Abstract
The Akt substrate of 160 kDa (AS160) is phosphorylated on Akt substrate (PAS) motifs in response to insulin and contraction in skeletal muscle, regulating glucose uptake. Here we discovered a dissociation between AS160 protein expression and apparent AS160 PAS phosphorylation among soleus, tibialis anterior, and extensor digitorum longus muscles. Immunodepletion of AS160 in tibialis anterior muscle lysates resulted in minimal depletion of the PAS band at 160 kDa, suggesting the presence of an additional PAS immunoreactive protein. By immunoprecipitation and mass spectrometry, we identified this protein as the AS160 paralog TBC1D1, an obesity candidate gene regulating GLUT4 translocation in adipocytes. TBC1D1 expression was severalfold higher in skeletal muscles compared with all other tissues and was the dominant protein detected by the anti-PAS antibody at 160 kDa in tibialis anterior and extensor digitorum longus but not soleus muscles. In vivo stimulation by insulin, contraction, and the AMP-activated protein kinase (AMPK) activator AICAR increased TBC1D1 PAS phosphorylation. Using mass spectrometry on TBC1D1 from mouse skeletal muscle, we identified several novel phosphorylation sites on TBC1D1 and found the majority were consensus or near consensus sites for AMPK. Semiquantitative analysis of spectra suggested that AICAR caused greater overall phosphorylation of TBC1D1 sites compared with insulin. Purified Akt and AMPK phosphorylated TBC1D1 in vitro, and AMPK, but not Akt, reduced TBC1D1 electrophoretic mobility. TBC1D1 is a major PAS immunoreactive protein in skeletal muscle that is phosphorylated in vivo by insulin, AICAR, and contraction. Both Akt and AMPK phosphorylate TBC1D1, but AMPK may be the more robust regulator.
A defining pathology of type 2 diabetes is impaired insulin-stimulated glucose uptake in skeletal muscle. Skeletal muscle is the largest tissue in the human body by mass and is the chief site of insulin-stimulated glucose disposal. Insulin stimulation causes translocation of GLUT4 glucose transporters from intracellular regions to the plasma membrane and t-tubule system where they function to import glucose. In individuals with type 2 diabetes, insulin fails to stimulate adequate GLUT4 translocation, resulting in impaired glucose uptake and poor glucose tolerance.
Skeletal muscle is unique as an insulin-sensitive tissue because voluntary contraction during exercise causes GLUT4 translocation completely independent of insulin signaling (1, 2). Contraction-stimulated glucose uptake is preserved in the muscle of individuals with type 2 diabetes, thus demonstrating the existence of signaling pathways that circumvent defective components of the insulin signaling pathway (3). If and where insulin- and contraction-stimulated glucose uptake pathways converge have been topics of considerable interest. Recently, the Akt substrate of 160 kDa (AS160)2 was identified as a mediator of both insulin- and contraction-stimulated glucose uptake and, therefore, a potential nexus for convergent signaling (4, 5).
AS160 is a functional rab-GTPase-activating protein (rab-GAP) and is thought to restrain exocytotic GLUT4 translocation by keeping target rabs in an inactive, GDP-bound state (6-8). Phosphorylation of AS160 at Akt substrate motifs (RXRXX(S/T)) is proposed to inhibit AS160 activity or cause dissociation from GLUT4 vesicles. Presumably, removal or disruption of AS160 GAP activity permits target rabs to return to an active, GTP-bound state thereby initiating GLUT4 exocytotic trafficking (6). In support of this concept, serine to alanine mutations that abolish phosphorylation of AS160 on Akt substrate motifs impair insulin- and contraction-stimulated glucose uptake (4, 5, 8). This effect is completely negated by a point mutation disabling the AS160 GAP domain (4, 5), suggesting that the outcome of AS160 phosphorylation is removal or suppression of its GAP activity.
Phosphorylation of AS160 is increased by many stimuli including insulin-like growth factor-1, epidermal growth factor, and phorbol esters (9, 10). In adult skeletal muscle, insulin, the AMP-activated protein kinase (AMPK) activator 5-aminoimidizole-4-carboxymide 1-β-d-ribofuranoside (AICAR), and contraction stimulate AS160 phosphorylation concomitant with glucose uptake (11-13). Rodent studies suggest that in skeletal muscle, insulin-stimulated AS160 phosphorylation is regulated by Akt, AICAR-stimulated AS160 phosphorylation is regulated by AMPK, and contraction-stimulated AS160 phosphorylation is regulated chiefly by AMPK but also Akt (12, 14).
Phosphorylation of AS160 has been routinely measured by immunoblotting lysates or immunoprecipitates with a phospho-Akt-substrate (PAS) antibody that binds to phosphorylated Akt substrate motifs (11, 12, 14-21). Furthermore, AS160 was discovered by using the PAS antibody to immunoprecipitate proteins harboring phosphorylated Akt substrate motifs from insulin-stimulated 3T3-L1 adipocytes (17). However, the expression of Akt substrates may vary by tissue, and Akt substrates other than AS160 that also have a molecular weight near 160 may be detected by the PAS antibody.
In the current study we found a dissociation between AS160 expression and apparent insulin- and AICAR-stimulated AS160 phosphorylation among skeletal muscles of different fiber types, which suggested the presence of an insulin- and AICAR-regulated protein other than AS160. Using mass spectrometry, we discovered this protein to be TBC1D1, an AS160 paralog and severe obesity candidate gene in humans (22) recently reported to regulate GLUT4 translocation in adipocytes (23). We found that TBC1D1 is highly expressed in skeletal muscle but not white adipose tissue or heart. We demonstrate that in skeletal muscle, insulin, AICAR, and contraction directly regulate TBC1D1 phosphorylation. We also report the de novo identification of phosphorylation sites on endogenous TBC1D1 from mouse skeletal muscle and their comparative regulation by insulin and AICAR.
EXPERIMENTAL PROCEDURES
Animals—Protocols for animal use were reviewed and approved by the Institutional Animal Care and Use Committee of the Joslin Diabetes Center. Experimental work was conducted on male ICR mice, aged 8-10 weeks, purchased from Charles River Laboratories (Wilmington, MA). All mice were housed in a 12:12-h light:dark cycle and fed a standard laboratory diet and water ad libitum. Mice were restricted from food for ∼5 h before the start of experiments. Animal experiments started at ∼1 p.m.
In Vivo Insulin and AICAR Administration—For both insulin and AICAR experiments, mice were anesthetized by intraperitoneal injection of sodium pentobarbital (90-100 mg/kg). To elicit a maximal insulin response, mice were injected intraperitoneally with 1 unit of recombinant human insulin (humulin R, #HI 210, Lilly). Controls were injected with saline (0.9% NaCl) or not injected. No difference was observed between saline injected and non-injected controls. To activate AMPK, mice were injected subcutaneously with 1 mg/g AICAR (#A8129, Sigma Aldrich) dissolved in saline (50 mg/ml). Because of solubility and injection volume limitations, this was a practical maximum dosage. Controls were injected with saline (0.9% NaCl). Mice were euthanized by cervical dislocation, and muscles were immediately dissected and snap-frozen in liquid nitrogen.
In Situ Contraction—Mice were anesthetized by intraperitoneal injection of sodium pentobarbital (90-100 mg/kg). Peroneal nerves to both hind limbs were surgically exposed. One hind limb was subjected to electrical stimulation using a Grass S88 pulse generator (Grass Instruments, Quincy, MA) for 15 min (train rate, 2/s; train duration, 500 ms; pulse rate, 100 Hz; duration, 0.1 ms at 1-10 V), and the other hind limb served as a sham-operated control. Because of normal variation in the surgery and the resultant contact of the electrode with the nerve, voltage was manually adjusted so that muscles directly innervated by the peroneal nerve contracted with a full range of motion without the recruitment of extraneous motor groups. Mice were euthanized by cervical dislocation immediately after the cessation of contraction, and tibialis anterior muscles were immediately dissected and snap-frozen in liquid nitrogen.
