Abstract
Protein geranylgeranylation is critical for the function of a number of proteins such as RhoA, Rac, and Rab. Protein geranylgeranyltransferase I (GGTase-I) and Rab geranylgeranyltransferase (RabGGTase) catalyze these modifications. In this work, we first describe the identification and characterization of small molecule inhibitors of GGTase-I (GGTI) with two novel scaffolds from a library consisting of allenoate-derived compounds. These compounds exhibit specific inhibition of GGTase-I and act by competing with a substrate protein. Derivatization of a carboxylic acid emanating from the core ring of one of the GGTI compounds dramatically improves their cellular activity. The improved GGTI compounds inhibit proliferation of a variety of human cancer cell lines and cause G1 cell cycle arrest and induction of p21CIP1/WAF1. We also report the identification of novel small molecule inhibitors of RabGGTase. These compounds were identified first by screening our GGTI compounds for those that also exhibited RabGGTase inhibition. This led to the discovery of a common structural feature for RabGGTase inhibitors: the presence of a characteristic six-atom aliphatic tail attached to the penta-substituted pyrrolidine core. Further screening led to the identification of compounds with preferential inhibition of RabGGTase. These compounds inhibit RabGGTase activity by competing with the substrate protein. These novel compounds may provide valuable reagents to study protein geranylgeranylation.
Protein prenylation is a post-translational modification of proteins involving the addition of isoprenoids (1–5). Specifically, protein farnesylation involves the addition of a C15 farnesyl group to proteins ending with the C-terminal CAAX motif (where C is cysteine; A is an aliphatic amino acid; and X is usually serine, methionine, glutamine, cysteine, or alanine). Farnesylated proteins include Ras proteins, Rheb proteins, nuclear lamins, and Hdj2. Protein geranylgeranylation involves the addition of a longer isoprenoid, C20 geranylgeranyl group. Two different types of geranylgeranylation have been reported. Rho family proteins such as RhoA, Cdc42, and Rac as well as the γ-subunit of heterotrimeric G-proteins are geranylgeranylated at a cysteine within the CAAL motif (similar to the CAAX motif, but the C-terminal amino acid is leucine or phenylalanine) at their C termini. Rab proteins involved in protein transport across the secretory and endocytosis pathways are also geranylgeranylated. These proteins usually end with CC (two cysteines) or CXC at the C termini, and both cysteines are geranylgeranylated.
Recent studies highlight the physiological significance of protein geranylgeranylation. Knock-out mice specific for the β-subunit of geranylgeranyltransferase I (GGTase-I)2 have been established (6). Characterization of GGTase-I-deficient cells showed proliferation inhibition and accumulation of p21CIP1/WAF1, pointing to the significance of GGTase-I in cell proliferation and cell cycle progression (6). GGTase-I deficiency reduced oncogenic K-ras-induced lung tumor formation in mice, pointing to the significance of inhibiting GGTase-I to block tumor formation (6). Recent studies also showed that a number of geranylgeranylated proteins play important roles in tumorigenesis and metastasis. In addition to RhoA and Cdc42 proteins, RalA protein was recently found to be activated downstream of Ras in most pancreatic cancer cells harboring oncogenic K-ras mutation (7). RalB plays critical roles in the survival pathway (8). RhoC is overexpressed in metastatic cancer, and RhoC knock-out mice exhibit defects in metastasis (9, 10). Overexpression of Rab25 in breast and ovarian cancer cells has been reported, and this mutation is a determinant for the aggressiveness of these cancers (11, 12). Rab25 is also up-regulated in prostate cancer and transitional cell bladder cancer (11). Overexpression of other Rab proteins such as Rab5a and Rab7 in cancer has been reported (13, 14).
Protein geranylgeranylation is catalyzed by two types of enzymes. GGTase-I catalyzes monogeranylgeranylation of proteins such as Rho, Rac, and Cdc42. This enzyme is a heterodimer consisting of α- and β-subunits (15). Rab geranylgeranyltransferase (RabGGTase or GGTase-II) catalyzes digeranylgeranylation of Rab proteins (16, 17). This enzyme also contains α- and β-subunits, but contains an additional subunit, the Rab escort protein (REP) (16, 18). The REP subunit binds to the substrate Rab protein (19). The α- and β-subunits share homology with corresponding subunits of GGTase-I.
Small molecule inhibitors of GGTases (GGTIs) provide novel reagents to study geranylgeranylation. Development of peptidomimetic GGTI compounds derived from the CAAL peptide was pioneered by Sebti and co-workers (20–24); compounds (GGTI-298, GGTI-2154, etc.) exhibit cellular effects, including cell cycle arrest and apoptosis induction, and inhibit tumor growth in mice. However, development of non-peptidomimetic inhibitors lagged behind. Recently, the first non-peptidomimetic compound, GGTI-DU40, was reported (25). As for RabGGTase inhibitors, no effective inhibitors have been identified so far (26). 2-Hydroxy-2-phosphono-3-(3-pyridinyl)-propanoic acid has been shown to inhibit geranylgeranylation of Rab proteins; however, a high concentration (mm) of compound was used to achieve inhibition (27). Thus, further development of compounds that inhibit GGTase-I or RabGGTase is of importance, as they provide novel reagents to deepen our understanding of protein geranylgeranylation.
In this study, we first report the identification and characterization of novel inhibitors of GGTase-I that were identified from a chemical compound library that we constructed through diversity-oriented synthesis using allenoic acid-derived starting materials (28). A specific modification that improves their cellular activity has been identified. Second, we report the identification and characterization of novel compounds with preferential inhibition of RabGGTase. These compounds have a scaffold that is shared with GGTI but possess an extra structural feature.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture—NIH3T3 cells were maintained in Dulbecco's modified Eagle's medium (Cellgro, Herndon, VA) supplemented with 10% (v/v) fetal bovine serum (FBS; HyClone, Logan, UT), 2% l-glutamine, 1% penicillin, and 1% streptomycin stock solutions (Invitrogen). K562 cells were maintained in RPMI 1640 medium (Cellgro) supplemented with 10% (v/v) FBS and penicillin/streptomycin. PANC-1 cells were maintained in Dulbecco's modified Eagle's medium/F-12 (Invitrogen) supplemented with 10% (v/v) FBS and penicillin/streptomycin. MCF-7 cells were maintained in Eagle's minimum essential medium (Cellgro) supplemented with 10% (v/v) FBS and penicillin/streptomycin.
