Abstract
Annexin A2, a calcium-dependent phospholipid-binding protein, is abundantly expressed in alveolar type II cells where it plays a role in lung surfactant secretion. Nevertheless, little is known about the details of its cellular pathways. To identify annexin A2-regulated or associated proteins, we silenced endogenous annexin A2 expression in rat alveolar type II cells by RNA interference and assessed the change of the cellular transcriptome by DNA microarray analysis. The loss of annexin A2 resulted in the change of 61 genes. Thirteen of the selected genes (11 down-regulated and 2 up-regulated genes) were validated by real time quantitative PCR. When the loss of rat annexin A2 was rescued by overexpressing EGFP-tagged human annexin A2, six of seven selected targets returned to their normal expression level, indicating that these genes are indeed annexin A2-associated targets. One of the targets, Rab14, co-immunoprecipitated with annexin A2. Rab14 also co-localized in part with annexin A2 and lamellar bodies in alveolar type II cells. The silencing of Rab14 resulted in a decrease in surfactant secretion, suggesting that Rab14 may play a role in surfactant secretion.
The alveolar epithelium is composed of morphologically and functionally distinct type I and II cells. Type I cells provide the bulk of the surface area for gas exchange, whereas type II cells secrete lung surfactant to maintain mechanical stability of the alveoli (1–3). Lung surfactant is stored and secreted by exocytosis of lamellar bodies, which are specialized organelles found in type II cells. Exocytosis of the lamellar bodies is triggered by several intracellular signaling pathways. A key component of signaling involves the elevation of cytosolic Ca2+ concentration in type II cells (4, 5). Because lung surfactant secretion is regulated by Ca2+ signals, it is important to identify Ca2+-regulated proteins for exocytosis.
Annexins are a family of Ca2+-dependent phospholipid-binding proteins. More than 50 different annexin isoforms have been identified (6, 7). Each annexin consists of a short variable N-terminal segment and a conserved C-terminal core domain. They have unique Ca2+-binding sites, which enable them to bind with negatively charged lipids. Annexins participate in many membrane-related events, such as the organization of membrane domains, the linkages of membrane-cytoskeleton, exocytosis and endocytosis, and the regulation of ion fluxes (7). Annexin A2 is abundant in alveolar type II cells. Several studies have indicated that annexin A2 is involved in Ca2+-regulated exocytosis (8–10). In permeabilized chromaffin cells, the time-dependent loss of secretory capacity can be blocked by the addition of annexin A2 to the culture medium (9). Knockdown of annexin A2 reduces the Ca2+-evoked exocytosis of Weibel-Palade bodies in endothelial cells (10). In alveolar type II cells, accumulating evidence supports a role for annexin A2 in controlling the fusion of lamellar bodies with the plasma membrane and promoting surfactant secretion (11–14). Nevertheless, relatively little is known about the molecular mechanisms of annexin A2 action in this process, especially in relation to its targets and biological pathways.
In this study, the expression of annexin A2 in rat type II cells was silenced by adenovirus-mediated short hairpin RNA (shRNA)2 to identify annexin A2-related genes at a genomic level. The genes that were up- or down-regulated because of the loss of annexin A2 were identified by the significance of microarray (SAM) test. Thirteen selected genes were validated by real time quantitative PCR, and six were further confirmed by rescuing the loss of annexin A2 with overexpression of human annexin A2. Most of the genes have not been previously reported as annexin A2-related genes. Of great interest is Rab14, which physically interacts with annexin A2 and is functionally related to annexin A2-mediated surfactant secretion. This finding provides another piece to the puzzle of the mechanisms of lung surfactant secretion and will help guide future experiments in this field of research.
MATERIALS AND METHODS
Alveolar Type II Cell Isolation and Culture—Type II cells were isolated from 180-to 200-g Sprague-Dawley rats, according to the method of Dobbs et al. (15). The purities of type II cells preparations were >90% as determined by the modified Papanicolaou staining. The viability of these cells was over 92%. To maintain the phenotype, isolated type II cells were cultured in an air-liquid model as described before (16).
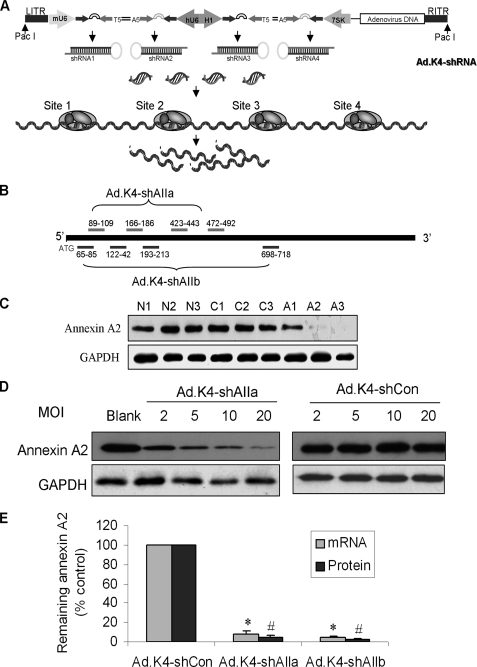
Construction of RNA Interference and Overexpression Vectors— A new pK4-shRNA vector was used to silence gene efficiently, in which the simultaneous expression of four different shRNA sequences was driven by four different polymerase III promoters (human U6, H1, 7SK, and mouse U6) in a single vector (Fig. 1A) as described previously (17). Two sets of four siRNA sequences against rat annexin A2 were selected for constructing the K4 vectors for silencing annexin A2 (Fig. 1B, Ad.K4-shAIIa and Ad.K4-shAIIb). One set of four siRNA sequences targeting to rat Rab14 at the positions 11–31, 111–131, 139–159, and 438–456 were used to silence Rab14 (Ad.K4-shRab14). The control vector, Ad.K4-shCon, contains four siRNAs without obvious homologies to mammalian genes (5′-AATTCTCCGAACGTGTCACGT-3′, 5′-GACAGCTAGGTTATCACGATC-3′, 5′-TGCGTTAGCTGCGTCAAGCAT-3′, and 5′-ACTTACTGTGCGTAGTTAGCC-3′).
FIGURE 1.