Preparation of Tissue Lysates—The following buffer was utilized to prepare lysates for immunoblotting, myosin heavy chain separation, measurement of citrate synthase activity, and immunoprecipitation: 50 mm Tris-HCl, 250 mm mannitol, 50 mm NaCl, 50 mm NaF, 1 mm EDTA, 1 mm EGTA, 5 mm sodium pyrophosphate, 5 mm β-glycerophosphate, 10% glycerol (v/v), 1% Nonidet P-40 substitute (v/v) (Calbiochem), 1 mm Na3VO4, 1 mm DTT, and Complete Mini protease inhibitor tablets (Roche Applied Science, #4693124), pH 7.4, at 4 °C. Frozen tissues were pulverized at liquid nitrogen temperature and homogenized with a Polytron (Brinkman Instruments, Westbury, NY) in ice-cold buffer. Lysates were centrifuged at 14,000 × g for 10 min. Lysate protein concentrations were determined by the Bradford assay (24).
Immunoblots—Lysates (30 μg protein) and immunoprecipitates were separated by SDS-PAGE before immunoblotting (25). Antibody-bound proteins were visualized on film using chemiluminescence detection reagents (PerkinElmer Life Sciences). Exposed film was scanned with an ImageScanner (GE Healthcare), and bands were quantitated by densitometry (Fluor-Chem 2.0; Alpha Innotech, San Leandro, CA). Commercially available primary antibodies were anti-AS160 (#07-741, Millipore, Billerica, MA), anti-GLUT4 (#400064, Calbiochem), anti-α-tubulin (#SC5286, Santa Cruz Biotechnology, Santa Cruz, CA), anti-PAS (#9611, Cell Signaling Technology, Danvers, MA), anti-phospho-Akt-Thr-308 (#9275, Cell Signaling Technology), and anti-phospho-AMPK-Thr 172 (#2535, Cell Signaling Technology). Serum-purified anti-TBC1D1 antibody was generated by Cell Signaling Technology by immunizing rabbits. To confirm that the anti-TBC1D1 antibody did not cross-react with AS160, TBC1D1 was immunoprecipitated from a tibialis anterior muscle lysate, and the pre-depletion lysate, supernatant, and immunoprecipitate were immunoblotted for TBC1D1 and AS160. Immunoprecipitations without antibody or lysate were included as additional controls and demonstrated that the immunodepletion and immunoprecipitation of TBC1D1 were dependent upon the anti-TBC1D1 antibody but that the anti-TBC1D1 antibody alone did not generate the TBC1D1 signal detected in the immunoprecipitate lane. To minimize the possibility of nonspecific interactions, the anti-TBC1D1 antibody preparations used to immunoprecipitate and immunoblot TBC1D1 were from different rabbits.
Immunoprecipitations—AS160 was immunoprecipitated with a goat polyclonal antibody made against the C-terminal region of AS160 (#ab5909, Abcam, Cambridge, MA, or #100-1313, Novus Biologicals, Littleton, CO). The long form of AS160 was immunoprecipitated with an antibody made against the splice exon of AS160. This antibody was kindly donated by Dr. Gustav Lienhard of Dartmouth Medical School, Hanover, NH. Protein G-agarose beads (#22851, Pierce) or protein G Fast Flow-Sepharose beads (#17-0618-01, GE Healthcare) were used to bind anti-AS160, anti-TBC1D1, or anti-PAS antibodies. Bead-antibody-protein complexes were washed 1× with lysis buffer, 1× or 2× with lysis buffer + 500 mm NaCl, and 1× with lysis buffer. Pellets were aspirated and spotted with 5-10 μl of 1 μg/μl bovine serum albumin before elution. Bovine serum albumin was utilized as a carrier protein to maximize the efficiency of immunoprecipitated protein elution. Proteins were eluted from protein G beads by adding Laemmli buffer (26) and heating for 5 min at 95 °C.
Myosin Heavy Chain Separation—Myosin heavy chain isoforms were separated as previously described (27) with slight modification. Before electrophoresis, β-mercaptoethanol (1 μl/ml) was added to a 12× upper running buffer consisting of 600 mm Tris (base), 900 mm glycine, and 0.6% SDS. Lower running buffer consisted of 50 mm Tris (base), 75 mm glycine, and 0.05% SDS. Soleus muscle (1.5 μg), tibialis anterior (TA) muscle (1 μg), and extensor digitorum longus (EDL) muscle (1 μg) lysates were prepared as above under “Preparation of Tissue Lysates” (this section of text) and were heated at 95 °C for 5 min in 2× Laemmli buffer at a final volume of 12 μl. After heating, 10 μl was loaded onto gels for separation. Gels were run at 100 V for 1 h and 150 V for ∼20 h. Temperature was maintained at 4-8 °C for the duration of the run. Gels were silver-stained with the SilverSNAP Stain Kit II (Pierce, 24612). Myosin heavy chain fraction was quantitated from images of scanned gels using the 1D-Multi function of AlphaEase FC software.
Citrate Synthase Activity—Citrate synthase activity was measured according to the method of Srere (28) with slight modification. Lysates were prepared as above under “Preparation of Tissue Lysates,” and citrate synthase activity was measured at room temperature on a 96-well plate with a final reaction volume of 200 μl.
Polymerase Chain Reaction—Total RNA was isolated from muscle and white adipose tissue with RNeasy Fibrous Tissue (#74704) and Lipid Tissue (#74804) mini kits according to the manufacturer's instructions (Qiagen, Valencia CA). RNA was reverse-transcribed to cDNA with Moloney murine leukemia virus reverse transcriptase (#M1701, Promega, Madison, WI). cDNA was amplified for 40 cycles using SYBR Green PCR Master Mix (#4309155, Applied Biosystems, Framingham, MA) on an ABI Prism 7000 sequence detection system (Applied Biosystems). Primer efficiencies and the relative differences in the starting quantity of each transcript were derived from the equations developed by Pfaffl (29). β-Actin was utilized as an internal standard. Primers utilized were: AS160 forward, 5′-CCATGAAGGAGGACTCCAAA-3′, and AS160 reverse, 5′-TCATGCAGGCTGAACTTGTC-3′; TBC1D1 forward, 5′-GTGAGGAAGAGGCGTTCAAG-3′, and TBC1D1 reverse, 5′-AGGTCTCGGTGGTAATCGTG-3′; β-actin forward, 5′-CAACGAGCGGTTCCGATG-3′, and β-actin reverse, 5′-GCCACAGGATTCCATACCCA-3′. To compare the relative abundances of AS160 and TBC1D1 splice variants, cDNA was amplified with exon-flanking primers for 30 cycles (98 °C × 10s, 61.7 °C × 20s, and 72 °C × 60s) with the Phusion Hot Start High-Fidelity DNA Polymerase (#F540-S, New England Biolabs). Primers utilized were: AS160 forward, 5′-GGCTTGGAAGTATGGACAGC-3′, and AS160 reverse, 5′-CATGGTGGGAGAGAGAGGAG-3′; TBC1D1 forward, 5′-GTGACTCAGAGGGCCACATT-3′, and TBC1D1 reverse, 5′-TGGCCACTCGAAGGAATATC-3′. Amplicons were separated by agarose gel electrophoresis and imaged with ethidium bromide staining under UV light.
In Vitro Phosphorylation of TBC1D1—TBC1D1 sufficient for 60 reactions was immunoprecipitated from a ∼6-mg pooled tibialis anterior muscle lysate. The following buffers were used for this procedure: enzyme buffer (50 mm Tris, 250 mm mannitol, 1 mm EDTA, 1 mm EGTA, 10% glycerol, 0.02% Brij-35, pH 7.4, at 4 °C) and kinase activation buffer (100 mm Hepes, 200 mm NaCl, 25 mm MgCl2, 2 mm EDTA, 20% glycerol, 2 mm DTT, 0.5 mm AMP, and 0.5 mm ATP, pH 7.0); DTT, AMP, and ATP were added just before use. Immunoprecipitated TBC1D1 was incubated with 10 ng of recombinant AMPK (#P47-10H, SignalChem, Richmond, BC, Canada) or recombinant Akt (#14-447, Millipore, Billerica, MA) for 0, 30, or 60 min, 10 ng of AMPK + 10 ng Akt for 60 min, or enzyme buffer without enzyme for 60 min at 37 °C. Before use, AMPK and Akt were diluted in enzyme buffer with 1 μg/μl bovine serum albumin and 2 mm DTT. Protein G-antibody-TBC1D1 complexes were aspirated and resuspended in enzyme buffer with 4 mm DTT and bovine serum albumin equivalent to 5 μg per reaction. Immunoprecipitated TBC1D1 suspension, kinase activation buffer, double distilled H2O, and diluted enzymes were combined at a ratio of 1:2:1:1. Reactions were stopped by adding 4× Laemmli buffer, and phosphorylation was analyzed by immunoblotting with the PAS antibody.
