycans recognized on endogenous functional receptors. Furthermore, enzymatic modification of endogenous receptors allowed us to explore the recognition of glycans by galectins on cell surfaces, without using genetically engineered modification, which can alter glycan expression in unpredictable ways. The importance of this is illustrated when considering that global alterations in glycosylation can have profound effects on glycoprotein trafficking, affecting surface half-life and total expression (58-60), making it difficult to easily translate differences in binding to possible differences in specificity as opposed to differences in cell surface receptor numbers. Furthermore, previous results suggest that specific modification of select ligands may be critical in conveying receptor specificity for Gal-1 (34), raising the question whether similar requirements exist for Gal-2 and Gal-3.
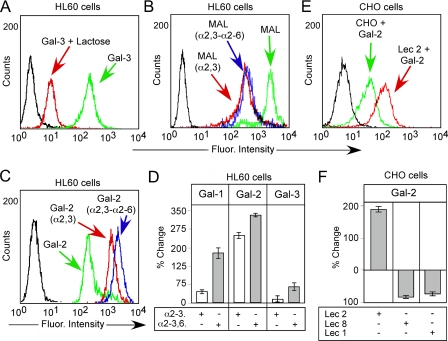
To examine binding of Gal-1, Gal-2, and Gal-3 toward HL60 cells, we first sought to determine whether binding of each galectin could be readily detected and whether this binding was carbohydrate-dependent. Gal-1, Gal-2, and Gal-3 each bound HL60 cells (Fig. 6, A and D). Importantly, binding was inhibited by lactose (Fig. 6A and data not shown), indicating that cell surface recognition was carbohydrate-dependent. On the glycan microarray, Gal-1 displayed no binding to α2-6-sialylated glycans, although it exhibited similar binding to either α2-3-sialylated or nonsialylated glycans. To test whether similar binding behavior might occur on the cell surface, cells were treated with either S. typhimurium neuraminidase, an α2-3-specific neuraminidase, or C. perfringens, an α2-3-α2-6-neuraminidase. Treatment of HL60 cells with either α2-3-neuraminidase or α2-3-α2-6-neuraminidase resulted in a comparable reduction in M. amurensis binding (Fig. 6B), indicating removal of α2-3-sialylated linkages following each treatment. Importantly, Gal-1 displayed a significantly greater increase in cell surface binding following treatment of cells with α2-3-α2-6-neuraminidase, compared with that with α2-3-neuraminidase alone (Fig. 6D). These results suggest that Gal-1 has a greater tolerance for the α2-3-sialyl LacNAc modification than α2-6-sialylation on cell surface glycans. By contrast, Gal-2 bound significantly better to cells treated with either neuraminidase (Fig. 6, C and D), suggesting that both α2-3- and α2-6-sialylation of endogenous ligands significantly inhibit binding. Unlike Gal-1 or Gal-2, Gal-3 demonstrated a less significant increase in binding following either treatment (Fig. 6D).
FIGURE 6.
Gal-1, Gal-2, and Gal-3 recognition of sialylated LacNAc on HL60 cells. A, HL60 cells were incubated with 10 μg/ml Gal-3 with or without 50 mm lactose as indicated followed by flow cytometric analysis. B, HL60 cells were treated with S. typhimurium neuraminidase, an α2-3-specific neuraminidase, or C. perfringens α2-3-, α2-6-neuraminidase for 12 h followed by staining with 10 μg/ml M. amurensis (MAL) as indicated followed by flow cytometric analysis. C, HL60 treated as in B were stained with Gal-2 followed by analysis using flow cytometry. D, quantification of flow cytometric data. Bars represent the percent change in cell surface binding when compared with the mean fluorescent intensity of nontreated cells. E, representative histogram of Gal-2 binding to CHO cells and Lec 2 cells as indicated. F, quantification of flow cytometric data of Gal-2 binding toward CHO cells. Bars represent the percent change in cell surface binding when compared with the mean fluorescent intensity wild type CHO cells ± S.D.
Although the binding of Gal-1 and Gal-3 toward CHO cells and mutant CHO cell derivatives (Lec mutants) has been extensively studied (61), the binding of Gal-2 toward CHO cells, which only generate α2-3-sialylated glycans, and Lec mutants has not been evaluated. Previous results demonstrated that Gal-1 and Gal-3 fail to increase binding toward Lec 2 CHO cell mutants, which fail to generate α2-3-sialylated glycans (61), consistent with the present findings. Although the functional consequence of galectin binding to CHO cells is unknown, we next sought to determine whether similar binding preferences observed toward HL60 cells by Gal-2 also occurred on CHO cells. Importantly, Gal-2 exhibited a significant increase in binding toward Lec 2 cells, which fail to generate α2-3-sialylated glycans, when compared with wild type CHO (Fig. 6, E and F), consistent with HL60 cells (Fig. 6, C and D), whereas it exhibited significantly reduced binding toward Lec 8 cells and Lec 1 cells (Fig. 6F), both of which fail to generate terminal LacNAc (61). These results strongly suggest that sialylation of either linkage reduces Gal-2 glycan recognition. Taken together, these results demonstrate that sialylation uniquely modulates the recognition of cell surface glycans by Gal-1, Gal-2, and Gal-3.
