Abstract
Osteoclasts are bone-resorbing cells, but they also secrete and respond to cytokines. Here, we test the hypothesis that osteoclasts secrete the lymphatic growth factor, VEGF-C, to increase their resorptive activity. Osteoclasts and osteoclast precursors were generated by culturing splenocytes with macrophage colony-stimulating factor and RANKL from wild-type, NF-κBp50-/-/p52-/-, and Src-/- mice. Expression of VEGFs was measured by real time reverse transcription-PCR, Western blotting, and immunostaining. The effect of VEGF-C signaling on osteoclast function was determined by osteoclastogenesis and pit assays. RANKL increased the expression of VEGF-C but not of other VEGFs in osteoclasts and their precursors. RANKL-induced VEGF-C expression was reduced in NF-κBp50-/-/p52-/- precursors or wild-type cells treated with an NF-κB inhibitor. VEGF-C directly stimulated RANKL-mediated bone resorption, which was reduced by the VEGF-C-specific receptor blocker, VEGFR3:Fc. Osteoclasts express VEGFR3, and VEGF-C stimulated Src phosphorylation in osteoclasts. VEGF-C-mediated bone resorption was abolished in Src-/- osteoclasts or cells treated with an Src inhibitor. We conclude that RANKL stimulates osteoclasts and their precursors to release VEGF-C through an NF-κB-dependent mechanism, indicating that VEGF-C is a new RANKL target gene in osteoclasts and functions as an autocrine factor regulating osteoclast activity.
Osteoclasts are bone-resorbing cells that play important roles in normal and pathologic bone remodeling. Osteoclast formation requires the expression of macrophage colony-stimulating factor (M-CSF)3 and receptor activator of nuclear factor κB (RANK) ligand (RANKL) by accessory cells, such as osteoblasts and bone marrow stromal cells, and of RANK, a member of the TNF receptor family, by osteoclast precursors (OCPs). RANKL and RANK interaction in OCPs triggers a cascade of signal transducing events and sequentially activates the transcription factors, NF-κB, AP-1, and NFATc1, leading to differentiation of OCPs to osteoclasts (1-3).
Recent studies demonstrate that osteoclasts are involved in more complex processes than simply resorption of bone. Osteoclasts can be activated to secrete factors that act through autocrine and paracrine mechanisms to contribute to inflammation and autoimmunity (4). For example, osteoclasts from patients with Paget's disease or giant cell tumors of bone produce higher levels of various cytokines and growth factors, including IL-6 and platelet-derived growth factor, than cells from normal bone (5). In co-cultures, myeloma cells and osteoclasts constitutively secrete the proangiogenic factors, VEGF-A (vascular endothelial growth factor-A) and osteopontin (6). In addition, platelet-derived growth factor-BB has been detected in the conditioned medium of the mouse macrophage/osteoclast precursor Raw 264.7 cells treated with RANKL (7). These findings indicate that osteoclasts also function as immunomodulators and affect the functions of other processes in addition to bone resorption (8, 9).
In the course of searching for new genes that osteoclasts express under pathologic conditions, we performed microarray analysis using mRNA from purified blood and bone marrow CD11b+/Gr-1-/lo OCPs from TNF transgenic (TNF-Tg) mice with established arthritis and wild-type (WT) mice. We detected significantly increased VEGF-C expression in cells from the arthritic mice (10). The VEGF gene family consists of VEGF-A, VEGF-B, VEGF-C, VEGF-D, and placental growth factor. VEGF-A, the prototype of VEGF ligands binds and activates two receptors, VEGF receptor 1 (VEGFR1) and VEGFR2 (11, 12), which are expressed by osteoclasts (13, 14). VEGF-A can substitute for M-CSF to support RANKL-induced osteoclastic bone resorption (15) through VEGFR1 signaling (16) and increases the area of bone resorption pits formed by mature osteoclasts (17). VEGF-A and placental growth factor also stimulate osteoclast recruitment (15, 18). The above studies proposed that VEGFs are released by cells other than osteoclasts, including blood vessel endothelium (13), osteoblasts (19), and tumor cells (6), and that they affect osteoclasts and OCPs through a paracrine mechanism. Whether or not osteoclasts themselves produce VEGFs that could function in an autocrine manner has not been studied to date.
VEGF-C has a unique role in lymphangiogenesis as a potent lymphatic endothelial cell growth factor through VEGFR3-mediated signaling. VEGF-C is highly expressed by numerous cancer cells, inflammatory macrophages, and synoviocytes (20-22), but factors that control VEGF-C expression have not been studied in detail. TNF, IL-1, and FGF stimulate VEGF-C expression in blood endothelium and fibroblastic cells (23-25), but to date only one publication has reported VEGF-C protein expression in osteoclast-like giant cells, and this was in immunohistochemically stained sections of a human pancreatic mucinous cystadenocarcinoma (26). The clinical or physiological significance of VEGF-C expression in this setting is unknown. Furthermore, little is known about the role of VEGF-C/VEGFR3 signaling in osteoclast formation and activation.
