Abstract
We report here that blocking the activity of the 26 S proteasome results in drastic changes in the morphology of the mitochondria and accumulation of intermembrane space (IMS) proteins. Using endonuclease G (endoG) as a model IMS protein, we found that accumulation of wild-type but to a greater extent mutant endoG leads to changes in the morphology of the mitochondria similar to those observed following proteasomal inhibition. Further, we show that wild-type but to a greater extent mutant endoG is a substrate for ubiquitination, suggesting the presence of a protein quality control. Conversely, we also report that wild-type but not mutant endoG is a substrate for the mitochondrial protease Omi but only upon inhibition of the proteasome. These findings suggest that although elimination of mutant IMS proteins is strictly dependent on ubiquitination, elimination of excess or spontaneously misfolded wild-type IMS proteins is monitored by ubiquitination and as a second checkpoint by Omi cleavage when the proteasome function is deficient. One implication of our finding is that in the context of attenuated proteasomal function, accumulation of IMS proteins would contribute to the collapse of the mitochondrial network such as that observed in neurodegenerative diseases. Another implication is that such collapse could be accelerated either by mutations in IMS proteins or by mutations in Omi itself.
The elimination of misfolded proteins represents an important mechanism for the maintenance of cellular viability. Such protein quality controls (PQC)4 involve the binding of a chaperone to the misfolded protein and its presentation to the ubiquitin-dependent proteasome degradation pathway (1, 2). Linkage of ubiquitin to a protein is a highly organized process involving the sequential action of a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin-ligase (E3) (3). Most of the regulation of the ubiquitination pathway occurs at the level of the ubiquitin ligase. This enzymatic cascade results in the attachment of a polyubiquitin chain onto specific lysine residues of a substrate. Although several types of polyubiquitin chains have now been reported, chains that are built on lysine 48 of ubiquitin serve as a signal for degradation by the 26 S proteasome. The 20 S catalytic core of the proteasome consists of three distinct proteolytic activities within a cylindrical barrel that is flanked by two 19 S regulatory caps. The proteasomes are localized both in the nucleus and in the cytoplasm, where they exist free or in association with organelles including the endoplasmic reticulum (4, 5).
PQC in the cytoplasm involves chaperones of the heat shock protein (hsp) family that act in concert with ubiquitin ligases (2). A similar PQC acting in the nucleus has recently been reported (6). Ubiquitin-dependent PQC was also found to monitor the folding of proteins present in the lumen of the endoplasmic reticulum, a result that was at first surprising since proteins that are translocated across the ER membrane were thought to be no longer accessible to the cytosolic ubiquitin-proteasome pathway (1, 7, 8). However, it was subsequently found that misfolded proteins in the ER lumen are recognized by luminal chaperones such as BiP and retrotranslocated into the cytosol for their ubiquitin-dependent degradation (9). Further, the formation of the ubiquitin chain appears to be required for the retrotranslocation process (9, 10), and a fraction of the proteasomes associates with the ER membrane to degrade these proteins as they emerge (11). The observation that inhibition of the proteasome activity results in the accumulation of misfolded proteins on the luminal side of the ER supports this model.
PQC acting in the mitochondria has also been described, but unlike other PQC, elimination of misfolded proteins from the mitochondria does not require ubiquitination but rather the action of the AAA-proteases (12, 13). The mitochondrion is a complex organelle that contains two membranes, the outer membrane and the inner membrane. The later forms invaginations called cristae and contains the matrix where mitochondrial DNA resides. The space between the inner and outer membranes is referred to as the intermembrane space (IMS) and contains several proteins. Most IMS proteins are best known for their roles in the initiation of apoptosis upon their release from the mitochondria and include among others cytochrome c, smac, HtrA2/Omi, and endonuclease-G (endoG).
Recently, the substrates of the AAA-proteases have been characterized and shown to include soluble proteins from the matrix and transmembrane proteins from the inner membrane (14). However, only one protein of the IMS was identified, raising questions about what regulates the folding quality of these proteins.
We report here that inhibition of the proteasome leads to the accumulation of IMS proteins in the mitochondria and that a ubiquitin-dependent protein quality control limits the import of mutant IMS proteins. In addition, we found that aberrant accumulation of mutated IMS proteins promotes the condensation of the mitochondria similarly to that observed upon inhibition of the proteasome. Taken together, our results suggest a novel role of the ubiquitin pathway in the maintenance of the mitochondria.