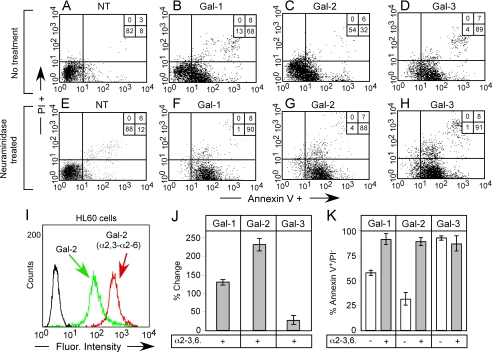
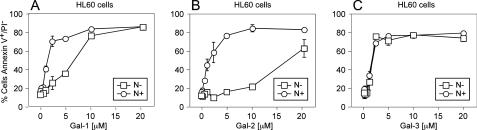
Although these general changes in cell surface binding corroborate the binding observed toward immobilized synthetic glycans, it is not clear whether these changes occur on the actual receptors through which Gal-1, Gal-2, and Gal-3 signal cellular responses, or whether these changes simply reflect arbitrary glycan recognition following enzymatic manipulation or genetic alterations in cell surface glycans. To test this, we removed terminal sialic acid on HL60 cells with A. ureafaciens α2-3-α2-6-neuraminidase, which allows removal of sialic acid at physiological pH. Treatment of HL60 cells with A. ureafaciens neuraminidase resulted in similar alterations in cell surface glycan recognition by Gal-1, Gal-2, and Gal-3 (Fig. 7, I and J), as observed following treatment with C. perfringens neuraminidase (Fig. 6D). To test the relationship of binding of galectins to their biological signaling activity, we examined their ability to induce surface exposure of PS. We previously showed that some galectins induce PS exposure independently of apoptosis, by Src-kinase-mediated pathways (20). We found that neuraminidase treatment of HL60 cells significantly enhanced their responses to Gal-1- and Gal-2-induced PS exposure (Fig. 7, B, C, F, G, and K). By contrast, treatment of HL60 cells with neuraminidase failed to enhance sensitivity to Gal-3-induced PS exposure (Fig. 7, D, H, and K). Importantly, binding of Gal-1, Gal-2, and Gal-3 to HL60 cells before and after treatment with neuraminidase correlated with HL60 sensitivity to each galectin (Fig. 7, J and K), demonstrating that these changes occur on the functional galectin counter receptors. To determine whether this altered sensitivity toward Gal-1 and Gal-2 occurred over a wide range of concentrations, we examined a dose response of HL60 toward Gal-1, Gal-2, and Gal-3 with or without pretreatment with neuraminidase. HL60 cells experienced the greatest enhancement in sensitivity toward Gal-2-induced PS exposure following treatment with neuraminidase, with ∼20 μm Gal-2 required to induce substantial PS exposure in nontreated and around ∼2 μm to induce similar responses in neuraminidase-treated cells (Fig. 8B). Neuraminidase-treated HL60 cells also showed enhanced PS exposure in response to Gal-1, although not to the same extent as Gal-2 (Fig. 8A). Interestingly, unlike Gal-1 and Gal-2, HL60 sensitivity toward Gal-3 was unaltered over all the concentrations tested (Fig. 8C). Gal-1-, Gal-2-, and Gal-3-induced agglutination paralleled the induction of PS exposure (data not shown), demonstrating that changes in cell surface recognition underscored alterations in sensitivity. Taken together, these results demonstrate that cell surface sialylation distinctly alters cellular signaling by Gal-1, Gal-2, and Gal-3.
FIGURE 7.
Desialylation differentially alters cellular sensitivity toward galectin-induced PS exposure. A-H, HL60 cells were either incubated with buffer control (A-D) or 100 milliunits of A. ureafaciens neuraminidase (E-H) for 1 h followed by treatment of cells with 20 μm Gal-1, Gal-2, or Gal-3. Cells were washed in 50 mm lactose, stained with annexin-V FITC and propidium iodide (PI), followed by flow cytometric analysis. Cells that were annexin-V-positive and propidium iodide-negative were considered positive for PS exposure. Numbers represent the percent of total cells found in each quadrant. I, HL60 cells were treated with 100 milliunits of A. ureafaciens neuraminidase for 1 h, followed by staining with 10 μg/ml Gal-2 and analysis by flow cytometry. J, quantification of Gal-1, Gal-2, and Gal-3 binding toward HL60 cells following treatment with A. ureafaciens neuraminidase. Bars represent the percent change in cell surface binding when compared with the mean fluorescent intensity of nontreated cells ± S.D. K, quantification of PS exposure (annexin-V+/PI-) on neuraminidase-treated (NT) or untreated cells following treatment with Gal-1, Gal-2, or Gal-3 as outlined in A as mean percentage ± S.D.
FIGURE 8.
Dose response of desialylated HL60 cells to Gal-1, Gal-2, and Gal-3. HL60 cells were incubated with either 100 milliunits of A. ureafaciens neuraminidase (circles) or buffer control (squares) for 1 h followed by treatment of cells with the indicated concentrations of Gal-1, Gal-2, or Gal-3 for 4 h. Cells were disengaged with 50 mm lactose and stained for PS exposure with annexin-V-FITC. The percent cells annexin V+/propidium iodide- are shown ± S.D.
Gal-1, Gal-2, and Gal-3 Display Differential Recognition of Cell Surface Poly(LacNAc)—The unique effects of sialylation on cell surface glycan recognition strongly suggested that Gal-1, Gal-2, and Gal-3 possess fundamentally different mechanisms of LacNAc recognition on the cell surface. Because Gal-1, Gal-2, and Gal-3 exhibited distinct modes of poly(LacNAc) interaction on the array, we next sought to determine whether distinct poly(LacNAc) interaction might also occur with cell surface ligands. We first treated HL60 cells with jack bean β-galactosidase to remove terminal galactose residues. Treatment of cells with β-galactosidase resulted in reduced RCA binding (Fig. 9A), indicating that many cell surface galactose residues were accessible to the enzyme. Importantly, a similar reduction in cell surface recognition by Gal-1 and Gal-2 occurred following β-galactosidase treatment (Fig. 9, B and E). However, the binding of Gal-3 to cells was less affected by β-galactosidase treatment (Fig. 9B). These results suggest that Gal-3 does not share the same requirement for terminal galactose residues on the cell surface as seen for Gal-1 and Gal-2.
FIGURE 9.
Gal-1, Gal-2, and Gal-3 recognize poly(LacNAc) glycans on HL60 cells. A, Gal-1, Gal-2, and Gal-3 binding toward HL60 cells following treatment with jack bean β-galactosidase with or without pretreatment of cells with A. ure-afaciens neuraminidase. B, Gal-1, Gal-2, and Gal-3 binding toward HL60 cells following treatment with either B. fragilis or E. freundii endo-β-galactosidase. Bars represent the percent change in cell surface binding when compared with the mean fluorescent intensity of nontreated cells ± S.D. C, confocal analysis of Gal-1, Gal-2, Gal-3, and LEA binding toward cell surface glycans on HL60 cells. D, confocal analysis of RCA-I binding toward cell surface glycans on HL60 cells or HL60 cells treated with 100 milliunits A.ureafaciens neuraminidase (dsHL60). E, confocal analysis of Gal-1, Gal-2, Gal-3 binding toward cell surface glycans on HL60 cells treated with 100 milliunits of A. ureafaciens neuraminidase (dsHL60). F, confocal analysis of Gal-2 and LEA binding toward cell surface glycans on HL60 cells treated with 100 milliunits of A. ureafaciens neuraminidase (dsHL60).
