Abstract
Within the yeast mitochondrial ATP synthase, subunit h is a small nuclear encoded protein belonging to the so-called “peripheral stalk” that connects the enzyme catalytic F1 component to the mitochondrial inner membrane. This study examines the role of subunit h in ATP synthase function and assembly using a regulatable, doxycycline-repressible subunit h gene to overcome the strong instability of the mtDNA previously observed in strains lacking the native subunit h gene. Yeast cells expressing less than 3% of subunit h, but still containing intact mitochondrial genomes, grew poorly on respiratory substrates because of a major impairment of ATP synthesis originating from the ATP synthase, whereas the respiratory chain complexes were not affected. The lack of ATP synthesis in the subunit h-depleted (δh) mitochondria was attributed to defects in the assembly/stability of the ATP synthase. A main feature of δh-mitochondria was a very low content (<6%) in the mitochondrially encoded Atp6p subunit, an essential component of the enzyme proton channel, which was in large part because of a slowing down in translation. Interestingly, depletion of subunit h resulted in dramatic changes in mitochondrial cristae morphology, which further supports the existence of a link between the ATP synthase and the folding/biogenesis of the inner mitochondrial membrane.
In the inner mitochondrial membrane, the F1F0-ATP
synthase synthesizes ATP using the H+ electrochemical gradient
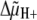 generated by the respiratory chain. This is a reversible enzyme that can also
hydrolyze ATP in a reaction coupled to a proton transport out of the
mitochondrial matrix. Historically, two different parts of this enzymatic
complex have been described as follows: (i) the water-soluble sector
F1, composed of α, β, γ, δ, and ε
subunits, which retains the ability to hydrolyze ATP in solution; (ii) the
F0 sector, which is embedded in the inner mitochondrial membrane
and composed, in yeast, of subunits 9, 6, 4, 8, f, OSCP, h, and
d. The ATP synthesis is only possible when the F1 is bound
to the F0 (1).
generated by the respiratory chain. This is a reversible enzyme that can also
hydrolyze ATP in a reaction coupled to a proton transport out of the
mitochondrial matrix. Historically, two different parts of this enzymatic
complex have been described as follows: (i) the water-soluble sector
F1, composed of α, β, γ, δ, and ε
subunits, which retains the ability to hydrolyze ATP in solution; (ii) the
F0 sector, which is embedded in the inner mitochondrial membrane
and composed, in yeast, of subunits 9, 6, 4, 8, f, OSCP, h, and
d. The ATP synthesis is only possible when the F1 is bound
to the F0 (1).
The ATP synthase complex can be described as a molecular rotary motor. The rotor, which is driven by the proton flux through the inner membrane, is composed of the subunit 9 oligomer (10 copies) and the central stalk subunits γ, δ, and ε. The stator is composed of nine major subunits. Subunits β, which hold the catalytic sites, are organized with subunits α in a soluble (αβ)3 hexamer. Subunits OSCP, 4, d, h, f, 8, and i form a peripheral stalk linked to the catalytic (αβ)3 hexamer and to subunit 6. Subunit 9 oligomer and subunit 6 interact in the membrane to form a proton-conducting pathway. The current model for ATP synthase energy coupling is the binding change mechanism (2), according to which affinity changes for substrates and products at the catalytic sites are coupled to proton transport via the rotation of the γ subunit inside the (αβ)3 hexamer (2). Evidence of this rotation during ATP hydrolysis or synthesis in the isolated F1 sector or the entire enzyme has been obtained by optical microscopy and fluorescence resonance energy transfer (3–6).
Subunits e, g, k, and i of the mitochondrial ATP synthase are referred to as “supernumerary” because they have no counterparts in bacteria. These subunits are not essential for cellular growth on nonfermentable sources and have only been found, for the first three, associated with ATP synthase dimers (7). The absence of subunit e and/or subunit g results in a strong alteration of the cristae morphology (8), indicating that the ATP synthase controls in some way the biogenesis of the inner mitochondrial membrane.
It has been proposed that the F1 sector biogenesis occurs independently of the F0 sector (9) and requires the presence of the general Hsp60p chaperone and at least three assembly factors (Atp12p, Atp11p, and Fmc1p) with specific actions in the hexamerization of the α-F1 and β-F1 proteins (10–12). Subunit 9 pre-formed oligomer in the inner mitochondrial membrane is believed to anchor the F1 sector to the membrane (13), forming an F1-910 complex to which subunits 8, 4, and other F0 components are assembled (14, 15). Finally, the two chaperones Atp10p and Atp23p were shown to be required for the assembly of subunit 6 (16–18), the last subunit incorporated to the complex.
In this work we focused on the role of subunit h within the ATP synthase. Subunit h is a 10.4-kDa water-soluble protein encoded by the nuclear gene ATP14. Although they share only 15% identity, yeast subunit h and bovine F6 subunit were shown to be functionally equivalent as evidenced by the ability of F6 to complement the absence of subunit h gene in Δatp14 yeast cells (where Δatp14 indicates deletion of the S. cerevisiae ATP14 gene, coding for ATP synthase subunit h) (19). Subunit h is part of the ATP synthase peripheral stalk, as shown by chemical cross-link studies that revealed its proximities to subunits α, f, d, and 4 (20, 21) and by electron microscopy analyses (22). Cross-links between two subunits h belonging to two ATP synthase monomers indicated that this subunit is located near or at the dimerization interface of this complex (23). Recently, a subcomplex of the peripheral stalk of the bovine ATP synthase was obtained by co-expression in Escherichia coli of large soluble domains of bovine subunits 4, d, and F6 (homologous to yeast subunit h). This sub-complex was purified and crystallized (24), showing that subunit F6 is located at the top of the peripheral stalk, wrapped along subunit 4 C terminus (25).
Further insight on the role of subunit h was sought through the construction of a yeast strain lacking the ATP14 gene (26). However, as for many other ATP synthase mutations, Δh yeast was genetically unstable in the form of more than 90% of ρ-/ρ0 cells, i.e. cells issued from large/complete deletions in the mtDNA. This hampered the analysis of the consequences of a lack in subunit h on the ATP synthase because of the secondary loss of several components of the mitochondrial energy-transducing system encoded by the mtDNA, among which are the two main subunits (Atp6p and Atp9p) of the ATP synthase proton channel (26).
To overcome this problem, we constructed a subunit h regulatable gene, under the control of a doxycycline repressible promoter (27). Such a method has already been successful when used for the study of several ATP synthase subunits (28, 29), among which is the δ subunit whose gene deletion results in populations exclusively composed of ρ-/ρ0 cells. In this way, we were able to generate yeast populations almost completely depleted in subunit h where most (>95%) of the cells were still ρ+. This allowed us to investigate in excellent conditions the specific consequences of a lack in subunit h. We show that subunit h depletion leads to a secondary loss in subunit 6 due in large part to a decrease of its synthesis, whereas most of the other enzyme subunits still show a rather good accumulation. In addition, we provide evidence that subunit h is critical for a correct maintenance of mitochondrial cristae morphology, which further supports the existence of a link between ATP synthase and the folding of the inner mitochondrial membrane.