Mass Spectrometry—For mass spectrometry experiments, AS160 and PAS-160 (TBC1D1) were immunoprecipitated from pooled tibialis anterior muscle lysates (∼40 mg, ∼5 mg/ml), subjected to SDS-PAGE, and stained with GelCode Blue Stain Reagent (#24592, Pierce). For PAS-160 identification and analysis at the Joslin Diabetes Center Proteomics Core, AS160 was simultaneously depleted with both the C-terminal and splice exon AS160 antibodies before TBC1D1 immunoprecipitation with the PAS antibody. Samples were reduced and alkylated with iodacetamide before SDS-PAGE. Samples were digested with trypsin, and peptides were analyzed by liquid chromatography-tandem mass spectrometry in an LTQ linear ion trap mass spectrometer (Thermofinnigan, San Jose, CA) by methods routinely utilized by the Joslin Diabetes Center Proteomics Core (30-32). To compare insulin- and AICAR-stimulated phosphorylation of TBC1D1, AS160 was depleted with the C-terminal AS160 antibody, phosphorylated TBC1D1 was immunoprecipitated with the PAS antibody, and samples were reduced and alkylated in-gel. Samples were digested with trypsin or chymotrypsin and analyzed by liquid chromatography-tandem mass spectrometry in an LTQ-Orbitrap mass spectrometer (Thermofinnigan) by methods routinely utilized by the Taplin Biological Mass Spectrometry Facility (33-35). In all cases data were analyzed with the sequest algorithm, and reported phosphopeptides were verified by manual inspection of spectra.
Data Analysis and Statistics—Data from immunoblots were normalized by setting the average of control or reference values to 1. For real-time PCR, AS160 cDNA levels in individual soleus muscles were set to 1, and data were normalized to AS160 in soleus muscle. Statistical analyses were completed using SigmaStat 3.5 (Systat, San Jose, CA) or Excel (Microsoft, Redmond, WA). Means were compared by t test, one-way analysis of variance (ANOVA) or two-way ANOVA. When differences between means were detected by one- or two-way analysis of variance, Fisher's least significance difference test was used for post hoc testing. When data failed tests for normality or equal variance, data were rank-transformed before analysis. Data are expressed as the means ± S.E. The differences between groups were considered significant when p < 0.05.
RESULTS
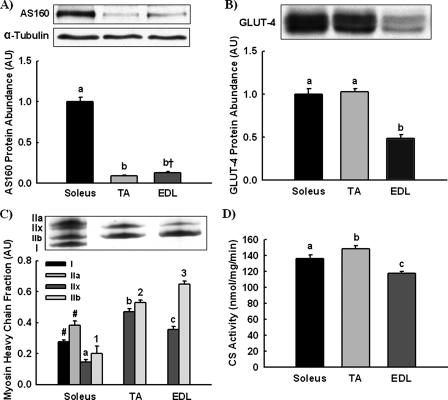
AS160 Expression—AS160 functions as a brake to restrain GLUT4 transporter exocytosis. Therefore, we compared AS160 and GLUT4 expression in muscles of different fiber types to test the hypothesis that muscles expressing higher levels of GLUT4 would express greater amounts of AS160. AS160 expression (Fig. 1A), GLUT4 expression (Fig. 1B), myosin heavy chain fraction (Fig. 1C), and citrate synthase activity (Fig. 1D) were measured in soleus, TA, and EDL muscle. AS160 expression was ∼10-fold greater in soleus muscle compared with tibialis anterior and EDL muscle. In contrast, GLUT4 expression was equal in soleus and tibialis anterior muscles. Myosin heavy chain analysis demonstrated that soleus, but not tibialis anterior or EDL muscle, expressed type I and IIa myosin heavy chain. No association was found with citrate synthase activity, a marker for mitochondrial content, which was greatest in tibialis anterior muscle followed by soleus and then EDL. Thus, AS160 expression was associated with myosin heavy chain type I and IIa, not GLUT4 expression or citrate synthase activity.
FIGURE 1.
AS160 protein abundance is greatest in soleus muscle. Relative AS160 protein (A) and GLUT4 protein abundances (B) were compared in soleus, TA, and EDL muscle by immunoblotting. α-Tubulin was utilized as a loading control for both AS160 and GLUT4 but is only shown for AS160 because quantitated images for AS160 and GLUT4 were from the same gels. C, myosin heavy chain fractions were determined by electrophoretic separation and silver staining. D, citrate synthase (CS) activity was measured by spectrophotometric assay. The data are expressed as means ± S.E. (n = 6-8). a-c, 1-3, groups within each panel not sharing a common letter are statistically different at p < 0.05; †, p = 0.052. Groups annotated by letters cannot be compared with groups annotated by numbers. #, types I and IIa myosin heavy chain were only detected in soleus muscle and excluded from the statistical analysis.
Insulin-stimulated PAS-160 Phosphorylation—Phosphorylation of AS160 at Akt substrate motifs (RXRXXS(S/T)) has been measured in numerous studies by immunoblotting with a PAS antibody (11, 12, 14-21). Because the PAS antibody can detect multiple phosphorylated proteins, the band detected by the PAS antibody at a molecular weight of 160 will be referred to as PAS-160.
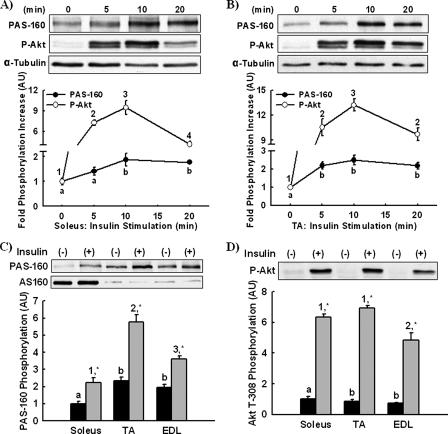
Insulin stimulates AS160 phosphorylation and increases glucose uptake by removing the brake effect of AS160 on GLUT4 exocytosis (8). We tested the hypothesis that maximal insulin-stimulated PAS-160 phosphorylation would be proportional to AS160 expression. Soleus and tibialis anterior muscles express the greatest and least amounts of AS160. Accordingly, we used these muscles to first determine the time courses of maximal PAS-160 and Akt Thr-308 phosphorylation by injecting mice with insulin for 5, 10, or 20 min. Blood glucose was not significantly decreased until the 20-min time point and not to the point of hypoglycemia (221 ± 7, 144 ± 6 mg/dl). In comparison to controls (time 0), PAS-160 and Akt Thr-308 phosphorylation in both soleus (Fig. 2A) and tibialis anterior (Fig. 2B) muscle were maximal at 10 min. Using lysates from the 10-min time point, we compared total PAS-160 phosphorylation among soleus, tibialis anterior, and EDL muscles (Fig. 2C). Surprisingly, PAS-160 phosphorylation was greatest in tibialis anterior muscle, not soleus muscle. Thus, AS160 expression and PAS-160 phosphorylation varied inversely. Akt Thr-308 phosphorylation in soleus and tibialis anterior muscles was similar, suggesting the disparity in PAS-160 phosphorylation was not to due to differences in phosphorylation-dependent Akt activity (Fig. 2D).
FIGURE 2.
Insulin-stimulated PAS-160 phosphorylation is greatest in TA muscle. The time courses of insulin-stimulated Akt substrate phosphorylation at a molecular weight of 160 (PAS-160) and Akt Thr-308 phosphorylation (P-Akt) in vivo were assessed by injecting mice with insulin intraperitoneally for 0 (control), 5, 10, or 20 min. PAS-160 and P-Akt were measured in soleus (A) and tibialis anterior muscle lysates (B) by immunoblotting. α-Tubulin was utilized as a loading control. Maximal PAS-160 (C) and P-Akt phosphorylation (D) between soleus, TA, and EDL muscles were compared by immunoblotting lysates from the 10-min time points. A paired immunoblot for AS160 is included in panel C. The data are expressed as the means ± S.E. (n = 6-8). a and b, 1-4, groups within each panel not marked by the same letter or number are statistically different. Groups annotated by letters cannot be compared with groups annotated by numbers. *, Insulin stimulation for a given muscle had a statistically significant effect on phosphorylation compared with basal, p < 0.05.