Materials—[3H]Farnesyl diphosphate (21.5 Ci/mmol) and [3H]geranylgeranyl diphosphate (GGPP) (23.0 Ci/mmol) were purchased from PerkinElmer Life Sciences. BMS-225975 was kindly provided by Dr. Veeraswamy Manne (Bristol-Myers Squibb Co.). GGTI-298 was purchased from Calbiochem. The prenyltransferases used are recombinant enzymes. GGTase-I, farnesyltransferase (FTase), RabGGTase, REP-1, and Rab7 were purchased from Jena Bioscience (Jena, Germany). Other chemicals were obtained from Sigma. The allenoate-derived compound library, including P3-E5 and P5-H6, were synthesized as described (28). Detailed methods for the synthesis of P63-F10, P63-C7, P63-E11, P62-A5, P62-C11, P62-E4, and modified P5-H6 compounds (P61-A2, P61-A5, P61-A6, P61-A7 and P61-B4) will be described elsewhere.3
In Vitro Enzyme Assays—GGTase-I and FTase activities were determined by following the incorporation of radiolabeled isoprenoid [3H]geranylgeranyl or [3H]farnesyl into substrate proteins. FTase or GGTase-I (50 nm) was used to initiate reactions containing 0.4 μm [3H]farnesyl diphosphate or 0.5 μm [3H]GGPP and 2 μm maltose-binding protein-tagged substrates (K-Ras4B for FTase and RhoA for GGTase-I) in 20 μlof buffer containing 50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 5 μm ZnCl2, 0.01% Triton X-100, and 1 mm dithiothreitol. Inhibitors were added at the indicated concentrations. The final Me2SO concentration was 2.5% for all samples. Reactions were carried out for 10 min at 30 °C. The reaction mixture was spotted onto a filter paper and treated with 10% trichloroacetic acid, followed by ethanol and acetone washing. The filter was counted using a scintillation counter. Kinetic assays in which the GGPP concentration was varied employed fixed concentrations of GGTase-I and RhoA, and reactions were carried out for 5 min. Similarly, fixed concentrations of GGTase-I and GGPP were used when the amount of RhoA was varied. For RabGGTase assays, the reaction contained the following components in 20 μl: 0.625 μlof[3H]GGPP (0.7 μm), 25 nm RabGGTase, 0.6 μm REP-1, 0.6 μm purified Rab7 or Ypt1 protein, 40 mm HEPES (pH 7.5), 150 mm NaCl, 5 mm dithiothreitol, 3 mm MgCl2, and 0.3% CHAPS. Reactions were carried out for 20 min at 37 °C, and the products were analyzed as described above for the GGTase-I reaction. Graphing and Michaelis-Menten analysis were performed using Prism Version 5 (GraphPad, San Diego, CA).
Inhibition of Geranylgeranylation in Cells—Inhibition of GGTase-I-catalyzed protein geranylgeranylation was assessed by examining the accumulation of unprenylated Rap1. To measure the level of unprenylated Rap1, cells were cultured in Dulbecco's modified Eagle's medium plus 10% (v/v) FBS overnight, and then Me2SO or appropriate inhibitors were added. Incubation was continued for 48 h. The cells were harvested and lysed in lysis buffer (20 mm Tris-HCl (pH 7.5), 150 mm
NaCl, 1 mm EDTA, 1% Nonidet P-40, and 1× protease inhibitor mixture). Whole cell lysates of NIH3T3 cells were electrophoresed on a 12% SDS-polyacrylamide gel, transferred to nitrocellulose membranes, and immunoblotted with antibody against the unprenylated form of Rap1 (goat, catalog no. sc-1482, Santa Cruz Biotechnology, Inc.), total Rap1 (catalog no. sc-65, Santa Cruz Biotechnology, Inc.) or actin (catalog no. A4700, Sigma). Actin was used as a loading control. Inhibition of Rab geranylgeranylation in cells (supplemental Fig. S2) was examined as described previously (29). Briefly, whole cell lysates were subjected to 4 m urea and 15% SDS-PAGE, followed by immunoblotting with antibody against Rab5b (catalog no. sc-598, Santa Cruz Biotechnology, Inc.) or actin. Subcellular fractionations (supplemental Fig. S3) were performed as described by Gomes et al. (30). Briefly, cells were treated with P49-F6 for 48 h. After osmotic lysis, cell debris were removed by centrifugation at 500 × g for 10 min, and the supernatant was subjected to ultracentrifugation at 100,000 × g for 60 min. The supernatant from the ultracentrifugation was collected as a soluble fraction. The pellet was collected as a membrane fraction. These fractions were subjected to electrophoresis on 10% SDS-polyacrylamide gels, followed by immunoblotting with antibody against Rab5b. RhoGDI (catalog no. sc-360, Santa Cruz Biotechnology, Inc.) and Na+/K+-ATPase (catalog no. A276, Sigma) were used as markers for soluble and membrane fractions, respectively.
Cell Viability and Cell Cycle Analysis—Cell viability was determined using the CCK-8 cell counting kit (Dojindo Molecular Technologies, Kumamoto, Japan) as described previously (31). Briefly, cells (5 × 103) were plated onto 96-well plates and treated with the appropriate inhibitor as indicated in the figure legends. Cell viability was calculated relative to the Me2SO control. The cell cycle profile was analyzed by flow cytometry as described previously (32).
Transcriptional Reporter Assays—For the p21CIP1/WAF1 promoter-luciferase assay, NIH3T3 cells were transfected with p21CIP1/WAF1 promoter-luciferase or vector plasmids (both plasmids were provided by Dr. Genhong Cheng) (33). Cells were treated with GGTI compounds. A Promega luciferase assay kit was used according to the manufacturer's protocol.
Statistical Analysis—The statistical significance of the difference was determined using Student's unpaired t test. A p value <0.05 was considered statistically significant.
