Abstract
The biosynthesis of 60 S ribosomal subunits in Saccharomyces cerevisiae requires Tif6p, the yeast homologue of mammalian eIF6. This protein is necessary for the formation of 60 S ribosomal subunits because it is essential for the processing of 35 S pre-rRNA to the mature 25 S and 5.8 S rRNAs. In the present work, using molecular genetic and biochemical analyses, we show that Hrr25p, an isoform of yeast casein kinase I, phosphorylates Tif6p both in vitro and in vivo. Tryptic phosphopeptide mapping of in vitro phosphorylated Tif6p by Hrr25p and 32P-labeled Tif6p isolated from yeast cells followed by mass spectrometric analysis revealed that phosphorylation occurred on a single tryptic peptide at Ser-174. Sucrose gradient fractionation and coimmunoprecipitation experiments demonstrate that a small but significant fraction of Hrr25p is bound to 66 S preribosomal particles that also contain bound Tif6p. Depletion of Hrr25p from a conditional yeast mutant that fails to phosphorylate Tif6p was unable to process pre-rRNAs efficiently, resulting in significant reduction in the formation of 25 S rRNA. These results along with our previous observations that phosphorylatable Ser-174 is required for yeast cell growth and viability, suggest that Hrr25p-mediated phosphorylation of Tif6p plays a critical role in the biogenesis of 60 S ribosomal subunits in yeast cells.
Eukaryotic translation initiation factor 6 (eIF6)3 was initially purified as a protein that can bind the 60 S ribosomal subunit and prevent its association with the 40 S ribosomal subunit (1–4). Based on this ribosomal subunit anti-association property, the protein was originally thought to be an initiation factor that functions to provide a pool of free ribosomal subunits required for initiation of protein synthesis (5). The protein was named eIF6, although a role in translation was not demonstrated in these earlier studies. To understand the function of this protein in translation, Si et al. (6) first cloned the human cDNA and then the yeast Saccharomyces cerevisiae gene (7) encoding functionally active eIF6, each of 245 amino acids. The two proteins are 72% identical. The yeast gene, designated TIF6, is a single copy gene that is essential for cell growth and viability (7). These properties of TIF6 allowed the construction of a conditional null allele by placing its expression under the control of the regulatable GAL10 promoter. Depletion of Tif6p in this yeast mutant strain inhibited the rate of in vivo protein synthesis (7). However, a more detailed analysis of the protein synthesis parameters in Tif6p-depleted cells showed that the reduced rate of protein synthesis was not due to a direct inhibition in initiation (7). Rather, the biogenesis of 60 S ribosomal subunits was severely inhibited. Similar observations were also reported by Sanvito et al. (8), who identified eIF6 from mammalian cells as a β4 integrin-interacting protein. Specifically, lack of Tif6p in yeast cells prevented the processing of pre-ribosomal RNA (pre-rRNA) to the mature 25 S and 5.8 S rRNAs, the constituents of the 60 S ribosomal particle (9). In agreement with these observations, Tif6p was found to be a constituent of a multiprotein assembly complex associated with 60 S pre-ribosomal particles (also known as the 66 S pre-ribosomal particles) in the nucleolus that are the intermediates in the biosynthesis of mature 60 S ribosomal subunits (7, 10).
In previous studies we have observed that in both mammalian and yeast cells, eIF6 (Tif6p) is phosphorylated, and we have identified casein kinase I α (CK Iα) as the protein kinase responsible for this modification in mammalian cell extracts (11). The sites of in vitro phosphorylation in mammalian eIF6 were identified as the serine residues at positions 174 (major site accounting for >90% of the total in vitro phosphorylation) and 175 (<10% phosphorylation). The serine residue at position 174 is present in a highly conserved consensus CK I sequence from yeast to mammals (11). Mutation of Ser-174 to alanine abolished phosphorylation of Tif6p in yeast cells by >75% and caused a loss of cell growth and viability (11). When both Ser-174 and Ser-175 were mutated to alanine, phosphorylation was virtually abolished (11). These observations suggested that phosphorylatable serine residues at Ser-174 and Ser-175 play an important regulatory role in the function of Tif6p in yeast cells. However, the protein kinase(s) responsible for phosphorylation of Tif6p in yeast has not been identified.
In the present work we have carried out molecular genetic and biochemical analyses to show that Hrr25p, an isoform of the budding yeast CK I (12) that is encoded by an essential gene HRR25, phosphorylates Tif6p at Ser-174 in vitro and in vivo. Tryptic phosphopeptide mapping shows that this is the only Tif6p site phosphorylated in yeast cells. We also show that a small but significant fraction of Hrr25p localizes along with Tif6p on 66 S pre-ribosomal particles. Furthermore, Hrr25p-mediated phosphorylation is also required for efficient processing of pre-rRNAs to form the mature rRNAs. These results suggest that constitutively active kinase Hrr25p plays an important regulatory role in coordinating the process of 60 S and 40 S ribosome biogenesis in yeast.
EXPERIMENTAL PROCEDURES
Media, Growth Conditions, and Genetic Methods—Yeast strains were grown at 30 °C in standard media as described previously (7). For pre-rRNA processing experiments using [methyl-3H]methionine pulse, synthetic complete medium lacking methionine and containing either 2% galactose (SGal) or 2% dextrose (SD) was used as the carbon source. These methionine-lacking media were designated SGal-Met and SD-Met, respectively. Yeast genetic methods as well as methods for isolation of plasmids and genomic DNAs, cloning, and bacterial transformation were carried out as described (13, 14).
Yeast Strains—The genotype of yeast strains used in this work are described in Table 1. The construction of yeast strains used in this study is as follows. For epitope-tagging of chromosomal genes at the 3′ ends of their open reading frames (ORFs), the PCR-based gene targeting method of Longtine et al. (15) was used. The construction of several other strains were as follows. For overexpression of hemagglutinin (HA)-tagged Hrr25p in yeast cells, the ORF of HRR25 was amplified by PCR from yeast genomic DNA using Pyrococcus DNA polymerase (Stratagene) using two primer sequences as follows; N terminus 5′-dccggatccatggacttaagagtaggaag-3′ having a BamHI overhang and C terminus 5′-dttgcggccgcttacaaccaaattgactggcca-3′ having a NotI overhang. The PCR product was used in a three-fragment ligation reaction with a NotI/SacI fragment derived from the vector pGEXYP1 and URA3-based CEN plasmid pRS316GAL digested at the BamHI and SacI sites. This reaction yielded the recombinant plasmid pGAL-HRR25-HA. The plasmid was transformed into a yeast strain KSY606 (Table 1) to generate a yeast strain, UBY616, which expressed HA-tagged Hrr25p when grown in a medium containing galactose as the carbon source. The strain PRY101 that expresses Myc-tagged Hrr25p from its genomic promoter and HA-tagged Tif6p from a CEN plasmid under the transcriptional control of its natural promoter was constructed by tagging the genomic copy of HRR25 of yeast strain KSY607 MATαtif6::HIS3, p[LEU2 TIF6-HA]) at its ORF 3′ end. In the strain KKY387 (16), a kind gift of Dr. Martha Cyert of Stanford University, a rapidly degradable form of Hrr25p (degron-tagged Hrr25p) is expressed from a galactose-inducible promoter in a plasmid, whereas the strain YPH499 is the isogenic parental wild-type strain in which Hrr25p is expressed from its own endogenous promoter (16). A chromosomal copy of TIF6 in these two strains was the Myc epitope tagged at the 3′ end to generate PRY102 and PRY103 strains, respectively. The strain PRY104 was constructed by tagging the genomic copy of TIF6 at its ORF 3′ end with a tandem affinity purification (TAP) cassette.
TABLE 1.