Here, we used primary OCPs derived from the spleen cells of WT and NF-κB-deficient mice and demonstrate that RANKL stimulated osteoclasts and OCPs to produce significantly increased amounts of VEGF-C. VEGF-C treatment or retroviral overexpression of VEGF-C in OCPs increased osteoclastic bone-resorbing activity but had no effect on osteoclast formation or survival. Disruption of VEGFR3 signaling by a VEGFR3:Fc chimeric protein reduced RANKL-mediated osteoclastic bone resorption, whereas interruption of VEGFR1 or VEGFR2 signaling had no effect. Our findings reveal that VEGF-C is a new target gene for RANKL in osteoclasts and that it acts as an autocrine factor positively regulating osteoclast function.
MATERIALS AND METHODS
Animals—WT (C57Bl/6), NF-κBp50-/-/p52-/- double knock-out (dKO) (C57Bl/6 × 129), and Src-/- (C57Bl/6) mice were used. NF-κB and Src knock-out mouse colonies were maintained and genotyped, as we described previously (27, 28). In experiments using knock-out mice, their WT littermates were used as controls. Except for NF-κB dKO mice, which were used at 3-4 weeks old due to early death of these animals, all other mice used were between 6 and 10 weeks old. The Institutional Animal Care and Use Committee of Rochester University approved all studies.
Reagents—Recombinant murine RANKL was purchased from R&D Systems Inc. (Minneapolis, MN), recombinant rat VEGF-C was from Calbiochem, fluorescein isothiocyanate-anti-mouse CD11b (M1/70) was from eBioscience (San Diego, CA), rabbit antibody to VEGF-C (H-190) and c-Src (N-16) were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), rabbit phosphospecific antibody to Src(phospho-Tyr418) was from BioSource and the Alexa Fluor 546 F(ab′)2 fragment of goat anti-rabbit IgG (H + L) antibody and To-Pro-3 iodide were purchased from Molecular Probes, Inc. (Carlsbad, CA). Murine RANK:Fc was provided by Dr. W. Dougall (Amgen Inc., Seattle, WA), and murine VEGF R2:Fc and VEGFR3:Fc were purchased fromR&D Systems Inc. VEGFR1:Fc was provided by ImClone Systems Inc. (New York). A small molecular NF-κB activation inhibitor was purchased from Calbiochem (catalog number 481406).
Real Time Quantitative RT-PCR—Total RNA was extracted using TRIzol reagent, and cDNA was synthesized using a RNA PCR Core Kit (Applied Biosystems, Branchburg, NJ). Quantitative PCR amplification was performed with gene-specific primers using an iCycler multiple-color real time PCR detection system (Bio-Rad), as we described previously (29). The primer sequences are listed in supplemental Table 1. For each sample, the relative levels were normalized to actin in the same sample.
Western Blot Analysis—Cells were lysed with mammalian protein extraction reagent (Pierce) containing a protease inhibitor mixture (Roche Applied Science). Conditioned media were concentrated with centrifugal devices (Pall Life Sciences, Ann Arbor, MI). Whole cell lysates (20 μg of protein) and concentrated conditioned media (80 μg of protein) were resolved by 12% SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted with anti-VEGF-C or anti-actin antibody (Sigma). Membranes were then washed and incubated with a horseradish peroxidase-conjugated secondary antibody (BioRad) and visualized by an enhanced chemiluminescence system (Amersham Biosciences) according to the manufacturer's instructions.
Immunofluorescence Staining—Cells were cultured on glass slides and fixed in cold methanol at -20 °C for 10 min. After washing with PBS, cells were incubated in 0.1% Triton and blocked with 1% bovine serum albumin plus PBS for 30 min. The fixed cells were stained with a mixture of fluorescein isothiocyanate-anti-CD11b and anti-VEGF-C antibody followed by Alexa Fluor 546 goat anti-rabbit IgG, and To-Pro-3 iodide.
Electrophoretic Mobility Shift Assay (EMSA)—Nuclear extract preparation and EMSA were performed, as described previously (30, 31). The following double-stranded oligonucleotides were used according to the published mouse VEGF-C promoter sequence (32) (only the top strands are shown): VEGF-C NF-κB binding sequence (WT), 5′-GCCCAGGGGGGTCCCCGGGAGG-3′; mutated VEGF-C in which the NF-κB binding sequence was mutated as indicated in underline (mut), 5′-GCCCAGGGGATTCTCCGGGAGG-3′; SP-1 binding sequence (control; Invitrogen), 5′-CGAGCCGGCCCCGCCCATC-3′. For competition assays, nuclear extracts and unlabeled oligonucleotides were preincubated for 15 min at room temperature with binding reaction buffer. The DNA-protein complex formed was then separated from free oligonucleotide on 5% native polyacrylamide gels.
Chromatin Immunoprecipitation (ChIP)—The ChIP assay was performed essentially according to the instructions in the Millipore EZ-ChIP™ kit. Briefly, Raw 264.7 cells were treated with RANKL (10 ng/ml) for 30 min and fixed in 1% formaldehyde for 10 min. The cross-linked cells were sonicated and immunoprecipitated with 1 μg of anti-NF-κB p65 or p50 antibodies (Santa Cruz Biotechnology) or mouse IgG overnight at 4 °C. Immune complexes were then recovered with Protein G-agarose beads, washed five times with the buffers provided with the kit, and then eluted with immunoprecipitation elution buffer (50 mm NaHCO3, 1% SDS) at room temperature. The samples were decross-linked, purified using spin columns, and resolved in sterile distilled water. PCRs were performed using 1 cycle at 95 °C for 1 min and 35 cycles at 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s, followed by further elongation at 72 °C for 10 min. PCR products were visualized on a 2% agarose gel with ethidium bromide staining. The primer pairs for the mouse VEGF-C promoter in the ChIP assays were 5′-GAGGGCAAAAGTTGCGAGC-3′ and 5′-GTGAGGCTGAGGTCCTCTCCT-3′, and these bind at -47 and +56 bp from the putative NF-κB binding sequence, respectively. The expected amplicon size is 103 bp.