RESULTS
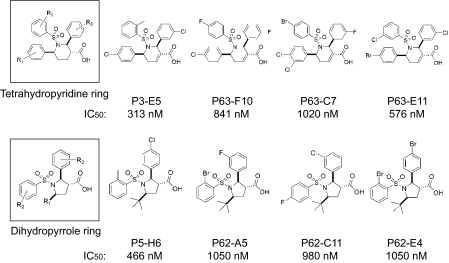
Allenoic Acid-based Chemical Compound Library and Identification of GGTase-I Inhibitors—Previously, we reported the construction of a library of allenoate-derived compounds and the identification of novel GGTI compounds (28). The library construction involved the use of allenoates as multireactive core molecules. Using a second set of building blocks (imines, aldehydes, and maleimides) that react with allenoates under similar reaction conditions (phosphine catalysis), we produced diverse compounds, including dihydropyrroles (34), tetrahydropyridines (35), bicyclic succinimides,4 dioxanylidenes (36), and α-pyrones (37). The identification of GGTI compounds was carried out first by screening a 171-compound pilot library using an in vitro assay with RhoA protein as a substrate. Scaffolds that initially showed activity were optimized by solid-phase split-and-pool combinatorial synthesis. This enabled us to identify two types of novel compounds: one group containing a tetrahydropyridine ring as its core scaffold and the other group having a dihydropyrrole ring as its core scaffold. Fig. 1 shows the structures and potencies of four representative compounds from each group, together with a general structure of each group.
FIGURE 1.
Novel core structures of GGTI and molecular structures of potent inhibitors. IC50 values for GGTase-I inhibition in vitro by compounds were measured as described under “Experimental Procedures.”
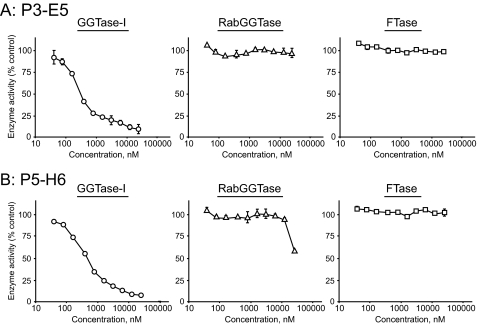
Two compounds with the highest potency in each group, P3-E5 and P5-H6, were further characterized. As described, P3-E5 and P5-H6 inhibit GGTase-I with IC50 values of 313 and 466 nm, respectively (Fig. 1). The specificity of GGTase-I inhibition by P3-E5 and P5-H6 was examined by assaying their ability to inhibit two closely related enzymes, FTase and RabGGTase. No significant inhibition of FTase activity by these compounds was observed even when the concentration was increased to 50,000 nm (Fig. 2). Similarly, P3-E5 showed little inhibition of RabGGTase even at 50,000 nm. P5-H6 showed little inhibition of RabGGTase up to 10,000 nm.
FIGURE 2.
Effect of P3-E5 (A) and P5-H6 (B) on the enzymatic activity of GGTase-I (left), RabGGTase (middle), and FTase (right). Varying concentrations of the compounds were added to each enzyme reaction. Data represent the mean ± S.D. of two measurements from two independent experiments.
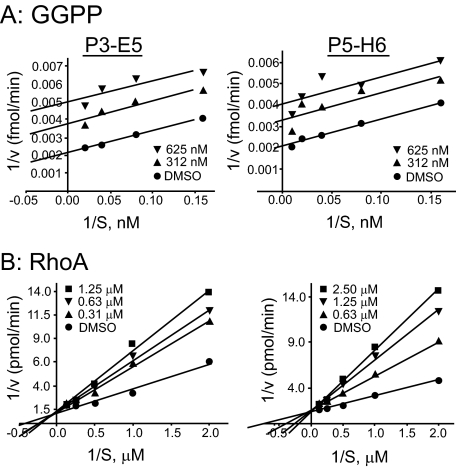
GGTIs Compete with the Substrate Protein—Michaelis-Menten analysis of the inhibition of GGTase-I is shown in Fig. 3. Fig. 3A shows the data derived from the results obtained using varying concentrations of GGPP, whereas Fig. 3B shows data derived from the results obtained using varying concentrations of the substrate protein RhoA. These results revealed that P3-E5 and P5-H6 are competitive inhibitors with respect to the substrate protein and uncompetitive inhibitors with respect to GGPP. P3-E5 and P5-H6 compete for binding of the protein substrate but not the isoprenoid substrate of GGTase-I. Ki values of 187 ± 13 and 408 ± 32 nm were calculated for P3-E5 and P5-H6, respectively.
FIGURE 3.
Kinetic analysis of GGTase-I inhibition. Shown are double-reciprocal plots obtained from substrate velocity curves for the inhibition of GGTase-I by P3-E5 (left) and P5-H6 (right). A, varying GGPP concentrations with a fixed RhoA protein concentration were used. B, varying RhoA protein concentrations with a fixed GGPP concentration were used. The amount of GGTI used is indicated. DMSO, Me2SO.
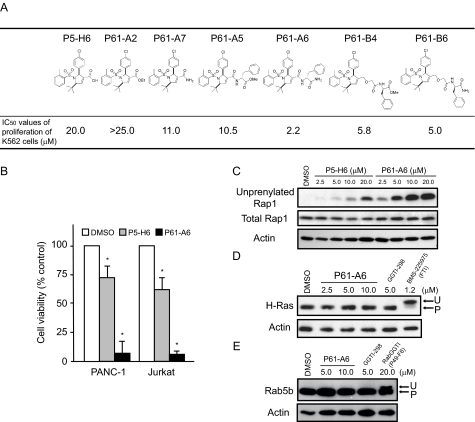
Improvement of the Cellular Activity of GGTI by Modification of P5-H6—We found that replacing a carboxyl group of the dihydropyrrole ring of P5-H6 significantly improved its cellular activity. This is shown in Fig. 4A; we synthesized a series of compounds with different moieties replacing the carboxyl group. The potency of these compounds to inhibit proliferation of K562 leukemic cells was examined, and their IC50 values are shown. The original P5-H6 compound containing a carboxylic acid moiety exhibited inhibition of proliferation of K562 cells with an IC50 value of 20 μm. Converting the free acid to an ethyl ester did not improve its cellular potency. However, amidation of this moiety led to improvement in its potency. Moreover, coupling of P5-H6 with an l-phenylalanine group resulted in significant improvement of cellular potency. Two compounds, P61-A6 and P61-B6, showed good potency with IC50 values of 2.2 and 5.0 μm in inhibiting proliferation of K562 cells, respectively. Comparison of the inhibitory activity of P5-H6 and P61-A6 in the pancreatic cancer cell line PANC-1 and Jurkat cells is shown in Fig. 4B.