To determine the extent to which poly(LacNAc) recognition may be important for galectin binding, we next treated cells with either B. fragilis or E. freundii endo-β-galactosidase; both can potentially cleave poly(LacNAc) glycans yet exhibit different preferences of cleavage on the cell surface. B. fragilis endo-β-galactosidase substantially cleaves poly(LacNAc) on tetraantennary N-glycans, although it has little activity toward poly(LacNAc) on triantennary N-glycans; by contrast, E. freundii endo-β-galactosidase efficiently cleaves poly(LacNAc) on N-glycans with fewer branches (62). Treatment of cells with E. freundii, but not B. fragilis, endo-β-galactosidase, resulted in reduced LEA binding (Fig. 9, C and F). LEA is a plant lectin that has strong specificity for poly(LacNAc) chains (57). Importantly, a similar reduction in glycan recognition by Gal-1, Gal-2, and Gal-3 also occurred following E. freundii treatment (Fig. 9, D and F). Similar results occurred if cells were pretreated with neuraminidase (data not shown). Taken together, these results demonstrate that although the requirement for terminal galactose residues may differ between Gal-1, Gal-2, and Gal-3, each of these galectins recognize poly(LacNAc)-containing glycans on HL60 cells.
We were curious as to whether the ligands for these three galectins were co-localized with LEA-binding sites. Dual staining and confocal microscopy revealed that Gal-1, Gal-2, and Gal-3 all co-localized with LEA binding on the cell surface (Fig. 9C). Unlike Gal-1, Gal-2, and Gal-3, RCA-1 displayed a uniform cell surface binding to HL60 cells (Fig. 9D), suggesting that Gal-1, Gal-2, and Gal-3 bind specific ligands that may reside within membrane microdomains. Because treatment of cells with neuraminidase enhanced binding and signaling by Gal-1 and Gal-2, we next sought to determine whether treatment of cells with neuraminidase might alter the discrete binding pattern exhibited by Gal-1, Gal-2, and Gal-3. Although RCA-I continued to exhibit a uniform staining pattern following neuraminidase treatment (Fig. 9D), Gal-1, Gal-2, and Gal-3 staining appeared unaltered (Fig. 9E). To determine whether Gal-1, Gal-2, and Gal-3 remained co-localized with LEA following neuraminidase treatment, cells were co-stained with LEA. Importantly, even after treatment with neuraminidase, Gal-2 and LEA remained predominantly co-localized (Fig. 9F), demonstrating that neuraminidase treatment likely exposes additional poly(LacNAc)-containing glycans, as opposed to simply increasing nonspecific binding to LacNAc-containing glycans over the entire cell surface. Similar results were observed for Gal-1 and Gal-3 (data not shown). Taken together, these results strongly suggest that poly(LacNAc)-containing glycans are components of the receptors for Gal-1, Gal-2, and Gal-3 on the cell surface.
In contrast to Gal-1, both Gal-2 and Gal-3 displayed significant increases in binding toward H antigen (blood group O), blood group A, and blood group B glycans, when compared with LacNAc on the solid phase array (Fig. 2, A-D). To further explore whether the predicted glycan specificity for Gal-1, Gal-2, and Gal-3 using the solid phase format represents cell surface glycan recognition, we examined the binding of these three galectins toward erythrocytes expressing different blood group glycans. Attempts to analyze Gal-1, Gal-2, and Gal-3 erythrocyte cell surface binding directly was impeded by the fragile nature of erythrocytes and the high propensity of galectins to agglutinate these cells, which caused massive fragmentation upon flow cytometric analysis (data not shown). Therefore, we evaluated the agglutination potential of Gal-1, Gal-2, and Gal-3 toward erythrocytes and compared this to binding toward HL60 cells. Although agglutination requires both binding and dimerization of each galectin, comparison of agglutination between several cell types for a particular galectin should give a relative correlation for binding between cell types decorated with different glycans. Using this approach, Gal-2 and Gal-3 showed more potent agglutination of erythrocytes when compared with binding to HL60 cells, and demonstrated a preference for blood group A and blood group B erythrocytes (Table 2). By contrast, Gal-1 exhibited similar agglutination toward erythrocytes as toward binding to HL60 cells, confirming that Gal-1 has no blood group preference (Table 2). These results demonstrate that Gal-2 and Gal-3 display preference for blood group cell surface glycans, corroborating the predicted specificity obtained from the glycan microarray and further demonstrating that Gal-1, Gal-2, and Gal-3 exhibit unique glycan recognition.
TABLE 2.
Cellular agglutination by Gal-1, Gal-2, and Gal-3
| O-RBCsa | A-RBCs | B-RBCs | HL60 | dsHL60 | |
|---|---|---|---|---|---|
| μm | μm | μm | μm | μm | |
| Gal-1 | 1 | 1 | 1 | 1 | 0.25 |
| Gal-2 | 0.13 | 0.06 | 0.06 | 20 | 0.5 |
| Gal-3 | 0.5 | 0.25 | 0.25 | 1 | 1 |
Cells were incubated with galectins in round bottom wells, as described under “Experimental Procedures.” Each concentration shown represents the lowest tested concentration in a serial dilution at which cell agglutination was visible. RBC means red blood cells.