Yeast strains used
| Strain | Genotype | Reference or source |
|---|---|---|
| W303 α | MATα leu2-3,112 his3-1, 15 ade2-1 1 trp-1 ura3-1 can1-100 | 13 |
| KSY603 | MATα leu2-3,112 his3-1, 15 ade2-1 1 trp-1 ura3-1 can1-100 tif6::HIS3 p[URA3 GAL10::UbTIF6] | 7 |
| KSY606 | MATα leu2-3, 112 his3-1,15 ade2-1 trp1-1 ura 3-1 can 1-100 tif6::HIS3, p[TRP1 TIF6-myc] | 11 |
| KSY607 | MATα leu2-3, 112 his3-1,15 ade2-1 trp1-1 ura 3-1 can 1-100 tif6::HIS3, p[LEU2 TIF6-HA] | 11 |
| UBY616 | MATα leu2-3, 112 his3-1, 15 ade2-1 trp1-1 ura3-1 can 1-100 tif6::HIS3 p[TRP1myc TIF6] p[URA3 GAL10::HRR25-HA] | This work |
| YPH499 | MATa ura3-52 lys2-801 ade2-101 trp-Δ63 his3-Δ200 leu2-Δ1 | 16 |
| KKY387 | MATa ura3-52 lys2-801 ade2-101 trp-Δ63 his3-Δ200 leu2-Δ1 hrr25Δ::loxP-kanMX-loxP p[LEU2 GAL10::3HA-HRR25degron] | 16 |
| PRY101 | MATα leu2-3, 112 his3-1, 15 ade2-1 trp1-1 ura3-1 can 1-100 tif6::HIS3, p[LEU2 TIF6-HA], HRR25-myc9::TRP1 | This work |
| PRY102 | MATa ura3-52 lys2-801 ade2-101 trp-Δ63 his3-Δ200 leu2-Δ1 TIF6-myc13::HIS3 | This work |
| PRY103 | MATa ura3-52 lys2-801 ade2-101 trp-Δ63 his3-Δ200 leu2-Δ1 hrr25Δ::loxP-kanMX-loxPp[LEU2 GAL4::3HA-HRR25degron] TIF6-myc13::HIS3 | This work |
| PRY104 | MATα leu2-3,112 his3-1, 15 ade2-1 1 trp-1 ura3-1 can 1-100 TIF6-TAP::KANr | This work |
| PRY105 | MATα leu2-3,112 his3-1, 15 ade2-1 1 trp-1 ura3-1 can 1-100 tif6::HIS3 p[URA3 GAL10::UbTIF6] p[TRP1 TIF6-GFP] | This work |
| PRY106 | MATα leu2-3,112 his3-1, 15 ade2-1 1 trp-1 ura3-1 can 1-100 tif6::HIS3 p[URA3 GAL10::UbTIF6] p[TRP1 tif6-GFP(S174A,S175A)] | This work |
Isolation of Hrr25p and Yck1p—All operations were carried out at 0–4 °C unless otherwise stated. Exponentially growing UBY616 cells (100-ml culture) in which Hrr25p is expressed as a HA-tagged protein were harvested. The washed cells were suspended in 1 ml of lysis buffer (50 mm Tris-HCl, pH 7.5, 10 mm KCl, 1 mm DTT, 10 mm MgCl2, 10% glycerol, and a mixture of protease inhibitors), lysed by vortexing with glass beads, and then centrifuged. The supernatant was incubated with 20 μl of a 50% suspension of agarose for about 90 min with gentle mixing in a rotator followed by centrifugation to remove the agarose beads. The supernatant was then incubated with 20 μl of agarose-conjugated anti-HA antibody (Santa Cruz Biotechnology, Berkeley, CA) for about 3 h with gentle mixing in a rotator. Immunocomplexes bound to agarose beads were isolated by centrifugation and washed sequentially with 0.5 ml of a immunoprecipitation buffer containing 20 mm Tris-HCl, pH 7.5, 2 mm DTT, 5 mm MgCl2, 1% Triton X-100, and 500 mm KCl (twice) and, second, with lysis buffer (once). HA-tagged Hrr25p bound to agarose beads was recovered by centrifugation and then suspended in 10 μl of the kinase buffer (20 mm Tris-HCl, pH 7.5, 100 mm KCl, 5 mm DTT, and 10 mm MgCl2) and used for phosphorylation of Tif6p.
Recombinant GST-tagged Hrr25p was isolated from Escherichia coli BL21/pLys cells expressing GST-Hrr25p from an expression plasmid KK196 (16), a kind gift of Dr. Martha Cyert. GST-Hrr25p was purified from lysates of these bacterial cells by affinity purification from glutathione-Sepharose beads as described by Kafadar et al. (16).
His7-Yck1p was isolated from A2085 yeast cells harboring the plasmid pBM4536 (17), a kind gift of Dr. Mark Johnston of Washington University School of Medicine, St. Louis, MO. Purification was carried out using Ni2+-NTA-agarose column as described (17).
Preparation of Hrr25p-depleted Yeast Cell-free Extracts—Exponentially growing cells (A600 = 0.2) of wild-type YPH499 (+Hrr25p) and the isogenic conditional mutant strain KKY387 in YPGal media were each filtered through a sterile 0.2-μm filter and then suspended in YPD media and allowed to grow for about 8 h (A600 of about 0.6) until there was no detectable Hrr25p in the mutant strain as determined by Western blot analysis of cell lysates. Harvested cells were lysed in buffer containing 20 mm Tris-HCl, pH 7.5, 100 mm KCl, 1 mm DTT, 10 mm MgCl2, and phosphatase inhibitors (50 mm NaF, 50 mm sodium molybdate) and used as a source of protein kinase for Tif6p phosphorylation. It should be noted that the growth of KKY387 cells begins to slow down 12–14 h after transfer to SD medium.
Coimmunoprecipitation of Tif6p with Hrr25p—A 100-ml culture of exponentially growing PRY101 yeast cells was harvested, washed twice with ice-cold water, and then suspended in 500 μl of a coimmunoprecipitation buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10% glycerol, 1 mm DTT, 10 mm MgCl2, and an aliquot of a mixture of protease inhibitors). Cells were disrupted by vortexing with glass beads and centrifuged. The glass beads were washed once with 500 μl of the coimmunoprecipitation buffer, and the supernatant obtained after centrifugation was mixed with the initial supernatant. The combined supernatant (about 1 ml) was incubated with 10 μl of agarose-conjugated anti-HA antibody (Santa Cruz) for about 4 h with gentle mixing at 4 °C. Immunocomplexes bound to agarose beads were isolated by centrifugation and washed with 0.5 ml of the coimmunoprecipitation buffer (twice). Proteins bound to the immunocomplexes were eluted by suspending the beads in 30 μl of 125 mm Tris-HCl, pH 6.8, 2% SDS, and 10% glycerol and incubating the suspension in a boiling water bath for 5 min. After centrifugation, the supernatant was treated with 100 mm DTT, cooled, and then subjected to SDS-PAGE (15% gel). The resolved proteins were transferred electrophoretically to a polyvinylidene difluoride membrane. The washed membrane was then analyzed by Western blotting using appropriate antibodies.
Assay for in Vitro Phosphorylation of Tif6p—Phosphorylation of Tif6p was carried out in reaction mixtures (30–50 μl each) containing 20 mm Tris-HCl, pH 7.5, 100 mm KCl, 5 mm DTT, 10 mm MgCl2, 100 μm [γ-32P]ATP (10,000 cpm/pmol), 15 pmol of recombinant His6-Tif6p, a mixture of phosphatase inhibitors (50 mm NaF, 50 mm sodium molybdate), and the indicated amounts of either yeast cell-free extracts or the purified kinase protein. In some experiments the nuclear or cytoplasmic fractions of yeast cell-free extracts replaced the total yeast cell-free extracts as the source of kinase. After incubation at 30 °C for 30 min, each reaction was treated with 450 μl of buffer I (20 mm potassium phosphate, pH 7.8, 0.5 m NaCl, 6 mm imidazole-HCl, pH 8.0, and 10% glycerol) followed by the addition of 15 μl of equilibrated Ni-NTA-agarose bead suspension. The solutions were mixed gently at 4 °C for 2 h then centrifuged. The agarose beads were washed twice with 1 ml of buffer I containing 50 mm imidazole. The washed beads containing bound His6-Tif6p were suspended in 50 μl of SDS-loading buffer, boiled, and then subjected to SDS-PAGE (either 15% gel or 4–15% gradient gel as indicated). For reactions using the purified kinase, the incubated reaction mixtures were directly treated with SDS-loading buffer, boiled, and then subjected to SDS-PAGE. In each case, after SDS-PAGE, the resolved proteins were transferred to a polyvinylidene difluoride membrane. The membrane was analyzed by autoradiography to determine the extent of phosphorylation of Tif6p. Subsequently, the recovery of Tif6p in each reaction was determined by Western blot analysis using anti-His antibody.
Phosphopeptide Mapping—The procedure for the preparation of tryptic phosphopeptides from immunocomplexes of in vivo 32P-labeled Tif6p was similar to that described previously (18). For two-dimensional separation of tryptic phosphopeptides by thin-layer chromatography, the samples were spotted 4 cm from the bottom edge of a 20 × 20-cm thin layer cellulose plate (Fisher) and electrophoresed at pH 1.9 in 2.5% formic acid, 17.8% acetic acid for 20 min at 16 °C at 1000 V. The plates were air-dried and then subjected to ascending chromatography in pyridine:acetic acid:1-butanol:H2O (50:15:75:60). The air-dried plates were subsequently subjected to autoradiography using phosphorimaging.