FIGURE 4.
Improved cellular activity of modified GGTI compounds. A, molecular structures of P5-H6 and modified P5-H6 compounds. K562 cells were treated with the modified compounds for 72 h, and the cell number was then counted as described under “Experimental Procedures.” IC50 values for cell viability relative to Me2SO (DMSO) were measured. B, inhibitory effect of 12.5 μm P5-H6 or P61-A6 on PANC-1 and Jurkat cell viability. Data represent the mean ± S.D. of two measurements from two independent experiments. *, p < 0.05 compared with the value for Me2SO. C–E, P5-H6 or P61-A6 treatment inhibits Rap1 geranylgeranylation in NIH3T3 cells. Whole cell lysates from cells treated with Me2SO, P5-H6, P61-A6, GGTI (GGTI-298), FTase inhibitor (BMS-225975), or RabGGTI (P49-F6) for 48 h were prepared and processed for immunoblot analysis using antibody against the unprenylated form of Rap1 (C, upper panel), total Rap1 (C, middle panel), H-Ras (D, upper panel), Rab5b (E, upper panel), or actin (C–E, lower panels). The immunoblots shown are representative of two independent experiments for each treatment.
The improved potency of these compounds to inhibit cell proliferation correlates with their increased ability to inhibit protein geranylgeranylation inside the cell. Results on this point using P5-H6 and P61-A6 are shown in Fig. 4C. In this experiment, inhibition of protein geranylgeranylation was evaluated using an antibody that specifically detects unprenylated Rap1. Treatment with P5-H6 or P61-A6 led to the appearance of the unprocessed Rap1 band in a dose-dependent manner. Note that the appearance of the Rap1 band was observed at 2.5 μm with P61-A6, whereas it was not seen with P5-H6, reflecting a significant improvement in the potency of P61-A6 to inhibit protein geranylgeranylation.
In contrast to its effects on GGTase-I, P61-A6 did not inhibit protein farnesylation. This was examined using the farnesylated protein H-Ras. Although FTase inhibitor (BMS-225975) slowed the mobility of H-Ras protein on SDS-polyacrylamide gel, no such mobility shift was observed with P61-A6 or another GGTI compound, GGTI-298. Similarly, P61-A6 did not inhibit geranylgeranylation of Rab5b, as a slow migrating band representing that of unmodified Rab5b was detected only after treatment with RabGGTase inhibitors (P49-F6; see below) and not with P61-A6 (Fig. 4E).
Although P61-A6 exhibited improved ability to inhibit geranylgeranylation in cells, its ability to inhibit the GGTase-I enzyme was less than that of P5-H6, as the IC50 value for enzyme inhibition was 1 μm. No significant inhibition of FTase or RabGGTase activity by the P61-A6 compound was observed even when the concentration was increased to 100,000 nm (data not shown).
GGTI Compounds Inhibit Proliferation of Various Human Cancer Cell Lines and Cause G1 Cell Cycle Arrest—As shown in supplemental Table S1, significant inhibition of proliferation of a variety of human cancer cell lines was observed. Thus, these GGTI compounds inhibit a broad range of human cancer cell lines.
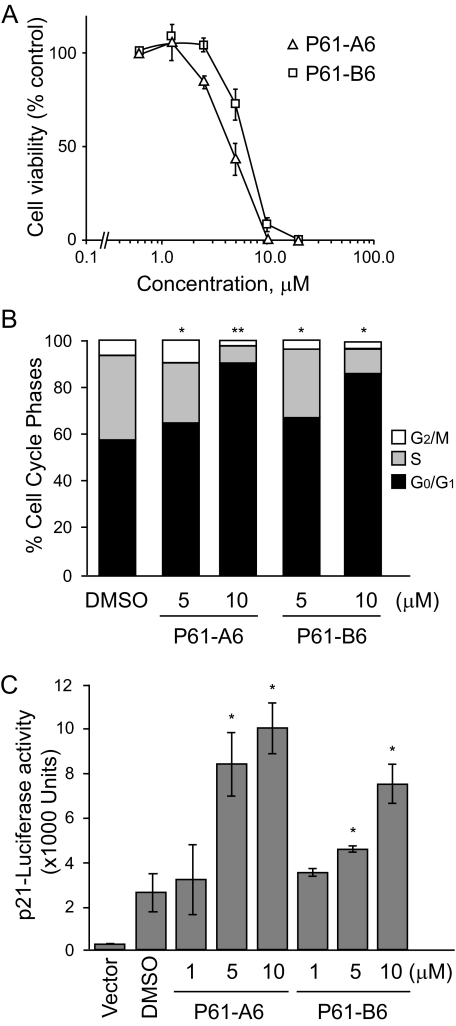
The inhibition of proliferation by GGTI appears to be due to the inhibition of cell cycle progression. As shown in Fig. 5A, treatment of the breast cancer cell line MCF-7 with P61-A6 or P61-B6 caused dose-dependent inhibition of proliferation. This was associated with a significant dose-dependent enrichment of G1 phase cells, whereas the percentage of S phase cells decreased (Fig. 5B and supplemental Fig. S1). Similar G1 enrichment was observed with a leukemic cell line, Jurkat; two pancreatic cancer cell lines, PANC-1 and MiaPaCa2; and another breast cancer cell line, MDA-MB-231 (supplemental Table S2).
FIGURE 5.
Effects of P61-A6 or P61-B6 on cell proliferation and cell cycle in MCF-7 cells. A, shown is the inhibition of proliferation of MCF-7 cells by P61-A6 and P61-B6. MCF-7 cells were treated with P61-A6, P61-B6, or Me2SO for 72 h. The cell number was counted as described under “Experimental Procedures.” Cell viability relative to the Me2SO control (100% value) is plotted. B, MCF-7 cells were treated with the indicated concentrations of P61-A6, P61-B6, or Me2SO (DMSO) for 48 h. Cell cycle profiles were monitored by flow cytometry. The percentages of cells in each phase of the cell cycle are indicated by different shades. C, NIH3T3 cells were transfected with p21CIP1/WAF1-luciferase or empty vector. Cells were treated with P61-A6 or P61-B6 compound at the indicated concentrations or with Me2SO for 48 h, and luciferase assay was performed. Data represent the mean ± S.D. of two measurements from two independent experiments. *, p < 0.05; **, p < 0.005 compared with the value for Me2SO.