Retroviral Vector and Infection—The coding sequence of human VEGF-C was amplified from MDA-231 human breast cancer cell line by PCR using forward primer 5′-CCTTGCGGCCGCCACCATGCACT-3′ (NotI) and reverse primer 5′ AAATCTTAAGTGTTTGGTCATTG (EcoRI) according to the published cDNA sequence (GenBank™ accession number X94216). After confirmation of the VEGF-C sequence by DNA sequencing, VEGF-C was cloned into the EcoRI site of the pMX-IRES-GFP vector provided by Dr. Matsuo Koichi. The retrovirus vector was transfected into the Plat-E retroviral packaging cell line, and viral supernatant was collected 48 h later for infection as we described previously (31, 33). pMXGFP plasmid was used as control vector.
Osteoclastogenesis and Bone Resorption Assays—Spleens were ground on a mesh to make single splenocyte suspensions. Red blood cells were lysed with NH4Cl (StemCell Technologies, Vancouver, Canada) on ice for 10 min and washed with medium twice and resuspended in α-minimal essential medium containing 10% fetal bovine serum. The cells were then cultured with 1:20 diluted conditioned medium containing M-CSF (from an M-CSF-producing cell line) (34) and RANKL with or without additional treatment to form mature osteoclasts. Alternatively, the cells were cultured with M-CSF for 3 days to generate OCPs, as we described previously (33), and then infected with retroviral supernatants and cultured with M-CSF and RANKL on bone slices to form mature osteoclasts. The cells were fixed and stained for tartrate-resistant acid (TRAP) activity to identify osteoclasts as TRAP+ cells containing at least three nuclei and then removed and stained with 0.5% toluidine blue to visualize resorption pits for bone resorption activity, as we described previously (31, 33).
Staining of Actin Rings—Osteoclasts cultured on bone slices were fixed with 3% paraformaldehyde for 5 min and permeabilized for 5 min in PBS containing 0.5% Triton X-100. After three washes with cold PBS, cells on bone slices were stained using rhodamine-conjugated phalloidin (Molecular Probes, Inc., Eugene, OR) for 30 min. Osteoclasts with actin rings were counted using a phase-contrast fluorescence microscope.
Statistics—Data are presented as mean ± S.D. of triplicates, and all experiments were performed at least twice with similar results. Statistical analyses were performed with Statview statistical software (SAS, Cary, NC). Differences between two groups were compared using unpaired Student's t test, and more than two groups were compared using one-way analysis of variance, followed by the Bonferroni/Dunnet test. p values less than 0.05 were considered to be statistically significant.
RESULTS
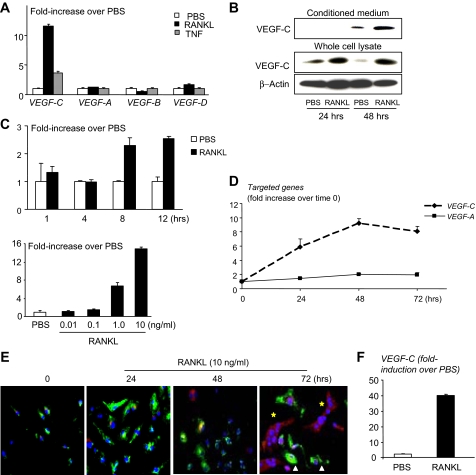
RANKL Stimulates Osteoclasts and OCPs to Produce VEGF-C—Recently, we found that OCPs from peripheral blood and bone marrow of TNF-Tg arthritic mice express high levels of VEGF-C compared with those from WT littermates (10). These mice also have increased expression of TNF, IL-1, and RANKL. To determine if these cytokines stimulate VEGF-C production by OCPs, we treated primary OCPs generated from WT splenocytes with RANKL, TNF, or IL-1. RANKL increased VEGF-C mRNA expression 12-fold, whereas TNF increased it only 3-4-fold (Fig. 1A). IL-1 increased VEGF-C expression to the same extent as TNF (data not shown). No effect on VEGF-A, -B, or -D expression was detected in the same samples (Fig. 1A). In conditioned media from these experiments, expression of a processed and functional form of VEGF-C protein (29 kDa) was increased at 48 h after RANKL treatment (Fig. 1B). Time and dose-response studies showed that RANKL, at the dose that is used routinely in osteoclastogenesis assays (10 ng/ml), increased VEGF-C mRNA levels, starting at 8 h (Fig. 1C). The lowest dose of RANKL that induced VEGF-C expression was 1 ng/ml (Fig. 1C), which is lower than the dose (10 ng/ml) used typically to induce maximum osteoclast formation. In preliminary experiments, we demonstrated that 1 ng/ml RANKL induced 4-fold fewer osteoclasts than 10 ng/ml RANKL on plastic culture wells.
FIGURE 1.