EXPERIMENTAL PROCEDURES
Materials—Digitonin (Sigma) was recrystallized twice in EtOH (30). All other chemicals were of reagent grade quality.
Strains and Media—The Saccharomyces cerevisiae strain SDC22 (MATα, arg8::HIS3, ade2-1, ura3-11,15, leu2-3,112, trp1-1, can1-100, ARG8m, rho+) was the wild type strain (28). Rich glucose (YPD), galactose (YPGal), or lactate (YPLac) media and synthetic complete medium used for growing yeast strains were prepared as described previously (28, 31).
Genetic and Molecular Biology Methods—DNA manipulations were done using standard methods (32). Bacteria and yeast were respectively transformed by electroporation and lithium chloride methods (33).
Construction of Mutant Strains—The coding sequence of subunit h was PCR-amplified from previously described pRS313 ATP14 (26). The resulting ATP14 amplicon was cloned into pCM189 (BamHI-NotI) downstream of the doxycycline-repressible promoter to give pRF1. pRF1-transformed strain, selected on synthetic complete medium lacking uracil, was named RFY1. In this strain, the ATP14 gene was disrupted at its locus with KanR gene using a PCR-based method as described previously (34). Cells that had integrated KanR at the ATP14 gene locus were selected on YPD plates containing 200 μg·ml-1 of geneticin and analyzed by PCR. A clone called RFY5-1 carrying the expected disruption of ATP14 gene, in a ρ+ state and containing pRF1 was retained for further analysis.
Quantification of the Amount of ρ-/ρ0 Cells upon Subunit h Depletion by Doxycycline Addition—The amount of ρ-/ρ0 cells was calculated from the difference between individual colonies growing on YPD versus residual individual colonies growing after replication on complete glycerol medium. In order not to take into account the cells resulting from plasmid loss, RFY5-1 cells (Δatp14/pCM189-ATP14) were primarily mated with ρ0 (ATP14) cells, so that the inability to grow on complete glycerol medium could only be attributed to a loss of or a deletion in mitochondrial DNA.
Biochemical Procedures—Cells were grown aerobically at 28 °C in a liquid complete medium containing 2% lactate or 2% galactose as carbon source and harvested in logarithmic growth phase (8.107 cells·ml-1). Mitochondria were prepared as described previously (35) and suspended in the isolation buffer (0.6 m mannitol, 2 mm EGTA, 10 mm Tris maleate, pH 6.8). Protein concentration was determined according to the Lowry method (36) in the presence of 5% (w/v) SDS with bovine serum albumin as standard protein.
Oxygen consumption rates were measured at 28 °C with a Clark-type oxygen electrode on freshly prepared mitochondria diluted to 0.3 mg·ml-1 in respiration buffer (0.65 m mannitol, 0.3 mm EGTA, 3 mm Tris-phosphate, 10 mm Tris-maleate, pH 6.75) as described (37). Variations in transmembrane potential (ΔΨ) were evaluated in the same conditions by measurement of rhodamine 123 (1 μg·ml-1) fluorescence quenching using λem = 530 nm and λex = 485 nm on a SAFAS (Monte-Carlo, Monaco) fluorescence spectrophotometer (38).
ATP synthesis was measured by bioluminescence using the luciferin/luciferase kit from PerkinElmer Life Sciences and an LKB-Wallac luminometer. ATP hydrolysis and oligomycin sensitivity were measured on Triton X-100-treated mitochondria (0.75 g·g-1 of proteins) as already described (39).
Protein Extracts—Whole cell protein extracts were prepared as described by Egner et al. (40). For SDS-PAGE analyses, mitochondrial proteins were precipitated with 3 m trichloroacetic acid prior to their solubilization in sample buffer containing 1% SDS. For CN-PAGE4 analyses, mitochondrial proteins were solubilized by digitonin at 5 mg of protein·ml-1 at a detergent to protein ratio varying from 0.5 to 2 g·g-1 in 150 mm potassium acetate, 2 mm 6-aminohexanoic acid, 10% (m/v) glycerol, 30 mm HEPES, pH 7.4, and protease inhibitor mixture.
Electrophoretic and Western Blot Analyses—SDS-PAGE was performed using Tris-Tricine 15% polyacrylamide slab gels according to Schägger and von Jagow (41). CN-PAGE experiments were carried out as described by Schägger (42). Mitochondrial digitonin extracts were centrifuged at 4 °C for 15 min at 40,000 × g. Forty microliters of supernatant were immediately loaded onto a 3–13% polyacrylamide slab gel. After electrophoresis, ATPase activity was revealed by incubating the gel in 5 mm ATP, 5 mm MgCl2, 0.05% (w/v) lead acetate, 50 mm glycine-NaOH, pH 8.6 (43). The bands of interest were cut, incubated in 1% (w/v) SDS for 1 h, and analyzed in a second dimension by SDS-PAGE (41).
Western blot analyses have been described previously (26). Proteins were electrotransferred onto nitrocellulose membranes (Membrane Protean BA83, Schleicher & Schuell). Primary antibodies were polyclonal rabbit antibodies. Secondary antibodies were peroxidase-conjugated goat anti-rabbit antibodies (Jackson ImmunoResearch). Western blots were revealed using the enhanced chemiluminescence method (Amersham Biosciences) on a CCD camera (GeneGnome, Syngene Bio-Imaging) or with photographic films (Hyperfilm, Amersham Biosciences). Quantification was done using ImageJ software.
Synthesis and Degradation of Mitochondrial Translation Products—The RFY5-1 strain was grown to early exponential phase (4 × 107 cells·ml-1) in 100 ml of liquid YPGal medium containing or not doxycycline (10 μg·ml-1). About 4 × 109 cells were harvested by centrifugation and washed twice with cold sterile water. Cells were resuspended in 5 ml of a low sulfate medium containing 1% galactose, supplemented with adenine, uracil, leucine, and tryptophan (50 mg·l-1 each) ± doxycycline (10 μg·ml-1) and incubated for 40 min at 28 °C. Cycloheximide (0.6 mg·ml-1) was added to the cell suspension. After a 5-min incubation (pulse labeling) at 28 °C, 2 mCi (74 MBq) of 35S in vitro cell labeling mix (Amersham Biosciences) was added, and the incubation was carried out for another 5 min at 28 °C. The labeling reaction was stopped by the addition of 400 μl of a Met + Cys solution (250 mm). For each time of chase, 1 ml of cell suspension was mixed with 1 ml of casamino acids 100 mg·ml-1, Na2SO4 2 mg·ml-1. Cells were harvested and washed twice. The pellet was resuspended in 1 ml of NaOH 1.85 m, β-mercaptoethanol 7.5% and incubated for 10 min at 4 °C. One millimeter of 50% trichloroacetic acid was added, and after a 10-min incubation at 4 °C, the solution was centrifuged for 5 min at 20,000 × g. The pellet was resuspended in 100 μl of 5% SDS, neutralized by addition of aliquots of Tris 1 m, incubated for 15 min at 42 °C, and centrifuged for 5 min at 20,000 × g. Protein concentration was determined on the supernatant, and 50 μg of each sample was separated on a 12.5% acrylamide gel, containing 4 m urea and 25% glycerol (44). The gel was dried out, and the incorporated radioactivity was counted using a Typhoon PhosphorImager (Fuji). For each sample, each subunit level was quantified relatively to the total incorporated radioactivity.