One of the proposed mechanisms of GGTI effects on cell cycle progression is the inhibition of RhoA, which negatively regulates expression of the Cdk inhibitor p21CIP1/WAF1. To investigate whether our GGTI compound induces p21CIP1/WAF1 expression, luciferase expression using the p21CIP1/WAF1 promoter (33) was measured. Transient expression systems with NIH3T3 cells were used to examine the ability of P61-A6 to induce p21CIP1/WAF1-luciferase expression. P61-A6 induced significant (4-fold) induction of luciferase activity versus Me2SO in a dose-dependent manner (Fig. 5C).
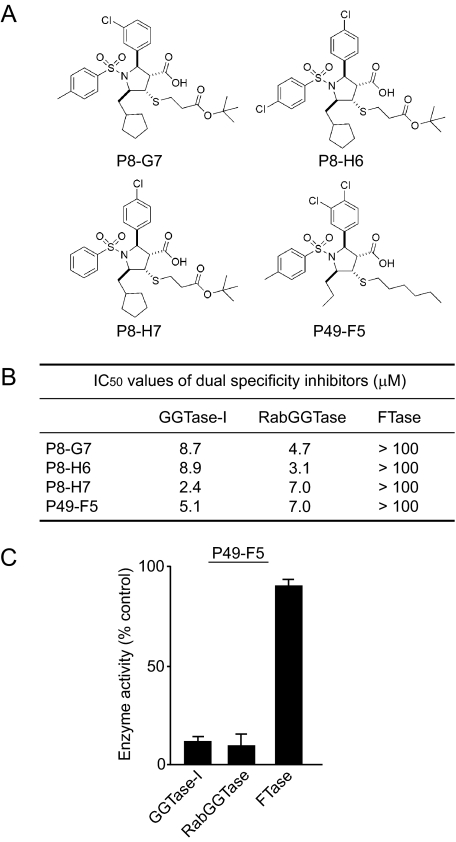
Identification of Dual Specificity Inhibitors of GGTase-I and RabGGTase—The characterization of our library of allenoate-derived compounds yielded novel compounds that have the ability to inhibit RabGGTase. The identification of these novel compounds was initially prompted by the observation that P5-H6 exhibited a slight inhibition of RabGGTase at concentrations >10,000 nm (Fig. 2B). This suggested to us that these GGTIs have the ability to act weakly on RabGGTase. If this were the case, we speculated that it might be possible to identify a subset of GGTI compounds that inhibit RabGGTase. With this reasoning, we examined our library of 3601 compounds related to P5-H6, listed all the compounds that exhibited GGTase-I inhibition, and screened them to see whether we could identify compounds that also inhibited RabGGTase. The assay was carried out using RabGGTase as described under “Experimental Procedures.” Of 428 compounds that exhibited GGTase-I inhibition (>50% inhibition at 50 μm), we found that 60 compounds also exhibited the ability to inhibit RabGGTase (>50% inhibition at 25 μm). Fig. 6 shows the four most potent compounds (P8-G7, P8-H6, P8-H7, and P49-F5) that exhibited inhibition of both GGTase-I and RabGGTase in the 2–9 μm range. On the other hand, these compounds did not inhibit FTase even at >100 μm (Fig. 6, B and C). Examination of their structures led us to notice a feature that is common to these dual specificity compounds; they all have a characteristic sixatom aliphatic tail attached to the penta-substituted pyrrolidine core via thioether linkage (putative RabGGTase inhibitor (RabGGTI) feature).
FIGURE 6.
Dual specificity inhibitors of GGTase-I and RabGGTase. A, molecular structures of dual specificity inhibitors. B, IC50 values for dual specificity inhibitors of GGTase-I, RabGGTase, and FTase. C, inhibitory effect of 25 μm P49-F5 on the in vitro activities of GGTase-I, RabGGTase, and FTase.
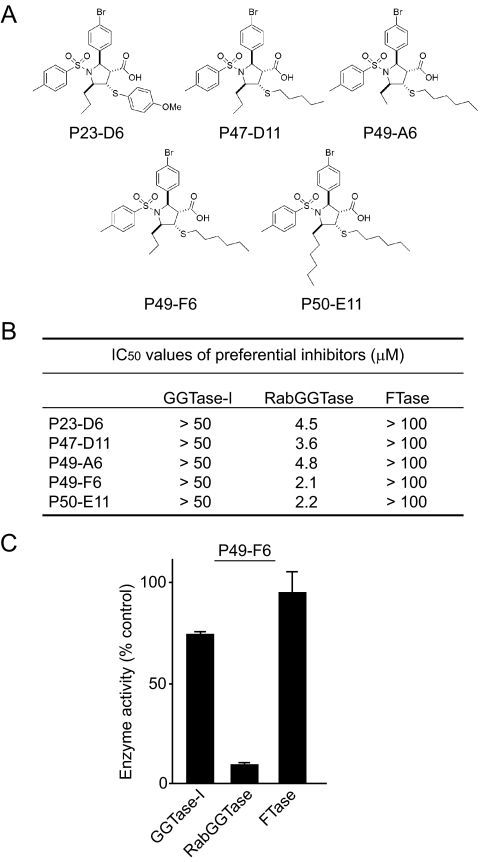
Identification of RabGGTIs—The finding of the putative RabGGTI feature is important, as this feature may be used to predict RabGGTI activity. We first examined our 3601-compound library and identified 524 compounds that have this feature. We then screened these compounds by carrying out RabGGTase assay. This resulted in the identification of 23 compounds that exhibited preferential inhibition of RabGGTase (<50% inhibition of GGTase-I at 50 μm and >50% inhibition of RabGGTase at 25 μm). Fig. 7 shows the structures and IC50 values for the five most potent preferential RabGGTIs, P23-D6, P47-D11, P49-A6, P49-F6, and P50-E11. Three of these compounds contain an n-hexylmercapto substituent at C-4 of the pyrrolidine ring. Additionally, we found n-pentyl thioether (P47-D11) and para-methoxyphenyl thioether (P23D6) appendages in the group of preferential RabGGTIs.
FIGURE 7.
Preferential RabGGTIs. A, molecular structures of RabGGTIs. B, IC50 values for preferential RabGGTIs against GGTase-I, RabGGTase, and FTase. C, inhibitory effect of 25 μm P49-F6 on the in vitro activities of GGTase-I, RabGGTase, and FTase.