Generation and confirmation of the RNA interference vectors. A, diagrammatic representation of the Ad.K4-shRNA vector showing the placement of the mU6, hU6, H1, and 7SK promoters in relation to the shRNA sequences. B, illustration of the Ad.K4-shAIIa and Ad.K4-shAIIb vectors showing the positions of the four siRNA sequences for each vector. C, alveolar type II cells were transduced with shRNA adenovirus or control adenovirus at an equal dose of 50 m.o.i. and cultured for 24, 48, or 72 h prior to harvesting. Equal amounts of protein (20 μg) were immunoblotted for annexin A2. The same membranes were re-probed for GAPDH to confirm equal loading. N1–3, blank control; C1–3, Ad.K4-Con vector; A1–3, Ad.K4-shAIIa vector. D, alveolar type II cells were transduced with shRNA adenovirus or control adenovirus at doses of 2, 5, 10, or 20 m.o.i. and cultured for 72 h prior to harvesting. The membranes were sequentially probed for annexin A2 and GAPDH. E, comparison of Ad.K4-shAIIa and Ad.K4shAIIb. Alveolar type II cells were transduced for 72 h with either control (K4-shCon) or silencing adenovirus (K4-shAIIa and K4-shAIIb) at an m.o.i. of 50. mRNA and protein levels were determined by RT-PCR and Western blot. GAPDH (for mRNA) and β-actin (for protein) were used as loading controls. Data were expressed as a percentage of K4-shCon. *, p < 0.05 versus Ad.K4shCon; #, p < 0.05 versus Ad.K4-shCon (n = 3).
To construct overexpression vectors, full-length human annexin A2 cDNA was amplified by PCR from the I.M.A.G.E. clone of 3535154 with the primer set of 5′-GAGGATCCATGTCTACTGTTCACGAA-3′ and 5′-GGACTAGTTCATCTCCACCACACA-3′. The full-length cDNA sequences of id1, p55cdc, retnla, and prdx6 were PCR-amplified from cDNA of rat type II cells with the corresponding primer sets as follows: 5′-GAGGATCCATGAAGGTCGCCAGTAGCAGTG-3′ and 5′-TGGACTAGTGATTTGCGACACAAGATGCG-3′ (for id1); 5′-GAGGATCCATGGCGCAGTTCGTGTTCGAGAGCGATTTG-3′ and 5′-GACTAGTGACCGGATGCCTTGGTGGATGAGGCTAC-3′ (for p55cdc); 5′-GAGGATCCATGAAGACTGCAACCTGTTC-3′ and 5′-TGGACTAGTGAGGACAGTTGGCAGCAG-3′ (for retnla); and 5′-GAAGATCTATGCCCGGAGGGCTGCTTCTC-3′ and 5′-GACTAGTGAAGGCTGGGGCGTATAACGGA-3′ (for prdx6). After double digestion with BamHI and SpeI, human annexin A2, rat id1, rat p55cdc, rat retnla, and rat prdx6 were cloned into our previously reported pENTR/CMV-EGFP vector through corresponding sites (18). The pEGFP-Rab14 overexpression vector was kindly provided by Dr. Richard H. Scheller (19). The expression of the EGFP fusion protein was verified by Western blot and fluorescence microscopy.
Adenovirus Generation—To construct the adenoviral vectors, the expression cassettes within the pENTR vectors were shuttled into an adenoviral vector, pAd/PL-DEST, through the Gateway recombination method (Invitrogen). After being linearized by PacI, the adenoviral plasmids were transfected into 293A cells for the generation of adenovirus. Viruses were purified by the Adeno-X mini purification kit (Clontech) and titered according to the EGFP signal on 293A transducted cells.
Adenovirus Transduction and RNA Preparation—Freshly isolated alveolar type II cells were mixed with adenovirus at different multiplicities of infection (m.o.i.) and cultured on an air-liquid model (16) for various days. Total RNA was extracted using TRI Reagent® (Molecular Research Center, Cincinnati, OH). For the rescue experiment, adenovirus expressing an EGFP-tagged human annexin A2 fusion protein (hAII-EGFP) was added to type II cells 1 day after the transduction of Ad.K4-shAIIa and continually cultured for an additional 2 days before the samples were collected.
Microarray Hybridization—The DNA microarray slides were prepared in-house using the Pan Rat 10K oligonucleotide set (MWG Biotec Inc., High Point, NC) that contains 6,221 known rat genes, 3,594 rat expressed sequence tags, and 169 Arabidopsis negative controls as described previously (20).
Two shRNA vectors (Ad.K4-shAIIa and Ad.K4-shAIIb) were used for the silencing of annexin A2. Ad.K4-shCon was used as the virus control. The blank control without virus transduction was applied to evaluate the potential effects of adenovirus on alveolar type II cells. Reference RNA (Ref) obtained from SuperArray Co. was used to monitor the change in gene expression level. A reference design was performed as follows: Ad.K4-shAIIa/Ref, Ad.K4-shAIIb/Ref, Ad.K4-shCon/Ref, and Blank/Ref, each with dye flipping. There were four biological replications. RNA (10 μg total) from each sample was reverse-transcribed into cDNA with Cy3 (green)- or Alexa 647 (red)-specific oligo(dT) primers from the 3DNA Array 50 kit (Genisphere, Hatfield, PA). The cDNA was purified with Microcom YM-30 columns (Millipore, Billerica, MA) and dissolved in 1× hybridization buffer (50% formamide, 6× SSC, and 0.2% SDS) at the concentration of 0.3 μg/μl.
The two-step microarray hybridization was carried out with the 3DNA 50 expression kit. Before hybridization, the slides were washed with 0.2% SDS once and with deionized water four times and then dried by centrifugation. The denatured two color-paired cDNA mixture was added to DNA microarray slides and hybridized at 42 °C for 48 h. After being washed, the slides were re-hybridized with Cy3- and Alexa 647-specific capture reagents at 42 °C for 2 h and scanned twice with a ScanArray Express scanner (PerkinElmer Life Sciences).
Microarray Data Analysis—The signal intensity for each spot was captured by Genepix 5.0. The ratios between each sample and reference cDNA were normalized by LOWESS normalization using the RealSpot software package developed in our laboratory (21). A quality index for each spot, based on signal intensity and signal-to-background ratio, was calculated by RealSpot. Data with a mean quality index of <1 were discarded. One class SAM statistical test was applied to the remaining genes using a cutoff q value of <0.05. The major functional categories were assigned using gene ontology.
Quantitative Real Time PCR—The validation of selected target genes with the QuantiTect SYBR Green PCR kit (Qiagen Inc., Valencia, CA) was performed. Each RNA sample was treated with 1 μg of DNase and was reverse-transcribed into cDNAs with 0.2 μg/μl (dT)17, 0.3 μg/μl random hexamer primer, and 200 units of Moloney murine leukemia virus reverse transcriptase (Invitrogen). The real time PCR thermal conditions for all genes were 95 °C for 15 min, followed by 40 cycles each at 95 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and 77 °C for 35 s (signal collection temperature). The primer pairs are listed in Table 1. The relative real time PCR quantification was based on the ΔΔCt method. The endogenous reference gene was GAPDH.
TABLE 1.