Northern Blot Analysis of Mitochondrial Transcripts—Mitochondrial RNAs were extracted, separated, and analyzed as described in Ref. 45 from RFY5-1 strain grown for 14 h to early exponential phase in liquid YPGal medium in the absence or presence of doxycycline (10 μg·ml-1). Specific DNA probes for ATP6 and 15 S rRNA were labeled with 32P. Blots were analyzed on a Typhoon PhosphorImager (Fuji) and quantified using ImageJ software.
Freezing and Freeze Substitution for Ultrastructural Analyses of Yeast Cells—The RFY5-1 strain was grown for 14 h to early exponential phase (107 cells·ml-1) in liquid YPGal medium in the absence or presence of doxycycline (10 μg·ml-1). The yeast pellets were placed on the surface of a copper EM grid (400 mesh) coated with Formvar. Each loop was very quickly submersed in pre-cooled liquid propane and kept at -180 °C in liquid nitrogen. The loops were then transferred to a pre-cooled solution of 4% osmium tetroxide in dry acetone in a 1.8-ml polypropylene vial and kept at -82 °C for 48 h (substitution fixation), warmed gradually to room temperature, and washed three times in dry acetone. Specimens were stained for 1 h with 1% (w/v) uranyl acetate in acetone at 4 °C in a dark room. Following another rinse in dry acetone, the loops were progressively infiltrated with araldite (epoxy resin, Fluka). Ultrathin sections were contrasted with lead citrate. Observations were done on a Philips Tecnai 12 electron microscope. Data presented in this study were analyzed and written with free softwares under GNU GPL license.
RESULTS
Depletion of Subunit h Severely Compromises the Respiratory Growth of Yeast—We have constructed the RFY5-1 strain by transformation of yeast cells with a plasmid where the coding sequence of ATP14 is under the control of a doxycycline-repressible promoter followed by deletion of the chromosomal ATP14 gene. In this way, one can block the expression of subunit h simply and quickly by the addition of doxycycline to the growth medium. In nonfermentable lactate medium, doubling time of RFY5-1 was 190 ± 16 min in the absence of doxycycline and increased to 824 ± 19 min in the presence of 10 μg·ml-1 doxycycline (data not shown). Progressive attenuation of subunit h after the addition of doxycycline was observed by Western blotting of whole cell protein extracts prepared at different culture times (Fig. 1A). After 24 h, i.e. less than 2 generations, about 6% of subunit h was still present in the RFY5-1 cells grown in the presence of doxycycline. When galactose, a fermentable carbon source, was used to grow RFY5-1, the doubling time was almost not modified by the addition of doxycycline, i.e. 150 and 170 min without and with doxycycline, respectively, and after 14 h, i.e. 3 generations, subunit h was virtually absent in whole cell extracts (Fig. 1B). However, when the cells grown for 14 h in galactose + doxycycline medium were transferred into lactate + doxycycline medium, a slow residual growth was still observed, whereas the same cells recovered a normal respiratory growth when doxycycline was not included in the lactate medium (Fig. 1D). After 75 h, the lactate + doxycycline culture reached a cell density similar to that of the culture made in absence of doxycycline. Thus, despite an apparent total cellular lack in subunit h, respiration-dependent growth was not completely abolished. However, when the Western blot analyses were performed with mitochondrial protein preparations instead of whole cell extracts, a low level of subunit h could be detected at about 3% compared with control mitochondrial proteins (Fig. 1E). The residual respiratory growth of RFY5-1 in the presence of doxycycline was thus likely because of a small transcriptional leak at the level of the doxycycline-repressible promoter.
FIGURE 1.
Subunit h depletion and percentage of ρ-/ρ0 cells in doxycycline-treated cells. A, cells were grown in nonfermentable YPLac 2% medium containing or not 10 μg·ml-1 doxycycline (Dox). Cellular extracts were prepared at different culture times. B, cells were grown in fermentable YPGal 2% medium containing or not 10 μg·ml-1 of doxycycline. Cellular extracts were prepared from cells harvested 500 min (∼3 generations) after doxycycline addition. A and B, 50 μg of proteins were submitted to SDS-PAGE and transferred onto nitrocellulose. The blots were probed with polyclonal antibodies raised against subunit h. C, cells were grown in fermentable YPGal 2% medium containing (closed circle) or not (open circle) doxycycline (10 μg·ml-1). Percentage of ρ-/ρ0 cells was quantified as described under “Experimental Procedures.” The arrow indicates the time at which cells were harvested prior to the mitochondrial preparation. D, RFY5-1 cells were grown for 14 h in YPGal 2% medium containing 10 μg·ml-1 of doxycycline. The complete subunit h depletion was verified in cell extracts by SDS-PAGE and Western blotting (not shown). Cells were then harvested and grown in a nonfermentable medium (YPLac 2%) containing (closed circle) or not (open circle) doxycycline (10 μg·ml-1). Cell growth was monitored by measuring absorbance at 550 nm. E, mitochondria were isolated from RFY5-1 strain grown in YPGal medium during 14 h without or with doxycycline (10 μg·ml-1). Fifty μg of proteins were submitted to SDS-PAGE and transferred onto nitrocellulose. The blots were probed with polyclonal antibodies raised against subunit h and porin as an internal standard.
Influence of a Depletion in Subunit h on the Stability of the mtDNA—In previous studies, cultures of Δatp14 yeast were found to contain a high number (>95%) of ρ-/ρ0 cells. The reason for this mtDNA instability is not known. The accumulation of ρ-/ρ0 cells by RFY5-1 strain after the addition of doxycycline (Fig. 1C) remained very low at least during 5–6 generations. At 25 h after the addition of doxycycline, i.e. after about 8–9 generations, the proportion of cells increased to about 20 versus 5% for the culture of RFY5-1 made in the absence of doxycycline. It might be that after a block in subunit h expression, mtDNA molecules are progressively lost over cell divisions, and ρ-/ρ0 cells appear after quite a large number of generations. The production of cultures of cells lacking the ATP14 gene requires at least 30 generations after the initial recombination event leading to the deletion of ATP14 from the chromosome. With the doxycycline-repressible subunit h gene, only a limited number of generations suffices to obtain large cultures almost completely depleted in subunit h. This system provides excellent conditions for the study of the consequences of a specific lack in subunit h. Throughout this study, doxycycline was used at a 10 μg·ml-1 concentration, and yeast grown in the presence of this concentration of doxycycline will be named δh, as opposed to Δh in which the nuclear ATP14 gene is disrupted.
Subunit h Is Essential for ATP Synthesis but Not for ATP Hydrolysis by the ATP Synthase—To further characterize the consequences of a lack in subunit h on oxidative phosphorylations, several parameters were investigated as follows: respiratory rates, ATP synthesis and hydrolysis activities, and mitochondrial electrical transmembrane potential. Measurements were done on mitochondria isolated from RFY5-1 strain grown 14 h in a galactose medium containing (δh-mitochondria) or not (control mitochondria) doxycycline (10 μg·ml-1).