Tandem Affinity Purification and Identification of Tif6p-associated RNA and Protein Components—The genomic copy of TIF6 of the yeast strain W303α (Table 1) was C-terminal-tagged with the TAP-tag cassette using PCR-based gene targeting methods (15). The resulting yeast strain, designated PRY104 (Table 1), expresses TAP-tagged Tif6p from its genomic promoter. This strain grew at a rate similar to the untagged wild-type strain. Immunoblot analysis of extracts of this strain showed a protein band whose size corresponded to Tif6p-TAP fusion protein. Furthermore, the Tif6p-TAP fusion protein sedimented in sucrose gradients primarily at the 60 S–66 S region (data not shown). Cells expressing Tif6p-TAP were grown in YPD at 30 °C to an A600 of about 1.0, and cell-free extracts were subjected to tandem affinity purification as described by Rigaut et al. (19). RNAs that co-purified with the Tif6p-TAP complex(s) were extracted from the final eluate with phenol/chloroform/isoamyl alcohol, precipitated with ethanol, and analyzed by Northern blot by using 32P-labeled deoxyoligonucleotide probes (9) that specifically detect each pre-rRNA species. Primer extension was carried out as described by Venema et al. (20). The probe used for detection of 35 S and 25.5 S pre-rRNAs by primer extension was (5′-ACACGCTGTATAGAGACTAGGC-3′) and (5′-CGCCTAGACGCTCTCTTCTTA-3′), respectively, as described by these investigators (20).
Other Materials and Methods—Recombinant yeast His6-Tif6p was purified from BL21 (DE3) cells carrying the ORF of yeast Tif6p in the plasmid pRSET-A by following a procedure similar to that described previously for the purification of His6-tagged mammalian eIF6 (11). The procedure involved affinity purification from a Ni-NTA column followed by gradient elution from an fast protein liquid chromatography-Mono Q column. The final preparation was >95% pure as judged by SDS-polyacrylamide gel electrophoresis followed by Coomassie Blue staining.
Nuclear and cytoplasmic extracts from exponentially growing yeast cells were prepared by an adaptation of the method of Evans and Engelke (21). The procedures used for pulse-chase labeling of pre-rRNAs with [methyl-3H]methionine and subsequent analysis of RNA by gel electrophoresis were as described (9). RNA was isolated from each immunocomplex by a phenol/chloroform extraction procedure as described (9). Purified yeast eIF5 (Tif5p) was a kind gift of Drs. Michael Acker and Jon Lorsch of the Johns Hopkins University of Medicine, Baltimore, MD. Anti-Pgk1p antibodies were purchased from Molecular Probes, whereas anti-Nop1p antibodies were obtained from EnCor Biotechnology Inc. Anti-Tcm1p (L3) antibodies and Northern probes JW71 and JW499 were kind gifts of Dr. Jonathan Warner of this institution.
RESULTS
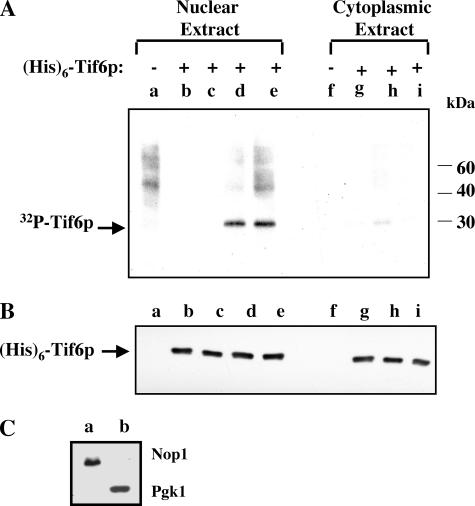
Hrr25p Phosphorylates Tif6p in Vitro—To identify the kinase responsible for phosphorylation of Tif6p in vitro and to characterize the phosphorylation reaction, yeast cell-free extracts were fractionated into nuclear and cytosolic fractions. The validity of the fractionation was verified by using antibodies for 3-phosphoglycerate kinase, Pgk1p (a marker for cytoplasmic proteins), and Nop1p (a marker for nucleolar proteins) (Fig. 1, panel C). Using bacterially expressed recombinant His6-Tif6p as the substrate and [γ-32P]ATP as the phosphoryl donor and increasing concentrations of either the nuclear or cytoplasmic extract as the source of the kinase, we observed that Tif6p was readily phosphorylated by the nuclear extract (Fig. 1, panel A, lanes d and e). In contrast, under the same conditions the relative efficiency of phosphorylation of Tif6p by the cytosolic fraction was low (Fig. 1A, lane h). A higher concentration of the cytosolic fraction was inhibitory in the phosphorylation reaction (Fig. 1A, lane i). The presence or absence of phosphatase inhibitors did not have any effect on the extent of phosphorylation (data not shown). Furthermore, the amount of His6-Tif6p recovered from each reaction was similar (Fig. 1, panel B).
FIGURE 1.
Tif6p is phosphorylated by a nuclear kinase in yeast cells. A, the preparation of reaction mixtures (50 μl each) containing either yeast nuclear or cytoplasmic extracts and the subsequent analysis of phosphorylation of Tif6p were as described under “Assay for In Vitro Phosphorylation of Tif6p” under “Experimental Procedures.” Where indicated, 15 pmol of recombinant His6-Tif6p were added to the reactions. The amount of nuclear and cytoplasmic extracts added were as follows: lane a, 8 μg; lane b, 1 μg; lane c, 2 μg; lane d, 4 μg; lane e, 8 μg; lane f, 20 μg; lane g, 10 μg; lane h, 20 μg; lane i, 50 μg. B, the recovery of His6-Tif6p in each reaction was determined by Western blot analysis using anti-His antibodies. C, nuclear (lane a) and cytoplasmic extracts (lane b) (30 μg each) were subjected to Western blot analysis using anti-Nop1p and anti-Pgk1p antibodies, respectively.
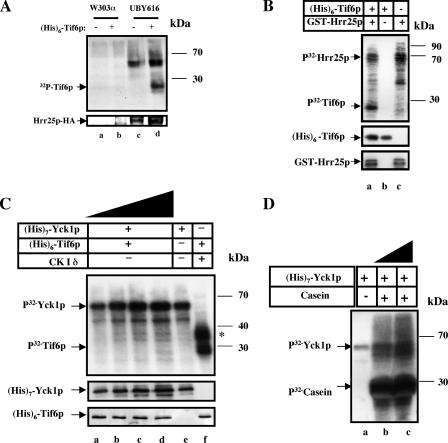
It has been reported previously that mammalian eIF6 is phosphorylated at Ser-174 by CK Iα (11). In view of our observation (11) that this serine residue is present in a highly conserved consensus CK I sequence (12) between yeast and mammals, we initially investigated whether a particular isoform(s) of yeast CK I can phosphorylate Tif6p in vitro. In S. cerevisiae there are four different isoforms of CK I, each encoded by a separate gene, YCK I, YCK 2, YCK 3, and HRR25 (12, 22–24). Among these isoforms, Hrr25p localizes throughout the cell (16), whereasYck1p, Yck2p, and Yck3p are prenylated and tightly bound to the plasma membrane (24, 25). To show if any isoform of yeast CK I is able to phosphorylate Tif6p in vitro, we purified Hrr25p, Yck1p, and Yck2p and used these purified proteins to phosphorylate recombinant Tif6p (Fig. 2). Tif6p was readily phosphorylated by both immunopurified Hrr25p (Fig. 2A) and by bacterially expressed recombinant GST-Hrr25p (Fig. 2B). In contrast, the other purified CK I isoform, Yck1p, was unable to phosphorylate Tif6p (Fig. 2C). Similar observations were made using β-galactosidase fusion protein of Yck2p (22, 23) as the source of the kinase (data not shown). Both Yck1p (Fig. 2D, lanes b and c) and Yck2p (data not shown) were, however, able to phosphorylate casein, a model in vitro substrate, for all isoforms of CK I, indicating that these kinases were enzymatically active. It should be noted that with all the isoforms of CK I tested, an additional higher molecular weight band (apparent Mr = 55,000–58,000) was observed in all reactions (Fig. 2). Presumably, this band represents the autophosphorylated form in each case. All the isoforms of yeast CK I including Hrr25p are known to autophosphorylate and migrate in SDS-PAGE with an apparent Mr of about 55,000–58,000 (12, 26).
FIGURE 2.