Primers used in real time PCR
| Gene name | Forward sequence (5′–3′) | Reversed sequence (5′–3′) |
|---|---|---|
| Annexin A2 | CACCTCCAGAAAGTGTTCGAAAG | GGCTTGTTCTGAATGCACTGAA |
| gapdh | AACTCCCTCAAGATTGTCAGCAA | CACAGTCTTCTGAGTGGCAGTGA |
| retnla | CCTGCTGGGATGACTGCTACT | TCAACGATTAAGCACAGGCAGTT |
| id-1 | GAACGTTCTGCTCTACGACATGA | GCAGTATCTCCACCTTGCTCACTT |
| rab14 | ACCTCACCAACCAAACACTGTA | ACATTCTCTCCCGTTTTTGCACT |
| atp1a1 | CTGGGAGGCTTCTTCACTTATT | GCTGTCCTCCACATCATTGAT |
| atp2a1 | TCTGGCTTCTCGGTTCCATCT | GACCATGAGCCACTGGGTAAAG |
| coq7 | GCAGGTCATTAAGCAATTTCGA | CGGCCTGGATAAGTCTCTTCAA |
| sp-c | AGCTCCAGGAACCTACTGCTACAT | AGGACTTGGCCTGGAAGTTCTT |
| dcx | CTAGCAGTCAGCTCTCAACACCTAA | GGAATCGCCAAGTGAATCAGA |
| fn1 | GGCAACAAATGATCTTTGAGGAA | CATCTACATTCGGCAGGTATGGT |
| gnb2 | CAGACATTCATAGGTCACGAATCG | AGGTCAAACAGGCGACATGTG |
| gts8 | AAGAAGTCAGTGCTCCTGTGTTGT | TCCAGGTTGCAGGAACTTCTTAA |
| map17 | CCCCAGGCTACAGGGAATGT | GCCTGAGATGGCTGTGATTCA |
| p55cdc | GGCAACAGGAGGAGGTACCA | ATGGAGCACACCTGGGAATG |
| prdx6 | GCTGCAGTTCCGTAGAAAGATTG | AATGTTTTGGTGCCATTTCCAT |
| ps20 | GATGGTCCGGAGGAAGTGTTAC | TGCAGGATGTGGCACTCATAG |
| psti | CTGCACAGTTCTTCTGAGTTTTGG | ATATGCAAATCCACCTACCTGCTAA |
| s100A1 | CACGGTGGCTTGTAACAACTTCT | TAGGGAGATACAGGTGGGATGAG |
| s100A6 | GACCGTAACAAGGATCAGGAAGTAA | CTGCCTTCCCATTTTATTTCAGA |
| sp4 | ACATTGAAGGGTGTGGGAAAGT | TTGCCACAAAACATCCAGTTG |
| tspan8 | AAATGCCAAGCTTTTGTCAGAAA | CAAATCTCAAGCCACAGCACTTA |
Western Blot—Adenovirus-treated or untreated type II cells were extracted using lysis buffer (10 mm Tris-HCl, pH 7.5, 1% Triton X-100, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin) on ice. Protein samples (20 μg) were separated on 10% SDS-PAGE and electroblotted to a nitrocellulose membrane. After being blocked with 5% fat-free milk in TTBS (20 mm Tris-HCl, pH 7.6, 150 mm NaCl, 0.1% Tween 20) for 1 h, the blot was incubated overnight at 4 °C with the following primary antibodies: rabbit anti-annexin A2 (Santa Cruz Biotechnology; 1:2,000 dilution); mouse monoclonal anti-annexin A2 (Zymed Laboratories Inc.; 1:1000 dilution); rabbit anti-Rab14 (kindly provided by Dr. Richard H. Scheller; 1:500 dilution); mouse monoclonal anti-GFP (Santa Cruz Biotechnology; 1:500 dilution); mouse monoclonal anti-GADPH (Ambion; 1:5000); rabbit polyclonal anti-β-actin (Sigma; 1:2000 dilution). Protein loading controls were done with either rabbit anti-β-actin antibody or anti-GADPH. The membrane was washed with TTBS and incubated with horseradish peroxidase-conjugated anti-mouse or rabbit IgG (1:1000–2,000 dilution) for 1 h. The membrane was developed with enhanced chemiluminescence reagents (Amersham Biosciences) and exposed to x-ray film.
Immunoprecipitation—To overexpress selected targets (Id1, Retnla, Prdx6, and Rab14) with an EGFP tag, 1 μg of each plasmid (pEGFP-Rab14, pENTR/CMV-Prdx6EGFP, pENTR/CMV-RetnlaEGFP, and pENTR/CMV-p55cdcEGFP) was transfected into 293A cells cultured on 6-well plates. Transfection was performed using Lipofectamine 2000 when the cells reached 80% confluence. Cells were washed with phosphate-buffered saline 48 h after transfection and lysed in 500 μl of M-PER mammalian protein extraction reagent (Pierce). Centrifugation at 14,000 rpm for 20 min was performed to remove the insoluble materials. Rabbit polyclonal anti-GFP antibodies (whole serum, 1 μl; Abcam) or control rabbit IgG was incubated with 250 μg of protein of cell lysate for 1 h at 4 °C, followed by incubation with 20 μl of protein A-agarose and 10 μl of protein G PLUS-agarose (Santa Cruz Biotechnology) overnight at 4 °C. For p65 immunoprecipitation, 250 μg of protein of cell lysate was directly incubated with 20 μl of NF-κB p65 (c-20) AC rabbit polyclonal IgG-conjugated agarose (Santa Cruz Biotechnology) overnight at 4 °C. The beads were pelleted at 800 × g for 5 min at 4 °C, then washed with cold phosphate-buffered saline four times, and boiled with 30 μl of 2× SDS sample buffer. The soluble fractions were detected by Western blotting.
Immunolocalization of Rab14 in Alveolar Type II Cells—Immunolocalization was performed exactly as described earlier (22). In brief, freshly isolated type II cells (0.5 × 106) were suspended in complete Dulbecco's modified Eagle's medium (10% fetal bovine serum; 1× nonessential amino acids and antibiotics) and cultured on sterile glass coverslips. Following overnight culture, the cells were washed with ice-cold phosphate-buffered saline and fixed with 4% paraformaldehyde. The cells were then permeabilized with 0.5% Triton X-100 for 20 min and blocked with 10% fetal bovine serum for 1 h before incubating with rabbit anti-Rab14 (1:100) and mouse anti-LB180 antibodies (1:500) or mouse anti-annexin A2 antibodies (1:100) overnight at 4 °C. Later the cells were washed and incubated with Alexa 488-conjugated goat anti-rabbit or Cy3-conjugated goat anti-mouse antibodies at a dilution of 1:250 for 1 h at room temperature. Following washing, the slides were mounted onto glass slides by adding anti-fade to prevent the quenching of the fluorophores. The proteins were visualized with a Nikon Eclipse E600 fluorescent microscope.
Lung Surfactant Secretion—Alveolar type II cells were cultured on an air-liquid model as described (16). Adenovirus (100 m.o.i.) was added on day 3. Lung surfactant secretion was performed on day 7 as described previously (23).