Respiration rates were measured with either NADH or ascorbate/TMPD as substrates (Fig. 2A). Because the amount of respiratory complexes could vary from one preparation to another, respiratory rates were expressed as a percentage of the maximal rate obtained in the presence of the membrane uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) and using ascorbate and 2,2,4-trimethyl-1,3-pentanediol (TMPD) as electron donors. Basal (state 4), uncoupled (NADH + CCCP), and maximal respiration (ascorbate/TMPD + CCCP) rates were relatively similar in mitochondria whether depleted or not in subunit h, showing that the depletion in subunit h did not increase the passive permeability to protons of the inner membrane and did not affect the expression of the respiratory chain complexes. Addition of ADP stimulated the respiration to state 3 in both mitochondrial preparations but to a much lesser extent in δh-mitochondria than in the control. In δh-mitochondria, the respiratory control was only 1.2 ± 0.1 versus 2.4 ± 0.2 in control mitochondria. Mitochondrial ATP synthesis was monitored using NADH as substrate in the presence of a saturating amount of ADP (Fig. 2B). In the control mitochondria, ATP synthesis rate was high (0.72 μmol ATP·min-1·mg-1 of proteins), giving an ATP/O ratio of 0.8. The addition of oligomycin, a specific inhibitor of the ATP synthase proton channel, inhibited ATP synthesis almost totally. In δh-mitochondria ATP synthesis activity was only 14% of the control rate, resulting in an ATP/O ratio of 0.3, and it was totally inhibited by oligomycin. Taken together, these results showed that subunit h is critical for ATP synthesis. The residual ATP synthesis is likely because of the low content in intact ATP synthases still present in the δh-mitochondria because of the inability of doxycycline to totally block the expression of subunit h (see above).
FIGURE 2.
Respiratory rates, ATP synthesis, and hydrolysis activities of mitochondria isolated from RFY5-1 strain grown with and without doxycycline. Mitochondria were prepared from RFY5-1 strain grown in YPGal medium without (open bars) and with doxycycline (dox) (closed bars) for 14 h as described under “Experimental Procedures.” Values are means determined from four different mitochondrial preparations. A, respiratory rates are expressed as percentage of highest rates obtained with ascorbate and TMPD. Mitochondria (0.3 mg·ml-1 proteins) were incubated in the respiratory buffer, and additions were 4 mm NADH (state 4), 4 mm NADH + 50 μm ADP (state 3), 4 mm NADH + 3 μm CCCP, or 15 mm ascorbate and 0.6 mm TMPD (Asc+TMPD). The maximal respiration rate measured using ascorbate and TMPD as electron donors were 2220 ± 190 and 1810 ± 270 nanoatoms O·min-1·mg-1 proteins for control and subunit h-depleted mitochondria respectively. B, ATP synthesis rates were measured by bioluminescence as described under “Experimental Procedures” after the addition of 7.5 mm of ADP. C, ATPase activities were measured as described under “Experimental Procedures.” Oligomycin (oligo) was used at a concentration of 33μg·mg-1 of protein.
ATPase activity was measured in both control and subunit h-depleted mitochondria (Fig. 2C). At pH 8.4, ATP hydrolysis rates measured in Triton X-100-solubilized mitochondria were 4.8 ± 0.3 μmol of Pi·min-1·mg-1 protein and 6.2 ± 0.6 μmol of Pi·min-1·mg-1 protein in control and δh-mitochondria, respectively. In the wild type mitochondria, 70% of this activity was oligomycin-sensitive, whereas only 10% of this activity was inhibited by oligomycin in the δh-mitochondria. These data indicate that, when subunit h is absent, the F1 sector is correctly assembled and fully functional, whereas the rest of the ATP synthase complex is not correctly assembled or coupled to the F1 sector.
The influence of a lack in subunit h was further assessed by monitoring the mitochondrial electrical transmembrane potential difference (ΔΨ) using fluorescence quenching of rhodamine 123. Fig. 3A, left panel, shows that in control mitochondria, as expected, ethanol addition induced an important fluorescence quenching because of the establishment of a ΔΨ by the respiratory chain complexes. ADP addition induced a decrease of this quenching because of both proton re-entry through the F1F0-ATP synthase and ΔΨ consumption by the ADP/ATP carrier. The transmembrane potential increased when most of the ADP had been transformed into ATP. Further additions of KCN and of the uncoupler CCCP led to the collapse of the membrane potential. In the δh-mitochondria, even though the addition of ethanol established a ΔΨ comparable with the one established in the control strain mitochondria, addition of ADP had almost no influence on ΔΨ (Fig. 3A, right panel), a finding consistent with the low state 3 respiration rate and the low ATP synthesis activity observed in these mitochondria (Fig. 2, A and B). We next examined the ability of δh-mitochondria to establish a ΔΨ when hydrolyzing ATP. Mitochondria were first energized with ethanol to remove the F1-ATPase natural inhibitor, IF1p (46), and ΔΨ was then collapsed following the addition of KCN. In control mitochondria, ATP induced a large and stable ΔΨ, which was reversed by DCCD addition, reflecting the specific proton pumping activity of the F1F0-ATPase (Fig. 3B, left panel). In the δh-mitochondria, ATP addition established a very slight ΔΨ that was partially sensitive to DCCD (Fig. 3B, right panel). These results show that a lack in subunit h very severely compromises the proton pumping activity of the F1F0-ATP synthase.
FIGURE 3.
ΔΨ establishment in control and subunit h-depleted mitochondria. Rhodamine 123-monitored ΔΨ variations were assayed, as described under “Experimental Procedures,” in isolated mitochondria (0.3 mg·ml-1) from RFY5-1 grown without (left panels) and with doxycycline (Dox) (right panels) in the YPGal medium. A, ΔΨ variations during oxidative phosphorylation. B, ΔΨ establishment by ATP hydrolysis. Additions were 0.3 mg·ml-1 mitochondria (m), 170 mm ethanol (EtOH), 50 μm ADP, 600 μm KCN, 400 μm ATP, 10 μg·mg-1 of proteins DCCD and 3 μm CCCP. a.u., arbitrary units.
Subunits 6 and f Are in Low Amounts in Subunit h-depleted Yeasts—To understand at the molecular level the severe decrease in mitochondrial ATP synthesis activity resulting from subunit h depletion, the assembly of the ATP synthase was studied in δh-mitochondria. For that purpose, we first carried out immunoblots of mitochondrial proteins to quantify the contents in the different enzyme subunits. Signals obtained for each subunit were quantified and normalized using the signal obtained for porin as an internal standard.