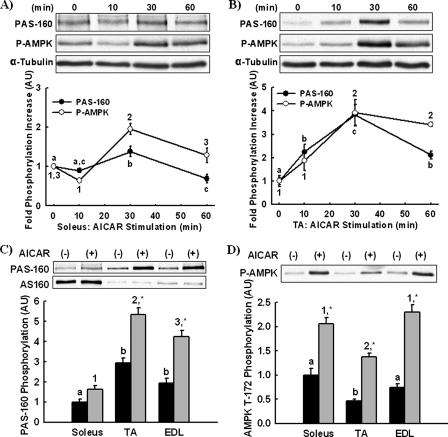
AICAR-stimulated PAS-160 Phosphorylation—AICAR-stimulated activation of the AMPK also causes PAS-160 phosphorylation (11, 12, 14). We determined the time course of AICAR-stimulated PAS phosphorylation and found that PAS-160 and AMPK Thr-172 phosphorylation in both soleus (Fig. 3A) and tibialis anterior (Fig. 3B) muscles were maximal at 30 min. Using lysates from the 30-min time point, we determined that AICAR-stimulated PAS-160 phosphorylation was greatest in tibialis anterior muscle not soleus muscle (Fig. 3C). AMPK Thr-172 phosphorylation was greater in soleus than tibialis anterior muscle, suggesting that the lower level of PAS-160 phosphorylation in soleus was not due to less activation of AMPK. Thus, AS160 expression and PAS-160 phosphorylation with both insulin and AICAR stimulation were completely dissociated.
FIGURE 3.
AICAR-stimulated PAS-160 phosphorylation is greatest in TA muscle. The time courses of AICAR-stimulated Akt substrate phosphorylation at a molecular weight of 160 (PAS-160) and AMPK Thr-172 phosphorylation (P-AMPK) in vivo were determined by injecting mice with AICAR subcutaneously for 0 (control), 10, 30, or 60 min. PAS-160 and P-AMPK were measured in soleus (A) and tibialis anterior muscle (B) lysates by immunoblotting. α-Tubulin was utilized as a loading control. Maximal PAS-160 (C) and P-AMPK (D) phosphorylation between soleus, TA, and EDL muscle was compared by immunoblotting lysates from the 30-min time points. A paired immunoblot for AS160 is included in panel C. The data are expressed as the means ± S.E. (n = 3-8). a-c, 1-3, groups within each panel not marked by the same letter or number are statistically different. Groups annotated by letters cannot be compared with groups annotated by numbers. *, AICAR stimulation for a given muscle had a statistically significant effect on phosphorylation compared with basal, p < 0.05.
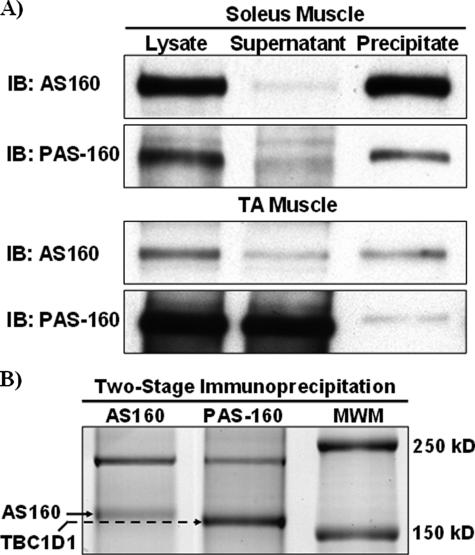
Identification of PAS-160 in Tibialis Anterior Muscle as TBC1D1—The disparate pattern of AS160 expression and PAS-160 phosphorylation between soleus and tibialis anterior muscles suggested two possibilities. First, the PAS-160 phosphorylation detected in tibialis anterior muscle could be originating from a protein other than AS160. Second, AS160 PAS phosphorylation could be regulated in a fiber-type-specific manner independent of AS160 expression. To address this question, we immunoprecipitated AS160 from soleus and tibialis anterior muscle lysates and compared immunoprecipitation and immunodepletion of AS160 and PAS-160 (Fig. 4A). In samples from soleus muscle, the PAS-160 signal in the pre-depletion lysate, supernatant, and immunoprecipitate corresponded with AS160. Conversely, in samples from tibialis anterior muscle, AS160 depletion did not result in significant PAS-160 depletion. This suggested that AS160 constituted only a small fraction of PAS-160 in tibialis anterior muscle.
FIGURE 4.
Identification of TBC1D1 as PAS-160 in TA muscle. A, to determine whether AS160 was the phospho-Akt-substrate detected at a molecular weight of 160 (PAS-160) in soleus and tibialis anterior muscle, AS160 was immunoprecipitated from insulin-stimulated soleus and tibialis anterior muscle lysates. Lysates, supernatants, and immunoprecipitated AS160 were immunoblotted (IB) for both AS160 and PAS-160. Gels for AS160 and PAS-160 immunoblots were loaded identically with volume equivalents of lysates and supernatants and a supra-proportional amount of AS160 immunoprecipitate to compensate for inefficient elution. AS160 and PAS-160 immunodepletion and immunoprecipitation patterns matched in soleus but not tibialis anterior muscle. B, to determine the identity of PAS-160 in tibialis anterior muscle, lysates were first immunodepleted of AS160; then the PAS antibody was used to immunoprecipitate the remaining PAS-160. AS160 and PAS-160 immunoprecipitates were subjected to SDS-PAGE and stained with Coomassie Blue. The AS160 and PAS-160 bands were excised from the gel and identified by mass spectrometry. The identity of AS160 was confirmed, and PAS-160 was identified as TBC1D1.
To determine the identity of PAS-160 in tibialis anterior muscle, lysates were first immunodepleted of AS160. Next, the PAS antibody was used to immunoprecipitate PAS-160 from the AS160-depleted supernatants. AS160 and PAS immunoprecipitates were subjected to SDS-PAGE, and the resulting gel was stained with Coomassie Blue (Fig. 4B). Staining with Coomassie Blue revealed that the protein immunoprecipitated by the PAS antibody was more abundant than AS160 and had slightly greater electrophoretic mobility compared with AS160. Using mass spectrometry, we confirmed the identity of AS160 and discovered the identity of the PAS immunoprecipitated protein to be TBC1D1.
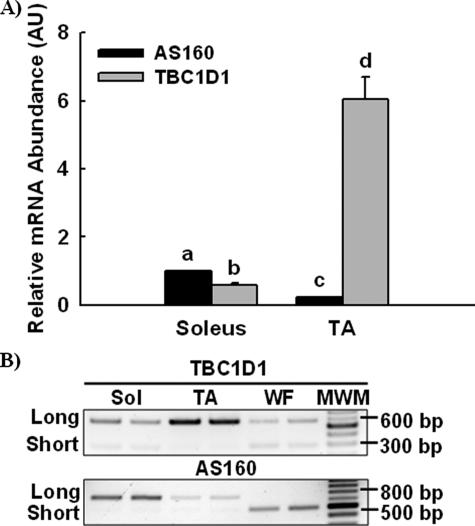
Characterization of TBC1D1 mRNA Expression—Little was known about TBC1D1, so we first characterized expression of TBC1D1 mRNA in comparison to AS160. Relative TBC1D1 and AS160 mRNA abundances in soleus and tibialis anterior muscles were compared by real-time PCR (Fig. 5A). AS160 mRNA expression in soleus muscle was greater than TBC1D1, whereas in tibialis anterior muscle TBC1D1 was many-fold greater than AS160. Sequencing databases indicated that TBC1D1 and AS160 each express a long and short splice variant. To determine the relative expression of the long and short TBC1D1 and AS160 splice variants in soleus, tibialis anterior, and white adipose tissue, we amplified TBC1D1 and AS160 by PCR with splice exon-flanking primers, separated amplicons by agarose gel electrophoresis, and imaged amplicons with ethidium bromide staining under ultraviolet light (Fig. 5B). The long form of TBC1D1 predominated in skeletal muscle, whereas both long and short forms were expressed similarly in white adipose tissue. Similar to TBC1D1, the long form of AS160 predominated in skeletal muscle. However, in contrast to TBC1D1, white adipose tissue only expressed the short form of AS160.