RESULTS
Annexin A2 Knockdown in Primary Alveolar Type II Cells with the Adenovirus-based Ad.K4-shRNA System—We have recently developed a new vector expressing multiple shRNAs that enhances silencing efficiency and reduces the amount of adenovirus required to achieve maximal silencing (17). This vector contains four different shRNA expression cassettes driven by the human U6, H1, 7SK promoters, and the mouse U6 promoter (Fig. 1A). To silence the rat annexin A2 gene, we constructed two adenoviral vectors, Ad.K4-shAIIa and Ad.K4-shAIIb (Fig. 1B), both containing four siRNA sequences targeting to the coding sequences of rat annexin A2 at different positions. The Ad.shCon vector expressing four different shRNAs without obvious homologies to mammalian DNA sequences was used as a negative control.
At an m.o.i. of 50, a time course analysis that tested the effects of the adenovirus at 24, 48, and 72 h showed that the amount of annexin A2 protein was reduced by >98% 72 h after transduction of Ad.K4-shAIIa (Fig. 1C, A1–A3). The control vector Ad.K4-shCon (Fig. 1C, C1–C3) as well as the blank control (N1–N3) had no effect on the expression of annexin A2. A dose-response experiment showed that the transduction of Ad.K4-shAIIa virus at an m.o.i. of 20 inhibited >95% of annexin A2 protein expression (Fig. 1D). The control vector showed no differences in the annexin A2 protein levels at all four doses. Ad.K4-shAIIb had a similar silencing efficiency as Ad.K4-shAIIb at both mRNA and protein levels using an m.o.i. of 50 (Fig. 1E). Based on these data, we used RNA samples prepared from type II cells 72 h after virus transduction at an m.o.i. of 50 for the DNA microarray.
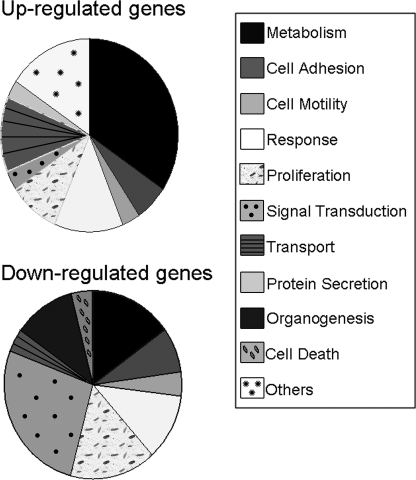
Changes in the Cellular Transcriptome upon RNAi-mediated Inhibition of Annexin A2 Expression—To evaluate the effect of annexin A2 knockdown on global gene expression in type II cells, two K4-shRNA vectors, Ad.K4-shAIIa and Ad.K4-shAIIb, were used to silence annexin A2. We reasoned that if K4-shRNAs can elicit a target-specific response, both should induce the same changes in the gene expression profiles. Ad.K4-shCon was used as a control to correct for potential off-target effects that could be induced by siRNAs in general or by the virus transduction. By analyzing a genome-wide microarray of ∼10,000 genes, we observed significant changes of 135 genes in Ad.K4-shAIIa and 185 genes in Ad.K4-shAIIb. Of them, 61 genes are commonly changed in both Ad.K4-shAIIa and Ad.K4-shAIIb. These genes were catalogued according to their gene ontology information. The pie chart in Fig. 2 shows the percentage of the genes in major functional categories for up- and down-regulated genes.
FIGURE 2.
Functional categories of the identified annexin A2-associated genes. The pie chart shows major functional categories of up-regulated and down-regulated genes based on gene ontology information. Response genes include immune response, inflammatory response, and response to stress.
Table 2 lists the commonly changed genes that were up- or down-regulated by more than 2-fold. Nineteen genes were down-regulated and seven genes were up-regulated in the absence of annexin A2. The identified set included genes that are involved in diverse processes, including cell adhesion, membrane fusion, fatty acid metabolism, and antioxidant defense. There are several genes related to calcium iron binding and intracellular transport, indicating that those targets are functionally close to annexin A2 in type II cells.
TABLE 2.
Commonly changed genes (> 2-fold) in annexin A2-silenced type II cells
| GenBank™ GeneID | Symbol | Gene name | -Fold change |
|---|---|---|---|
| NM_019905 | anxa2 | Annexin A2 | –5.24 |
| NM_053333 | retnla | Resistin-like molecule α | –4.33 |
| S68809 | s100a1 | s100a1 protein | –4.16 |
| NM_019143 | fn1 | Fibronectin 1 | –2.87 |
| NM_053485 | s100a6 | s100a6 protein | –2.74 |
| AF037272 | ps20 | Wap four-disulfide core domain protein | –2.66 |
| NM_012504 | atp1a1 | ATPase Na+/K+-transporting α1 polypeptide | –2.56 |
| NM_012761 | sp4 | Sp4 transcription factor | –2.47 |
| D11325 | psTI | Pancreatic secretory trypsin inhibitor | –2.45 |
| NM_017342 | sftpc | Surfactant associated protein C | –2.36 |
| NM_012797 | id1 | Inhibitor of DNA binding 1 helix-loop-helix protein | –2.34 |
| X81449 | keratin 19 | Keratin 19 | –2.32 |
| NM_031834 | sult1a1 | Sulfotransferase family 1A | –2.32 |
| NM_133526 | tm4sf3 | Transmembrane 4 superfamily member 3 | –2.27 |
| U46149 | coq7 | COQ7 | –2.18 |
| X62660 | Glutathione transferase subunit 8 | –2.15 | |
| AF397193 | gnb2 | G-protein β2 subunit | –2.07 |
| NM_053589 | rab14 | Small GTP-binding protein RAB14 | –2.05 |
| NM_053576 | prdx6 | Peroxiredoxin 6 | –2.04 |
| AF402772 | map17 | Membrane-associated protein MAP17 | –2.00 |
| S39779 | preproET-3 | Preproendothelin-3 | 2.03 |
| NM_058213 | atp2a1 | ATPase, Ca2+-transporting, fast twitch 1 | 2.10 |
| NM_053379 | dcx | Doublecortin gene | 2.16 |
| U05341 | p55cdc | p55CDC | 2.18 |
| M58653 | grp-ca | Glutamine/glutamic acid-rich protein isoform calcium | 2.29 |
| AF016180 | go-vn3 | Putative pheromone receptor | 2.43 |
| NM_012556 | fabp1 | Fatty acid-binding protein 1 | 2.51 |
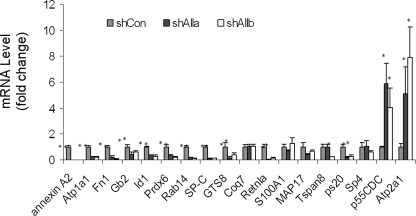
Real Time PCR Verification—To examine the reliability of the microarray data, 18 genes (16 down-regulated and 2 up-regulated) were chosen from Table 2 for validation. These 18 genes were chosen based on their possible association with annexin A2-mediated cellular functions. The RNAs from the same samples used in the DNA microarray were used for quantitative real time PCR analysis. As shown in Fig. 3, 13 of 18 genes were validated by real time PCR, including 11 that were down-regulated (annexin A2, atp1a1, fn1, gb2, id1, prdx6, rab14, sp-c, gts8, retnla, and ps20) and 2 that were up-regulated (atp2a1 and p55cdc).