RANKL increases expression of VEGF-C by osteoclasts. WT splenocytes were cultured with M-CSF-containing medium (1:20) for 3 days to generate OCPs. A, OCPs were treated with RANKL or TNF (10 ng/ml) for 24 h, and the expression levels of VEGF-A, -B, -C, and -D mRNA were examined by real time RT-PCR. The -fold increase in RANKL-treated over PBS-treated cells was calculated. Values are the means plus S.D. of triplicate loadings. B, Raw 264.7 cells were serum-starved overnight and treated with RANKL. VEGF-C protein expression was examined by Western blot analysis. C, OCPs were treated with RANKL for various times (upper graph) or with different amounts of RANKL for 24 h (lower graph). Expression of VEGF-C mRNA was measured by RT-PCR. D, OCPs were cultured on glass slides with M-CSF and RANKL. The expression of VEGF-C and VEGF-A mRNA was examined at various times by real time RT-PCR. E, cells were stained with FTIC-anti-CD11b (green) and rabbit anti-VEGF-C followed by anti-rabbit Alexa Fluor 546 antibody (red). TO-PRO-3 iodide was used for nuclear staining (blue). Pictures were taken under a confocal microscope at power 10. F, WT spleen cells were cultured with RANKL and M-CSF for 5 days to generate osteoclasts. When mature osteoclasts were observed under an inverted microscope, RANKL was removed from the culture medium overnight. Cells were then retreated with RANKL (10 ng/ml) or PBS for 24 h. The expression levels of VEGF-C mRNA were determined by real time RT-PCR. Values are the means plus S.D. of triplicate loadings.
We next examined the expression pattern of VEGF-C mRNA and protein in WT cells during RANKL-induced osteoclastogenesis. These increased with time and peaked at 48-72 h when mature osteoclasts had formed (Fig. 1, D and E). In contrast, expression levels of VEGF-A, the predominant angiogenic VEGF family member (11), were unchanged during osteoclast differentiation (Fig. 1D). To examine the distribution of VEGF-C protein and its relationship with expression of CD11b, a surface marker of OCPs and macrophages, we performed double immunocytochemical staining of cells cultured on glass slides with anti-CD11b and anti-VEGF-C antibodies using a confocal microscope (Fig. 1E). As reported previously (35), CD11b protein was expressed on the surface of OCPs, and this expression disappeared when OCPs became mature multinucleated osteoclasts. In contrast, VEGF-C protein expression increased progressively in the cytoplasm of these mononuclear cells as they differentiated into osteoclasts. Mononuclear OCPs also express VEGF-C in their cytoplasm while they have CD11b staining on the cell surface (Fig. 1E).
To determine if RANKL stimulates mature osteoclasts to express VEGF-C, we cultured OCPs with RANKL and M-CSF and removed RANKL from the cultures overnight when multinucleated osteoclasts were visible under an inverted microscope. The cells were then treated with RANKL for a further 24 h. Under these conditions, RANKL-induced VEGF-C mRNA expression was increased further (Fig. 1F).
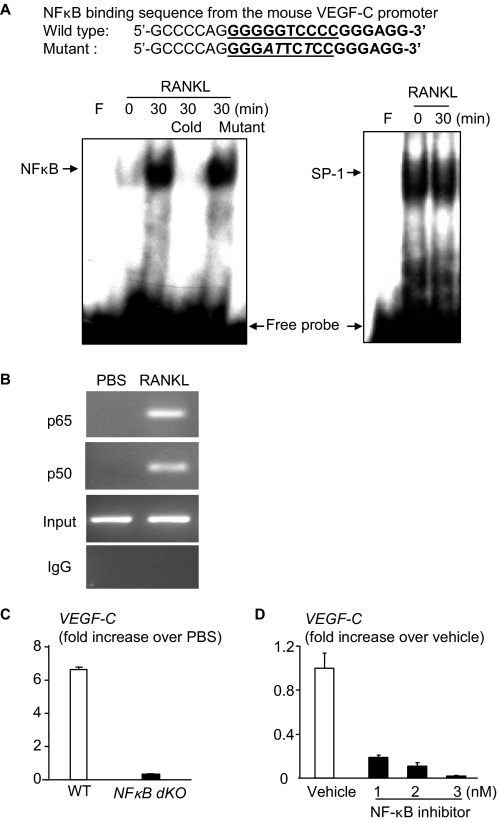
NF-κB Regulates RANKL-induced Expression of VEGF-C by OCPs—To investigate the mechanisms by which RANKL induces VEGF-C expression, we focused on NF-κB signaling, because it plays an essential role in osteoclastogenesis down-stream from RANKL/RANK (3) and because the -108/-99 region of the mouse VEGF-C promoter has a putative NF-κB binding sequence (32). First, we searched our mRNA microarray data from RANKL-treated OCPs from NF-κB dKO mice and WT littermates and found that VEGF-C expression was reduced in NF-κB dKO cells compared with WT cells (data not shown). We then performed Western blot and EMSA assays using Raw264.7 cells and an oligonucleotide probe synthesized according to the putative NF-κB recognition sequence localized in the -108/-99 region of the mouse VEGF-C promoter. RANKL treatment stimulated nuclear translocation of NF-κB p65 and p50 proteins, as anticipated (data not shown), and binding of nuclear protein to the NF-κB probe (Fig. 2A). This nuclear binding was abolished by an excess of unlabeled WT but not by an oligonucleotide in which the NF-κB binding sequence has been mutated to prevent binding (Fig. 2A). To determine if RANKL stimulates binding of NF-κB protein to the VEGF-C promoter in intact cells, we performed a ChIP assay using the same experimental conditions used in the EMSA and demonstrated that in RANKL-treated cells, NF-κB-containing proteins bound to the NF-κB binding sequence of the VEGF-C promoter (Fig. 2B). To further confirm involvement of NF-κB in RANKL-induced VEGF-C expression, splenocytes from NF-κB dKO and WT mice were cultured with M-CSF for 3 days to generate OCPs and then were treated with RANKL for 24 h. Similar to the microarray data, RANKL-induced VEGF-C expression in the dKO cells was significantly reduced compared with WT cells (Fig. 2C). We also treated WT OCPs with a recently developed nonspecific NF-κB inhibitor (31, 36) and found that it also inhibited RANKL-induced VEGF-C expression in a dose-dependent manner (Fig. 2D).