FIGURE 5.
TBC1D1 and AS160 mRNA expression and relative distribution of splice variants. A, relative TBC1D1 and AS160 mRNA abundances were compared by isolating RNA from TA and soleus muscle, reverse transcribing RNA to cDNA, and then amplifying cDNA by real-time PCR. The data are expressed as the means ± S.E. (n = 8). a-d, groups within each panel not sharing a common letter or number are statistically different, p < 0.05. B, relative expression of TBC1D1 and AS160 splice variants within soleus muscle, tibialis anterior muscle, and white fat were compared by isolating RNA, reverse transcribing RNA to cDNA, amplifying TBC1D1 and AS160 cDNA by PCR with splice exon-flanking primers, separating amplicons by agarose gel electrophoresis, and imaging with ethidium bromide staining under ultraviolet light. Replicates produced similar results.
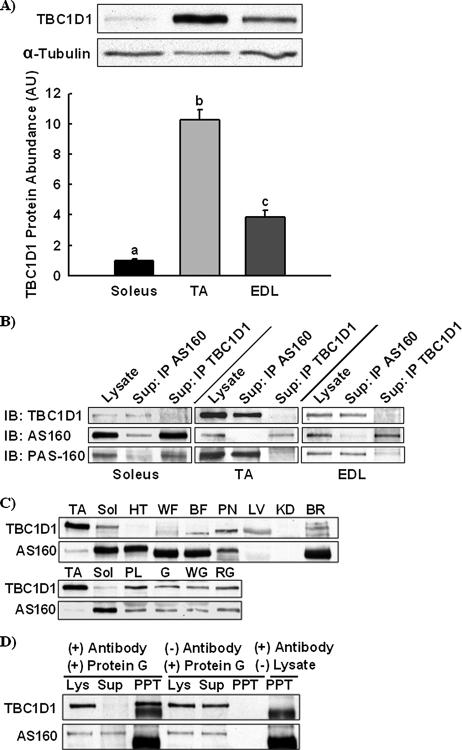
Characterization of TBC1D1 Protein Expression—We developed an anti-TBC1D1 antibody to directly measure TBC1D1 protein expression by immunoblotting. We compared relative TBC1D1 protein expression in soleus, tibialis anterior, and EDL muscles (Fig. 6A). TBC1D1 protein expression was highest in tibialis anterior muscle (more than 10-fold greater than soleus muscle) followed by EDL and then soleus. Thus, TBC1D1, not AS160 expression, was proportional to insulin- and AICAR-stimulated PAS-160 phosphorylation. To determine the relative contributions of TBC1D1 and AS160 to the PAS-160 signal in soleus, tibialis anterior, and EDL muscles, we immunodepleted muscle lysates of AS160 and TBC1D1. Supernatants depleted of either AS160 or TBC1D1 were immunoblotted for PAS-160 in comparison to starting lysates (Fig. 6B). Immunodepletion of TBC1D1 from tibialis anterior and EDL, but not soleus muscle, resulted in nearly complete PAS-160 depletion. In soleus muscle, immunodepletion of AS160, but not TBC1D1, resulted in nearly complete PAS-160 depletion. Thus, PAS-160 may be almost exclusively AS160 in soleus and TBC1D1 in tibialis anterior. Accordingly, the time courses of PAS-160 phosphorylation in soleus (Figs. 2A and 3A) and tibialis anterior (Figs. 2B and 3B) muscle may represent the time courses of AS160 and TBC1D1 PAS phosphorylation, respectively.
FIGURE 6.
TBC1D1 is the major PAS-160 in skeletal muscle. A, relative TBC1D1 protein abundance in soleus, TA, and EDL muscle was compared by immunoblotting. α-Tubulin was utilized as a loading control. The data are expressed as means ± S.E. (n = 8). a-c, groups within each panel not sharing a common letter or number are statistically different, p < 0.05. B, AS160 and TBC1D1 were immunoprecipitated (IP) from soleus, TA, and EDL muscle lysates. Pre-depletion lysates and depleted supernatants were immunoblotted (IB) for TBC1D1, AS160, and phospho-Akt substrate at a molecular weight of 160 (PAS-160). C, TBC1D1 and AS160 protein expression in TA, soleus muscle (sol), heart (HT), white adipose tissue (WA), brown adipose tissue (BA), pancreas (PN), liver (LV), kidney (KD), brain (BR), plantaris muscle (P), whole gastrocnemius muscle (G), white gastrocnemius muscle (WG), and red gastrocnemius muscle (RG) were compared by immunoblotting. Replicates produced similar results. D, to confirm that the anti-TBC1D1 antibody did not cross-react with AS160, TBC1D1 was immunoprecipitated from a tibialis anterior muscle lysate, and the pre-depletion lysate, supernatant, and immunoprecipitate (PPT) were immunoblotted for TBC1D1 and AS160. Gels for TBC1D1 and AS160 immunoblots were loaded identically. Immunoprecipitations without antibody or lysate were included as additional controls and demonstrated that the immunodepletion and immunoprecipitation of TBC1D1 were dependent upon the anti-TBC1D1 antibody but that the anti-TBC1D1 antibody alone did not generate the TBC1D1 signal detected in the immunoprecipitate lane.
Next, using tibialis anterior and soleus muscle as reference points, we compared TBC1D1 and AS160 protein among multiple tissues and muscles (Fig. 6C). TBC1D1 and AS160 were detected at different molecular weights among different tissues indicating the tissue-specific distribution of splice variants. AS160 expression was similar among soleus, heart, white adipose tissue, brown adipose tissue, and brain and significantly lower in pancreas and other muscles, with none detected in liver or kidney. TBC1D1 expression was highest in muscle, with low levels in white adipose tissue and brown adipose tissue and none detected in heart. In muscle, TBC1D1 expression was greatest in tibialis anterior followed by plantaris and red gastrocnemius. Soleus muscle had the lowest level of TBC1D1 among muscles but still expressed significantly more than white adipose tissue and heart. Because insulin-stimulated GLUT4 translocation is regulated by a similar mechanism in skeletal muscle, heart, and adipose tissue, this raises the possibility that TBC1D1 may have a specialized function in skeletal muscle, potentially regulating insulin-independent mechanisms of glucose transport. Fig. 6D shows that the TBC1D1 antibody does not cross-react with AS160.
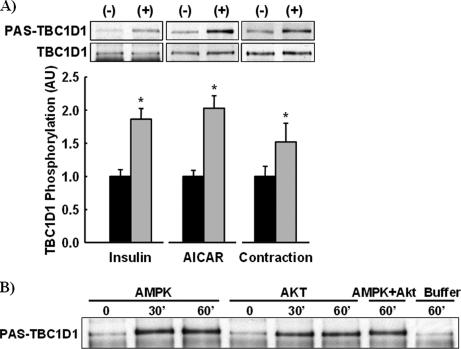
Insulin, AICAR, and Muscle Contraction Increase TBC1D1 Phosphorylation—To test the hypothesis that stimuli that increase glucose transport in muscle increase TBC1D1 phosphorylation, we immunoprecipitated TBC1D1 from insulin-, AICAR-, and contraction-stimulated tibialis anterior muscle and immunoblotted immunoprecipitates with the PAS antibody (Fig. 7A). All three stimuli significantly increased TBC1D1 PAS phosphorylation. The finding that contraction-stimulated TBC1D1 PAS phosphorylation was slightly less than with insulin and AICAR may be due to the fact that contracted muscles were snap-frozen ∼45 s after cessation of contraction. This could allow for some dephosphorylation, whereas AICAR- and insulin-stimulated signaling would persist after dissection until freezing. Interestingly, AICAR and contraction, but not insulin, induced a slight upward band shift on immunoblots, demonstrating a greater decrease in TBC1D1 electrophoretic mobility. This suggests that AICAR and contraction may stimulate TBC1D1 phosphorylation at multiple sites.