EXPERIMENTAL PROCEDURES
Cell Culture, Plasmids, and Transfections—HEK293T were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics (Invitrogen). Wild-type and mutant endonuclease-G were cloned in the pEGFP-N1 plasmid. All transfections were performed using the FuGENE™ 6 system, as described by the manufacturer (Roche Applied Science).
Immunoprecipitation and Immunoblot Analysis—For protein extractions, cells were washed three times in ice-cold PBS and lysed in 200 μl of ice-cold lysis buffer (50 mm Tris, pH 7.5, 250 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, 50 mm NaF, 0.2 mm Na3VO4, 1 g/ml leupeptin, 1 g/ml pepstatin, 100 g/ml phenylmethylsulfonyl fluoride, 1 mm dithiothreitol). Lysates were clarified by centrifugation at 13,000 rpm for 20 min at 4 °C, and the protein concentration of the supernatant was assayed using Bio-Rad protein assay (Bio-Rad). Immunoprecipitations were performed as described previously (15), and for immunoblot analysis, proteins were separated by SDS-PAGE electrophoresis, transferred to nitrocellulose membrane (PerkinElmer Life Sciences), and probed with the following antibodies: rabbit anti-PHB2 (Upstate Biotechnology), mouse anti-FLAG (Sigma), mouse anti-cytochrome c (BD Biosciences), mouse anti-tubulin antibody, 12G10 (University of Iowa), mouse monoclonal anti-Myc antibody 9E10 to detect Myc-ubiquitin, mouse monoclonal anti-HA 12CA5 to detect HA-ubiquitin, rabbit anti-endonulceaseG (Calbiochem, PC684), and rabbit anti-GFP (Santa Cruz Biotechnology, cs-8334). Immunoblots were developed by ECL (Amersham Biosciences).
Immunofluorescence Staining—HEK293T cells were plated in 6-well tissue culture plates containing a sterile coverslip at a density of 2.5 × 105 cells/well and were transfected with GFP-endoG. Cells were washed three times and fixed in 3.7% paraformaldehyde-PBS for 15 min at room temperature. The cells were then permeabilized using PBS containing 0.1% Triton X-100, 0.5% bovine serum albumin for 5 min at room temperature. A blocking solution of 2% bovine serum albumin, PBS was applied to the cells for 15 min at room temperature. Mouse anti-endoG (1:200, BD Pharmingen) was used, and anti-mouse Alexa Fluor 594 or anti-rabbit Alexa Fluor 488 (Molecular probes) was used as secondary antibody at a dilution of 1/1000 and was incubated for 1 h at room temperature. Following antibody incubations, cells were washed with 1 ml of 0.05% Triton X-100, PBS for 2 min and then rinsed in PBS. Coverslips were removed from wells and mounted on glass slides, in Citifluor AF1 (Alltech) mounting solution. Images were taken on a Nikon eclipse microscope equipped with a DIC L 10X and a confocal laser scanning microscope (Zeiss LSM 510 Meta) and processed using Spot advanced program 3.5.8 (Diagnostic Instruments Inc.). All experiments were done at room temperatures.
RESULTS
Inhibition of the 26 S Proteasome Promotes the Accumulation of IMS Proteins and Electron-dense Mitochondria—To investigate the potential role of the ubiquitin pathway in the monitoring of IMS proteins, we first inhibited the proteasome using LLnL and analyzed the effect of such treatment on cytochrome c by immunofluorescence. We found that the staining intensity of cytochrome c increased when compared with untreated cells (supplemental Fig. S1). As LLnL also inhibits the activity of calpain, the role of the proteasome in this observation was further tested using a more specific proteasome inhibitor, MG132. Treatment with MG132 also led to the accumulation of cytochrome c, and further, led to the clustering of the staining in a perinuclear location (supplemental Fig. S1). To determine whether this effect was specific to cytochrome c, immunofluorescence of another intermembrane space protein, namely endoG, was also performed in the presence or absence of proteasome inhibitors. We found that as it was observed with cytochrome c, the levels of endoG increased upon treatment with proteasome inhibitor and further that the staining also displayed a clustered pattern (Fig. 1A). The same clustering was observed using antibody against other IMS proteins, namely Omi (supplemental Fig. S2), and further, the same observation was reported by another group using immunofluorescence against PINK (16); therefore, inhibition of the proteasome leads to the clustering of all IMS proteins tested.