Most of the essential ATP synthase subunits were present in δh-mitochondria in slightly decreased quantities (Fig. 4) and always accounted for more than 70% of the signals found in control mitochondria. This included F1 sector subunits (α, β, γ, δ, and ε subunits) and peripheral stalk subunits such as subunits 4, OSCP, and d. Subunit 9 level was also not significantly affected. In contrast, the accumulation levels of subunit 6 (6.7 ± 4%) and of subunit f (19.5% ± 9%) were strongly decreased. The lack in subunit 6 suffices to explain the drastic loss in ATP synthesis in the subunit h-depleted cells. Concerning the nonessential ATP synthase components, subunits i, e, and g were decreased by 40–60% and subunit k by about 80%. In the four different subunit h-depleted mitochondrial preparations tested, the amount of subunit h never exceeded 3% of the amount present in control mitochondria.
FIGURE 4.
Quantification of the different subunits of the F1F0-ATP synthase in RFY5-1 strain grown in the absence or presence of doxycycline. A and B, mitochondria isolated from RFY5-1 strain grown in YPGal medium in the absence or presence of doxycycline (Dox) (10 μg·ml-1) were prepared as described under “Experimental Procedures.” A, mitochondrial proteins (50 μg) were separated by Tris-Tricine SDS-PAGE, and the presence of the different subunits was revealed by Western blotting using appropriate antibodies. B, obtained signals were quantified. For each subunit, the relative quantity is the ratio between the signal obtained for mitochondria isolated from RFY5-1 strain grown in the presence of doxycycline (10 μg·ml-1) and the signal obtained for mitochondria isolated from the same strain grown in the absence of the antibiotic and normalized to the ratio obtained for porin. This ratio is expressed in percentage (relative quantity (%) = 100 × ((signal RFY5-1 + Dox for subunit x)/(signal RFY5-1-Dox for subunit x)) × ((signal RFY5-1 + Dox for porin)/(signal RFY5-1-Dox for porin))). Measurements were obtained from four different mitochondrial preparations.
Subunit 6 Synthesis in Subunit h-depleted Mitochondria—The low content in subunit 6 in the subunit h-depleted mitochondria could be due either to an increased rate of degradation or a slower rate of synthesis. To distinguish between these two possibilities, we performed pulse-chase labeling experiments, in which cytosolic protein synthesis was inhibited by cycloheximide. Subunit 6 was still synthesized in doxycycline-treated cells but to a much lower level than in control cells. As shown on Fig. 5, A–C, after a 5-min labeling, the quantity of neo-synthesized subunit 6 in the subunit h-depleted cells was decreased by 72 ± 1% compared with the control cells, whereas the other mtDNA-encoded proteins still showed a rather good synthesis. Chase experiments were performed to measure the degradation rates of the neo-synthesized proteins. The half-life time of subunit 6 was 47.7 ± 0.14 min and 53.6 ± 0.15 min in control and δh-cells, respectively (Fig. 5D). Thus, the low level of subunit 6 in δh-mitochondria was mostly because of a decreased rate of synthesis.
FIGURE 5.
Synthesis and degradation of mitochondrial translation products. A, RFY5-1 strain grown in the absence (-dox) or in the presence (+dox) of doxycycline (dox) (10 μg·ml-1) was labeled in vivo with 35S-labeled (methionine + cysteine) for 5 min in the presence of cycloheximide to inhibit cytosolic protein synthesis. Excesses of unlabeled methionine and cysteine were added, and samples were taken after the indicated times of chase at 28 °C as described under “Experimental Procedures.” Proteins were separated on a 12.5% gel containing 4 m urea and 25% glycerol. The gel was dried and analyzed with a PhosphorImager. B, for a better resolution of Atp6p, the contrast of the gel area containing Cox3p and Atp6p was enhanced. C, at time 0 of the chase, the level of each subunit was quantified relatively to the total radioactivity incorporated. The ratios between signals obtained for growth in the presence and absence of doxycycline were calculated for each subunit. D, at each time of the chase, Atp6p level was quantified relatively to the total radioactivity incorporated and expressed as a percentage of signal obtained at time 0 of the chase. (Gray circles, absence of doxycycline in the growth medium. Black circles, presence of doxycycline in the growth medium). a.u., arbitrary units.
We then performed Northern blot analyses of total cellular RNAs isolated from RFY5-1 strain grown 14 h in a galactose medium containing (δh-cells) or not (control cells) doxycycline (10 μg·ml-1). The RNAs were hybridized with probes specific to ATP6 and 15 S rRNA transcripts. For each strain, quantification of ATP6 RNA was normalized to 15 S rRNA. These data showed a 35% decrease in the amount of ATP6 RNA in the subunit h-depleted cells (Fig. 6). Thus, the down-regulation of subunit 6 in δh yeast was in large part because of a slowing down in translation.
FIGURE 6.
Northern blot analysis of mitochondrial transcripts isolated from RFY5-1 strain grown with or without doxycycline. Approximately 15 μg of mitochondrial RNA of RFY5-1 grown in the absence (-Dox) or in the presence (+Dox) of doxycycline (Dox) (10 μg·ml-1) were separated on a 1% agarose/formaldehyde gel. The RNAs were blotted onto Nytran and hybridized separately to probes specific to ATP6 and 15S rRNA transcripts. For each strain, the different RNAs were normalized to 15 S rRNA.
Subunit h Depletion Destabilizes the Supramolecular Structure of the Yeast F1F0-ATP Synthase—The influence of a lack in subunit h on the ATP synthase assembly was further examined by CN-PAGE analysis of digitonin-extracted mitochondrial complexes. The presence of the ATP synthase in gel was revealed by its ATPase activity (Fig. 7A). For the control mitochondria, the ATP synthase was, as expected, found as monomers, dimers, and/or oligomers depending on the digitonin/protein ratio used for solubilization of the mitochondria. For the subunit h-depleted mitochondria, the major ATP hydrolysis signal was that of free F1 sector. At a high digitonin to protein ratio (2 g·g-1), a signal of weaker intensity, possibly corresponding to ATP synthase dimers was also detected in extracts from δh-mitochondria. This was confirmed by a second dimension SDS-PAGE followed by Western blot analysis with antibodies against subunits 6 and h (Fig. 7B). Subunits 6 and h were also detected, although to a lesser extent, at a position corresponding to monomeric ATP synthase indicating that a small amount of monomeric enzyme was also present in the δh-mitochondria. The presence of low amounts of fully assembled F1F0 complexes in the δh-mitochondria is because of a small transcriptional leak at the level of the regulatable subunit h gene promoter that could not be completely blocked by doxycycline (see above). A low molecular weight complex showing ATPase activity, possibly corresponding to unassembled F1 subunits, was detected on CN-PAGE of extracts from δh-mitochondria (Fig. 7). Altogether, these analyses indicated that subunit h is required for a correct assembly of the other ATP synthase subunits and/or for the stability of the complex.
FIGURE 7.
Assembly and oligomerization state of subunit h-depleted ATP synthase. A, mitochondria from RFY5-1 strain grown in the absence or in the presence of doxycycline (Dox) were solubilized at different digitonin/protein ratio. After centrifugation, the mitochondrial complexes were separated by CN-PAGE. The gel was incubated with ATP-Mg2+ and lead acetate to reveal the ATPase activity. For representation purposes, the figure is the negative of the original gel. B, samples from δh-mitochondria 2 g·g-1 digitonin extract were separated by CN-PAGE in duplicate: the 1st lane was revealed by ATPase activity (upper part of the B); the 2nd lane was separated in a second dimension by SDS-PAGE, analyzed by Western blotting, and revealed by anti-6 and anti-h polyclonal antibodies (lower part of B). (dim., dimer; mon., monomer).