As shown in Fig. 7 (B and C), all these compounds inhibited RabGGTase with IC50 values of 2–5 μm, whereas the inhibition of GGTase-I required >50 μm. No inhibition of FTase was observed with these compounds at 100 μm. Inhibition of Rab geranylgeranylation in cells was examined using Rab5b protein. In this experiment, we examined the mobility of Rab5b proteins (supplemental Fig. S2A). P49-F6, one of the preferential RabGGTIs, caused the appearance of a slow migrating Rab5b protein that represents an unprenylated form (29). On the other hand, P49-F6 did not cause a mobility shift of H-Ras and did not induce the appearance of unprenylated Rap1 (supplemental Fig. S2B), suggesting that it does not inhibit FTase or GGTase-I. We also examined the intracellular localization of Rab5b proteins (supplemental Fig. S3). Treatment with P49-F6 resulted in an increase of Rab5b protein in the soluble fraction.
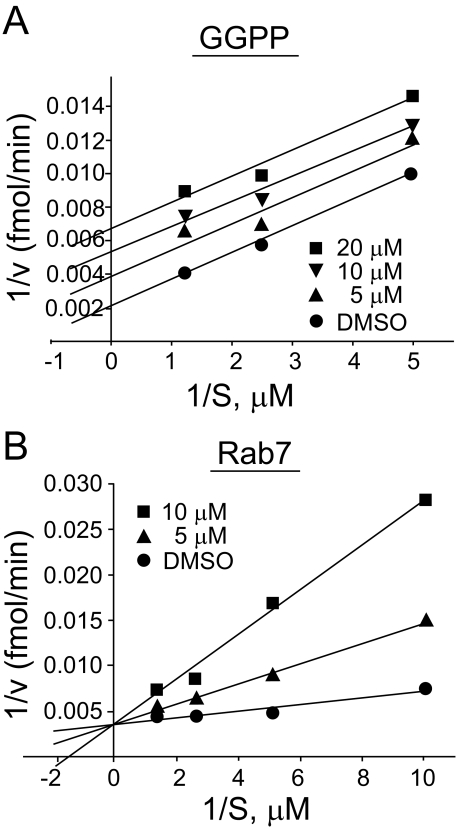
RabGGTI Competes with the Substrate Protein—Kinetic analysis was carried out to examine whether our RabGGTI compounds compete with the substrate protein. RabGGTase consists of a tightly bound core complex, the α- and β-subunits, and the third REP subunit. Therefore, these subunits were first mixed together in the presence of a low concentration of GGPP; the concentration of each substrate was altered; and the effect of inhibition by RabGGTI was examined. As shown in Fig. 8B, the RabGGTI compound P49-F6 inhibited RabGGTase activity with respect to the substrate protein Rab7. On the other hand, increasing the concentration of GGPP did not influence the inhibition by RabGGTI (Fig. 8A), suggesting that they are uncompetitive inhibitors with respect to GGPP. A Ki value of 1.36 ± 0.38 μm was calculated from these experiments.
FIGURE 8.
Characterization of P49-F6 inhibition of RabGGTase. In vitro RabGGTase assay was carried out with varying concentration of GGPP (A) or Rab7 (B). Each reaction mixture contained recombinant RabGGTase and recombinant REP-1 protein. They were mixed with a low concentration of GGPP (0.125 μm) to form an active enzyme first. Duplicate reactions were carried out as described under “Experimental Procedures.” The amount of RabGGTI used is indicated. DMSO, Me2SO.
DISCUSSION
In this work, we first reported the identification of novel small molecule inhibitors of GGTase-I from the library of allenoate-derived compounds. Two novel scaffolds were identified. Our compounds are non-peptidomimetic inhibitors that compete with the substrate protein. Specific inhibition of protein geranylgeranylation but not protein farnesylation was established with the purified enzymes as well as with treated cells. Other non-peptidomimetic GGTI compounds have been reported (25); however, 2–20 μm was used to exert cellular activity with tissue culture cells. Our compounds represent an important addition to the growing list of GGTI compounds.
We found that derivatization of the carboxyl group of the dihydropyrrole ring of our initial compound, P5-H6, resulted in a dramatic increase in cellular potency. In particular, changing the carboxyl group to the corresponding amide by coupling it with l-phenylalanine was effective. On the other hand, converting the free carboxylic acid into the ethyl ester did not lead to any improvement. Similarly, converting to a methyl ester did not result in improvement either.4 These results suggest that the improvement was not simply due to the removal of the charge but that the phenylalanine moiety exerted an additional effect. The improvement of the cellular activity of our GGTI compound correlated with an increase in the ability to inhibit protein geranylgeranylation, as detected by the appearance of unprenylated Rap1 protein. On the other hand, the modification did not improve the potency of these compounds to inhibit GGTase-I enzyme. Therefore, the improvement of cellular activity may reflect increased cellular uptake or stability of the compound.
Our GGTI compounds exhibit inhibition of proliferation of human cancer cell lines, including leukemic, pancreatic cancer, and breast cancer cell lines. One of the hallmarks of GGTI is that this class of inhibitors causes cell cycle arrest at the G1 phase (38, 39). Our inhibitors exhibited significant G1 arrest with the human cancer cell lines examined (Fig. 5B and supplemental Table S2). In particular, dramatic G1 arrest was observed with a breast cancer cell line MCF-7. In addition, our GGTI compounds induced p21CIP1/WAF1 expression, as observed using a luciferase reporter assay. These results are in-line with the idea that our GGTI inhibits RhoA, which acts as a negative regulator of p21CIP1/WAF1 expression (39–41). These results also agree with the recent observation of the accumulation of p21CIP1/WAF1 in GGTase-I-deficient cells from knock-out mice (6).
In this study, we have also reported the identification of a novel type of RabGGTIs. These compounds share the scaffold with one of the GGTI compounds but possess an extra hydrophobic tail emanating from the core ring. Only a handful of RabGGTIs have been identified in the past (26). Commonly used inhibitors are bisphosphonate-type compounds; however, the inhibition of RabGGTase requires approximately mm concentrations of the compounds. In contrast, our compounds inhibit RabGGTase with a single μm concentration. Although we did observe inhibition of geranylgeranylation of Rab protein in cells, further modification of the compounds was needed to improve their cellular activity. Recent studies suggest that RabGGTIs may be valuable as anticancer drugs. A study using small interfering RNA showed that the inhibition of RabGGTase leads to apoptosis induction in human cancer cells (42). Elevated levels of RabGGTase are detected in a number of human cancers (42). Furthermore, FTase inhibitor compounds that inhibit RabGGTase induce mislocalization of Rab protein and apoptosis (42).