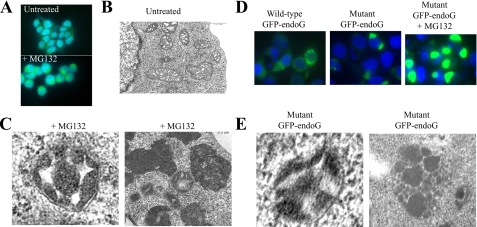
FIGURE 1.
Inhibition of the proteasome or accumulation of mutant EndoG promotes mitochondrial condensation. A, HEK293T cells were treated with or without the proteasome inhibitor MG132 for 18 h, and the levels of endogenous endoG was determined by immunofluorescence. B, electron microscopy of the mitochondria of untreated HEK293T cells. C, HEK293T cells were treated with MG132 for 18 h, and the mitochondria were visualized by electron microscopy. The panel on the left shows a wheel-like morphology, whereas the panel on the right shows dark condensed mitochondria. D, HEK293T cells were transfected with either wild-type or mutant GFP-endoG for 24 h with and without proteasome inhibitor added for the last 18 h. The numbers of transfected cells with an even mitochondrial distribution versus a mitochondrial aggregation pattern were scored. E, HEK293T cells were transfected with mutant endoG for 24 h, and the mitochondria were visualized by electron microscopy.
We next determined the effect of proteasome inhibition on the mitochondria rather than on the staining pattern of individual IMS proteins using electron microscopy. This analysis revealed that upon proteasome inhibition, the mitochondria became filled with electron-dense material when compared with mitochondria in untreated cells (Fig. 1, B and C). These results support the notion that upon inhibition of the proteasome, proteins accumulate in the mitochondria, causing the opacity, and are consistent with the accumulation of IMS proteins observed by immunofluorescence. Further, these results raised the possibility that a proteasome-dependent PQC normally acts to limit the import of excess or misfolded mitochondrial proteins. In agreement with this possibility, the accumulation of mitochondrial proteins and the appearance of electron-dense mitochondria was also reported following treatment with an inhibitor of the chaperone hsp90 (17).
To begin investigating the possibility of a PQC, HEK293T cells were transfected with wild-type GFP-endonuclease G or a mutant form of endonuclease G (endoG-N174) in the absence of proteasome inhibitor. We found that accumulation of wild-type endonuclease G led to a normal mitochondrial staining in 78% of the transfected cells (Fig. 1D), whereas in 22% of the cells, GFP-endoG staining showed a perinuclear clustering similar to those observed with endogenous endoG in the presence of proteasome inhibitor. When mutant endoG was overexpressed, the percentage of cells with clustered staining pattern increased to 58% of cells (Fig. 1D). We then tested the effect of proteasome inhibitor on the staining pattern of mutant endoG. We found that upon proteasomal stress, 100% of transfected cells show GFP-positive clustering with increased staining intensity (Fig. 1C). The accumulation of mutant endoG in the mitochondria was confirmed using fractionation of the mitochondria and cytosol. As expected, endoG localized to the mitochondria, and further, treatment with LLnL led to an increased in the levels of endoG in the mitochondria fraction but not in the cytosol (supplemental Fig. S3). We next tested the effect of accumulation of mutant endoG on the morphology of the mitochondria by electron microscopy. We found that overexpression of endoG-N174 also led to the accumulation of electron-dense mitochondria (Fig. 1E) similar to those observed upon treatment with proteasome inhibitor (Fig. 1C). However, the opacity of the mitochondria was not as severe as the opacity observed following proteasome inhibition, suggesting that although accumulation of IMS proteins such as endoG is likely to contribute to the appearance of electron dense material, upon proteasomal stress, stabilization of other proteins also contribute to this phenotype. In agreement with this possibility, inhibition of hsp90 was reported to lead to the accumulation of mitochondrial proteins from the matrix and inner membrane (17).