FIGURE 3.
Real time PCR verification of microarray data. To verify the microarray data, 18 genes were chosen from the list in Table 2 based on their possible association with annexin A2 or surfactant-related functions. The same RNA samples for the microarray analysis were used for real time RT-PCR. GAPDH was used as an endogenous reference gene. *, p < 0.05 versus shCon (n = 4, each performed in duplicate).
Coq7, S100A1, MAP17, Tspan8, and Sp4 were not significantly changed as seen by real time RT-PCR, but the microarray assay indicated that those genes were expressed significantly higher in the control type II cells than annexin A2-depleted type II cells. For Tspan8, the expression level decreased in one of the treatments as revealed by real time PCR. The inconsistent results of five genes were likely because of low mRNA abundance, high variation among samples, or differences in data normalization (locally weighted regression and scatterplot smoothing or LOWESS versus housekeeping gene). Four genes (prepro-et-3, psti, dcx, and grp-ca) were inconclusive because of the poor amplification (data not shown).
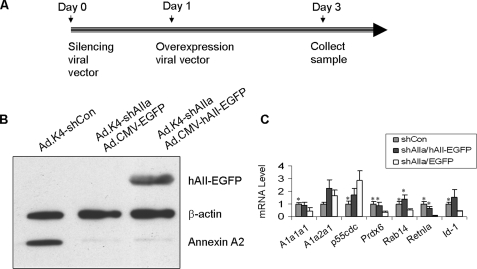
Rescue of Silencing by the Overexpression Construct—To confirm the specificity of the RNAi phenotype, annexin A2 silencing was rescued by overexpressing an RNAi-resistant human annexin A2 construct to see whether the RNAi effect could be reversed. One approach to generate an RNAi-resistant construct is to create silent point mutations in the target site of the coding sequence or to select an siRNA targeted to the sequence in the 3′-untranslated region of a gene. Another way is to use a cross-species DNA construct if these are mismatches in the target sites among species. 2–5 base mismatches exist in each of the four siRNA sequences of the Ad.K4-shAIIa vector between rat and human annexin A2. We have previously shown that the transduction of the Ad.K4-shAIIa vector into human A549 cells had little effect on the expression of human annexin A2 (17), indicating that human annexin A2 can be used to rescue the loss of annexin A2 caused by Ad.K4-shAIIa in rat cells. We used the adenoviral vector, Ad.CMV-hAII-EGFP, that contains the human annexin A2 fused with an EGFP tag under the control of the cytomegalovirus promoter. The C-terminal EGFP tag makes it easily to distinguish overexpressed annexin A2 from endogenous annexin A2. In our experimental design (Fig. 4A), the rescuing viral vector was added to the cells 1 day after the knockdown of annexin A2. The culture continued for additional 2 days prior to sample collection. We found that Ad.K4-shAIIa viral vector silenced >95% of endogenous rat annexin A2 but had no effect on the expression of the hAII-EGFP fusion protein as examined by Western blot (Fig. 4B). Real time PCR analysis revealed that the expression of Prdx6, Rab14, and Retnla returned to the control level after rescuing the loss of endogenous annexin A2 by the hAII-EGFP fusion protein (Fig. 4C). Other genes were variable or partially reversed to the control level. Because the targets confirmed by the rescue experiment may be functionally regulated or associated by annexin A2, we selected these targets for a physical interaction study.
FIGURE 4.
Rescue of rat annexin A2 silencing by overexpression of human annexin A2. A, diagram of the experimental design used for the rescue experiment. Alveolar type II cells were transduced with the control vector (Ad.K4-shCon), Ad.K4-shAIIa silencing vector combined with the Ad.CMV-EGFP vector or Ad.K4-shAIIa silencing vector combined with the Ad.CMV-hAII-EGFP overexpression vector. The silencing vector was added at Day 0 (the day of plating), and the overexpression vector was added at Day 1. The cells were collected at Day 3. B, Western blot. Equal amounts of protein (20 μg) from each treatment were immunoblotted for the EGFP-tagged human annexin A2 (hAII-EGFP), endogenous annexin A2, or β-actin using anti-annexin A2 and anti-β-actin antibodies. C, real time PCR of seven genes chosen from Table 2. The primer sequences used to amplify the respective genes are listed in Table 1. The mRNA level is presented as a ratio over shCon. Gray bar, control cells treated with shCon; black bar, the cells treated with the shAIIa-hAII-EGFP vector; and white bar, the cells treated with the shAIIa-EGFP vector. *, p < 0.05 versus shAIIa/EGFP.
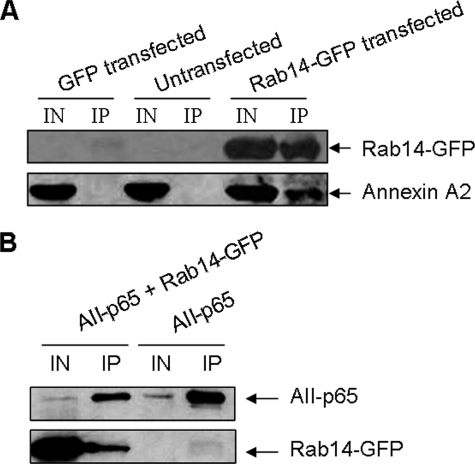
Identification of Annexin A2-interacting Proteins by Immunoprecipitation—To assess the possibility that annexin A2 directly interacts with any of the identified targets by microarray analysis, co-immunoprecipitation experiments were performed. pEGFP-Rab14 was transiently transfected into 293A cells and immunoprecipitated using anti-GFP antibodies. pEGFP expressing EGFP alone was used as a control. Anti-GFP antibodies pulled down Rab14-EGFP in the pEGFP-Rab14-transfected cells, and endogenous annexin A2 was co-precipitated with it (Fig. 5A). In the pEGFP-transfected cells and untransfected cells, no annexin A2 appeared in the immunoprecipitates. Immunoprecipitation using the control IgG showed no GFP or annexin A2 expression as expected (data not shown). Similar results were performed with pENTR/CMV-Prdx6EGFP, pENTR/CMV-RetnlaEGFP, or pENTR/CMV-p55CDCEGFP. Little annexin A2 was detected in the immunoprecipitates using anti-GFP antibodies (data not shown).
FIGURE 5.
Physical interaction of annexin A2 and Rab14 by co-immunoprecipitation. A, 293A cells were transfected with pEGFP-Rab14 or pEGFP for 48 h. The cell lysate was immunoprecipitated (IP) with rabbit polyclonal anti-GFP antibodies and detected by Western blot using mouse monoclonal anti-GFP and anti-annexin A2 antibodies. IN, input. B, 293A cells were transfected with pEGFP-Rab14 and/or pCMV-AII-p65 for 48 h. The cell lysate was immunoprecipitated (IP) with anti-p65-agarose and analyzed by Western blot using anti-GFP and anti-annexin A2 antibodies. IN, input.