FIGURE 2.
Involvement of NF-κB in RANKL-induced VEGF-C expression. Raw264.7 cells were treated with RANKL (10 ng/ml) for 30 min. A, the binding of nuclear extracts to the NF-κB binding sequence of the VEGF-C probe was determined by EMSA. The specificity of binding was confirmed by using 50-fold more unlabeled WT or a mutated probe in which the NF-κB binding sequence was mutated. An SP-1 probe was used as a loading control. B, cell lysates were immunoprecipitated using anti-p65 or anti-p50 antibodies and subjected to a ChIP assay. The PCR was performed using primers that bind at -47 and +56 from the NF-κB binding sequence. IgG immunoprecipitated samples were used as negative controls. Preimmunoprecipitated samples were used as loading controls (Input lanes). C, OCPs generated from NF-κB dKO mice and WT littermates were treated with RANKL for 24 h. The expression of VEGF-C mRNA was measured by real time RT-PCR as in Fig. 1A. D, WT OCPs were treated with RANKL with or without an NF-κB inhibitor for 24 h, and expression of VEGF-C was determined by real time RT-PCR. The -fold decrease in expression in the NF-κB inhibitor-treated over vehicle-treated cells was calculated.
VEGF-C Increases the Bone Resorptive Activity of Osteoclasts—To determine if VEGF-C plays a direct role in osteoclast generation, activation, or survival, we cultured WT splenocytes on plastic dishes with RANKL and M-CSF in the presence or absence of VEGF-C. VEGF-C had no significant effect on basal or RANKL-induced osteoclast formation (data not shown). To examine if VEGF-C affects osteoclastic bone resorption on bone slices, we modified our typical culture protocol and used 10-fold lower concentrations of RANKL and M-CSF, which we determined in preliminary studies induced minimal resorption. Under these culture conditions, VEGF-C had no effect on osteoclast numbers cultured on either plastic plates (data not shown) or bone slices, but it significantly increased the osteoclast-induced resorption pit area in a dose-dependent manner (Fig. 3A). To confirm this observation, we constructed a retroviral construct, which contains the human VEGF-C cDNA followed by eGFP cDNA (pMX-hVEGF-C-IRSE-eGFP). Infection of WT OCPs with this hVEGF-C retrovirus on plastic plates induced high expression of hVEGF-C, which was confirmed by real time RT-PCR (Fig. 3B) and GFP fluorescence (Fig. 3C). Similar to the effect of exogenous VEGF-C treatment in Fig. 3A, overexpression of VEGF-C significantly increased the resorption pit area but had no effect on osteoclast numbers (Fig. 3D) or survival (supplemental Fig. 1). Furthermore, we consistently observed that TRAP-stained osteoclasts that had been generated on plastic dishes with low doses of RANKL and M-CSF in the presence of VEGF-C appeared to have thicker cell membranes than PBS-treated controls (Fig. 3E, arrow).
FIGURE 3.
VEGF-C increases osteoclastic bone resorption and actin ring formation. A, WT OCPs were cultured on bone slides in suboptimal concentrations of M-CSF (1:200) and RANKL (0.5 ng/ml) with or without VEGF-C for 10 days. The number of TRAP+ osteoclasts and the area of resorption pits on bone slices were assessed. Values are the means plus S.D. of four bone slices. B, WT OCPs were infected with hVEGF-C retrovirus supernatant and cultured on plastic dishes. The expression of the hVFGF-C transgene was determined by real time RT-PCR. C, infected cells were visualized by eGFP fluorescence under a fluorescent microscope, showing that more than 80% infected cells are GFP+. D, infected cells were cultured on bone slices in suboptimal concentrations of M-CSF and RANKL. The number of TRAP+ osteoclasts and the area of resorption pits were assessed as in A. E, WT OCPs were cultured in suboptimal concentrations of M-CSF and RANKL with or without VEGF-C. Pictures of TRAP stained osteoclasts on plastic show a clear continuous cell membrane structure in VEGF-C-treated cells (arrows) compared with cells without VEGF-C, which have incompletely defined cell membranes. F, osteoclasts cultured on bone slices were fixed and stained with rhodamine-conjugated phalloidin to observe actin rings (arrows). G, the number of osteoclasts with completely formed actin rings was counted under a fluorescent microscope. Values are the means plus S.D. of four bone slices. *, p < 0.05 versus PBS-treated or GFP virus-infected cells.