DISCUSSION
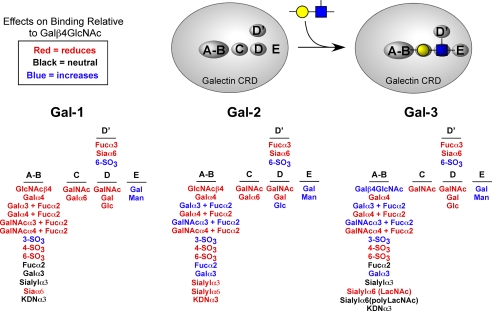
In our studies we have explored the glycan specificity of the galectins Gal-1, Gal-2, and Gal-3 using a combined approach of a solid phase assay system with glycan microarrays compared with cell surface binding experiments. Defining the dose response of Gal-1, Gal-2, and Gal-3 allowed a greater understanding of differences in glycan recognition and specificity between the galectins. These results demonstrate that each galectin exhibits significant differences in glycan binding specificity but also show overlapping recognition of some glycans (Fig. 10 and Table 3). These results provide new insights into both the overlapping and unique biological effects induced by these proteins. Taken together, our results, which provide relative apparent affinities for a wide variety of glycans (Table 3), suggest a new functional map of the CRD for each galectin. This map illustrates the impact of modification of the basic LacNAc core on glycan recognition by Gal-1, Gal-2, and Gal-3 and suggests that unique subsites exist with each CRD. Modification of the LacNAc core can either enhance, permit, or preclude modified LacNAc recognition by each respective galectin (Fig. 10). The binding modes of the different glycans suggest that each galectin may be viewed to have within its carbohydrate recognition domain a number of subsites, which we have indicated as A/B, C, D, D′, and E.
FIGURE 10.
Representative model of the CRD for Gal-1, Gal-2, and Gal-3 to illustrate the effect of LacNAc substitution on glycan recognition. Red indicates that the specific addition of the specified structure and linkage at the respective site reduces or abolishes recognition by the indicated galectin. Black refers to those modifications that had no effect on glycan recognition. Blue represents those modifications that produced more favorable binding than LacNAc alone.
TABLE 3.
Half-maximal binding of Gal-1, Gal-2, and Gal-3 to glycans on the microarray, derived from the dose-response curves
| >10 μma | ~5 μm | ~1 μm | ~0.5 μm | <0.1 μm | |
|---|---|---|---|---|---|
| Gal-1 | Extended core 2 | (LacNAc)3 | Extended core 4 | LacNAc2 NG | |
| LacNAc | Sialylα3LacNAc2 NG | SO3LacNAc | |||
| H antigen | Fucα2(LacNAc)3 | ||||
| GalαLacNAc | Sialylα3(LacNAc)3 | ||||
| BGB | LacNAc-Lex-Lex | ||||
| Gal-2 | SO3LacNAc | LacNAc2 NG | BGA | ||
| H antigen | |||||
| GalαLacNAc | |||||
| BGB | |||||
| Fucα2(LacNAc)3 | |||||
| Gal-3 | LacNAc2 NG | BGB | Fucα2(LacNAc)3 | ||
| SO3LacNAc | BGA | Sialylα3(LacNAc)3 | |||
| Sialylα3LacNAc2 NG | (LacNAc)3 | GlcNAc(LacNAc)2 | |||
| GalαLN | Sialylα6(LacNAc)2 |
The combined use of the glycan microarray coupled with the evaluation of cell surface binding provided a useful strategy to predict the nature of endogenously expressed glycans recognized by Gal-1, Gal-2, and Gal-3 on functional receptors. We chose to use glycan modification of fixed cells rather than manipulating the entire glycome of the cell; such manipulations could alter receptor trafficking and surface half-life in unpredictable ways (58-60). Using this approach, one of the most striking examples of differential glycan recognition, both on the array and on the cell surface, was the differential recognition of sialylated glycans by Gal-1, Gal-2, and Gal-3. For example, the presence of terminal sialic acid on glycans in either α2-3 or α2-6 linkage to galactose blocked their recognition by Gal-2 on the microarray. Similarly, Gal-2 binding to HL60 cells was significantly enhanced following treatment of cells with either α2-3- or α2-3-α2-6-neuraminidase. By contrast, Gal-1 bound well to α2-3- but not α2-6-sialylated glycans on the microarray, which was consistent with our finding that treatment of HL-60 cells with α2-3-α2-6-neuraminidase enhanced recognition of HL60 cells much more than α2-3-neuraminidase alone. Although Gal-3 failed to recognize α2-6-sialylated N-glycans lacking poly(LacNAc), it did bind to α2-6-sialylated poly(LacNAc) on the microarray, suggesting that sialylation of poly(LacNAc) does not alter its binding by Gal-1. Similarly, Gal-3 exhibited little change in cell surface binding following treatment with either neuraminidase, suggesting that sialylation is not a key regulator of cellular sensitivity toward Gal-3. This finding is consistent with previous results demonstrating that α2-3 cell surface sialylation fails to significantly alter either Gal-1 or Gal-3 binding to CHO cells (61), whereas a reduction in α2-3-sialylation significantly enhanced Gal-2 binding toward CHO cells.
Changes in cell binding following neuraminidase treatment correlated with altered cellular sensitivity to each galectin, indicating that sialylation can regulate the functional receptors utilized by Gal-1, Gal-2, and Gal-3. Consistent with this, recent results demonstrate that α2-3- and α2-6-sialylation fails to alter T cell sensitivity toward Gal-3; by contrast, α2-6-sialylation, but not α2-3-sialylation, blocks T cells from responding to Gal-1 (63). These results demonstrate that receptor sialylation can significantly and uniquely alter cellular sensitivity toward Gal-1, Gal-2, or Gal-3.
Differential recognition of poly(LacNAc) likely underscores the disparate effects of sialylation on glycan recognition and signaling by Gal-1, Gal-2, and Gal-3. Unlike Gal-3, Gal-1 and Gal-2 require the terminal LacNAc unit for poly-(LacNAc) recognition, making modifications of the terminal Gal relevant in glycan recognition. By contrast, although Gal-3 did not bind to the α2-6-sialylated LacNAc2 N-glycan, which contains single terminal LacNAc units on each branch, Gal-3 bound well to α2-6-sialyl poly(LacNAc). The ability of Gal-3 to bind poly(LacNAc) in the absence of a terminal, nonreducing, and available Gal residue strongly suggests that Gal-3 binds α2-6-sialyl poly(LacNAc) as a result of recognizing internal, as opposed to terminal, LacNAc units.