Modification of the Cristae Morphology in Subunit h-depleted Mitochondria—It has been reported previously that the F1F0-ATP synthase dimerization/oligomerization is involved in the generation of mitochondrial cristae morphology (8, 29). Yeast mutants devoid of either subunit e or g were found to form onion-like structures inside their mitochondria (8) and (Fig. 8C) instead of the small and poorly defined cristae seen in wild type cells (Fig. 8A). Progressive attenuation of subunits e and g expression leads to a morphology intermediate between that of the wild type and onion-like structures, under the form of parallel sheets that cross the whole organelle (29) (Fig. 8D). Interestingly the δh-mitochondria (Fig. 8B) exhibited a morphology very similar to that of mitochondria exhibiting a 60% depletion in subunit e or g. This finding indicates that the ATP synthase subunit h is crucial to a proper biogenesis/folding of the inner mitochondrial membrane.
FIGURE 8.
Alteration of inner structures of mitochondria from RFY5-1 strain grown in the presence of doxycycline. RFY5-1 cells grown in YPGal medium in the absence (A) or in the presence (B) of doxycycline (10 μg·ml-1) for 14 h were analyzed by transmission electron microscopy as described under “Experimental Procedures.” C, Δtim11 cells grown in a YPLac medium. Picture taken from Ref. 8. D, TetO-TIM11 cells were grown in the presence of doxycycline (20 μg·ml-1) for 6 h. Picture taken from Ref. 29. Each picture is representative of the mitochondria observed in the cells. Bars indicate 100 nm.
DISCUSSION
Previous studies on the topology of subunit h in the F1F0-ATP synthase complex revealed the central position of this subunit within the ATP synthase peripheral stalk (20). Biochemical and structural data highlighted the central position of subunit h (F6) in the peripheral stalk that may confer to this protein an important role in the physical coupling between F1 and F0 and in the peripheral stalk stability (20, 22, 25).
To better understand the role of subunit h in ATP synthase assembly and function, we have constructed yeast cells in which the expression of the subunit h gene can be modulated exogenously, through a doxycycline-repressible promoter. With this system we were able to generate populations almost completely depleted in subunit h with only a minimal number, about 5%, of ρ-/ρ0 cells, thus creating correct conditions to analyze the consequences of a specific lack in subunit h.
Depletion of subunit h was found to severely compromise the growth of yeast in nonfermentable media, because of a major impairment in ATP synthesis by the ATP synthase, although the respiratory complexes were not affected. The lack in ATP synthesis in the subunit h-depleted (δh) mitochondria was attributed to defects in the assembly/stability of the ATP synthase. A main feature of δh-mitochondria was a very low content (<6%) in the Atp6p subunit, an essential component of the enzyme proton channel. Subunit f was also strongly reduced, whereas the other essential subunits were all present in much less reduced amounts. State 4 respiration rate in the δh-mitochondria was similar to that of control mitochondria, indicating that the absence of subunit h did not increase the normal permeability to protons of the inner mitochondrial membrane. Moreover, uncoupled and maximal respirations were not affected by subunit h depletion, indicating a normal function of the respiratory chain. Interestingly, depletion of subunit h resulted in dramatic changes in mitochondrial morphology, which further supports the idea that the ATP synthase has a critical role for the proper folding/biogenesis of the inner mitochondrial membrane.
Modification of the Mitochondrial Cristae Morphology in Subunit h-depleted Yeasts—The ATP synthase exists in the inner mitochondrial membrane as homodimers, and there is evidence that ATP synthase dimers associate with each other to form large oligomers. The subunits e, g, k, and i, which are nonessential for the ATP synthesis process, are associated with the ATP synthase peripheral stalk. Among these subunits, e and g were shown to be critical for the enzyme dimerization/oligomerization process (7, 8, 29). Remarkably, yeast strains lacking subunits e and/or g exhibit dramatic changes in mitochondrial morphology, characterized by numerous digitations and onion-like structures instead of the small and poorly defined cristae typical of wild type cells (8). These observations indicate that the supramolecular organization of the ATP synthase is required for the proper generation of mitochondrial cristae, and that the enzyme dimerization and/or oligomerization interface is a key link between ATP synthase and mitochondrial morphology. According to Allen (47), ATP synthase dimers would be responsible for a local deformation of the inner membrane that is amplified by association of additional dimeric complexes leading to cristae formation. However, evidence for this is still lacking.
Mitochondria in δh-yeast exhibited a morphology intermediate between wild type and onion-like structures with the presence of parallel sheets that cross the whole mitochondrial sections. Interestingly, an identical morphology had been observed in yeast cells partially depleted in subunits e and/or g (29). In this study, attenuation of subunits e and g was achieved using the same doxycycline-repressible gene expression system that we used here to block subunit h synthesis (29). The strong similarities in mitochondrial morphology in cells almost totally depleted in subunit h and cells partially depleted in subunits e and/or g, indicate that in both cell types something similar occurs that prevents a correct folding of the inner membrane. A main consequence of a 60–65% depletion in subunits e or g is that substantial amounts of monomeric ATP synthase accumulate in the inner membrane (29), whereas in wild type cells most of the ATP synthase complexes were under oligomeric state. It is possible that beyond a certain accumulation threshold in monomeric ATP synthase, the inner membrane cannot fold properly. We therefore assumed that partial F1F0 assemblies are present despite the absence of subunit h and still have the capacity to influence the organization of the inner membrane. In this respect, it should be noted that in mutants defective in the assembly of the ATP synthase F1 component and where many F0 subunits are missing because of an increased turnover of unassembled F0 subunits, there were no cristae at all, i.e. in these mutants the inner membrane did not form invaginations and remained parallel to the outer membrane (48). The putative assemblies that form in subunit h-depleted cells and that might be responsible for the observed disorganization of the inner membrane are certainly more complex than the sole F1 sector. Indeed in cells lacking mtDNA, where assembled F1 particles are still present despite the absence of F0 (49), the mitochondrial morphology is not very different from that seen in F1 assembly defective mutants,5 which indicates that some key determinant exists also in the F0 for the formation of cristae, as predicted in Allen's hypothesis. The rather good accumulation of most of the essential ATP synthase subunits in the δh-mitochondria is consistent with the assumption that partial F1F0 assemblies can form in absence of subunit h. That CN-PAGE analysis of δh-mitochondria digitonin extracts failed to detect any ATP synthase subcomplex other than F1 does not preclude that a lack in subunit h has a more limited impact on the assembly of incomplete ATP synthase subcomplexes in vivo. This is a reasonable view with regard to a recent study of yeast cells lacking the mitochondrial gene encoding the ATP synthase subunit 6 (45). It is generally assumed, based on the study of many ATP synthase assembly defective mutants, that subunit 6 is the last one to be inserted into the complex, i.e. the other enzyme subunits can still assemble in a Δatp6 mutant. However, only trace amounts of Atp6p-less F1F0 assemblies could be detected by CN-PAGE of Δatp6 mitochondria, indicating that such complexes are very fragile and easily disrupted upon extraction with mild detergent (digitonin) and/or during electrophoretic migration (45). Thus, when both Atp6p and subunit h are missing, it might become impossible to preserve in CN-PAGE the integrity of assemblies made by the other ATP synthase subunits. However, based on the mitochondrial morphology data discussed above, there is little doubt that some structure containing the F1 and part of the F0 forms in vivo when expression of the sole subunit h is blocked, and this structure is likely what causes the abnormal folding of the inner membrane in the δh-mitochondria.