FIGURE 7.
Insulin, AICAR, and contraction regulate TBC1D1 phosphorylation in skeletal muscle. A, TBC1D1 was immunoprecipitated from insulin-, AICAR-, and contraction-stimulated tibialis anterior muscle lysates and immunoblotted for both TBC1D1 and PAS phosphorylation. TBC1D1 phosphorylation was calculated by normalizing PAS-phosphorylated TBC1D1 to total TBC1D1. The data are expressed as the means ± S.E. (n = 6-8). *, stimulation caused a statistically significant increase in TBC1D1 phosphorylation compared with controls, p < 0.05. B, immunoprecipitated TBC1D1 was phosphorylated in vitro by AMPK and Akt for 0, 30, or 60 min, by AMPK and Akt combined for 60 min, and buffer alone for 60 min. Replicates produced similar results.
AMPK and Akt Phosphorylate TBC1D1—Insulin stimulation activates Akt, AICAR stimulation activates AMPK, and muscle contraction activates both AMPK and Akt. These phenomena suggested that AMPK and Akt may be TBC1D1 kinases. To test this hypothesis, TBC1D1 was immunoprecipitated and incubated in vitro with recombinant AMPK, Akt, or AMPK plus Akt for 30 or 60 min. Incubation with AMPK, Akt, or AMPK and Akt combined resulted in similar increases in TBC1D1 PAS phosphorylation (Fig. 7B). No differences were observed between 30- and 60-min incubations, indicating maximal TBC1D1 PAS phosphorylation was achieved by 30 min. TBC1D1 phosphorylation by AMPK, but not Akt, caused a distinct upward shift in electrophoretic mobility similar to AICAR and contraction stimulation, which activate AMPK in vivo (Fig. 7A). Together, these findings suggest that AMPK may phosphorylate TBC1D1 at multiple sites in vivo and in vitro.
AICAR and Insulin Differentially Regulate TBC1D1 Phosphorylation—We utilized mass spectrometry to identify TBC1D1 phosphorylation sites in skeletal muscle and test the hypothesis that AICAR and insulin stimulation result in differential phosphorylation of TBC1D1. AICAR- and insulin-stimulated tibialis anterior muscle lysates were first pre-cleared of AS160 by immunoprecipitation. Next, TBC1D1 was immunoprecipitated with the PAS antibody to enrich phosphorylated TBC1D1. Thus, phosphorylation was compared between AICAR- and insulin-stimulated TBC1D1 but not unstimulated TBC1D1. TBC1D1 phosphorylation sites were identified using an LTQ-Orbitrap mass spectrometer at Ser-231, Thr-253, Thr-499, Thr-590, Ser-621, Ser-660, and Ser-700 (Table 1).
TABLE 1.
TBC1D1 phosphorylation sites identified by mass spectrometry AICAR- and insulin-stimulated tibialis anterior muscle lysates were immunodepleted of AS160. Phosphorylated TBC1D1 was then immunoprecipitated from the supernatants using the PAS antibody and analyzed for phosphorylation by tandem liquid chromatography-tandem mass spectrometry using an LTQ-Orbitrap mass spectrometer. Relative ion peak intensities (RPI) for phosphopeptides were calculated by dividing the ion peak intensity for a phosphopeptide by that of its cognate nonphosphopeptide. The ratio of relative peak ion intensities (RPI ratio) was calculated by dividing AICAR relative peak ion intensities by insulin relative peak ion intensities. Although semiquantitative, the 10–54-fold greater relative peak ion intensities observed with AICAR-stimulated phosphorylation of Ser-231, Ser-660, and Ser-700 strongly suggest that AICAR stimulates greater phosphorylation of these sites than insulin. Thr-253 phosphorylation may be greater with insulin, and phosphorylation of Ser-621 may be similar between AICAR and insulin. Thr-499 phosphorylation was only observed with AICAR stimulation but appears to be phosphorylated at very low levels. We did not achieve coverage of the TBC1D1 PAS site, Thr-590, during the comparative analysis, perhaps due to modification of the epitope by the PAS antibody. Phosphorylation of Thr-590 was separately confirmed using an LTQ-linear ion trap mass spectrometer. All phosphopeptides were verified by manual inspection of spectra. S/T* denotes phosphorylation. N/A, not applicable.
| Phosphorylation site | Phosphopeptide | RPI AICAR | RPI insulin | RPI ratio |
|---|---|---|---|---|
| Ser-231 | SFS*QPGLR | 5.5 | 0.20 | 28 |
| KSFS*QPLGR | 34 | 0.64 | 54 | |
| Thr-253 | QDASLRRSST*F | 1.3 | 9.6 | 0.14 |
| Thr-499 | SLT*ESLESILSR | 0.0036 | N/A | N/A |
| Thr-590 | ANT*LSHFPVECPAPPEPAQSSPGVSQR | N/A | N/A | N/A |
| Ser-621 | YHS*VSTETPHER | 3.0 | 2.3 | 1.3 |
| Ser-660 | LNPSAS*SPNFFK | 0.83 | 0.023 | 37 |
| Ser-700 | LHSSSS*VPNFLK | 56 | 5.3 | 10 |
| KLHSSSS*VPNFLK | 43 | 0.83 | 52 |
The ratio of relative peak ion intensities was computed to semiquantitatively compare the effects of AICAR and insulin on specific phosphorylation sites. Peptides and their cognate phosphopeptides have similar ionization and detection efficiencies (36). Thus, the phosphorylation of a specific site can be semiquantitatively compared between different samples by normalizing the ion intensities of the phosphopeptides of interest to those of their cognate nonphosphopeptides, which serve as an internal standard. Sano et al. (5) previously used a similar approach to characterize insulin-stimulated phosphorylation of AS160. Our data suggest that relative to insulin, AICAR increased phosphorylation of Ser-231, Ser-660, and Ser-700 and that Thr-253 phosphorylation may be greater in insulin-treated samples. However, in contrast to the other phosphorylation sites we identified, Thr-253 is not conserved in humans. Thr-499 phosphorylation was only detected with AICAR stimulation, and Ser-621 phosphorylation was similar between AICAR and insulin. Although combined amino acid coverage between AICAR- and insulin-stimulated TBC1D1 exceeded 97%, we did not initially achieve coverage of the TBC1D1 PAS site, Thr-590, perhaps due to modification of the epitope by the PAS antibody. Because immunoblots with the PAS antibody demonstrated that AICAR and insulin regulate Thr-590 phosphorylation similarly, we did not pursue a comparative analysis but confirmed phosphorylation of Thr-590 on insulin-stimulated TBC1D1 using an LTQ-linear ion trap mass spectrometer. Future studies with phospho-specific antibodies will be required to quantitatively assess the regulation of these sites.
DISCUSSION
TBC1D1 (tre-2/USP6, BUB2, cdc16 domain family member 1) is a member of the TBC1 Rab-GTPase family and an AS160 paralog. TBC1D1 and AS160 are 47% identical and share several structural features. Both proteins contain two full or partial N-terminal phosphotyrosine binding domains, a splice-exon, a calmodulin binding domain, and a C-terminal Rab-GAP domain. The TBC1D1 and AS160 Rab-GAP domains exhibit identical Rab specificities (23). A key difference between TBC1D1 and AS160 is the number of Akt substrate motifs (RXRXX(S/T)). AS160 harbors six Akt substrate motifs, and insulin stimulation increases phosphorylation of five (5). Of these, only Thr-642 (human Thr-649) is conserved in TBC1D1. The paralogous TBC1D1 site is Thr-590 (human Thr-596). Several studies have demonstrated that phosphorylation of AS160 Akt substrate motifs regulates GLUT4 translocation and glucose uptake (4, 5, 8, 10, 37-39), but the function of TBC1D1 is less characterized.