The ability of Gal-1, Gal-2, and Gal-3 to recognize poly(Lac-NAc) is consistent with several previous studies (28, 34, 35, 54-56) and suggests that poly(LacNAc) may serve as the key glycan ligand for galectin-mediated effects. Indeed, previous studies demonstrated that HL60 cells possess few poly(Lac-NAc) glycans sensitive to E. freundii endo-β-galactosidase (64), suggesting that a specific poly(LacNAc) modification may serve as a functional glycan ligand to convey receptor specificity for Gal-1, Gal-2, and Gal-3 among many possible cell surface glycans containing LacNAc. Consistent with this, Gal-1, Gal-2, and Gal-3 co-localized with LEA, in restricted membrane microdomains that were identified by their binding of cholera toxin to the glycosphingolipid GM1.3 Neuraminidase, which exposed binding sites for RCA, failed to alter Gal-1, Gal-2, and Gal-3 cell surface localization or co-localization with LEA, suggesting that poly(LacNAc) glycans promote specific interactions with Gal-1, Gal-2, and Gal-3 within the functional receptor(s).
Although biochemical approaches to assessing glycan binding properties of GBPs can provide useful information, the presentation of cell surface glycans may differ sufficiently from presentation in these artificial formats such that binding may not reflect actual binding specificity toward cells surface glycans. For example, although Gal-1 exhibits high binding toward poly(LacNAc) glycans on cell surface glycans, many studies using solution-based platforms fail to demonstrate any preference of poly(LacNAc) in solution (21, 34), demonstrating that cell surface presentation uniquely promotes preferential binding. This may result from conformational constraints only relevant following immobilization of the nonreducing ends of the glycans. Similarly, although the present platform provided specificity information that was largely corroborated by cell surface binding in this study and in previous studies, such as the high affinity of Gal-1 for sulfated LacNAc (53, 65) and poly(LacNAc) (34, 35) and Gal-3 for blood group antigens and poly(LacNAc) (21-27), the presentation of many glycans on the microarray, including poly(LacNAc), only reflect terminal glycan modifications in the absence of their context as extensions and modifications of N-glycans, O-glycans, or glycolipids where they are normally presented on the cell surface. Therefore, the reduced binding of Gal-2 to poly(LacNAc) on the microarray, in comparison to the binding of Gal-2 to poly(LacNAc) on HL60 cells, may reflect a more specific requirement of Gal-2 for poly(LacNAc) presentation in the context of cell surface glycans. Such results demonstrate that although the current glycan microarray provides unique insights into the specificity of GBPs, the development of glycan microarrays from glycans harvested from natural sources will greatly facilitate the full elucidation of the binding requirements of GBPs. Furthermore, with a greater understanding of the carbohydrate binding requirements and specificity of Gal-1, Gal-2, and Gal-3 toward HL60 cell ligands, the identification of the functional glycoprotein receptors through which these galectins signal will be greatly facilitated. Studies of the functional receptor will likely provide key insight into the functional glycans recognized by Gal-1, Gal-2, and Gal-3 and the mechanisms by which they preferentially recognized these glycans.
The ability of Gal-2 and Gal-3 to recognize blood group antigens corroborates some earlier findings for Gal-3 (21, 25-28) and raises important questions concerning the functional consequence of this interaction. Interestingly, we did not detect the expression of any blood group antigens in HL60 cells using blood group-specific monoclonal antibodies,4 and our results suggest that poly(LacNAc)-containing ligands, rather than blood group antigens, comprise the major ligands on these cells for Gal-1, Gal-2, and Gal-3. Furthermore, although Gal-1 exhibited similar agglutination potential toward HL60 cells and erythrocytes, Gal-2 and Gal-3 exhibited much higher agglutination toward erythrocytes. These results suggest that Gal-2 and Gal-3 recognize erythrocyte glycans containing blood group antigens compared with non-blood group ligands on HL60 cells. Blood group antigens are thought to elicit immune responses that underlie the formation of anti-blood group antigen antibodies (66-70). Thus, Gal-2 and Gal-3 may be important in innate immune mechanisms in recognizing blood group-related glycans. Consistent with this, recent studies suggest a role for Gal-3 in innate immune recognition of several pathogens, including Candida albicans (69), Leishmania major (68, 71) and Schistosoma mansoni (72, 73). Given the relationship between galectins and the innate immunity (74), future studies will continue to examine the effect and modulation galectins impose on the innate immune system.
In addition to providing further understanding of galectin recognition of glycans, our study also potentially resolves some questions concerning previously suggested glycan ligands for Gal-1 and Gal-3. For example, several reports suggested that Gal-1 is a negative growth regulator of neuroblastoma cells, possibly through interactions with the ganglioside GM1 (50, 75). Follow-up studies demonstrated that Gal-1 exhibited binding to GM1 in solution at very high concentrations (51). However, in the present study, Gal-1 failed to recognize GM1 at any concentration on the glycan microarray, although cholera toxin subunit B readily bound GM1 in the same assay system, corroborating an earlier study (21). Thus, in the cells studied it is unlikely that GM1 is a functional receptor for Gal-1. Additional studies implicate Gal-1 and Gal-3 in mechanisms of neoplastic metastasis (76, 77), possibly through interactions with the disaccharide T antigen that is the basic unit of core 1 O-glycans (78, 79). However, in our study Gal-1 and Gal-3 did not bind significantly to core 1 O-glycan, suggesting that other neoplastic glycans may be responsible for mediating these effects. However, unique presentation of these glycan in vivo may be required for proper galectin recognition. Future studies will continue to evaluate the endogenous glycans responsible for galectin-mediating functions in vivo.
In summary, our results provide significant clarification and additional insight into the specificity of Gal-1, Gal-2, and Gal-3 for glycan ligands. The glycan array largely predicted the binding preferences of Gal-1, Gal-2, and Gal-3 toward cell surface glycans, illustrating the utility of the array in elucidating the carbohydrate binding preferences of Gal-1, Gal-2, and Gal-3. These results also provide additional biochemical understanding for the overlapping functions of each of these proteins, while also providing an explanation for the functionally unique and often opposing roles of these galectin family members in vivo (3, 4).