We next determined the effect of expression of endoG-N174 where the mitochondria localization signal was deleted (endoG-N174ΔMLS). We found that endoG-N174ΔMLS localized evenly throughout the cytoplasm and nucleus, and as expected, did not localize to the mitochondria (supplemental Fig. S4, a and b). Further, deletion of the mitochondrial signal completely abolished the ability of endoG-N174 to promote clustering despite the fact that its expression level was equal to those of endoG-N174 (supplemental Fig. S4c). This result indicates that overexpression of mutant endoG does not lead to protein aggresomes and that import in the mitochondria is required for the increased opacity and clustering of the mitochondria to be observed. Taken together, these results suggest that the activity of the proteasome is required to limit the accumulation of proteins in the IMS and that upon inhibition of the proteasome, such accumulation contributes to the collapse of the mitochondria.
IMS Proteins Are Monitored by a Ubiquitin-dependent Protein Quality Control Prior to Their Mitochondrial Import—To further explore the possibility of a proteasome-dependent PQC for IMS proteins, we next tested whether wild-type and mutant endoG are substrates for ubiquitination. We reasoned that if a PQC is monitoring such proteins, the ubiquitination of mutant endoG should be more drastic than wild-type endoG since such PQC do not only prevent the presence of excess and unassembled proteins but also promote the selective elimination of misfolded proteins. We found that although wild-type endoG was a substrate for ubiquitination, the ubiquitination of mutant endoG was drastically increased when compared with wild-type endoG (Fig. 2A). The same result was obtained with two additional mutants (supplemental Fig. S5). The increased ubiquitination of mutant versus wild-type version of endoG therefore argues that the ubiquitination is not simply due to overexpression but rather is consistent with the presence of a PQC of IMS proteins that selectively target misfolded proteins.
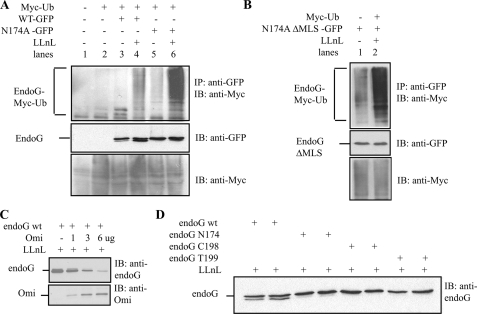
FIGURE 2.
Ubiquitin (Ub)-dependent protein quality control and Omi collaborate in the regulation of endoG. A, HEK293T cells were transfected with the indicated plasmids, and the proteasome inhibitor LLnL was added 18 h before harvesting. GFP-endoG was immunoprecipitated (IP) using anti-GFP antibody, and the immunoblot (IB) was developed using anti-Myc to detect ubiquitinated endoG. WT, wild type. B, HEK293T cells were transfected with the indicated plasmids, and the proteasome inhibitor LLnL was added 18 h before harvesting. GFP-endoG was immunoprecipitated using anti-GFP antibody, and the immunoblot was developed using anti-Myc to detect ubiquitinated endoG. C, HEK293T cells were transfected with the indicated plasmids, and the proteasome inhibitor LLnL was added 18 h before harvesting. The levels of GFP-endoG and Omi were detected by immunoblot using anti-endoG and Omi antibodies, respectively. D, HEK293T cells were transfected with the indicated plasmids, and the proteasome inhibitor LLnL was added 18 h before harvesting. The levels of GFP-endoG were detected by immunoblot using anti-endoG antibody.
We next aimed at addressing whether the ubiquitination take place in the cytosol prior to their import in the mitochondria, or as is the case in the endoplasmic reticulum, required import first and then retrotranslocation and ubiquitination (9, 11). To distinguish between these two possibilities, the mitochondrial localization signal (MLS) of mutant endoG was deleted, and the level of its ubiquitination was tested. We found that deletion of the MLS did not affect the ubiquitination of mutant endoG (Fig. 2B), suggesting that the ubiquitination does not require import into the IMS and does not involve retrotranslocation. Rather, this observation suggests that misfolded IMS proteins are ubiquitinated and eliminated in the cytoplasm prior to their import into the mitochondria. Therefore, our data suggest that the accumulation of IMS proteins in the mitochondria following proteasome inhibition is due to the lack of the elimination of the misfolded IMS proteins and their import in the mitochondria.