Osteoclasts resorb bone by reorganizing their cytoskeleton to form ruffled borders and actin-rich rings on the bone surface. To examine if VEGF-C influences actin ring formation by resorbing osteoclasts, we generated osteoclasts on bone slices using suboptimal concentrations of RANKL and M-CSF with or without VEGF-C and stained them with rhodamine-conjugated phalloidin to visualize actin-rich rings. Cells treated with low doses of RANKL and M-CSF had poorly formed actin rings, which were dotlike and discontinuous, unlike those in osteoclasts treated with optimal doses of these cytokines (Fig. 3F). The addition of VEGF-C to the cultures treated with suboptimal cytokine doses restored the actin ring dotlike pattern to normal (Fig. 3F). The number of osteoclasts with normal, intact actin rings was significantly greater in VEGF-C-treated compared with non-VEGF-C-treated cells (Fig. 3G).
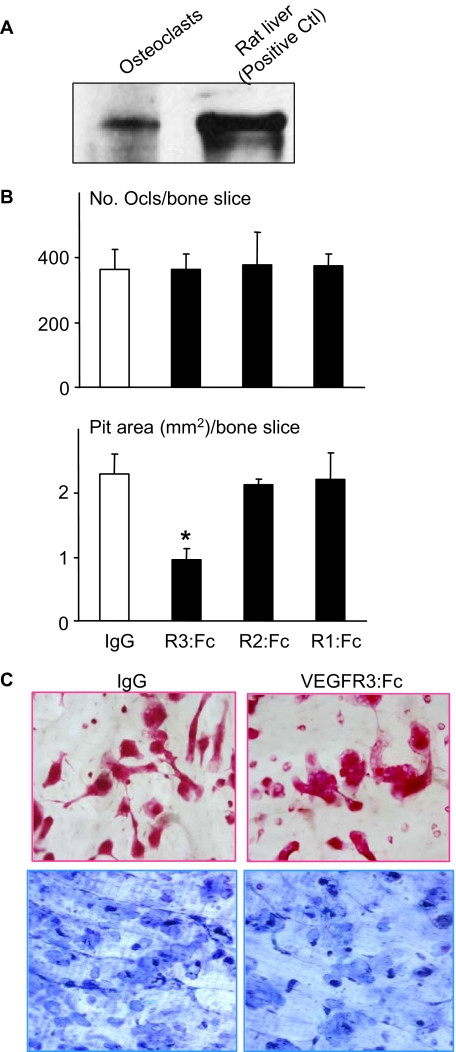
Blockade of VEGF-C Signaling Inhibits RANKL-mediated Osteoclastic Bone Resorption—The findings that RANKL stimulates osteoclasts to produce VEGF-C and that VEGF-C increases osteoclastic bone resorption led us to speculate that VEGF-C induced by RANKL may function in an autocrine manner to promote osteoclast activity. To test this hypothesis, we first demonstrated that osteoclasts express the VEGF-C-specific receptor, VEGFR3, using Western blot analysis (Fig. 4A). To determine if osteoclasts have functional VEGFR3 signaling, we treated osteoclasts with a VEGFR3-specific mutant VEGF-C (VEGF-C C156S) which is an agonist that binds specifically to VEGFR3 and no other VEGFRs in lymphatic endothelial cells (20). We found that this mutant agonist induced bone resorption (resorption pit area/bone slice (mm2): 2.0 ± 0.3 versus 0.75 ± 0.15 in PBS controls, p < 0.05). We then cultured WT OCPs with optimal doses of RANKL and M-CSF to induce resorption in the presence of various VEGF receptor antagonists, including VEGFR1:Fc, VEGFR2:Fc, or VEGFR3: Fc. VEGFR3:Fc significantly reduced RANKL-induced osteoclastic bone resorption by 60%, whereas neither VEGFR1:Fc nor VEGFR2:Fc had any inhibitory effect in the same experiments (Fig. 4). None of the VEGF receptor antagonists had any effect on osteoclast numbers.
FIGURE 4.
Blockade of VEGF-C/VEGFR-3 signaling reduces RANKL-mediated osteoclastic bone resorption. A, expression of VEGFR3 was examined by Western blot analysis using WT osteoclast cell lysates. Rat liver extracts (Santa Cruz Biotechnology) were included as a positive control. B, WT OCPs were cultured with M-CSF (1:20) and RANKL (10 ng/ml) on bone slices with or without VEGFR3:Fc, VEGFR2:Fc, VEGFR1:Fc (1 μg/ml), or control IgG isotype for 10 days. The number of TRAP+ osteoclasts and the area of resorption pits on bone slices were assessed. Values are the means plus S.D. of four bone slices. *, p < 0.05 versus IgG-treated cells. C, pictures were taken with a ×10 objective lens showing TRAP+ osteoclasts and resorption pits in IgG- or VEGFR3:Fc-treated cells.
VEGF-C Activates Osteoclastic Bone Resorption through Src Signaling—Src, p38, and Erk mediate VEGFR signaling in endothelial and cancer cells (38, 39). We and others have demonstrated that Src expression is essential in osteoclasts for the cytoskeletal reorganization that they require for bone resorption (40) (41). To examine the involvement of Src in VEGF-C-mediated osteoclast bone resorption, we treated osteoclasts derived from Src-/- cells with VEGF-C and observed no resorption pits (Fig. 5A). Since this may not show a VEGF-C-dependent mechanism because no resorption occurred in Src-/- osteoclasts even in the absence of VEGF-C, we used an Src inhibitor that we demonstrated previously to reduce bone resorption (42) and PTH-induced hypercalcemia (43). We found that it also prevented VEGF-C-induced osteoclastic bone resorption (Fig. 5B). Furthermore, VEGF-C as well as the VEGFR3-specific VEGF-C mutant increased phosphorylated but not total Src protein expression in osteoclasts (Fig. 5C).