Acknowledgments
We thank Sandy Cummings for technical assistance. We also thank Drs. Tongzhong Ju, Baoyun Xia, and Jamie Heimburg-Molinaro for helpful discussions.
This work was supported by National of Institutes of Health Grant HL085607, by resources from the Consortium for Functional Glycomics (Core D and Core H) funded by NIGMS Grant GM62116 from the National Institutes of Health, and by Swedish Research Council Grant 15390 (to H. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Gal, human galectin; βME, β-mercaptoethanol; LacNAc, N-acetyllactosamine (Galβ1GlcNAc); poly(LacNAc), poly-N-acetyllactosamine (Galβ1-4GlcNAc)n; SPR, surface plasmon resonance; CRD, carbohydrate recognition domain; GBP, glycan-binding protein; PBS, phosphate-buffered saline; PS, phosphatidylserine; FITC, fluorescein isothiocyanate; CHO, Chinese hamster ovary; LEA, L. esculentum agglutinin; RCA-I, R. communis agglutinin-I; NG, N-glycan.
S. R. Stowell and R. D. Cummings, unpublished data.
C. Arthur, S. R. Stowell, and R. D. Cummings, unpublished data.
References
- 1.Camby, I., Le Mercier, M., Lefranc, F., and Kiss, R. (2006) Glycobiology 16 R137-R157 [DOI] [PubMed] [Google Scholar]
- 2.Dumic, J., Dabelic, S., and Flogel, M. (2006) Biochim. Biophys. Acta 1760 616-635 [DOI] [PubMed] [Google Scholar]
- 3.Leffler, H., Carlsson, S., Hedlund, M., Qian, Y., and Poirier, F. (2004) Glycoconj. J. 19 433-440 [DOI] [PubMed] [Google Scholar]
- 4.Ilarregui, J. M., Bianco, G. A., Toscano, M. A., and Rabinovich, G. A. (2005) Ann. Rheum. Dis. 64 Suppl. 4, iv, 96-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubinstein, N., Ilarregui, J. M., Toscano, M. A., and Rabinovich, G. A. (2004) Tissue Antigens 64 1-12 [DOI] [PubMed] [Google Scholar]
- 6.Frigeri, L. G., Zuberi, R. I., and Liu, F. T. (1993) Biochemistry 32 7644-7649 [DOI] [PubMed] [Google Scholar]
- 7.La, M., Cao, T. V., Cerchiaro, G., Chilton, K., Hirabayashi, J., Kasai, K., Oliani, S. M., Chernajovsky, Y., and Perretti, M. (2003) Am. J. Pathol. 163 1505-1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sano, H., Hsu, D. K., Yu, L., Apgar, J. R., Kuwabara, I., Yamanaka, T., Hirashima, M., and Liu, F. T. (2000) J. Immunol. 165 2156-2164 [DOI] [PubMed] [Google Scholar]
- 9.Jeng, K. C., Frigeri, L. G., and Liu, F. T. (1994) Immunol. Lett. 42 113-116 [DOI] [PubMed] [Google Scholar]
- 10.Baggiolini, M., and Clark-Lewis, I. (1992) FEBS Lett. 307 97-101 [DOI] [PubMed] [Google Scholar]
- 11.Rabinovich, G. A., Sotomayor, C. E., Riera, C. M., Bianco, I., and Correa, S. G. (2000) Eur. J. Immunol. 30 1331-1339 [DOI] [PubMed] [Google Scholar]
- 12.Hsu, D. K., Yang, R. Y., Pan, Z., Yu, L., Salomon, D. R., Fung-Leung, W. P., and Liu, F. T. (2000) Am. J. Pathol. 156 1073-1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozaki, K., Inoue, K., Sato, H., Iida, A., Ohnishi, Y., Sekine, A., Sato, H., Odashiro, K., Nobuyoshi, M., Hori, M., Nakamura, Y., and Tanaka, T. (2004) Nature 429 72-75 [DOI] [PubMed] [Google Scholar]
- 14.Hahn, H. P., Pang, M., He, J., Hernandez, J. D., Yang, R. Y., Li, L. Y., Wang, X., Liu, F. T., and Baum, L. G. (2004) Cell Death Differ. 11 1277-1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sturm, A., Lensch, M., Andre, S., Kaltner, H., Wiedenmann, B., Rosewicz, S., Dignass, A. U., and Gabius, H. J. (2004) J. Immunol. 173 3825-3837 [DOI] [PubMed] [Google Scholar]
- 16.Stillman, B. N., Hsu, D. K., Pang, M., Brewer, C. F., Johnson, P., Liu, F. T., and Baum, L. G. (2006) J. Immunol. 176 778-789 [DOI] [PubMed] [Google Scholar]
- 17.Almkvist, J., Dahlgren, C., Leffler, H., and Karlsson, A. (2002) J. Immunol. 168 4034-4041 [DOI] [PubMed] [Google Scholar]
- 18.Almkvist, J., Faldt, J., Dahlgren, C., Leffler, H., and Karlsson, A. (2001) Infect. Immun. 69 832-837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaoka, A., Kuwabara, I., Frigeri, L. G., and Liu, F. T. (1995) J. Immunol. 154 3479-3487 [PubMed] [Google Scholar]
- 20.Karmakar, S., Cummings, R. D., and McEver, R. P. (2005) J. Biol. Chem. 280 28623-28631 [DOI] [PubMed] [Google Scholar]
- 21.Hirabayashi, J., Hashidate, T., Arata, Y., Nishi, N., Nakamura, T., Hirashima, M., Urashima, T., Oka, T., Futai, M., Muller, W. E., Yagi, F., and Kasai, K. (2002) Biochim. Biophys. Acta 1572 232-254 [DOI] [PubMed] [Google Scholar]
- 22.Brewer, C. F. (2004) Glycoconj. J. 19 459-465 [DOI] [PubMed] [Google Scholar]