Specificity of in vitro phosphorylation of Tif6p by Hrr25p, an isoform of yeast casein kinase I. In A, B, and C, 15 pmol of His6-Tif6p were incubated with [γ-32P]ATP and a yeast CK I isoform in standard phosphorylation reaction mixtures. The source of kinase was as follows. A, immunopurified HA-tagged Hrr25p from UBY616 cells and a parallel mock immunocomplex isolated from an isogenic wild-type strain w303α in which Hrr25p was not epitope tagged, as indicated. B, purified recombinant GST-Hrr25p (10 pmol). C, increasing concentrations of His7-Yck1p, purified from yeast cells were added as follows. Lane a, 2.5 pmol; lane b, 5 pmol; lane c, 10 pmol; lane d, 15 pmol; lane e, 5 pmol; mammalian CK1 (New England Biolabs) was used as a positive control in lane f. D, purified His7-Yck1p (7.5 pmol in lanes a and b and 15 pmol in lane c) was used to phosphorylate 4.7 μg of casein that replaced His6-Tif6p as the kinase substrate. In each panel, omission of either the substrate or the kinase is indicated. After incubation at 30 °C for 30 min, the reactions were subjected to SDS-PAGE, and the resolved proteins were transferred to a polyvinylidene difluoride membrane. The membrane was subjected to autoradiography. The slower migrating band (apparent Mr = 56,000) present in reactions containing no substrate presumably represents autophosphorylated Hrr25p or Yck1p as indicated. The recovery of Hrr25p (A and B) or Yck1p (C) determined by Western blot analysis is shown in the lower panel of each experiment. The asterisk in C indicates the autophosphorylated product of mammalian CK Iδ (apparent Mr = 35,000).
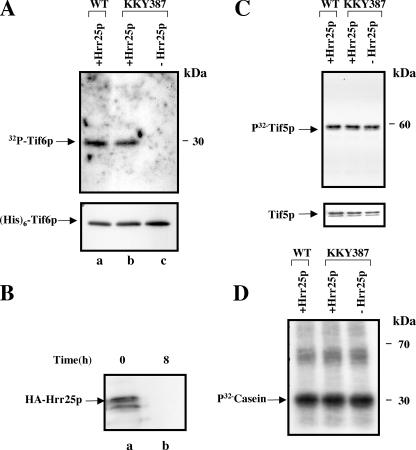
Tif6p Is Phosphorylated by Hrr25p in Yeast Cell Lysates—Additional confirmation that Hrr25p is indeed involved in phosphorylation of Tif6p in yeast cells came from comparison of the ability of the cell-free extracts isolated from two isogenic yeast strains YPH499 and KKY387 that differ in their conditional expression of Hrr25p (Table 1). In the strain KKY387, the genomic copy of HRR25 was deleted, and the essential function of HRR25 provided from a 2-μm plasmid in which Hrr25p was expressed from a galactose-inducible GAL10 promoter as an N-terminal-ubiquitinylated HA-tagged fusion protein (16). The presence of this N-terminal module (degron) results in rapid degradation of Hrr25p when the strain is grown in YPD (glucose) medium. In contrast, in the isogenic wild-type strain YPH499, Hrr25p is expressed from its genomic promoter as an untagged protein.
To investigate the role of Hrr25p in the phosphorylation of Tif6p, exponentially growing cultures of YPH499 and KKY387 growing in YPGal(galactose) medium were each divided into two parts. One part of each culture was maintained in YPGal medium, whereas the other part was shifted to a YPD medium and allowed to grow for 8 h to deplete Hrr25p from KKY387 cells (Fig. 3, panel B). Cell-free extracts prepared from each condition were then used as the source of protein kinase for in vitro phosphorylation of Tif6p (Fig. 3). Depletion of Hrr25p in the conditional mutant strain KKY387 resulted in severe inhibition of 32P incorporation into Tif6p (Fig. 3A, compare lanes b and c). Longer exposures failed to show any band corresponding to Tif6p in the Hrr25p-depleted cell extract lane. In contrast, extracts prepared from the isogenic wild-type strain YPH499, which contains the wild-type copy of HRR25, were active in phosphorylation of Tif6p (Fig. 3A, lane a) when subjected to the same YPGal to YPD media shift. It should be noted that this effect on Tif6p phosphorylation by Hrr25p is specific since phosphorylation of either yeast eIF5 (Tif5p) that is phosphorylated by casein kinase II (18) or the model substrate casein, which is known to be phosphorylated by all the isoforms of CK I as well as by CK II, is not significantly affected by the depletion of Hrr25p from yeast cells (Fig. 3, C and D, respectively). These results show that Hrr25p phosphorylates Tif6p in yeast cell lysates.
FIGURE 3.
Hrr25p phosphorylates Tif6p in yeast cell lysates. A, recombinant His6-Tif6p (15 pmol) was incubated in standard phosphorylation reaction with [γ-32P]ATP (10,000 cpm/pmol) and 250 μg of yeast extracts prepared from either wild-type (YPH 499) cells expressing Hrr25p (lane a) or the conditional mutant KKY387 cells grown either in SGal medium (lane b) expressing Hrr25p or in SD medium (lane c) depleted of detectable Hrr25p. Formation of 32P-labeled Tif6p was subsequently analyzed by SDS-PAGE followed by autoradiography as described under “Experimental Procedures” (upper panel). Recovery of His6-Tif6p in each reaction was estimated by Western blot analysis using anti-His antibody (lower panel). B, Western blot analysis of HA-Hrr25p of cell lysates prepared from KKY387 cells grown in YPGal medium (lane a) and following the shift to YPD medium and grown for 8 h (lane b). In lane a, the faster migrating band presumably represents the degradation product of Hrr25p. C, Tif5p (yeast eIF5), one of the known CK II protein kinase substrates (15 pmol), replaced His6-Tif6p as the kinase substrate. D, the model CK I and CK II substrate, casein (4.7μg), replaced His6-Tif6p as the kinase substrate.
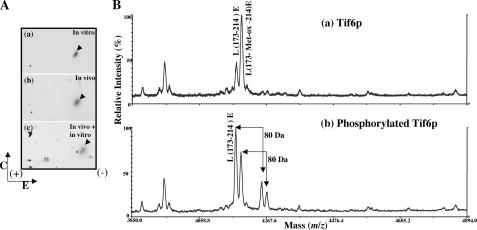
Hrr25p Phosphorylates Tif6p in Vivo and in Vitro at the Same Site—The sites in Tif6p phosphorylated in vivo and in vitro were determined by two-dimensional phosphopeptide mapping. 32P-Labeled Tif6p isolated from KSY607 yeast cells (Table 1) was digested with excess TPCK-trypsin, and the resulting phosphopeptides were separated by a combination of high voltage electrophoresis and thin layer chromatography. One major phosphopeptide was detected (Fig. 4A, panel b), suggesting that phosphorylation of Tif6p occurred primarily within one tryptic peptide. In a separate experiment, phosphoamino acid analysis of the same sample of Tif6p showed that serine was the only amino acid residue phosphorylated (data not shown). When recombinant His6-Tif6p was phosphorylated in vitro by immunopurified HA-tagged Hrr25p and the resulting 32P-labeled Tif6p was subjected to two-dimensional tryptic phosphopeptide mapping, the same distinct phosphopeptide observed in vivo was resolved (Fig. 4A, panel a). When the tryptic peptides obtained from 32P-labeled Tif6p isolated from yeast cells were mixed with those obtained from Tif6p labeled in vitro with immunopurified HA-Hrr25p, an identical phosphopeptide was resolved (Fig. 4A, panel c). These results suggest that Hrr25p phosphorylates Tif6p in vivo in yeast cells and that phosphorylation occurs at one or more serine residues present in the same tryptic peptide.
FIGURE 4.
Identification of the phosphopeptide generated from Tif6p labeled with 32P in vivo in yeast cells and in vitro by purified Hrr25p and determination of the phosphorylation site(s). A, HA-tagged Tif6p, labeled with [32P]orthophosphate in yeast cells, was isolated by immunoprecipitation and subjected to SDS-PAGE followed by electrotransfer to a polyvinylidene difluoride membrane. 32P-Labeled Tif6p was located by autoradiography, excised from the membrane, and treated with TPCK-trypsin. The isolated tryptic peptides (100 cpm each) were analyzed by two-dimensional phosphopeptide mapping as described under “Experimental Procedures.” a, 32P-labeled Tif6p phosphorylated in vitro by immunoprecipitated Hrr25p kinase; b, 32P-labeled Tif6p isolated from yeast cells by immunoprecipitation; c, aliquots of 32P-labeled tryptic peptides obtained from a and b were mixed (50 cpm from each panel). The direction of electrophoresis (E) and chromatography (C) are indicated. The sample application origin is indicated by a + sign within the chromatogram. B, MALDI-TOF mass spectra of the Glu-C digested unphosphorylated Tif6p (a) and phosphorylated Tif6p (b). Unphosphorylated and phosphorylated Tif6p were excised from the gel, reduced with 10 mm DTT, alkylated with 55 mm iodoacetamide, and then digested with Glu-C at room temperature overnight. The digestion products were analyzed by MALDI-TOF to identify the phosphopeptides with a PerSeptive MALDI-TOF DE-STR mass spectrometer. In a two ions were observed at 4173.8 and 4189.8 Da corresponding to the mass (m/z) of peptide L173–214E and its oxidized form L173-Met-ox-214E. Two ions corresponding to phosphorylated L173–214E and L173-Met-ox-214E at 4254.2 and 4270.2 Da were observed only from the Glu-C digestion of phosphorylated Tif6p sample in (b).