Omi Cleaves endoG Following Proteasome Inhibition—The analysis of wild-type and mutant endoG revealed that wild-type but not mutant endoG migrates as a doublet specifically in response to proteasome inhibition (Fig. 2A), suggesting that a protease may be activated following proteasome inhibition. During the course of this study, the mitochondrial protease Omi, which localized to the intermembrane space along with endoG, was reported to become activated following phosphorylation by PINK kinase in response to a variety of stresses including proteasome inhibition (18). We therefore tested whether Omi may be responsible for the cleavage of endoG following LLnL treatment. We found that transfection of Omi reduced the level of endoG, suggesting that the lower band of the doublet represents a partial cleavage product that retains the epitope but that Omi is able to degrade endoG completely (Fig. 2C). The specificity of Omi-mediated cleavage toward wild-type endoG was confirmed using two additional mutants of endoG. In all mutants tested, no cleavage was observed when compared with wild-type endoG (Fig. 2D). This result suggests that although excess or spontaneously misfolded wild-type endoG may be targeted for degradation by ubiquitination, when the proteasome function is inhibited, cleavage by Omi represents a second checkpoint to limit their accumulation in the mitochondria. However, elimination of mutant endoG is solely dependent on the ubiquitin pathway.
DISCUSSION
In this study, we present data supporting the presence of a PQC that monitors the folding state of proteins targeted to the IMS of the mitochondria. This PQC appears to act prior to the import into the IMS based on the observation that deletion of the mitochondrial targeting signal did not abolish the ubiquitination of mutant endoG. One alternative possibility, however, is that ΔMLS-endoG-N174 and endoGN-174 mutants are ubiquitinated by different ubiquitin ligases; ΔMLS-endoG-N174 is ubiquitinated by one ligase directly in the cytosol, whereas endoG-N174 is ubiquitinated by another ligase that would participate in its retrotranslocation. In support of this possibility, during the course of this work, Margineantu et al. (17) reported that the OSCP subunit of mitochondrial F1F0-ATPase is ubiquitinated and retrotranslocated. Therefore, whether endoG ubiquitination involves retrotranslocation remains unclear.
The accumulation of mitochondrial proteins following inhibition of the proteasome we describe here is also consistent with the study by Margineantu et al. (17), which reported similar findings using hsp90 inhibitors. Collectively, the findings reported here and the study by Margineantu et al. (17) strongly supports an important role for the ubiquitin pathway in the monitoring of mitochondrial protein quality.
During the course of this work, another study reported the activation of Omi following proteasome inhibition (18). Although several proteins have been reported to be substrates of Omi following its release from the mitochondria during apoptosis, the mitochondrial substrates of Omi following proteasomal stress remain uncharacterized. We report here that endoG is a substrate for Omi cleavage following proteasome inhibition. These results not only identify endoG as substrate for the mitochondrial function of Omi but also highlight that mutation in endoG render endoG resistant to such regulation.
These findings have several implications. Since altered function of the proteasome has been tightly linked to the progression of several neurodegenerative diseases, the first implication of our findings is that in the situation where proteasome function is defective, accumulation of mitochondrial proteins may cause a collapse of the mitochondrial network. A second implication is that such a collapse may be accelerated by the acquisition of mutation in IMS proteins such as endoG since such mutants are more efficient at inducing mitochondrial clustering and are resistant to the Omi-mediated checkpoint. A third implication is that mutation in Omi itself, which as been reported in Parkinson disease, may also contribute to the accelerated collapse of the mitochondrial network by eliminating the Omi checkpoint.
Collectively, the picture that emerges from our findings and that of others is that mitochondrial proteins are regulated by the ubiquitin-dependent protein quality control, possibly similarly to the one acting at the endoplasmic reticulum. Further, the proteasome also affects the activity of the protease Omi, which may represent an important second checkpoint to limit the accumulation of mitochondrial proteins. The observation that Omi and PINK (16) are both mutated in Parkinson disease supports this model. Whether other IMS proteins are mutated in neurodegenerative diseases and whether the combined defects in proteasome and such mutations correlate with accelerated progression of the disease await further investigations. If so, our findings may help differentiate patients with high risk of rapid progression from those suffering from a slow and less aggressive progression.