- 23.Sato, S., and Hughes, R. C. (1992) J. Biol. Chem. 267 6983-6990 [PubMed] [Google Scholar]
- 24.Solomon, J. C., Stoll, M. S., Penfold, P., Abbott, W. M., Childs, R. A., Hanfland, P., and Feizi, T. (1991) Carbohydr. Res. 213 293-307 [DOI] [PubMed] [Google Scholar]
- 25.Sparrow, C. P., Leffler, H., and Barondes, S. H. (1987) J. Biol. Chem. 262 7383-7390 [PubMed] [Google Scholar]
- 26.Leffler, H., and Barondes, S. H. (1986) J. Biol. Chem. 261 10119-10126 [PubMed] [Google Scholar]
- 27.Knibbs, R. N., Agrwal, N., Wang, J. L., and Goldstein, I. J. (1993) J. Biol. Chem. 268 14940-14947 [PubMed] [Google Scholar]
- 28.Feizi, T., Solomon, J. C., Yuen, C. T., Jeng, K. C., Frigeri, L. G., Hsu, D. K., and Liu, F. T. (1994) Biochemistry 33 6342-6349 [DOI] [PubMed] [Google Scholar]
- 29.Perillo, N. L., Pace, K. E., Seilhamer, J. J., and Baum, L. G. (1995) Nature 378 736-739 [DOI] [PubMed] [Google Scholar]
- 30.Ahmad, N., Gabius, H. J., Andre, S., Kaltner, H., Sabesan, S., Roy, R., Liu, B., Macaluso, F., and Brewer, C. F. (2004) J. Biol. Chem. 279 10841-10847 [DOI] [PubMed] [Google Scholar]
- 31.Massa, S. M., Cooper, D. N., Leffler, H., and Barondes, S. H. (1993) Biochemistry 32 260-267 [DOI] [PubMed] [Google Scholar]
- 32.Nieminen, J., Kuno, A., Hirabayashi, J., and Sato, S. (2007) J. Biol. Chem. 282 1374-1383 [DOI] [PubMed] [Google Scholar]
- 33.Blixt, O., Head, S., Mondala, T., Scanlan, C., Huflejt, M. E., Alvarez, R., Bryan, M. C., Fazio, F., Calarese, D., Stevens, J., Razi, N., Stevens, D. J., Skehel, J. J., van Die, I., Burton, D. R., Wilson, I. A., Cummings, R., Bovin, N., Wong, C. H., and Paulson, J. C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 17033-17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leppanen, A., Stowell, S., Blixt, O., and Cummings, R. D. (2005) J. Biol. Chem. 280 5549-5562 [DOI] [PubMed] [Google Scholar]
- 35.Stowell, S. R., Dias-Baruffi, M., Penttila, L., Renkonen, O., Nyame, A. K., and Cummings, R. D. (2004) Glycobiology 14 157-167 [DOI] [PubMed] [Google Scholar]
- 36.Ahmad, N., Gabius, H. J., Sabesan, S., Oscarson, S., and Brewer, C. F. (2004) Glycobiology 14 817-825 [DOI] [PubMed] [Google Scholar]
- 37.Bochner, B. S., Alvarez, R. A., Mehta, P., Bovin, N. V., Blixt, O., White, J. R., and Schnaar, R. L. (2005) J. Biol. Chem. 280 4307-4312 [DOI] [PubMed] [Google Scholar]
- 38.Stowell, S. R., Karmakar, S., Stowell, C. J., Dias-Baruffi, M., McEver, R. P., and Cummings, R. D. (2007) Blood 109 219-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukumori, T., Takenaka, Y., Yoshii, T., Kim, H. R., Hogan, V., Inohara, H., Kagawa, S., and Raz, A. (2003) Cancer Res. 63 8302-8311 [PubMed] [Google Scholar]
- 40.Dias-Baruffi, M., Zhu, H., Cho, M., Karmakar, S., McEver, R. P., and Cummings, R. D. (2003) J. Biol. Chem. 278 41282-41293 [DOI] [PubMed] [Google Scholar]
- 41.van Liempt, E., Bank, C. M., Mehta, P., Garcia-Vallejo, J. J., Kawar, Z. S., Geyer, R., Alvarez, R. A., Cummings, R. D., Kooyk, Y., and van Die, I. (2006) FEBS Lett. 580 6123-6131 [DOI] [PubMed] [Google Scholar]
- 42.Nam, H. J., Gurda-Whitaker, B., Yee Gan, W., Ilaria, S., McKenna, R., Mehta, P., Alvarez, R. A., and Agbandje-McKenna, M. (2006) J. Biol. Chem. 281 25670-25677 [DOI] [PubMed] [Google Scholar]
- 43.Carlsson, S., Oberg, C. T., Carlsson, M. C., Sundin, A., Nilsson, U. J., Smith, D., Cummings, R. D., Almkvist, J., Karlsson, A., and Leffler, H. (2007) Glycobiology 17 663-676 [DOI] [PubMed] [Google Scholar]
- 44.Baenziger, J. U., and Fiete, D. (1979) J. Biol. Chem. 254 9795-9799 [PubMed] [Google Scholar]
- 45.Cummings, R. D., and Kornfeld, S. (1982) J. Biol. Chem. 257 11230-11234 [PubMed] [Google Scholar]
- 46.Novogrodsky, A., Lotan, R., Ravid, A., and Sharon, N. (1975) J. Immunol. 115 1243-1248 [PubMed] [Google Scholar]
- 47.Lotan, R., Skutelsky, E., Danon, D., and Sharon, N. (1975) J. Biol. Chem. 250 8518-8523 [PubMed] [Google Scholar]
- 48.Kornfeld, R., and Kornfeld, S. (1985) Annu. Rev. Biochem. 54 631-664 [DOI] [PubMed] [Google Scholar]
- 49.Shibuya, N., Goldstein, I. J., Broekaert, W. F., Nsimba-Lubaki, M., Peeters, B., and Peumans, W. J. (1987) J. Biol. Chem. 262 1596-1601 [PubMed] [Google Scholar]
- 50.Kopitz, J., von Reitzenstein, C., Burchert, M., Cantz, M., and Gabius, H. J. (1998) J. Biol. Chem. 273 11205-11211 [DOI] [PubMed] [Google Scholar]