FIGURE 5.
c-Src is involved in VEGF-C-mediated osteoclast bone resorption. A, OCPs from WT or Src-/- mice were cultured on bone slices for 8 days and treated with RANKL, M-CSF, and VEGF-C. B, WT OCPs were cultured with M-CSF, RANKL, and VEGF-C with or without an Src inhibitor on bone slices for 8 days. The area of resorption pits on bone slices was assessed. Values are the means plus S.D. of four bone slices. *, p < 0.05 versus vehicle-treated cells or WT cells. #, p < 0.05 versus PBS-treated cells. C, WT OCPs were cultured with M-CSF and RANKL for 5 days to form osteoclasts. Cells were cultured in serum-free medium for 4 h and treated with VEGF-C or VEGFR3 agonist, VEGF-C (C156S), for 30 min. Expression of phospho-Src and total Src protein was determined by Western blot analysis.
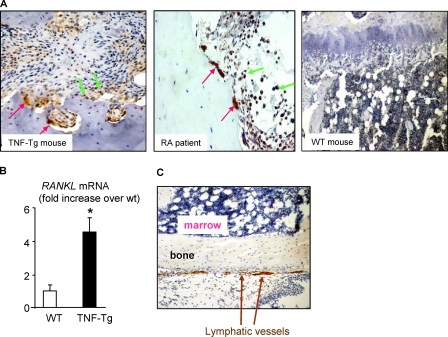
VEGF-C Expression Is Increased in Osteoclasts in Arthritic Joints—To determine if VEGF-C expression by osteoclasts is increased in vivo, we performed immunostaining of tissue sections from joints of mice with TNF-induced arthritis (TNF-Tg mice) and from patients with rheumatoid arthritis in which RANKL expression typically is increased (44). A strong signal for VEGF-C was observed in mature osteoclasts and some mononuclear cells with an anti-VEGF-C antibody (Fig. 6A). However, using the same dilution of VEGF-C antibody, we did not detect VEGF-C expression in osteoclasts from WT mice (Fig. 6A). To examine if RANKL expression is increased in joint samples from the TNF-Tg mice to account for increased VEGF-C expression by osteoclasts, we compared RANKL mRNA expression in joints of TNF-Tg mice with their WT littermates and found that joints of TNF-Tg mice have a high level of RANKL expression (Fig. 6B). In addition, we also used antibodies to the specific lymphatic endothelial cell markers, lymphatic vessel endothelial receptor 1 (Fig. 6C) and podoplanin (data not shown), and demonstrated that there are no lymphatic channels in the bone marrow cavities of mice.
FIGURE 6.
Increased VEGF-C and RANKL expression in samples from arthritic joints. A, joint sections from TNF-Tg arthritic mice (n = 3), patients with rheumatoid arthritis (n = 3), or long bones from WT littermates of TNF-Tg mice (n = 4) were immunostained with anti-VEGF-C antibody. Strong VEGF-C-positive staining was observed in mature osteoclasts (red arrows) and mononuclear cells (green arrows). B, total RNA was extracted from ankle joints of TNF-Tg mice and WT littermates. Expression of RANKL mRNA was determined by real time RT-PCR. The -fold increase in TNF-Tg mice over WT mice was calculated. Similar results were obtained from four pairs of TNF-Tg mice and WT littermates. C, paraffin sections of mouse long bone were immunostaining with anti-lymphatic vessel endothelial receptor 1 (LYVE-1) antibody to identify lymphatic vessels. The image (×10 power) indicates that LYVE-positive lymphatic vessels localize within the connective tissues outside of bone marrow cavity (arrows).
DISCUSSION
In this study, we have demonstrated that RANKL induces osteoclasts to express the lymphatic growth factor, VEGF-C, and that VEGF-C by binding to its receptor, VEGFR3, on osteoclasts directly increases osteoclastic bone resorption without affecting osteoclast formation or survival. This effect of RANKL was much greater than that of TNF or IL-1, two cytokines that, like RANKL, are highly expressed at sites of inflammation in bone, such in the inflamed joints of patients with rheumatoid arthritis and in TNF-Tg mice with inflammatory arthritis. These findings identify VEGF-C as a novel target of RANKL signaling in osteoclasts that functions by an autocrine mechanism to amplify the effects of RANKL on osteoclast function.
We observed that when osteoclasts were cultured on plastic plates with low concentrations of RANKL and M-CSF, their cells membranes appear thin and their actin rings appeared discontinuous and dotlike. VEGF-C treatment increased the thickness of the cell membranes and resulted in actin rings appearing normal and continuous (Fig. 3E). We think that these morphologic observations are relevant, since VEGF-C activates Src through phosphorylation (Fig. 5C) and affects actin ring formation (Fig. 3, F and G). Thus, it is likely that the increased thickness of the cell membranes reflects a more robust actin ring structure and that VEGF-C may increase osteoclastic bone resorption by a direct autocrine mechanism in osteoclasts: improved osteoclast binding to matrix. This direct action is unlike that of other VEGFs, which are produced by accessory cells in bone and stimulate osteoclasts through a paracrine mechanism (13, 18).