Our data demonstrate that TBC1D1 is the overall major phospho-Akt substrate detectable at a molecular weight of 160 (PAS-160) in tibialis anterior and EDL muscle. In comparison to the tibialis anterior and soleus muscle, the levels of TBC1D1 and AS160 in the plantaris and gastrocnemius muscles were similar to those of the EDL muscle where TBC1D1 is clearly the dominant PAS-160, suggesting that TBC1D1 is also the major PAS-160 in these muscles (Figs. 1A and 6, A-C). TBC1D1 expression in muscle, like AS160, was not associated with GLUT4 protein or citrate synthase activity, a marker of mitochondrial content. Interestingly, TBC1D1 expression in soleus, tibialis anterior, and EDL muscle tracked with myosin heavy chain type IIx content in skeletal muscle and was not found at all in heart muscle.
AICAR may stimulate greater PAS phosphorylation of TBC1D1 than AS160. The time courses of insulin-stimulated PAS-160 phosphorylation in soleus (AS160) and tibialis anterior (TBC1D1) muscles were similar. However, AICAR-stimulated PAS-160 phosphorylation in soleus muscle was several-fold less than in tibialis anterior muscle. This suggests AMPK may play a greater role in PAS phosphorylation of TBC1D1 than for AS160. Alternatively, PAS sites specifically regulated by AMPK may be less efficiently detected than those regulated by insulin. Further investigation will be needed to fully characterize determinants of TBC1D1 and AS160 phosphorylation and expression in different muscles.
Two studies suggest that TBC1D1 functions as a regulator of fuel homeostasis. First, genetic linkage analyses found that a TBC1D1 R125W missense variant contributes to severe obesity in humans (22). Further analyses suggested that the R125W allele requires interaction with an unidentified gene to have this effect. Second, Roach et al. (23) recently demonstrated that overexpression of wild type TBC1D1 in 3T3-L1 adipocytes severely impaired insulin-stimulated GLUT4 translocation. They found that insulin stimulation increased PAS antibody-detectable phosphorylation of TBC1D1. However, mutation of Thr-596 (mouse Thr-590) to Ala completely abolished TBC1D1 PAS phosphorylation without causing a further decrement in GLUT4 translocation. This suggests that PAS phosphorylation of TBC1D1 may not be the major mode of regulation.
Another potential mechanism of TBC1D1 regulation is through phosphorylation by AMPK. The greater comparative phosphorylation of TBC1D1 Ser-231, Ser-660, and Ser-700 by AICAR is consistent with direct phosphorylation by AMPK. Ser-231 and Ser-700 are consensus matches for the AMPK phosphorylation motif (Φ(Xβ)XX(S/T)XXXΦ; Φ= Met, Val, Leu, Ile, or Phe, and β = Arg, Lys, or His) (40), and Ser-660 is a consensus match save the lack of one basic residue at either the -3 or -4 position. Thr-590, the TBC1D1 PAS site phosphorylated by AMPK in vitro, is also one residue away from a consensus match. Furthermore, our observation that phosphorylation of TBC1D1 with AMPK but not Akt induced an electrophoretic mobility shift provides additional evidence for a greater comparative regulation by AMPK. Thus, the major mode of TBC1D1 regulation may be phosphorylation by AMPK.
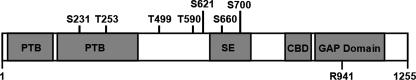
The splice variable region of TBC1D1 may have important regulatory functions. The mouse long variant of TBC1D1 contains a central 94-amino acid sequence (631-724) that is absent in the short variant. Interestingly, the AICAR-regulated phosphorylation sites at Ser-660 and Ser-700 are both within the splice variable region (Fig. 8). Roach et al. (23) found that overexpression of the wild type TBC1D1 short form in 3T3 L1 adipocytes reduced GLUT4 translocation by ∼80%, whereas in a previous study this group found that overexpression of wild type AS160 had no effect on translocation (5). Perhaps the absence of Ser-660 and Ser-700 in the overexpressed short form of wild type TBC1D1 caused the constitutive inhibitory effect on GLUT4 translocation. Alternatively, the robust overexpression of exogenous TBC1D1 compared with the minimal amounts of endogenous TBC1D1 expressed in adipose tissue may have led to inhibition of GLUT4 translocation.
FIGURE 8.
Diagram of TBC1D1 structure. TBC1D1 harbors two phosphotyrosine binding domains (PTB), a splice-exon (SE), a calmodulin-binding domain (CBD), seven identified phosphorylation sites, and a rab-GTPase activating domain (GAP domain). Arg-941 is the arginine residue necessary for catalytic activity.
Using an in vivo overexpression technique, we previously demonstrated that phosphorylation of AS160 regulates skeletal muscle glucose uptake (4, 41). We utilize the mouse tibialis anterior for this procedure because of its ideal size and accessibility. Because of the very high expression level of TBC1D1 in tibialis anterior muscle, this technique may be ineffective for functional characterization of TBC1D1 phosphorylation sites. Future studies, perhaps utilizing transgenic mice will be required to characterize the effect of TBC1D1 phosphorylation on skeletal muscle function. Because of the structural similarity of TBC1D1 to AS160, identical Rab specificity, regulation of glucose uptake in adipocytes, and unequivocal regulation by stimuli that regulate glucose uptake in skeletal muscle, we think that TBC1D1, like AS160, may function to regulate glucose uptake in skeletal muscle. A comparatively greater regulation of TBC1D1 by AMPK concomitant with the high expression of TBC1D1 in muscle is consistent with a role for TBC1D1 in AMPK-mediated glucose uptake in skeletal muscle during exercise. Nonetheless, it is also possible that TBC1D1 has no role in skeletal muscle glucose uptake and, instead, regulates another insulin, AICAR, and contraction-sensitive process.
In conclusion, we demonstrate that TBC1D1 expression is highest in skeletal muscle, that insulin, AICAR, and contraction regulate TBC1D1 phosphorylation, and that Akt and AMPK directly phosphorylate TBC1D1 in vitro. Using mass spectrometry, we compared phosphorylation of TBC1D1 by insulin and AICAR. Our data suggest that AICAR stimulated multisite phosphorylation of TBC1D1 by activating AMPK. To our knowledge, this is the first application of mass spectrometry for comprehensive de novo identification of phosphorylation sites on an endogenous mouse skeletal muscle protein.
Acknowledgments
We thank Ross Tomaino and Steve Gygi from the Taplin Biological Mass Spectrometry Facility for expert mass spectrometry service, James D. Casaletto and Lauren E. Peter for technical assistance, and Dr. Gustav Lienhard for the generous donation of AS160 exon-specific antibody and many helpful discussions.
This research was supported by National Institutes of Health Grants AR42238 and AR45670 (to L. J. G.), Individual Kirschstein National Research Service Award F32 DK075851 (to E. B. T.), Joslin Diabetes Center Diabetes and Endocrinology Research Center Grant DK36836, Canadian Institutes of Health Research Fellowship MFE-83802, and Canadian Diabetes Association Incentive Award PF-3-07-2255-DA (to D. A.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: AS160, Akt substrate of 160 kDa; AICAR, 5-aminoimidazole-4-carboxamide 1-β-d-ribofuranoside; AMPK, AMP-activated protein kinase; GAP, GTPase-activating protein; PAS, phospho-Akt substrate; PAS-160, phospho-Akt-substrate detectable at a molecular weight of 160; TBC1D1, tre-2/USP6, BUB2, cdc16 domain family member 1; AU, arbitrary units; DTT, dithiothreitol; TA, tibialis anterior; EDL, extensor digitorum longus.