Supplementary Material
Acknowledgments
Electron microscopy was performed by the shared facility at Mount Sinai, which is supported from the National Institutes of Health Shared Resources Grant 5R24 CA095823-04, National Science Foundation Major Research Instrumentation Grant DBI-9724504, and National Institutes of Health Shared Instrumentation Grant 1 S10RRO 9145-01.
This work was supported, in whole or in part, by the National Institutes of Health RO1 Grant CA109482 (to D. G.). This work was also supported by the Samuel Waxman Cancer Research Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains five supplemental figures.
Footnotes
The abbreviations used are: PQC, protein quality control(s); IMS, intermembrane space; endoG, endonuclease G; ER, endoplasmic reticulum; PBS, phosphate-buffered saline; HA, hemagglutinin; GFP, green fluorescent protein; MLS, mitochondrial localization signal; hsp, heat shock protein; LLnL, N-acetyl-l-leucyl-l-leucyl-l-norleucinal; PINK, PTEN-induced putative kinase 1.
References
- 1.Kostova, Z., and Wolf, D. H. (2003) EMBO J. 22 2309–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldberg, A. L. (2003) Nature 426 895–899 [DOI] [PubMed] [Google Scholar]
- 3.Ciechanover, A. (2005) Cell Death Differ. 12 1178–1190 [DOI] [PubMed] [Google Scholar]
- 4.Enenkel, C., Lehmann, A., and Kloetzel, P. M. (1998) EMBO J. 17 6144–6154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivett, A. J. (1998) Curr. Opin. Immunol. 10 110–114 [DOI] [PubMed] [Google Scholar]
- 6.Gardner, R. G., Nelson, Z. W., and Gottschling, D. E. (2005) Cell 120 803–815 [DOI] [PubMed] [Google Scholar]
- 7.Sitia, R., and Braakman, I. (2003) Nature 426 891–894 [DOI] [PubMed] [Google Scholar]
- 8.Romisch, K. (2005) Annu. Rev. Cell Dev. Biol. 21 435–456 [DOI] [PubMed] [Google Scholar]
- 9.Plemper, R. K., Bohmler, S., Bordallo, J., Sommer, T., and Wolf, D. H. (1997) Nature 388 891–895 [DOI] [PubMed] [Google Scholar]
- 10.Jarosch, E., Taxis, C., Volkwein, C., Bordallo, J., Finley, D., Wolf, D. H., and Sommer, T. (2002) Nat. Cell Biol. 4 134–139 [DOI] [PubMed] [Google Scholar]
- 11.Plemper, R. K., and Wolf, D. H. (1999) Mol. Biol. Rep. 26 125–130 [DOI] [PubMed] [Google Scholar]
- 12.Langer, T., Kaser, M., Klanner, C., and Leonhard, K. (2001) Biochem. Soc. Trans. 29 431–436 [DOI] [PubMed] [Google Scholar]
- 13.Arnold, I., and Langer, T. (2002) Biochim. Biophys. Acta 1592 89–96 [DOI] [PubMed] [Google Scholar]
- 14.Augustin, S., Nolden, M., Muller, S., Hardt, O., Arnold, I., and Langer, T. (2005) J. Biol. Chem. 280 2691–2699 [DOI] [PubMed] [Google Scholar]
- 15.Radke, S., Pirkmaier, A., and Germain, D. (2005) Oncogene 24 3448–3458 [DOI] [PubMed] [Google Scholar]
- 16.Muqit, M. M., Abou-Sleiman, P. M., Saurin, A. T., Harvey, K., Gandhi, S., Deas, E., Eaton, S., Payne Smith, M. D., Venner, K., Matilla, A., Healy, D. G., Gilks, W. P., Lees, A. J., Holton, J., Revesz, T., Parker, P. J., Harvey, R. J., Wood, N. W., and Latchman, D. S. (2006) J. Neurochem. 98 156–169 [DOI] [PubMed] [Google Scholar]
- 17.Margineantu, D. H., Emerson, C. B., Diaz, D., and Hockenbery, D. M. (2007) PLoS ONE 2 e1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plun-Favreau, H., Klupsch, K., Moisoi, N., Gandhi, S., Kjaer, S., Frith, D., Harvey, K., Deas, E., Harvey, R. J., McDonald, N., Wood, N. W., Martins, L. M., and Downward, J. (2007) Nat. Cell Biol. 9 1243–1252 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.