By contrast to subunit h-depleted cells, mitochondria in Δatp6 cells exhibit cristae that are not very different from those seen in wild type yeast (45) It thus appears that the altered mitochondrial morphology in subunit h-depleted cells is not because of the secondary loss of Atp6p but rather to the absence of subunit h itself. As the dimerization and/or oligomerization of the ATP synthase seems to be a key determinant for cristae formation and as subunit h was located near or at the dimerization interface (23), a reasonable view is that subunit h contributes in some way to the establishment of contacts between ATP synthase monomers, either directly or by allowing the dimer-specific subunits e and g to interact efficiently with the enzyme.
The strong decrease in subunit f content (80%) observed in subunit h-depleted yeast is an interesting result. Subunits h and f are known to be in close proximity within the ATP synthase peripheral stalk (20). Subunit h could facilitate the insertion of subunit f into the ATP synthase or stabilize the interactions of subunit f with its partner subunits. The decrease in the content of subunit f in δh-mitochondria might be due to an increased turnover of unassembled subunit f. Subunit k was also strongly reduced to about 20% of its normal content. Little information is available on this small soluble subunit. It is located in the intermembrane space, associated to the dimeric form of ATP synthase in detergent extracts, and its presence in the complex requires both subunits e and g (7, 50). Thus the observed decreased level of subunit k might not be a direct consequence of subunit h depletion but, more likely, might be due to the decrease in subunits e and g.
Subunit h Is Required for Subunit 6 Assembly—A main consequence of the subunit h depletion is the almost complete disappearance of subunit 6, whereas the other essential ATP synthase subunits, with the exception of subunit f, still show a somewhat good accumulation in mitochondria. As already mentioned above, the assembly of the ATP synthase is believed to end with the insertion of subunit 6. A key argument is that subunit 6 fails to accumulate in most mutants where one of the essential ATP synthase subunits, or a protein required for the assembly of the enzyme, is missing. Interestingly, the absence of subunit 6 in δh-yeast was in a large part because of a slowing down in synthesis. This is an unusual observation contrasting with most of the other ATP synthase assembly defective mutants where the absence of subunit 6 results from an increased turnover (12, 16–18, 49). This finding suggests that in addition to a post-translational regulation, subunit 6 might also be regulated at the level of synthesis. There is a number of organellar genes, in chloroplasts and mitochondria, that have been shown to be the target of a regulatory scheme, referred to as CES for Control by Espistasy of Synthesis (51), in which the synthesis of the proteins encoded by these genes is modulated in response to the state of assembly of the complexes to which they belong. A well documented example is the yeast mitochondrial COX1 gene encoding the Cox1p subunit of the respiratory complex IV. With a few exceptions, Cox1p translation was found to be selectively slowed down in mutants unable to assemble the complex IV (52). The down-regulation of Cox1p involves Mss51p, which is required to activate Cox1p translation. Mss51p remains bound to the new Cox1p polypeptide until it enters into the cytochrome c oxidase assembly pathway with the result that Cox1p synthesis is coupled to cytochrome c oxidase assembly. The recent discovery of a specific subunit 6 translation activator, Atp22p, gives support to the idea that the synthesis of subunit 6 might be modulated in response to the assembly of the ATP synthase (44, 53). The down-regulation of subunit 6 synthesis consecutive to a block in subunit h synthesis might be a means to prevent the formation of incomplete Atp6p-containing assemblies that could dissipate the mitochondrial membrane potential. The existence of a tight control on yeast subunit 6 is further supported by the discovery of two proteins, Atp10p and Atp23p, that were shown to exert specific actions in the assembly of Atp6p (17, 18).
Acknowledgments
We thank Jacques Vaillier for helpful technical assistance; Patrick Paumard, Théodore Weimann, Marie-France Giraud, and Alain Dautant for helpful discussions and critical reading of the manuscript, and Christine Schwimmer for English revisions.
This work was supported in part by the CNRS, Université Victor Ségalen, Bordeaux 2, and Etablissement Public Régional d'Aquitaine. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CN-PAGE, clear native PAGE; CCCP, carbonyl cyanide m-chlorophenylhydrazone; DCCD, N,N′-dicyclohexylcarbodiimide; TMPD, 2,2,4-trimethyl-1,3-pentanediol; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
J.-P. di Rago, unpublished data.
References
- 1.Velours, J., and Arselin, G. (2000) J. Bioenerg. Biomembr. 32 383-390 [DOI] [PubMed] [Google Scholar]
- 2.Boyer, P. D. (1993) Biochim. Biophys. Acta 1140 215-250 [DOI] [PubMed] [Google Scholar]
- 3.Duncan, T. M., Bulygin, V. V., Zhou, Y., Hutcheon, M. L., and Cross, R. L. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 10964-10968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noji, H., Yasuda, R., Yoshida, M., and Kinosita, K. J. (1997) Nature 386 299-302 [DOI] [PubMed] [Google Scholar]
- 5.Sabbert, D., Engelbrecht, S., and Junge, W. (1996) Nature 381 623-625 [DOI] [PubMed] [Google Scholar]
- 6.Zhou, Y., Duncan, T. M., Bulygin, V. V., Hutcheon, M. L., and Cross, R. L. (1996) Biochim. Biophys. Acta 1275 96-100 [DOI] [PubMed] [Google Scholar]
- 7.Arnold, I., Pfeiffer, K., Neupert, W., Stuart, R. A., and Schagger, H. (1998) EMBO J. 17 7170-7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paumard, P., Vaillier, J., Coulary, B., Schaeffer, J., Soubannier, V., Mueller, D. M., Brèthes, D., di Rago, J. P., and Velours, J. (2002) EMBO J. 21 221-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orian, J. M., Hadikusumo, R. G., Marzuki, S., and Linnane, A. W. (1984) J. Bioenerg. Biomembr. 16 561-581 [DOI] [PubMed] [Google Scholar]