References
- 1.Richter, E. A., Nielsen, J. N., Jorgensen, S. B., Frosig, C., Birk, J. B., and Wojtaszewski, J. F. (2004) Proc. Nutr. Soc. 63 211-216 [DOI] [PubMed] [Google Scholar]
- 2.Jessen, N., and Goodyear, L. J. (2005) J. Appl. Physiol. 99 330-337 [DOI] [PubMed] [Google Scholar]
- 3.Kennedy, J. W., Hirshman, M. F., Gervino, E. V., Ocel, J. V., Forse, R. A., Hoenig, S. J., Aronson, D., Goodyear, L. J., and Horton, E. S. (1999) Diabetes 48 1192-1197 [DOI] [PubMed] [Google Scholar]
- 4.Kramer, H. F., Witczak, C. A., Taylor, E. B., Fujii, N., Hirshman, M. F., and Goodyear, L. J. (2006) J. Biol. Chem. 281 31478-31485 [DOI] [PubMed] [Google Scholar]
- 5.Sano, H., Kane, S., Sano, E., Miinea, C. P., Asara, J. M., Lane, W. S., Garner, C. W., and Lienhard, G. E. (2003) J. Biol. Chem. 278 14599-14602 [DOI] [PubMed] [Google Scholar]
- 6.Sano, H., Eguez, L., Teruel, M. N., Fukuda, M., Chuang, T. D., Chavez, J. A., Lienhard, G. E., and McGraw, T. E. (2007) Cell Metab. 5 293-303 [DOI] [PubMed] [Google Scholar]
- 7.Miinea, C. P., Sano, H., Kane, S., Sano, E., Fukuda, M., Peranen, J., Lane, W. S., and Lienhard, G. E. (2005) Biochem. J. 391 87-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeigerer, A., McBrayer, M. K., and McGraw, T. E. (2004) Mol. Biol. Cell 15 4406-4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geraghty, K. M., Chen, S., Harthill, J. E., Ibrahim, A. F., Toth, R., Morrice, N. A., Vandermoere, F., Moorhead, G. B., Hardie, D. G., and MacKintosh, C. (2007) Biochem. J. 407 231-241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thong, F. S., Bilan, P. J., and Klip, A. (2007) Diabetes 56 414-423 [DOI] [PubMed] [Google Scholar]
- 11.Bruss, M. D., Arias, E. B., Lienhard, G. E., and Cartee, G. D. (2005) Diabetes 54 41-50 [DOI] [PubMed] [Google Scholar]
- 12.Kramer, H. F., Witczak, C. A., Fujii, N., Jessen, N., Taylor, E. B., Arnolds, D. E., Sakamoto, K., Hirshman, M. F., and Goodyear, L. J. (2006) Diabetes 55 2067-2076 [DOI] [PubMed] [Google Scholar]
- 13.Treebak, J. T., Birk, J. B., Rose, A. J., Kiens, B., Richter, E. A., and Wojtaszewski, J. F. (2007) Am. J. Physiol. Endocrinol. Metab. 292 715-722 [DOI] [PubMed] [Google Scholar]
- 14.Treebak, J. T., Glund, S., Deshmukh, A., Klein, D. K., Long, Y. C., Jensen, T. E., Jorgensen, S. B., Viollet, B., Andersson, L., Neumann, D., Wallimann, T., Richter, E. A., Chibalin, A. V., Zierath, J. R., and Wojtaszewski, J. F. (2006) Diabetes 55 2051-2058 [DOI] [PubMed] [Google Scholar]
- 15.Bouzakri, K., Karlsson, H. K., Vestergaard, H., Madsbad, S., Christiansen, E., and Zierath, J. R. (2006) Diabetes 55 785-791 [DOI] [PubMed] [Google Scholar]
- 16.Deshmukh, A., Coffey, V. G., Zhong, Z., Chibalin, A. V., Hawley, J. A., and Zierath, J. R. (2006) Diabetes 55 1776-1782 [DOI] [PubMed] [Google Scholar]
- 17.Kane, S., Sano, H., Liu, S. C., Asara, J. M., Lane, W. S., Garner, C. C., and Lienhard, G. E. (2002) J. Biol. Chem. 277 22115-22118 [DOI] [PubMed] [Google Scholar]
- 18.Karlsson, H. K., Ahlsen, M., Zierath, J. R., Wallberg-Henriksson, H., and Koistinen, H. A. (2006) Diabetes 55 1283-1288 [DOI] [PubMed] [Google Scholar]
- 19.Karlsson, H. K., Hallsten, K., Bjornholm, M., Tsuchida, H., Chibalin, A. V., Virtanen, K. A., Heinonen, O. J., Lonnqvist, F., Nuutila, P., and Zierath, J. R. (2005) Diabetes 54 1459-1467 [DOI] [PubMed] [Google Scholar]
- 20.Karlsson, H. K., Zierath, J. R., Kane, S., Krook, A., Lienhard, G. E., and Wallberg-Henriksson, H. (2005) Diabetes 54 1692-1697 [DOI] [PubMed] [Google Scholar]
- 21.Peck, G. R., Ye, S., Pham, V., Fernando, R. N., Macaulay, S. L., Chai, S. Y., and Albiston, A. L. (2006) Mol. Endocrinol. 20 2576-2583 [DOI] [PubMed] [Google Scholar]
- 22.Stone, S., Abkevich, V., Russell, D. L., Riley, R., Timms, K., Tran, T., Trem, D., Frank, D., Jammulapati, S., Neff, C. D., Iliev, D., Gress, R., He, G., Frech, G. C., Adams, T. D., Skolnick, M. H., Lanchbury, J. S., Gutin, A., Hunt, S. C., and Shattuck, D. (2006) Hum. Mol. Genet. 15 2709-2720 [DOI] [PubMed] [Google Scholar]
- 23.Roach, W. G., Chavez, J. A., Miinea, C. P., and Lienhard, G. E. (2007) Biochem. J. 403 353-358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradford, M. M. (1976) Anal. Biochem. 72 248-254 [DOI] [PubMed] [Google Scholar]
- 25.Towbin, H., Staehelin, T., and Gordon, J. (1979) Proc. Natl. Acad. Sci. U. S. A 76 4350-4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. (1970) Nature 227 680-685 [DOI] [PubMed] [Google Scholar]
- 27.Bamman, M. M., Clarke, M. S., Talmadge, R. J., and Feeback, D. L. (1999) Electrophoresis 20 466-468 [DOI] [PubMed] [Google Scholar]
- 28.Srere, P. (1969) Methods Enzymol. 13 3-6 [Google Scholar]
- 29.Pfaffl, M. W. (2001) Nucleic Acids Res. 29 2002-2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishida-Takahashi, R., Rosario, F., Gong, Y., Kopp, K., Stancheva, Z., Chen, X., Feener, E. P., and Myers, M. G., Jr. (2006) Mol. Cell. Biol. 26 4063-4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao, B. B., Hansen, H., Chen, H. C., and Feener, E. P. (2006) Biochem. J. 397 337-344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feener, E. P., Rosario, F., Dunn, S. L., Stancheva, Z., and Myers, M. G., Jr. (2004) Mol. Cell. Biol. 24 4968-4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villen, J., Beausoleil, S. A., Gerber, S. A., and Gygi, S. P. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1488-1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greer, E. L., Oskoui, P. R., Banko, M. R., Maniar, J. M., Gygi, M. P., Gygi, S. P., and Brunet, A. (2007) J. Biol. Chem. 282 30107-30119 [DOI] [PubMed] [Google Scholar]
- 35.Beausoleil, S. A., Villen, J., Gerber, S. A., Rush, J., and Gygi, S. P. (2006) Nat. Biotechnol. 24 1285-1292 [DOI] [PubMed] [Google Scholar]
- 36.Steen, H., Jebanathirajah, J. A., Rush, J., Morrice, N., and Kirschner, M. W. (2006) Mol. Cell. Proteomics 5 172-181 [DOI] [PubMed] [Google Scholar]
- 37.Bai, L., Wang, Y., Fan, J., Chen, Y., Ji, W., Qu, A., Xu, P., James, D. E., and Xu, T. (2007) Cell Metab. 5 47-57 [DOI] [PubMed] [Google Scholar]
- 38.Eguez, L., Lee, A., Chavez, J. A., Miinea, C. P., Kane, S., Lienhard, G. E., and McGraw, T. E. (2005) Cell Metab. 2 263-272 [DOI] [PubMed] [Google Scholar]
- 39.Ramm, G., Larance, M., Guilhaus, M., and James, D. E. (2006) J. Biol. Chem. 281 29174-29180 [DOI] [PubMed] [Google Scholar]
- 40.Dale, S., Wilson, W. A., Edelman, A. M., and Hardie, D. G. (1995) FEBS Lett. 361 191-195 [DOI] [PubMed] [Google Scholar]
- 41.Kramer, H. F., Taylor, E. B., Witczak, C. A., Fujii, N., Hirshman, M. F., and Goodyear, L. J. (2007) Diabetes 56 2854-2862 [DOI] [PubMed] [Google Scholar]