To identify the peptide(s) containing the phosphorylated serine residues, recombinant Tif6p was maximally phosphorylated in vitro with the purified kinase, and the 32P-labeled Tif6p was isolated by SDS-PAGE. Gel slices containing either the phosphorylated or unphosphorylated Tif6p (50 pmol each) were reduced, alkylated, and digested with Glu-C protease, which cleaves at the C termini of glutamic acid residues of the protein. Each digestion product was analyzed by MALDI-TOF with a Perseptive MALDI-TOF DE-STR mass spectrometer. From Glu-C digestion products of unphosphorylated Tif6p, two ions were observed at 4173.8 and 4189.8 Da corresponding to masses of the peptide Leu-173–Glu-214 and its methionine residue oxidized-form, respectively. In contrast, from the digestion products of phosphorylated Tif6p, two new ions at 4254.2 and 4270.2 Da corresponding to the addition of a phosphate onto the peptide Leu-173–Glu-214 and its methionine residue-oxidized form, respectively, were observed. These two ions were absent in the products obtained from the digests of the unphosphorylated Tif6p (Fig. 4B, compare the upper and lower panels).
To determine the phosphorylation site(s), the products of Glu-C digestion were further cleaved by trypsin, resulting in the formation of two peptide ions 1666.99 and 1747.01 corresponding to the peptide Leu-173–Arg-188 and its phosphorylation form. This result demonstrates that the phosphorylation site is either Ser-174 or on Ser-175. Using a combination of chemical derivation that converts the phosphoserine-containing peptides to propanylcysteine-containing peptides and tandem mass spectrometry, we identified that Ser-174 was the major phosphorylation site (data not shown).
These results are in agreement with our previous observation (11) that mutation of Ser-174 to alanine leads to >75% abolition of phosphorylation of Tif6p in vivo in yeast cells and caused loss of cell growth and viability. In contrast, mutation of Ser-175 alone inhibited phosphorylation only marginally (about 10–15% inhibition) and did not cause loss of viability of yeast cells (11). Thus, failure to detect phosphorylation at Ser-175 by mass spectrometry may be due to inefficient phosphorylation at this site. Alternatively, and perhaps more likely, in wild-type Tif6p, Ser-174, which is in the consensus CK I site, is the only site that is phosphorylated by Hrr25p. When this site is not phosphorylated, the adjacent Ser-175 is inefficiently phosphorylated.
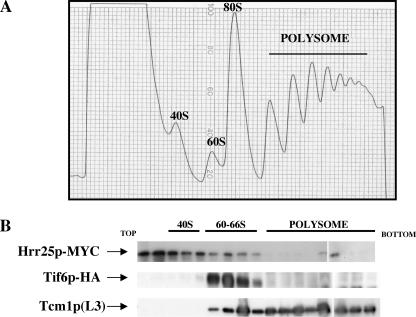
Hrr25p and Tif6p Bind to the 66 S Preribosomal Particles—It has been reported previously that Hrr25p is stably bound to both the 90 S and the pre-40 S particles (27) and is required for the synthesis of mature 40 S ribosomes (28). In contrast, Tif6p is known to be primarily associated with the 60 S-66 S ribosomal particles (7, 10). In view of our observation that Hrr25p phosphorylates Tif6p both in vitro and in vivo, the question naturally arises of whether a stable interaction of Tif6p and Hrr25p occurs on any of the pre-ribosomal particles. For this purpose, we constructed a strain PRY101 (Table 1) in which Tif6p was expressed as a HA-tagged protein, whereas Hrr25p was expressed as a Myc-tagged protein from their respective genomic loci. Cell-free extracts of exponentially growing culture of this strain were subjected to sucrose density gradient centrifugation. Polysome-ribosome fractions (Fig. 5, panel A) were then analyzed for the association of the HA-Tif6p and Myc-Hrr25p by Western blot analysis. As shown in Fig. 5, panel B, whereas Tif6p was localized primarily in the 60 S-66 S region as reported previously (7, 10), Hrr25p was localized mainly at the top of the gradient as well as in the 40 S region in agreement with the results previously reported (27). However, a small fraction of Hrr25p appeared to sediment in the 60 S-66 S region along with Tif6p.
FIGURE 5.
Binding of Myc-Hrr25p and HA-Tif6p to ribosomal particles. A, cell-free extracts prepared from PRY101 cells expressing Myc-tagged Hrr25p and HA-tagged Tif6p were loaded onto 11 ml of 7–47% sucrose density gradient and subjected to centrifugation as described previously (7). B, the Myc-tagged Hrr25p and HA-tagged Tif6p in the gradient fractions were detected by Western blotting using anti-Myc antibodies and anti-HA-antibodies, respectively. The positions of free 60 S particles as well as of 80 S monosomes and polysomes were detected by Western blotting using anti-Tcm1p (60 S ribosomal protein L3) antibodies.
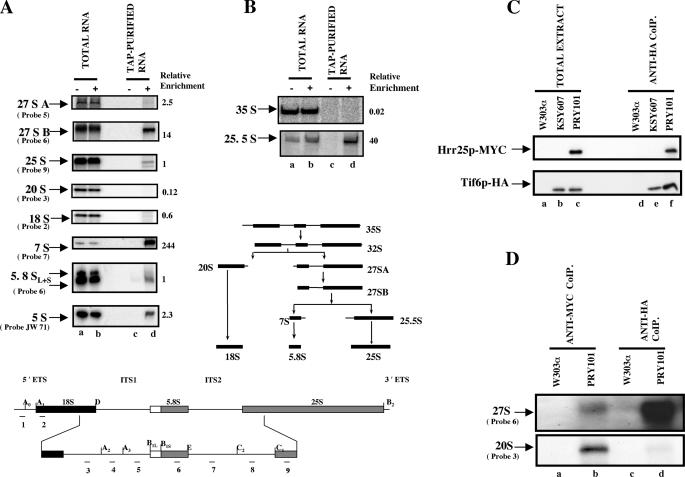
To determine the ribosomal particles on which Tif6p is associated, we used TAP protocol (19) to purify proteins and pre-rRNA(s) associated with the Tif6p-TAP purified complex from lysates of PRY104 yeast cells expressing Tif6p-TAP. The RNA that copurified with the Tif6p-TAP complex was subjected to Northern and primer extension analysis followed by phosphorimaging quantification. The RNA isolated from whole-cell extracts and untagged strains was also analyzed as controls. The results presented in Fig. 6, A and B, show that there was a selective enrichment of 27 SB, 25.5 S, and 7 S pre-rRNAs in the Tif6p-TAP-purified complex. However, no 35 S pre-ribosomal RNA was detected in the purified complex (Fig. 6, panel B). Based on the relative enrichment of 27 SB, 25.5 S and 7 S pre-rRNAs, we suggest that Tif6p is predominantly associated with two late 66 S pre-ribosomal particles, one containing 27 SB pre-rRNA and the other with more mature pre-60 S ribosomal particles containing 25.5 S and 7 S pre-rRNA(s). However, the absence of 35 S pre-rRNA(s) in the Tif6p-TAP associated complex suggests that Tif6p is not present in the 90 S particle (Fig. 6B).
FIGURE 6.