To further confirm the interaction of annexin A2 with Rab14, we constructed a pCMV-AII-p65 plasmid that contains the DNA insert encoding full-length rat annexin A2 (AII) protein and the transcriptional activation domain of the mouse NF-κB gene (p65, amino acids 364–550). We co-transfected 293A cells with the plasmids, pEGFP-Rab14, and pCMV-AII-p65. AII-p65 was immunoprecipitated using anti-p65 antibody. Immunoblotting was performed using anti-GFP antibodies to detect Rab14-EGFP and anti-annexin A2 antibodies to detect AII-p65, respectively. As shown in Fig. 5B, EGFP-Rab14 co-immunoprecipitated with AII-p65 in the AII-p65 and EGFP-Rab14-transfected cells but not in the AII-p65-transfected cells. Together, these immunoprecipitation experiments indicate that Rab14 physically interacts with annexin A2.
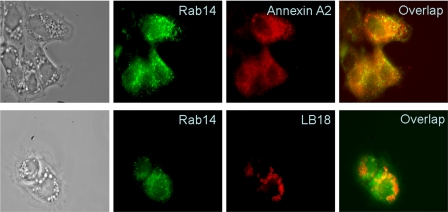
Co-localization of Rab14 and Annexin A2 in Type II Cells—To further explore the relationship between annexin A2 and Rab14, we performed localization studies in primary alveolar type II cells. Immunofluorescence studies indicate that Rab14 co-localized partially with annexin A2 in primary alveolar type II cells (Fig. 6). Furthermore, Rab14 also co-localized partially with the lamellar body membrane protein, LB-180, indicating that Rab14 is also present in some lamellar bodies (Fig. 6).
FIGURE 6.
Immunofluorescence of Rab14. Overnight cultured type II cells were double-labeled with rabbit polyclonal anti-Rab14 and mouse monoclonal anti-annexin A2 or anti-LB180 and examined by a fluorescence microscope.
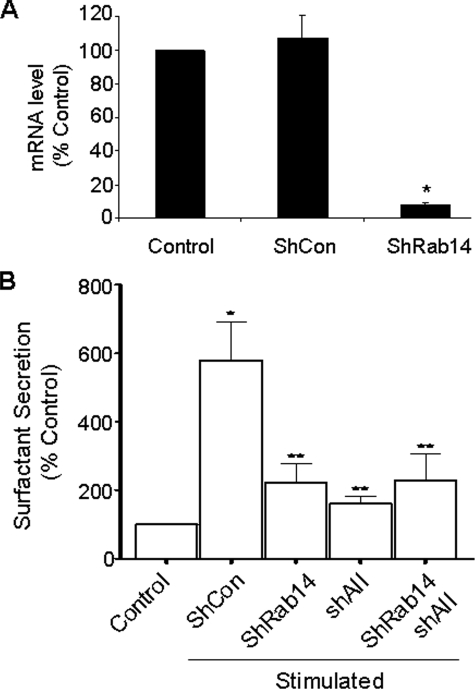
Lung Surfactant Secretion and Rab14—To address the functional roles of Rab14 in lung surfactant secretion, we constructed an adenoviral vector, Ad.K4-shRab14, which expresses four siRNAs specifically targeting to the coding regions of Rab14 at the positions of 11–31, 111–131, 139–159, and 438–456. Real time PCR analysis revealed that the transduction of alveolar type II cells with the Ad.K4-shRab14 resulted in ∼97% reduction in Rab14 mRNA expression (Fig. 7A). The Ad.K4-shRab14 also inhibited lung surfactant secretion (Fig. 7B). The silencing of annexin A2 with the Ad.K4-shAII also caused a decrease of lung surfactant secretion. Furthermore, the combination of Ad.K4-shRab14 and Ad.K4-shAII had a similar effect as the individual vector alone. Lack of the additive effects suggests that annexin A2 and Rab14 may act at the same pathway.
FIGURE 7.
Effect of Rab14 silencing on lung surfactant secretion. A, silencing of Rab14 in type II cells analyzed by real time PCR. 18 S rRNA was used to normalize the data. Data shown are means ± S.E. from three independent cell preparations, each assayed in duplicate. *, p < 0.05 versus shCon. B, lung surfactant secretion. Alveolar type II cells were cultured on an air-lipid interface. Adenovirus (m.o.i. 100) was added on day 3, and the cells were labeled with [3H]choline on day 6. The cells were stimulated with 1 mm ATP and 0.1 μm phorbol 12-myristate 13-acetate for 2 h on Day 7, and lung surfactant secretion was assayed. Data shown are means ± S.E. from three independent cell preparations. *, p < 0.01 versus Control; **, p < 0.05 versus shCon.
DISCUSSION
Annexins are multifunctional proteins involved in diverse cellular processes such as the control of intracellular calcium, vesicle trafficking, exocytosis and inhibition of phospholipase A2 and blood coagulation (6, 7). Annexin A2 is abundant in alveolar type II cells and has been implicated in the regulation of lung surfactant secretion. Evidence providing support for a role of annexin A2 in lung surfactant secretion is as follows. 1) Annexin A2 promotes in vitro fusion of lamellar bodies with the plasma membrane (11, 12). 2) Modification of annexin A2 by nitration or S-nitrosylation can eliminate its liposome aggregation activity (24, 25). 3) Depletion of annexin A2 from alveolar type II cell cytosol by immunoprecipitation almost completely inhibits the cytosol-induced fusion of lamellar bodies with the plasma membrane (11). 4) Knockdown of annexin A2 by RNA interference significantly reduces lung surfactant secretion (23). It is clear that annexin A2 promotes membrane fusion during the exocytosis of lamellar bodies; however, the interacting proteins and cellular pathways through which annexin A2 accomplishes the membrane fusion remain to be elucidated.
DNA microarray analysis has been widely used to identify informative signatures for uncovering important molecules and/or pathways. We therefore decided to use this method in association with an RNAi strategy to investigate the possible targets affected by the loss of annexin A2. Because it is difficult to transfect DNA vector-based shRNA into primary type II cells, we utilized an adenoviral system to deliver the shRNA vector. To reduce the possibility of off-target effects, we utilized our recently developed novel method (17) to construct a DNA vector expressing multiple shRNAs (K4-shRNA), in which four different shRNAs are transcribed from the same construct under the control of different promoters. Compared with the single shRNA vector method, the K4-shRNA vector significantly improves the silencing ability (17). Importantly, this construct design reduces the adenovirus required to silence a gene maximally and thus may reduce virus-related off-target and inflammatory responses. To further reduce off-target effects, we used two silencing vectors carrying different siRNA sequences targeted to rat annexin A2. Although two vectors had similar silencing efficiency, significantly changed genes because of the treatment of the two vectors were different (135 versus 185). Of these, 61 were commonly changed and were likely related to the loss of annexin A2. Thirteen of 18 genes were verified by real time PCR and 6 were further verified by rescue experiments, indicating that those targets functionally relate to annexin A2. Most of these genes were not known to be related with annexin A2 previously.