We have shown that our RabGGTI compounds inhibit the enzyme by competing with the substrate protein. Because our RabGGTI compounds are derived from GGTI compounds, RabGGTI and GGTI share the same scaffold. This may be the reason that both our GGTI and RabGGTI work by competing with the substrate protein. GGTase-I and RabGGTase share similar active-site structures (16, 26); both enzymes have a core structure that consists of α- and β-subunits. In addition, the corresponding subunits in these enzymes share significant homology. It will be interesting to examine whether these inhibitors bind to similar pockets in these enzymes. On the other hand, we found that RabGGTI compounds possess a structural feature that is unique to this group of inhibitors. They contain a characteristic long aliphatic tail attached to the penta-substituted pyrrolidine core. It will be interesting to understand how this feature contributes to the inhibition of RabGGTase. One possibility is that the aliphatic tail fits into a pocket or interferes with an enzymatic process that is specific to RabGGTase but not to GGTase-I. The significance of lipidbinding pockets of REP for the RabGGTase reaction has been suggested (18, 43). Further characterization of our GGTI and RabGGTI should lead to increased understanding of the possible differences and similarities in the active sites of the two enzymes.
In summary, we have reported three different types of inhibitors of protein geranylgeranylation. First, we reported novel GGTIs. They inhibit proliferation of a variety of human cancer cell lines and cause G1 cell cycle arrest. Second, we reported novel RabGGTIs. Interestingly, those compounds possess structural features that define this class of inhibitors. In the course of the identification of RabGGTI compounds, we also identified dual specificity inhibitors that inhibit both GGTase-I and RabGGTase. Use of these novel compounds may lead to further understanding of the roles of protein geranylgeranylation. In addition, they may provide lead compounds for the development of anticancer drugs based on the inhibition of protein geranylgeranylation.
Supplementary Material
Acknowledgments
We thank Alan Ikeda and Kathleen Sakamoto for discussion on the effect of GGTI on leukemic cells and Richard Finn for discussion on breast cancer cells.
This work was supported by National Institutes of Health Grants CA32737 (to F. T.) and GM071779 (to O. K.) and by a grant from the Susan E. Riley Foundation. The flow cytometric analysis done in the UCLA Flow Cytometry Core Facilities was supported in part by National Institutes of Health Grants CA16042 and AI28697. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1 and S2.
Footnotes
The abbreviations used are: GGTase-I, geranylgeranyltransferase I; RabGGTase, Rab geranylgeranyltransferase; REP, Rab escort protein; GGTI, geranylgeranyltransferase I inhibitor; FBS, fetal bovine serum; GGPP, geranylgeranyl diphosphate; FTase, farnesyltransferase; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; RabGGTI, Rab geranylgeranyltransferase inhibitor.
H. D. G. Fiji, M. Watanabe, L. Chan, F. Tamanoi, and O. Kwon, manuscript in preparation.
S. Castellano, H. D. G. Fiji, and O. Kwon, unpublished data.
References
- 1.Tamanoi, F., and Sigman, D. S. (eds) (2001) The Enzymes, Vol. 21, Academic Press, San Diego
- 2.Zhang, F. L., and Casey, P. J. (1996) Annu. Rev. Biochem. 65 241–270 [DOI] [PubMed] [Google Scholar]
- 3.Glomset, J. A., Gelb, M. H., and Farnsworth, C. C. (1990) Trends Biochem. Sci. 15 139–142 [DOI] [PubMed] [Google Scholar]
- 4.Gelb, M. H., Brunsveld, L., Hrycyna, C. A., Michaelis, S., Tamanoi, F., Van Voorhis, W. C., and Waldmann, H. (2006) Nat. Chem. Biol. 10 518–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox, A. D., and Der, C. J. (1992) Curr. Opin. Cell Biol. 4 1008–1016 [DOI] [PubMed] [Google Scholar]
- 6.Sjogren, A. K., Andersson, K. M., Liu, M., Cutts, B. A., Karlsson, C., Wahlstrom, A. M., Dalin, M., Weinbaum, C., Casey, P. J., Tarkowski, A., Swolin, B., Young, S. G., and Bergo, M. O. (2007) J. Clin. Investig. 117 1294–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim, K. H., Baines, A. T., Fiordalisi, J. J., Shipitsin, M., Feig, L. A., Cox, A. D., Der, C. J., and Counter, C. M. (2005) Cancer Cell 6 533–545 [DOI] [PubMed] [Google Scholar]
- 8.Chien, Y., and White, M. A. (2003) EMBO Rep. 4 800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, E. A., Golub, T. R., Lander, E. S., and Hynes, R. O. (2000) Nature 406 532–535 [DOI] [PubMed] [Google Scholar]
- 10.Hakem, A., Sanchez-Sweatman, O., You-Ten, A., Duncan, G., Wakeham, A., Khokha, R., and Mak, T. W. (2005) Genes Dev. 19 1974–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, K. W., Lahad, J. P., Kuo, W. L., Lapuk, A., Yamada, K., Auersperg, N., Liu, J., Smith-McCune, K., Lu, K. H., Fishman, D., Gray, J. W., and Mills, G. B. (2004) Nat. Med. 10 1251–1256 [DOI] [PubMed] [Google Scholar]
- 12.Cheng, K. W., Lahad, J. P., Gray, J. W., and Mills, G. B. (2005) Cancer Res. 65 2516–2519 [DOI] [PubMed] [Google Scholar]