Association of Tif6p and Hrr25p with pre-66 S ribosomal particles. The RNA isolated from the TAP-Tif6p-purified complex as well as from whole cell-free extracts (as described under “Experimental Procedures”) was subjected to Northern analysis (A) using 32P-labeled deoxyoligonucleotide probes complementary to different regions of 35 S pre-rRNA, shown in the lower panel of A and B, that specifically detects each pre-rRNA intermediate. B, primer extension analysis was carried out to detect 35 S and 25.5 S pre-rRNAs. +, cells expressing TAP-tagged Tif6p; –, cells expressing untagged wild-type cells. A schematic diagram of the processing intermediates of 35 S pre-rRNA to mature 25 S and 18 S rRNAs is also shown. After autoradiography by phosphorimaging, the band intensity of each RNA species was calculated using the ImageQuant software. Relative enrichment of each species was calculated as described by Harnpicharnchai et al. (10). This consisted of initially calculating the ratio of band intensity of each pre-rRNA species in the TAP-purified fractions to that for the same RNA species in total RNA fractions and then dividing this ratio by the ratio obtained for 25 S rRNA as described in Harnpicharnchai et al. (10). It should be noted that because the 35 S and 25.5 S pre-rRNAs were assayed by primer extension whereas other pre-rRNA species were assayed by Northern analysis, their relative enrichments cannot be directly compared. C, Hrr25p and Tif6p coimmunoprecipitate with the 66 S preribosomal particles. Cell-free extracts prepared from W303α (lane a), KSY607 expressing HA-tagged Tif6p (lane b), and PRY101 (lane c), expressing both HA-Tif6p and Myc-Hrr25p, were immunoprecipitated with anti-HA antibodies under conditions of coimmunoprecipitation as described under “Experimental Procedures.” The washed immunocomplexes (Anti-HA CoIP) as well as aliquots of each cell extract (Total extract), before immunoprecipitation, were analyzed by Western blotting using anti-HA antibodies to detect HA-tagged Tif6p (lower panel) or anti-Myc antibodies to detect Myc-tagged Hrr25p (upper panel). D, extracts from PRY101 and W303α cells were immunoprecipitated under conditions of coimmunoprecipitation with either anti-Myc antibodies (lanes a and b) to precipitate Myc-tagged Hrr25p-containing complexes or with anti-HA antibodies (lanes c and d) to precipitate HA-Tif6p-containing complexes. Each RNA sample, isolated from the washed beads by phenol/chloroform extraction, was then analyzed by Northern blotting using an appropriate DNA probe that specifically detects 27 SA+B pre-rRNAs (Probe 6) or 20 S pre-rRNA (Probe 3).
To determine whether Hrr25p could be associated with the 66 S pre-ribosomal particles, we investigated whether Tif6p and Hrr25p can be coimmunoprecipitated from cell extracts. For this purpose, extracts of PRY101 expressing HA-tagged Tif6p and Myc-tagged Hrr25p were immunoprecipitated with anti-HA antibody. Analysis of the immunocomplex by SDS-PAGE followed by immunoblotting with anti-HA and anti-Myc antibodies demonstrated that both Tif6p and Hrr25p were present in the immunocomplex (Fig. 6C, lane f). Similar results were obtained when the coimmunoprecipitation reaction was carried out with anti-Myc antibodies (data not shown). When the RNA samples isolated from the immunocomplex, obtained using either anti-HA or anti-Myc antibodies, were analyzed by Northern blot hybridization using appropriate DNA probes, the presence of 27 S pre-rRNA was readily detected (Fig. 6D, lanes d and b, respectively). These results indicate that Hrr25p that was coimmunoprecipitated with Tif6p was associated with the 66 S pre-ribosomal particles. In contrast, whereas 20 S pre-rRNA was detected when Hrr25p was immunoprecipitated with anti-Myc antibodies, no 20 S pre-rRNA was detected in the immunocomplex precipitated with HA-Tif6p (Fig. 6D, lanes b and d), indicating that unlike Hrr25p, Tif6p does not associate with the pre-40 S ribosomal particles. It should be noted that the amount of 27 S pre-rRNA immunoprecipitated with HA-tagged Tif6p was significantly higher than that immunoprecipitated with Myc-tagged Hrr25p. These results suggest that although a majority of cellular Tif6p is associated with 66 S preribosomal particles, only a small fraction of Hrr25p associates with these 66 S particles at any one time.
Hrr25p Is Required for Pre-rRNA Processing and Consequently for Ribosome Biogenesis—An important function of Tif6p in yeast cells is its association with the 66 S preribosomal particles (7, 10) and its participation in the processing of 27 S pre-rRNA to mature 25 S and 5.8 S rRNAs (9). In view of our observations presented here that Hrr25p is the kinase responsible for phosphorylating Tif6p in yeast cells, we investigated whether depletion Hrr25p, which should give rise to unphosphorylated Tif6p in yeast cells, inhibited the processing of pre-rRNA.
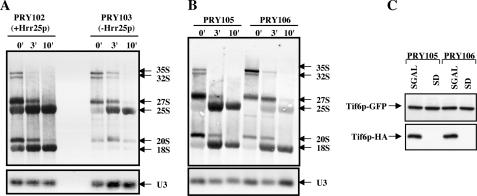
For this purpose exponentially growing cultures of the yeast strain PRY103 ([hrr25Δ::loxP-KanMX-loxP p[LEU2 GAL10::3HA-HRR25 degron TIF6-myc13::H1S3]) in SGal-Met medium containing a conditional Hrr25p expression system and the corresponding isogenic wild-type strain PRY102 ([HRR25 TIF6-myc13::H1S3]) in SGal-Met media were each transferred to SD-Met (glucose) medium, and the cells were allowed to grow for 8 h to deplete Hrr25p from the PRY103 cells. Cells were then pulse-labeled with [3H-methyl] methionine for 2.5 min and chased for 3 and 10 min with an excess of unlabeled methionine. [Because 25 S and 18 S rRNAs are highly and specifically methylated, the processing of pre-rRNAs can be readily monitored by labeling with [methyl-3H]methionine (29)]. Analysis of the total 3H-RNA samples isolated from each batch of cells by formaldehyde agarose gel electrophoresis followed by fluorography showed that in the wild-type PRY102 cells, the 35 S pre-rRNA and the 27 S and 20 S processing intermediates were rapidly chased into mature 25 S and 18 S rRNAs, as expected (Fig. 7, Panel A). In contrast, in Hrr25p-depleted mutant PRY103 cells, the rates of formation of all pre-rRNA intermediates were slower. There was a significant decrease in the conversion of 35 S pre-rRNA to 27 S and 20 S pre-rRNAs and the formation of 18 S mature rRNA was severely inhibited. This observation is in agreement with that of Schafer et al. (28) who reported that Hrr25p-mediated phosphorylation of several pre-40 S ribosomal proteins is essential for the formation of 18 S rRNA and consequently for 40 S ribosome biosynthesis. More importantly, in these Hrr25p-depleted cells, the formation of mature 25 S rRNA was also inhibited, albeit to a lesser extent than that of 18 S rRNAs (Fig. 7, Panel A). One striking aspect is that in Hrr25p-depleted cells, the processing of 35 S pre rRNA leads to significant loss of the RNA rather than the production of 27 S, 18 S and 25 S mature RNA species suggesting degradation of the unprocessed pre-RNAs that presumably accumulates in the absence of Hrr25p. Similar degradation of unprocessed pre-rRNAs were observed previously (9).
FIGURE 7.
Effect of lack of phosphorylation of Tif6p on its ability to function in pre-rRNA processing. A, the haploid yeast strains PRY102 (HRR25 TIF6-myc13::HIS3) and PRY103(hrr25Δ::loxP-kanMX-loxPp[LEU2 GAL10::3HA-HRR25degron]TIF6-myc113::HIS3), expressing the wild-type and the conditional mutant alleles of HRR25, respectively, were each grown at 30 °C in SGal-Met medium until the A600 reached 0.2. Each culture was then transferred to SD-Met (glucose) medium and allowed to grow for about 8 h to an A600 = 0.6. Under these conditions, Hrr25p was no longer detectable in cell lysates prepared from PRY103 cells (data not shown). Each batch of cells (5-ml culture) was pulse-labeled with 300 μCi of [methyl-3H]methionine for 2.5 min and then chased for 0, 3, and 10 min with an excess of unlabeled methionine (1 mg/ml). Total RNA was isolated from each batch of cells, and 3 μg of each RNA sample were subjected to a 1.2% agarose-6%formaldehyde gel electrophoresis and transferred to a Hybond-N+ nylon membrane (Amersham Biosciences). The membrane was UV-cross-linked and then baked for 20 min at 80 °C. The gel was then cut into two parts. The upper part of the gel was sprayed with En3Hance (PerkinElmer Life Sciences), dried, and subjected to fluorography at –70 °C for 10 days. The lower part of the membrane was subjected to Northern analysis using 32P-labeled deoxyoligonucleotide probe, designated JW 499 in Vilardell et al. (30), to detect U3 SnoRNA, which served as the loading control. B, effect of mutation of Ser-174 and Ser-175 of Tif6p on its ability to function in pre-rRNA processing. Exponentially growing cultures of PRY105 (tif6::HIS3 p[URA3 GAL10::Ub-HA-TIF6]p[TRP1TIF6-GFP]) and PRY106 (tif6::HIS3 p[URA3 GAL10::Ub-HA-TIF6]p[TRP1tif6-GFP]) in SGal-Met media (A600 = 0.5) were each transferred to equal volumes of SD-Met media and allowed to grow for 120 min to deplete HA-Tif6px from yeast cells as described under immunoblot analysis in C. Each culture was then subjected to [methyl-3H]methionine pulse-chase followed by analysis of [3H] rRNA formed as described under A above.