There are several possibilities for the association of the identified genes with annexin A2: (a) a direct physical interaction. Our co-immunoprecipitation studies revealed Rab14 as such a protein. The loss of annexin A2 may lead to the dissociation of Rab14 with annexin A2, and thus send a feedback signal for turning off Rab14 gene expression. (b) Annexin A2 is a substrate for pp60Src and insulin receptor kinase (26, 27). The defect of the signal transduction because of the absence of annexin A2 may change the expression of transcriptional factors such as Id1 and the genes that these factors control. (c) Annexin A2 is also involved in ion fluxes (28). Changes in cellular ion composition may regulate some of the ion channels such as Atp1a1 and Atp2a1. (d) Annexin A2 has been shown to bind with RNAs (29). Under certain conditions, nuclear annexin A2 was detected (30). Therefore, annexin A2 may directly regulate RNA transport and gene expression.
Rab proteins are the large family of the low molecular weight GTPases. More than 60 Rab proteins have been identified and implicated in the regulation of vesicle transport, including exocytosis (31). Rab proteins act as molecular switches for flipping GTP- and GDP-bound conformations. Because of the cooperative nature of Rab proteins, it is important to identify their interacting components. Several proteins interacting with the GTP-bound form of Rab proteins have been identified, but in many cases their functions are unknown. Strikingly, these proteins are structurally different from each other and are implicated in various cellular processes, suggesting that individual Rab proteins interacting with multiple effectors may control different steps in membrane trafficking events (32–35).
In comparison with the established roles of soluble NSF attachment protein receptor proteins, which mediate membrane fusion through the formation of the soluble NSF attachment protein receptor complex, Rabs act on a more upstream process in the initial physical contact between a vesicle and its target. It is believed that Rab proteins recruit distinct cytosolic effector proteins to the membrane and thus dock secretory granules onto the plasma membrane (36). This has been particularly well studied for yeast exocytosis. The elimination of a compartment-specific Rab blocks the downstream membrane fusion and leads to the accumulation of transport vesicles (37, 38). Deletion of Sec4p is lethal and also causes accumulation of small transport vesicles in the cytoplasm (39, 40). In mammalian cells, Rab proteins play a central role in constitutive and regulated exocytosis. Rab3A is one of the most notable and well characterized Rabs and is active in the regulated secretory pathway in the neuron (41). Involvement of Rab3A in insulin exocytosis was successively confirmed in vivo using Rab3A knock-out mice (42). Rab27 and Rab10 are also known to regulate organelle movement and regulate exocytosis in a wide variety of secretory cells (43, 44).
Rab proteins involved in the exocytosis of lamellar bodies in alveolar type II cells are unclear. One study identified Rab3D in type II cells, but only 25% of Rab3D is associated with lamellar bodies (45). Another study showed that Rab38 mRNA and protein were specifically located in alveolar type II cells and bronchial epithelial cells. However, Rab38 was not present in lamellar bodies (46, 47). Rab14 is a recently discovered new member and is uniquely expressed in all tissue (19). Rab14 is mainly localized to biosynthetic (rough endoplasmic reticulum, Golgi, and trans-Golgi network) and endosomal compartments (early endosomal vacuoles and associated vesicles), but a significant portion of Rab14 is also found in the plasma membrane of normal rat kidney cells (19). However, its function remains unknown. Our study indicated that the loss of annexin A2 down-regulated Rab14 expression. They interacted with each other physically. Furthermore, both proteins were partially co-localized in type II cells. Also, some Rab14 were present on lamellar bodies. Importantly, the loss of Rab14 reduced surfactant secretion. These results suggest that Rab14 may collaborate with annexin A2 in the secretion of surfactant in type II cells.
Acknowledgments
We thank Dr. Richard H. Scheller (Stanford University) for kindly providing Rab14 clone and anti-Rab14 antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL-052146, R01 HL-071628, and R01 HL-083188 (to L. L.). This work was also supported by a Center for Veterinary Health Sciences seed grant (to D. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: shRNA, short hairpin RNA; siRNA, short interfering RNA; m.o.i., multiplicity of infection; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; RT, reverse transcription; SAM, significance of microarray; GFP, green fluorescent protein; EGFP, enhanced GFP; RNAi, RNA interference.
References
- 1.Dietl, P., and Haller, T. (2005) Annu. Rev. Physiol. 67 595-621 [DOI] [PubMed] [Google Scholar]
- 2.Fehrenbach, H. (2001) Respir. Res. 2 33-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreeva, A. V., Kutuzov, M. A., and Voyno-Yasenetskaya, T. A. (2007) Am. J. Physiol. 293 L259-L271 [DOI] [PubMed] [Google Scholar]
- 4.Ichimura, H., Parthasarathi, K., Lindert, J., and Bhattacharya, J. (2006) Am. J. Physiol. 291 L596-L601 [DOI] [PubMed] [Google Scholar]
- 5.Haller, T., Auktor, K., Frick, M., Mair, N., and Dietl, P. (1999) Am. J. Physiol. 277 L893-L900 [DOI] [PubMed] [Google Scholar]
- 6.Gerke, V., and Moss, S. E. (2002) Physiol. Rev. 82 331-371 [DOI] [PubMed] [Google Scholar]
- 7.Gerke, V., Creutz, C. E., and Moss, S. E. (2005) Nat. Rev. Mol. Cell Biol. 6 449-461 [DOI] [PubMed] [Google Scholar]