- 10.Ackerman, S. H., and Tzagoloff, A. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 4986-4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ackerman, S. H. (2002) Biochim. Biophys. Acta 1555 101-105 [DOI] [PubMed] [Google Scholar]
- 12.Lefebvre-Legendre, L., Vaillier, J., Benabdelhak, H., Velours, J., Slonimski, P. P., and di Rago, J. P. (2001) J. Biol. Chem. 276 6789-6796 [DOI] [PubMed] [Google Scholar]
- 13.Stock, D., Leslie, A. G., and Walker, J. E. (1999) Science 286 1700-1705 [DOI] [PubMed] [Google Scholar]
- 14.Hadikusumo, R. G., Meltzer, S., Choo, W. M., Jean-Francois, M. J., Linnane, A. W., and Marzuki, S. (1988) Biochim. Biophys. Acta 933 212-222 [DOI] [PubMed] [Google Scholar]
- 15.Paul, M. F., Velours, J., Arselin de Chateaubodeau, G., Aigle, M., and Guérin, B. (1989) Eur. J. Biochem. 185 163-171 [DOI] [PubMed] [Google Scholar]
- 16.Tzagoloff, A., Barrientos, A., Neupert, W., and Herrmann, J. M. (2004) J. Biol. Chem. 279 19775-19780 [DOI] [PubMed] [Google Scholar]
- 17.Osman, C., Wilmes, C., Tatsuta, T., and Langer, T. (2007) Mol. Biol. Cell 18 627-635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng, X., Neupert, W., and Tzagoloff, A. (2006) Mol. Biol. Cell 18 617-626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velours, J., Vaillier, J., Paumard, P., Soubannier, V., Lai-Zhang, J., and Mueller, D. M. (2001) J. Biol. Chem. 276 8602-8607 [DOI] [PubMed] [Google Scholar]
- 20.Fronzes, R., Chaignepain, S., Bathany, K., Giraud, M. F., Arselin, G., Schmitter, J. M., Dautant, A., Velours, J., and Brèthes, D. (2003) Biochemistry 42 12038-12049 [DOI] [PubMed] [Google Scholar]
- 21.Soubannier, V., Rusconi, F., Vaillier, J., Arselin, G., Chaignepain, S., Graves, P. V., Schmitter, J. M., Zhang, J. L., Mueller, D., and Velours, J. (1999) Biochemistry 38 15017-15024 [DOI] [PubMed] [Google Scholar]
- 22.Rubinstein, J. L., Dickson, V. K., Runswick, M. J., and Walker, J. E. (2005) J. Mol. Biol. 345 513-520 [DOI] [PubMed] [Google Scholar]
- 23.Fronzes, R., Weimann, T., Vaillier, J., Velours, J., and Brèthes, D. (2006) Biochemistry 45 6715-6723 [DOI] [PubMed] [Google Scholar]
- 24.Silvester, J. A., Dickson, V. K., Runswick, M. J., Leslie, A. G. W., and Walker, J. E. (2006) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62 530-533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dickson, V. K., Silvester, J. A., Fearnley, I. M., Leslie, A. G. W., and Walker, J. E. (2006) EMBO J. 25 2911-2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arselin, G., Vaillier, J., Graves, P. V., and Velours, J. (1996) J. Biol. Chem. 271 20284-20290 [DOI] [PubMed] [Google Scholar]
- 27.Gari, E., Piedrafita, L., Aldea, M., and Herrero, E. (1997) Yeast 13 837-848 [DOI] [PubMed] [Google Scholar]
- 28.Duvezin-Caubet, S., Caron, M., Giraud, M. F., Velours, J., and di Rago, J. P. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13235-13240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arselin, G., Vaillier, J., Salin, B., Schaeffer, J., Giraud, M. F., Dautant, A., Brèthes, D., and Velours, J. (2004) J. Biol. Chem. 279 40392-40399 [DOI] [PubMed] [Google Scholar]
- 30.Kun, E., Kirsten, E., and Piper, W. N. (1979) Methods Enzymol. 55 115-118 [DOI] [PubMed] [Google Scholar]
- 31.Chateaubodeau, G. A., Guérin, M., and Guérin, B. (1976) Biochimie (Paris) 58 601-610 [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 33.Gietz, D., St Jean, A., Woods, R. A., and Schiestl, R. H. (1992) Nucleic Acids Res. 20 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guldener, U., Heck, S., Fielder, T., Beinhauer, J., and Hegemann, J. H. (1996) Nucleic Acids Res. 24 2519-2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Law, R. H., Manon, S., Devenish, R. J., and Nagley, P. (1995) Methods Enzymol. 260 133-163 [DOI] [PubMed] [Google Scholar]
- 36.Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951) J. Biol. Chem. 193 265-275 [PubMed] [Google Scholar]
- 37.Rigoulet, M., and Guérin, B. (1979) FEBS Lett. 102 18-22 [DOI] [PubMed] [Google Scholar]
- 38.Emaus, R. K., Grunwald, R., and Lemasters, J. J. (1986) Biochim. Biophys. Acta 850 436-448 [DOI] [PubMed] [Google Scholar]
- 39.Somlo, M. (1968) Eur. J. Biochem. 5 276-284 [DOI] [PubMed] [Google Scholar]
- 40.Egner, R., Mahe, Y., Pandjaitan, R., and Kuchler, K. (1995) Mol. Cell. Biol. 15 5879-5887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schagger, H., and von Jagow, G. (1987) Anal. Biochem. 166 368-379 [DOI] [PubMed] [Google Scholar]
- 42.Wittig, I., and Schagger, H. (2005) Proteomics 5 4338-4346 [DOI] [PubMed] [Google Scholar]
- 43.Grandier-Vazeille, X., and Guérin, M. (1996) Anal. Biochem. 242 248-254 [DOI] [PubMed] [Google Scholar]
- 44.Zeng, X., Hourset, A., and Tzagoloff, A. (2007) Genetics 175 55-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rak, M., Tétaud, E., Godard, F., Sagot, I., Salin, B., Duvezin-Caubet, S., Slonimski, P. P., Rytka, J., and di Rago, J. P. (2007) J. Biol. Chem. 282 10853-10864 [DOI] [PubMed] [Google Scholar]
- 46.Venard, R., Brèthes, D., Giraud, M. F., Vaillier, J., Velours, J., and Haraux, F. (2003) Biochemistry 42 7626-7636 [DOI] [PubMed] [Google Scholar]
- 47.Allen, R. D. (1995) Protoplasma 189 1-8 [Google Scholar]
- 48.Lefebvre-Legendre, L., Salin, B., Schaëffer, J., Brèthes, D., Dautant, A., Ackerman, S. H., and di Rago, J. P. (2005) J. Biol. Chem. 280 18386-18392 [DOI] [PubMed] [Google Scholar]
- 49.Ackerman, S. H., and Tzagoloff, A. (2005) Prog Nucleic Acids Res. Mol. Biol. 80 95-133 [DOI] [PubMed] [Google Scholar]
- 50.Brunner, S., Everard-Gigot, V., and Stuart, R. A. (2002) J. Biol. Chem. 277 48484-48489 [DOI] [PubMed] [Google Scholar]
- 51.Drapier, D., Rimbault, B., Vallon, O., Wollman, F. A., and Choquet, Y. (2007) EMBO J. 26 3581-3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barrientos, A., Zambrano, A., and Tzagoloff, A. (2004) EMBO J. 23 3472-3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Helfenbein, K. G., Ellis, T. P., Dieckmann, C. L., and Tzagoloff, A. (2003) J. Biol. Chem. 278 19751-19756 [DOI] [PubMed] [Google Scholar]