- 51.Siebert, H. C., Andre, S., Lu, S. Y., Frank, M., Kaltner, H., van Kuik, J. A., Korchagina, E. Y., Bovin, N., Tajkhorshid, E., Kaptein, R., Vliegenthart, J. F., von der Lieth, C. W., Jimenez-Barbero, J., Kopitz, J., and Gabius, H. J. (2003) Biochemistry 42 14762-14773 [DOI] [PubMed] [Google Scholar]
- 52.Reed, R. A., Mattai, J., and Shipley, G. G. (1987) Biochemistry 26 824-832 [DOI] [PubMed] [Google Scholar]
- 53.Allen, H. J., Ahmed, H., and Matta, K. L. (1998) Glycoconj. J. 15 691-695 [DOI] [PubMed] [Google Scholar]
- 54.Zhou, Q., and Cummings, R. D. (1990) Arch. Biochem. Biophys. 281 27-35 [DOI] [PubMed] [Google Scholar]
- 55.Woo, H. J., Shaw, L. M., Messier, J. M., and Mercurio, A. M. (1990) J. Biol. Chem. 265 7097-7099 [PubMed] [Google Scholar]
- 56.Cooper, D. N., Massa, S. M., and Barondes, S. H. (1991) J. Cell Biol. 115 1437-1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merkle, R. K., and Cummings, R. D. (1987) J. Biol. Chem. 262 8179-8189 [PubMed] [Google Scholar]
- 58.Zhang, P., Tian, X., Chandra, P., and Brouwer, K. L. (2005) Mol. Pharmacol. 67 1334-1341 [DOI] [PubMed] [Google Scholar]
- 59.Hatch, N. E., Hudson, M., Seto, M. L., Cunningham, M. L., and Bothwell, M. (2006) J. Biol. Chem. 281 27292-27305 [DOI] [PubMed] [Google Scholar]
- 60.Jiang, J., Suppiramaniam, V., and Wooten, M. W. (2006) Neurosignals 15 266-282 [DOI] [PubMed] [Google Scholar]
- 61.Patnaik, S. K., Potvin, B., Carlsson, S., Sturm, D., Leffler, H., and Stanley, P. (2006) Glycobiology 16 305-317 [DOI] [PubMed] [Google Scholar]
- 62.Hokke, C. H., Bergwerff, A. A., Van Dedem, G. W., Kamerling, J. P., and Vliegenthart, J. F. (1995) Eur. J. Biochem. 228 981-1008 [DOI] [PubMed] [Google Scholar]
- 63.Toscano, M. A., Bianco, G. A., Ilarregui, J. M., Croci, D. O., Correale, J., Hernandez, J. D., Zwirner, N. W., Poirier, F., Riley, E. M., Baum, L. G., and Rabinovich, G. A. (2007) Nat. Immunol. 8 825-834 [DOI] [PubMed] [Google Scholar]
- 64.Viitala, J., and Finne, J. (1984) Eur. J. Biochem. 138 393-397 [DOI] [PubMed] [Google Scholar]
- 65.Moiseeva, E. P., Williams, B., and Samani, N. J. (2003) Biochim. Biophys. Acta 1619 125-132 [DOI] [PubMed] [Google Scholar]
- 66.Springer, G. F., and Horton, R. E. (1969) J. Clin. Investig. 48 1280-1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oriol, R., Le Pendu, J., and Mollicone, R. (1986) Vox Sang. 51 161-171 [DOI] [PubMed] [Google Scholar]
- 68.Pelletier, I., Hashidate, T., Urashima, T., Nishi, N., Nakamura, T., Futai, M., Arata, Y., Kasai, K., Hirashima, M., Hirabayashi, J., and Sato, S. (2003) J. Biol. Chem. 278 22223-22230 [DOI] [PubMed] [Google Scholar]
- 69.Kohatsu, L., Hsu, D. K., Jegalian, A. G., Liu, F. T., and Baum, L. G. (2006) J. Immunol. 177 4718-4726 [DOI] [PubMed] [Google Scholar]
- 70.Yi, W., Shao, J., Zhu, L., Li, M., Singh, M., Lu, Y., Lin, S., Li, H., Ryu, K., Shen, J., Guo, H., Yao, Q., Bush, C. A., and Wang, P. G. (2005) J. Am. Chem. Soc. 127 2040-2041 [DOI] [PubMed] [Google Scholar]
- 71.Pelletier, I., and Sato, S. (2002) J. Biol. Chem. 277 17663-17670 [DOI] [PubMed] [Google Scholar]
- 72.Oliveira, F. L., Frazao, P., Chammas, R., Hsu, D. K., Liu, F. T., Borojevic, R., Takiya, C. M., and El-Cheikh, M. C. (2007) J. Leukocyte Biol. 82 300-310 [DOI] [PubMed] [Google Scholar]
- 73.Breuilh, L., Vanhoutte, F., Fontaine, J., van Stijn, C. M., Tillie-Leblond, I., Capron, M., Faveeuw, C., Jouault, T., van Die, I., Gosset, P., and Trottein, F. (2007) Infect. Immun. 75 5148-5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sato, S., and Nieminen, J. (2004) Glycoconj. J. 19 583-591 [DOI] [PubMed] [Google Scholar]
- 75.Kopitz, J., von Reitzenstein, C., Andre, S., Kaltner, H., Uhl, J., Ehemann, V., Cantz, M., and Gabius, H. J. (2001) J. Biol. Chem. 276 35917-35923 [DOI] [PubMed] [Google Scholar]
- 76.Glinsky, V. V., Glinsky, G. V., Glinskii, O. V., Huxley, V. H., Turk, J. R., Mossine, V. V., Deutscher, S. L., Pienta, K. J., and Quinn, T. P. (2003) Cancer Res. 63 3805-3811 [PubMed] [Google Scholar]
- 77.Glinsky, V. V., Glinsky, G. V., Rittenhouse-Olson, K., Huflejt, M. E., Glinskii, O. V., Deutscher, S. L., and Quinn, T. P. (2001) Cancer Res. 61 4851-4857 [PubMed] [Google Scholar]
- 78.Sangeetha, S. R., and Appukuttan, P. S. (2005) Int. J. Biol. Macromol. 35 269-276 [DOI] [PubMed] [Google Scholar]
- 79.Appukuttan, P. S. (2002) J. Mol. Recognit. 15 180-187 [DOI] [PubMed] [Google Scholar]