- 13.Croizet-Berger, K., Daumerie, C., Couvreur, M., Courtoy, P. J., and van den Hove, M. F. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 8277–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, H., Dai F., Yu, L., She, X., Zhao, Y., Jiang, J., Chen, X., and Zhao, S. (2002) Gene Expr. 10 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor, J. S., Reid, T. S., Terry, K. L., Casey, P. J., and Beese, L. S. (2003) EMBO J. 22 5963–5974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung, K. F., Baron, R., and Seabra, M. C. (2006) J. Lipid Res. 47 467–475 [DOI] [PubMed] [Google Scholar]
- 17.Zhang, H., Seabra, M. C., and Deisenhofer, J. (2000) Structure (Lond.) 8 241–251 [DOI] [PubMed] [Google Scholar]
- 18.Pylypenko, O., Rak, A., Reents, R., Niculae, A., Sidorovitch, V., Cioaca, M. D., Bessolitsyna, E., Thoma, N. H., Waldmann, H., Schlichting, I., Goody, R. S., and Alexandrov, K. (2003) Mol. Cell 11 483–494 [DOI] [PubMed] [Google Scholar]
- 19.Rak, A., Pylypenko, O., Niculae, A., Pyatkov, K., Goody, R. S., and Alexandrov, K. (2004) Cell 117 749–760 [DOI] [PubMed] [Google Scholar]
- 20.Sebti, S. M., and Adjei, A. A. (2004) Semin. Oncol. 31 28–39 [DOI] [PubMed] [Google Scholar]
- 21.Lerner, E. C., Qian, Y., Hamilton, A. D., and Sebti, S. M. (1995) J. Biol. Chem. 270 26770–26773 [DOI] [PubMed] [Google Scholar]
- 22.Vasudevan, A., Qian, Y., Vogt, A., Blaskovich, M. A., Ohkanda, J., Sebti, S. M., and Hamilton, A. D. (1999) J. Med. Chem. 42 1333–1340 [DOI] [PubMed] [Google Scholar]
- 23.Sun, J., Qian, Y., Hamilton, A. D., and Sebti, S. M. (1998) Oncogene 16 1467–1473 [DOI] [PubMed] [Google Scholar]
- 24.Sun, J., Ohkanda, J., Coppola, D., Yin, H., Kothare, M., Busciglio, B., Hamilton, A. D., and Sebti, S. M. (2003) Cancer Res. 63 8922–8929 [PubMed] [Google Scholar]
- 25.Peterson, Y. K., Kelly, P., Weinbaum, C. A., and Casey, P. J. (2006) J. Biol. Chem. 281 12445–12450 [DOI] [PubMed] [Google Scholar]
- 26.El Oualid, F., Cohen, L. H., van der Marel, G. A., and Overhand, M. (2006) Curr. Med. Chem. 13 2385–2427 [DOI] [PubMed] [Google Scholar]
- 27.Coxon, F. P., Helfrich, M. H., Larijani, B., Muzylak, M., Dunford, J. E., Marshall, D., McKinnon, A. D., Nesbitt, S. A., Horton, M. A., Seabra, M. C., Ebetino, F. H., and Rogers, M. J. (2001) J. Biol. Chem. 276 48213–48222 [DOI] [PubMed] [Google Scholar]
- 28.Castellano, S., Fiji, H. D., Kinderman, S. S., Watanabe, M., Leon, P., Tamanoi, F., and Kwon, O. (2007) J. Am. Chem. Soc. 129 5843–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanford, J. C., Foster, L., Kapadia, Z., and Wessling-Resnick M. (1995) Anal. Biochem. 224 547–556 [DOI] [PubMed] [Google Scholar]
- 30.Gomes, A. Q., Ali, B. R., Ramalho, J. S., Godfrey, R. F., Barral, D. C., Hume, A. N., and Seabra, M. C. (2003) Mol. Biol. Cell 14 1882–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, J., Liong, M., Zink, J. I., and Tamanoi, F. (2007) Small 3 1341–1346 [DOI] [PubMed] [Google Scholar]
- 32.Kato-Stankiewicz, J., Hakimi, I., Zhi, G., Zhang, J., Serebriiskii, I., Guo, L., Edamatsu, H., Koide, H., Menon, S., Eckl, R., Sakamuri, S., Lu, Y., Chen, Q. Z., Agarwal, S., Baumbach, W. R., Golemis, E. A., Tamanoi, F., and Khazak, V. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 14398–14403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo, W., Baluda, M. A., and Park, N. H. (1997) Oncogene 15 1143–1149 [DOI] [PubMed] [Google Scholar]
- 34.Zhu, X., Henry, C. E., and Kwon, O. (2005) Tetrahedron 61 6276–6282 [Google Scholar]
- 35.Zhu, X., Lan, J., and Kwon, O. (2003) J. Am. Chem. Soc. 125 4716–4717 [DOI] [PubMed] [Google Scholar]
- 36.Zhu, X., Henry, C. E., Wang, J., Dudding, T., and Kwon, O. (2005) Org. Lett. 7 1387–1390 [DOI] [PubMed] [Google Scholar]
- 37.Zhu, X., Schaffner, A., Li, R. C., and Kwon, O. (2005) Org. Lett. 7 2977–2980 [DOI] [PubMed] [Google Scholar]
- 38.Vogt, A., Sun, J., Qian, Y., Hamilton, A. D., and Sebti, S. M. (1997) J. Biol. Chem. 272 27224–27229 [DOI] [PubMed] [Google Scholar]
- 39.Sun, J., Qian, Y., Chen, Z., Marfurt, J., Hamilton, A. D., and Sebti, S. M. (1999) J. Biol. Chem. 274 6930–6934 [DOI] [PubMed] [Google Scholar]
- 40.Adnane, J., Bizouarn, F. A., Qian, Y., Hamilton, A. D., and Sebti, S. M. (1998) Mol. Cell. Biol. 18 6962–6970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olson, M. F., Paterson, H. F., and Marshall, C. J. (1998) Nature 394 295–299 [DOI] [PubMed] [Google Scholar]
- 42.Lackner, M. R., Kindt, R. M., Carroll, P. M., Brown, K., Cancilla, M. R., Chen, C., de Silva, H., Franke, Y., Guan, B., Heuer, T., Hung, T., Keegan, K., Lee, J. M., Manne, V., O'Brien, C., Parry, D., Perez-Villar, J. J., Reddy, R. K., Xiao, H., Zhan, H., Cockett, M., Plowman, G., Fitzgerald, K., Costa, M., and Ross-Macdonald, P. (2005) Cancer Cell 7 325–336 [DOI] [PubMed] [Google Scholar]
- 43.Pylypenko, O., Rak, A., Durek, T., Kushnir, S., Dursina, B. E., Thomae, N. H., Constantinescu, A. T., Brunsveld, L., Watzke, A., Waldmann, H., Goody, R. S., and Alexandrov, K. (2006) EMBO J. 25 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.