To show that the reduction in the formation of 25 S rRNA was due to lack of Hrr25p-mediated Tif6p phosphorylation, we carried out [methyl-3H]]methionine pulse-chase experiments in yeast cells under conditions where only phosphorylation of Tif6p by Hrr25p is blocked without affecting phosphorylation of any other cellular substrates. For this purpose, we constructed a yeast strain (tif6::HIS3 p[URA3 GALI0::Ub-HA-TIF6] p[TRP1tif6-GFP]) expressing both the wild-type and phosphomutant form of tif6p (SI74A, S175A) from two separate CEN plasmids. The resulting strain PRY106 expressed the wild-type Tif6p from a GAL10 promoter as an N-terminal-ubiquitinylated HA-tagged fusion protein and the mutant tif6p (S174A,S175A) as a C-terminal GFP-tagged fusion protein from its endogenous promoter. As a control, we constructed another yeast strain PRY105 (tif6p::HIS3 p[URA3 GAL10::Ub-HA-TIF6]p[TRP1 TIF6-GFP]) which is isogenic to the strain PRY106 except that it expresses wild-type Tif6p from its endogenous promoter instead of the phosphomutant form of Tif6p.
Exponentially growing cultures of both PRY105 and PRY106 strains in SGal-Met media were each transferred to SD-Met (glucose) media, and the cells were allowed to grow for 2 h to deplete HA-tagged Tif6p from both the PRY105 and PRY106 cells (Fig. 7C). Under these conditions, the expression of GFP-tagged Tif6p phosphomutant in the PRY106 cells as well as that of GFP-tagged wild-type Tif6p in PRY105 cells remained uninhibited (Fig. 7C). Each batch of cells was then pulse-labeled with [methyl-3H]]methionine for 2.5 min and chased for 0, 3, and 10 min with an excess of unlabeled methionine. Analysis of total [3H]RNA samples showed that in control PRY105 cells, the pre-rRNAs were rapidly chased into 25 S and 18 S rRNAs, as expected. In contrast, in PRY106 cells expressing only the unphosphorylatable Ala-mutant Tif6p, the formation of 25 S rRNA was significantly inhibited without affecting the level of 18 S rRNA (Fig. 7B). Taken together, the experiments presented in Fig. 7, A and B, suggest strongly that Hrr25p-mediated phosphorylation plays an essential role for the optimal formation of both mature 25 S rRNA and 18 S rRNA.
DISCUSSION
Ribosome biogenesis in eukaryotic cells is a complex process that occurs primarily in the nucleolus where four ribosomal rRNAs are formed, modified, and processed during their assembly with 78 (in yeast) and 79 (in mammals) ribosomal proteins into mature 40 S and 60 S ribosomal subunits (31, 32). In S. cerevisiae, where the process has been best characterized, the 18 S rRNA of the 40 S subunit and the 25 S and 5.8 S rRNAs of the 60 S subunit are transcribed from a 9.1-kilobase rDNA transcription unit by RNA polymerase I as a single large precursor RNA known as the 35 S pre-ribosomal RNA (pre-rRNA) (see Fig. 6 for RNA processing intermediates). Immediately after synthesis of this pre-rRNA, many ribosomal proteins as well as a large number of transacting non-ribosomal proteins associate with the 35 S pre-rRNA to form the 90 S ribonucleoprotein particle. These transacting non-ribosomal proteins are required for pre-rRNA processing, pre-rRNA modification, and ribosome assembly. Tif6p, the yeast homologue of mammalian eIF6, is one of these essential 60 S ribosomal assembly proteins that associates with the 66 S pre-ribosomal particles in the nucleolus (7, 10) and is required for the processing of 27 SB pre-rRNA to the mature 25 S and 5.8 S rRNAs leading to formation of mature 60 S ribosomal particles (11).
In this study we show that Hrr25p, an isoform of yeast CK I, phosphorylates Tif6p at a single tryptic peptide both in vitro and in vivo. This peptide contains the only known CK I phosphorylation motif (D/E)nXXST located at serine residue 174 (Fig. 4). Of the isoforms of yeast CK I we analyzed, only Hrr25p can efficiently phosphorylate Tif6p (Fig. 2). Depletion of Hrr25p from yeast cells by a conditional mutant yeast strain confirms that Hrr25p is the kinase responsible for Tif6p phosphorylation in vivo (Fig. 3). Phosphopeptide mapping and mass spectrometric analysis show that Ser-174 is the major site of phosphorylation of Tif6p by Hrr25p (Fig. 4). These results are in agreement with our previous finding that mutation of Tif6p at Ser-174 to alanine drastically reduced phosphorylation (>75%) and caused loss of cell growth and viability (11). When both Ser-174 and Ser-175 were mutated to alanine, phosphorylation of Tif6p was completely abolished (11).
In recent reports (27, 28), Hrr25p has been shown to stably associate with both the 90 S and the pre-40 S particles. The bound Hrr25p appears to phosphorylate two pre-40 S ribosomal assembly proteins, Enp1p and Ltv1p, as well as a 40 S ribosomal protein Rps3p in the pre-40 S ribosomal particles (28). Phosphorylation of these proteins appears to be required for the maturation of the pre-40 S particles. As discussed before, Tif6p is an essential 60 S ribosome assembly protein that associates with the 66 S pre-ribosomal particles. Analysis of the Tif6p-associated RNA components obtained by affinity purification of lysates of yeast cells expressing TAP-tagged Tif6p showed that Tif6p primarily associates with the two late 66 S pre-ribosomal particles (Fig. 6). Moreover, when we investigated the interaction between Tif6p and Hrr25p by coimmunoprecipitation, a small fraction of cellular Hrr25p was found to be associated with the 66 S pre-ribosomal particles to which Tif6p was also bound (Fig. 6). The presence of both Hrr25p and Tif6p on the same 66 S pre-ribosomal particles suggests that phosphorylation of Tif6p may occur on the 66 S preribosomal particles.
An important question that emerges is whether phosphorylation of Tif6p plays a role in the processing of 35 S pre-rRNA to form mature 18 S, 25 S, and 5.8 S rRNAs. Pulse-chase experiments presented to measure the rate of rRNA processing reactions clearly show that depletion of Hrr25p from yeast cells results in severe inhibition in the processing of 35 S pre rRNA to intermediate 27 S and 20 S pre-rRNAs. Subsequent processing of 20 S pre-rRNA to 18 S rRNA is also strongly inhibited, in agreement with the Northern analysis carried out by Schafer et al. (28). In contrast, Hrr25p-depleted yeast cells showed formation of 25 S rRNA, albeit at a much lower level than wild-type cells expressing Hrr25p. Additionally, in cells expressing only mutant Tif6p (S174A,S175A), where only Hrr25-mediated phosphorylation of Tif6p is prevented without affecting phosphorylation of any other cellular proteins, formation of 25 S rRNA is also severely inhibited without affecting the formation of 18 S rRNA (Fig. 7B). Reports published from several laboratories (33, 34) have shown that Tif6p is required for nuclear export of pre-60 S ribosomal particles for final maturation in the cytoplasm. Thus, the possibility exists that lack of phosphorylation of Tif6p affects nuclear export of Tif6p-bound pre-60 S ribosomal particles. This results in inhibition of nucleocytoplasmic recycling of Tif6p that is presumably required for continued nucleolar 60 S biogenesis.
Both mammalian and yeast cells contain multiple isoforms of CK I that associate with different cellular compartments (12). These isoforms have a highly conserved catalytic domain in their N- and C-terminal domains. These non-catalytic domains contribute to the discrete cellular localization of each isoform and are essential for their respective functions (12). Many of the mammalian isoforms of CK I have been shown to be involved in specific regulatory functions, e.g. during embryonic morphogenesis (35), Wnt signaling pathway (36, 37), mRNA metabolism (38), cell cycle regulation (39), and circadian rhythm in Drosophila (40). Of the four isoforms of CK I in yeast, Hrr25p has emerged as a major regulatory kinase affecting a number of discrete biological processes. This kinase has been shown to be involved in transcriptional response to DNA damage and repair (41, 42), vesicle budding in the secretory pathway from the endoplasmic reticulum (43), cell survival during stress response through phosphorylation of stress-responsive transcription factor, Crz1p (16), and monopolar attachment of sister kinetochores at meiosis 1 (44). We now show that Hrr25p is also responsible for phosphorylation of Tif6p, an essential 60 S ribosomal biogenesis protein. This observation along with that of Schafer et al. (28) showing Hrr25p also mediates phosphorylation of several proteins of the pre-40 S ribosomal particles suggests that Hrr25p plays an important dual role in the biogenesis of both 40 S and 60 S ribosomal subunits. It is tempting to speculate that the constitutively active kinase Hrr25p might play a pivotal role in coordinating the process of 60 S and 40 S ribosome biogenesis in yeast.