- 8.Chasserot-Golaz, S., Vitale, N., Umbrecht-Jenck, E., Knight, D., Gerke, V., and Bader, M. F. (2005) Mol. Biol. Cell 16 1108-1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ali, S. M., Geisow, M. J., and Burgoyne, R. D. (1989) Nature 340 313-315 [DOI] [PubMed] [Google Scholar]
- 10.Knop, M., Aareskjold, E., Bode, G., and Gerke, V. (2004) EMBO J. 23 2982-2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chattopadhyay, S., Sun, P., Wang, P., Abonyo, B., Cross, N. L., and Liu, L. (2003) J. Biol. Chem. 278 39675-39683 [DOI] [PubMed] [Google Scholar]
- 12.Liu, L., Wang, M., Fisher, A. B., and Zimmerman, U. J. P. (1996) Am. J. Physiol. 270 L668-L676 [DOI] [PubMed] [Google Scholar]
- 13.Liu, L., Tao, J. Q., Li, H. L., and Zimmerman, U. J. P. (1997) Arch. Biochem. Biophys. 342 322-328 [DOI] [PubMed] [Google Scholar]
- 14.Liu, L. (1999) Cell. Signal. 11 317-324 [DOI] [PubMed] [Google Scholar]
- 15.Dobbs, L. G., Gonzalez, R., and Williams, M. C. (1986) Am. Rev. Respir. Dis. 134 141-145 [DOI] [PubMed] [Google Scholar]
- 16.Mason, R. J., Lewis, M. C., Edeen, K. E., McCormick-Shannon, K., Nielsen, L. D., and Shannon, J. M. (2002) Am. J. Physiol. 282 L249-L258 [DOI] [PubMed] [Google Scholar]
- 17.Gou, D., Weng, T., Wang, Y., Wang, Z., Zhang, H., Gao, L., Chen, Z., Wang, P., and Liu, L. (2007) J. Gene Med. 9 751-763 [DOI] [PubMed] [Google Scholar]
- 18.Gou, D., Narasaraju, T., Chintagari, N., Jin, N., Wang, P., and Liu, L. (2004) Nucleic Acids Res. 32 e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Junutula, J. R., De Maziere, A. M., Peden, A. A., Ervin, K. E., Advani, R. J., van Dijk, S. M., Klumperman, J., and Scheller, R. H. (2004) Mol. Biol. Cell 15 2218-2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, Z., Chen, J. W., Weng, T., Jin, N., and Liu, L. (2006) BMC Genomics 7 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen, Z., and Liu, L. (2005) Physiol. Genomics 21 284-291 [DOI] [PubMed] [Google Scholar]
- 22.Chintagari, N. R., Jin, N., Wang, P., Narasaraju, T. A., Chen, J., and Liu, L. (2006) Am. J. Respir. Cell Mol. Biol. 34 677-687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gou, D., Wang, P., Jin, N., and Liu, L. (2004) Annexins 1 31-36 [Google Scholar]
- 24.Liu, L., Enright, E., Sun, P., Tsai, S. Y., Mehta, P., Beckman, D. L., and Terrian, D. M. (1998) Eur. J. Biochem. 269 4277-4286 [DOI] [PubMed] [Google Scholar]
- 25.Rowan, W. H., III, Sun, P., and Liu, L. (2002) Biochemistry 41 1409-1420 [DOI] [PubMed] [Google Scholar]
- 26.Karasik, A., Pepinsky, R. B., Shoelson, S. E., and Kahn, C. R. (1988) J. Biol. Chem. 263 11862-11867 [PubMed] [Google Scholar]
- 27.Hubaishy, I., Jones, P. G., Bjorge, J., Bellagamba, C., Fitzpatrick, S., Fujita, D. J. X., and Waisman, D. M. (1995) Biochemistry 34 14527-14534 [DOI] [PubMed] [Google Scholar]
- 28.Nilius, B., Gerke, V., Prenen, J., Szucs, G., Heinke, S., Weber, K., and Droogmans, G. (1996) J. Biol. Chem. 271 30631-30636 [DOI] [PubMed] [Google Scholar]
- 29.Filipenko, N. R., MacLeod, T. J., Yoon, C. S., and Waisman, D. M. (2004) J. Biol. Chem. 279 8723-8731 [DOI] [PubMed] [Google Scholar]
- 30.Eberhard, D. A., Karns, L. R., Vandenberg, S. R., and Creutz, C. E. (2001) J. Cell Sci. 114 3155-3166 [DOI] [PubMed] [Google Scholar]
- 31.Bock, J. B., Matern, H. T., Peden, A. A., and Scheller, R. H. (2001) Nature 409 839-841 [DOI] [PubMed] [Google Scholar]
- 32.Echard, A., Jollivet, F., Martinez, O., Lacapere, J. J., Rousselet, A., Janoueix-Lerosey, I., and Goud, B. (1998) Science 279 580-585 [DOI] [PubMed] [Google Scholar]
- 33.Kato, M., Sasaki, T., Ohya, T., Nakanishi, H., Nishioka, H., Imamura, M., and Takai, Y. (1996) J. Biol. Chem. 271 31775-31778 [DOI] [PubMed] [Google Scholar]
- 34.Ren, M., Zeng, J., De Lemos-Chiarandini, C., Rosenfeld, M., Adesnik, M., and Sabatini, D. D. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 5151-5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stenmark, H., Vitale, G., Ullrich, O., and Zerial, M. (1995) Cell 83 423-432 [DOI] [PubMed] [Google Scholar]
- 36.Zerial, M., and McBride, H. (2001) Nat. Rev. Mol. Cell Biol. 2 107-117 [DOI] [PubMed] [Google Scholar]
- 37.Segev, N., Mulholland, J., and Botstein, D. (1988) Cell 52 915-924 [DOI] [PubMed] [Google Scholar]
- 38.Benli, M., Doring, F., Robinson, D. G., Yang, X., and Gallwitz, D. (1996) EMBO J. 15 6460-6475 [PMC free article] [PubMed] [Google Scholar]
- 39.Goud, B., Salminen, A., Walworth, N. C., and Novick, P. J. (1988) Cell 53 753-768 [DOI] [PubMed] [Google Scholar]
- 40.Salminen, A., and Novick, P. J. (1987) Cell 49 527-538 [DOI] [PubMed] [Google Scholar]
- 41.Schluter, O. M., Khvotchev, M., Jahn, R., and Sudhof, T. C. (2002) J. Biol. Chem. 277 40919-40929 [DOI] [PubMed] [Google Scholar]
- 42.Yaekura, K., Julyan, R., Wicksteed, B. L., Hays, L. B., Alarcon, C., Sommers, S., Poitout, V., Baskin, D. G., Wang, Y., Philipson, L. H., and Rhodes, C. J. (2003) J. Biol. Chem. 278 9715-9721 [DOI] [PubMed] [Google Scholar]
- 43.Khvotchev, M. V., Ren, M., Takamori, S., Jahn, R., and Sudhof, T. C. (2003) J. Neurosci. 23 10531-10539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tolmachova, T., Anders, R., Stinchcombe, J., Bossi, G., Griffiths, G. M., Huxley, C., and Seabra, M. C. (2004) Mol. Biol. Cell 15 332-344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Weeren, L., de Graaff, A. M., Jamieson, J. D., Batenburg, J. J., and Valentijn, J. A. (2004) Am. J. Respir. Cell Mol. Biol. 30 288-295 [DOI] [PubMed] [Google Scholar]
- 46.Osanai, K., Iguchi, M., Takahashi, K., Nambu, Y., Sakuma, T., Toga, H., Ohya, N., Shimizu, H., Fisher, J. H., and Voelker, D. R. (2001) Am. J. Pathol. 158 1665-1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osanai, K., Takahashi, K., Nakamura, K., Takahashi, M., Ishigaki, M., Sakuma, T., Toga, H., Suzuki, T., and Voelker, D. R. (2005) Biol. Chem. 386 143-153 [DOI] [PubMed] [Google Scholar]