Acknowledgments
We are indebted to Dr. Michael C. Keogh of Albert Einstein College of Medicine for critically reading the manuscript and for many helpful suggestions during the course of this work. We also thank Dr. Robyn Moir, Dr. Neelam Desai, and Dr. Ian Willis of Albert Einstein College of Medicine for considerable help in the preparation of yeast nuclear and cytoplasmic extracts. Finally, we are grateful to Dr. Jonathan Warner of Albert Einstein College of Medicine for many helpful discussions during the course of our work.
This research was supported by National Institutes of Health Grant GM15399 and by NCI, National Institutes of Health Cancer Core Support Grant P30CA13330. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: eIF, eukaryotic (translation) initiation factor; CK I, casein kinase I; ORF, open reading frame; Ni-NTA, nickel-nitrilotriacetic acid; HA, hemagglutinin; DTT, dithiothreitol; YPD, yeast extract/peptone/dextrose; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; TAP, tandem affinity purification; GFP, green fluorescent protein; pre-rRNA, pre-ribosomal RNA; TPCK, l-1-tosylamido-2-phenylethyl chloromethyl ketone.
References
- 1.Russell, D. W., and Spremulli, L. L. (1979) J. Biol. Chem. 254 8796–8800 [PubMed] [Google Scholar]
- 2.Russell, D. W., and Spremulli, L. L. (1980) Arch. Biochem. Biophys. 201 518–526 [DOI] [PubMed] [Google Scholar]
- 3.Valenzuela, D. M., Chaudhuri, A., and Maitra, U. (1982) J. Biol. Chem. 257 7712–7719 [PubMed] [Google Scholar]
- 4.Raychaudhuri, P., Stringer, E. A., Valenzuela, D. M., and Maitra, U. (1984) J. Biol. Chem. 259 11930–11935 [PubMed] [Google Scholar]
- 5.Merrick, W. C., and Hershey, J. W. B. (1996) in Translational Control (Hershey, J. W. B., Matthews, M. B., and Sonenberg, N., eds) pp. 31–69, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 6.Si, K., Chaudhuri, J., Chevesich, J., and Maitra, U. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 14285–14290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Si, K., and Maitra, U. (1999) Mol. Cell. Biol. 19 1416–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanvito, F., Piatti, S., Villa, A., Bossi, M., Lucchini, G., Marchisio, P. C., and Biffo, S. (1999) J. Cell Biol. 144 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basu, U., Si, K., Warner, J. R., and Maitra, U. (2001) Mol. Cell. Biol. 21 1453–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harnpicharnchai, P., Jakovljevic, J., Horsey, E., Miles, T., Roman, J., Rout, M., Meagher, D., Imai, B., Guo, Y., Brame, C. J., Shabanowitz, J., Hunt, D. F., and Woolford, J. L., Jr. (2001) Mol. Cell 8 505–515 [DOI] [PubMed] [Google Scholar]
- 11.Basu, U., Si, K., Deng, H., and Maitra, U. (2003) Mol. Cell. Biol. 23 6187–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross, S. D., and Anderson, R. A. (1998) Cell Signal 10 699–711 [DOI] [PubMed] [Google Scholar]
- 13.Rose, M. D., Winston, F., and Heiter, P. (1989) Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 14.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 15.Longtine, M. S., McKenzie, A., III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998) Yeast 14 953–961 [DOI] [PubMed] [Google Scholar]
- 16.Kafadar, K. A., Zhu, H., Snyder, M., and Cyert, M. S. (2003) Genes Dev. 17 2698–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moriya, H., and Johnston, M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 1572–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maiti, T., Bandyopadhyay, A., and Maitra, U. (2003) Yeast 20 97–108 [DOI] [PubMed] [Google Scholar]
- 19.Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. (1999) Nat. Biotechnol. 17 1030–1032 [DOI] [PubMed] [Google Scholar]
- 20.Venema, J., Planta, R. J., and Raue, H. A. (1998) Methods Mol. Biol. 77 257–270 [DOI] [PubMed] [Google Scholar]
- 21.Evans, C. F., and Engelke, D. R. (1990) Methods Enzymol. 181 439–450 [DOI] [PubMed] [Google Scholar]
- 22.Robinson, L. C., Hubbard, E. J., Graves, P. R., DePaoli-Roach, A. A., Roach, P. J., Kung, C., Haas, D. W., Hagedorn, C. H., Goebl, M., Culbertson, M. R., and Carlson, M. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson, L. C., Menold, M. M., Garrett, S., and Culbertson, M. R. (1993) Mol. Cell. Biol. 13 2870–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vancura, A., Sessler, A., Leichus, B., and Kuret, J. (1994) J. Biol. Chem. 269 19271–19278 [PubMed] [Google Scholar]
- 25.Wang, X., Hoekstra, M. F., DeMaggio, A. J., Dhillon, N., Vancura, A., Kuret, J., Johnston, G. C., and Singer, R. A. (1996) Mol. Cell. Biol. 16 5375–5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeMaggio, A. J., Lindberg, R. A., Hunter, T., and Hoekstra, M. F. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 7008–7012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schafer, T., Strauss, D., Petfalski, E., Tollervey, D., and Hurt, E. (2003) EMBO J. 22 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schafer, T., Maco, B., Petfalski, E., Tollervey, D., Bottcher, B., Aebi, U., and Hurt, E. (2006) Nature 441 651–655 [DOI] [PubMed] [Google Scholar]
- 29.Udem, S. A., and Warner, J. R. (1972) J. Mol. Biol. 65 227–242 [DOI] [PubMed] [Google Scholar]
- 30.Vilardell, J., Chartrand, P., Singer, R. H., and Warner, J. R. (2000) RNA 6 1773–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venema, J., and Tollervey, D. (1999) Annu. Rev. Genet. 33 261–311 [DOI] [PubMed] [Google Scholar]
- 32.Kressler, D., Linder, P., and de La Cruz, J. (1999) Mol. Cell. Biol. 19 7897–7912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tschochner, H., and Hurt, E. (2003) Trends Cell Biol. 13 255–263 [DOI] [PubMed] [Google Scholar]
- 34.Menne, T. F., Goyenechea, B., Sanchez-Puig, N., Wong, C. C., Tonkin, L. M., Ancliff, P. J., Brost, R. L., Costanzo, M., Boone, C., and Warren, A. J. (2007) Nat. Genet. 39 486–495 [DOI] [PubMed] [Google Scholar]
- 35.McKay, R. M., Peters, J. M., and Graff, J. M. (2001) Dev. Biol. 235 378–387 [DOI] [PubMed] [Google Scholar]
- 36.McKay, R. M., Peters, J. M., and Graff, J. M. (2001) Dev. Biol. 235 388–396 [DOI] [PubMed] [Google Scholar]
- 37.Peters, J. M., McKay, R. M., McKay, J. P., and Graff, J. M. (1999) Nature 401 345–350 [DOI] [PubMed] [Google Scholar]
- 38.Gross, S. D., Loijens, J. C., and Anderson, R. A. (1999) J. Cell Sci. 112 2647–2656 [DOI] [PubMed] [Google Scholar]
- 39.Gross, S. D., Simerly, C., Schatten, G., and Anderson, R. A. (1997) J. Cell Sci. 110 3083–3090 [DOI] [PubMed] [Google Scholar]
- 40.Rosbash, M., Allada, R., McDonald, M., Peng, Y., and Zhao, J. (2003) Novartis Found. Symp. 253 223–232; discussion 252–225, 102–229, 232–227 passim [PubMed] [Google Scholar]
- 41.Hoekstra, M. F., Liskay, R. M., Ou, A. C., DeMaggio, A. J., Burbee, D. G., and Heffron, F. (1991) Science 253 1031–1034 [DOI] [PubMed] [Google Scholar]
- 42.Murakami, A., Kimura, K., and Nakano, A. (1999) J. Biol. Chem. 274 3804–3810 [DOI] [PubMed] [Google Scholar]
- 43.Ho, Y., Mason, S., Kobayashi, R., Hoekstra, M., and Andrews, B. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petronczki, M., Matos, J., Mori, S., Gregan, J., Bogdanova, A., Schwickart, M., Mechtler, K., Shirahige, K., Zachariae, W., and Nasmyth, K. (2006) Cell 126 1049–1064 [DOI] [PubMed] [Google Scholar]