It is becoming increasingly clear that autocrine regulation of formation and function is an important aspect of osteoclast activity. For example, TNF is produced by osteoclasts, and it stimulates osteoclasts to release IL-1 (45). An IL-1 antagonist partially blocks TNF-induced osteoclast formation, consistent with an autocrine mechanism (45). We have found that osteoclast precursors interact with bone matrix to trigger their own differentiation by producing IL-1 (46). Osteoclasts also negatively control their formation through RANKL-induced production of interferon β (47). Here we show that VEGF-C is another example of osteoclast autoregulation.
VEGF-C has been identified recently as a lymphatic endothelial cell growth factor (48), and we found that osteoclasts express VEGF-C. Thus, one possible additional function of VEGF-C expression by osteoclasts in bone might be to enhance growth of lymphatic channels in bone marrow. However, using antibodies to the specific lymphatic endothelial cell markers, lymphatic vessel endothelial receptor 1 (Fig. 6C) and podoplanin (data not shown), we found that there are no lymphatic channels in the bone marrow cavities of mice. Although it is known that there are no capillary beds in bone marrow and that these are replaced by a sinusoidal small vascular system, we believe that this is the first definitive evidence that the lymphatic system is also absent from bone marrow. In contrast to these findings within bone, we found remarkably enlarged lymphatic vascular networks in the soft tissues around inflamed joints of TNF-Tg arthritic mice (10), where RANKL levels are high and osteoclasts strongly express VEGF-C (Fig. 6). Thus, osteoclast-derived VEGF-C may play a more important role in pathologic conditions where RANKL and inflammatory cytokines are highly expressed, such as in arthritic joints, through two distinct mechanisms. One is to increase osteoclastic bone resorption, and another is to increase lymphatic channel numbers and presumably lymph flow away from inflamed joints. It remains to be determined if osteoclasts play a significant role in lymphatic vessel formation during development, but the absence of osteoclasts and lymph nodes in mice deficient in RANKL, RANK, or NF-κB p50/p52 (4, 35) suggests that this merits further study.
We found that NF-κB mediates RANKL-induced VEGF-C expression. This is not surprising, because NF-κB regulates transcription of many genes. The specificity of our findings is the involvement of NF-κB in RANKL-mediated VEGF-C expression. The importance of this finding is 2-fold. One is to link NF-κB, RANKL, VEGF-C, and osteoclastic bone resorption together in the context of joint inflammation, where each individual factor is known to be up-regulated. Another is related to recent reports of expression of VEGF-C by dendritic cells (37). Since dendritic cells are RANK-expressing cells and respond to RANKL, NF-κB-mediated VEGF-C expression by RANKL may also apply to dendritic cells.
In summary, we have demonstrated that the lymphatic growth factor, VEGF-C, is a new RANKL target gene in osteoclasts, and it stimulates bone resorption through a VEGFR3-mediated pathway. Thus, osteoclast-induced VEGF-C may have dual functions; it up-regulates osteoclast activation and stimulates lymphangiogenesis through autocrine and paracrine mechanisms, respectively.
Supplementary Material
Acknowledgments
We thank Dr. Toshio Kitamura for the Plat-E cell line, Dr. Sunao Takeshita for the M-CSF-producing cell line, Dr. Matsuo Koichi for the pMX-IRES-GFP vector, and Xiaoyun Zhang for technical assistance with the histology.
This work was supported, in whole or in part, by National Institutes of Health Grants PHS AR48697 and AR53586 (to L. X.) and AR43510 (to B. F. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
Footnotes
The abbreviations used are: M-CSF, macrophage colony-stimulating factor; TNF, tumor necrosis factor; OCP, osteoclast precursor; IL, interleukin; TNF-Tg, TNF transgenic; WT, wild type; dKO, double knock-out; RT, reverse transcription; PBS, phosphate-buffered saline; EMSA, electrophoretic mobility shift assay; ChIP, chromatin immunoprecipitation; TRAP, tartrate-resistant acid; GFP, green fluorescent protein; eGFP, enhanced green fluorescent protein.
References
- 1.Wada, T., Nakashima, T., Hiroshi, N., and Penninger, J. M. (2006) Trends Mol. Med. 12 17-25 [DOI] [PubMed] [Google Scholar]
- 2.Asagiri, M., and Takayanagi, H. (2007) Bone 40 251-264 [DOI] [PubMed] [Google Scholar]
- 3.Boyce, B. F., Yamashita, T., Yao, Z., Zhang, Q., Li, F., and Xing, L. (2005) J. Bone Miner. Metab. 23 (suppl.) 11-15 [DOI] [PubMed] [Google Scholar]
- 4.Takayanagi, H. (2007) Nat. Rev. Immunol. 7 292-304 [DOI] [PubMed] [Google Scholar]
- 5.Mills, B. G., and Frausto, A. (1997) Calcif. Tissue Int. 61 16-21 [DOI] [PubMed] [Google Scholar]
- 6.Tanaka, Y., Abe, M., Hiasa, M., Oda, A., Amou, H., Nakano, A., Takeuchi, K., Kitazoe, K., Kido, S., Inoue, D., Moriyama, K., Hashimoto, T., Ozaki, S., and Matsumoto, T. (2007) Clin. Cancer Res. 13 816-823 [DOI] [PubMed] [Google Scholar]
- 7.Kubota, K., Sakikawa, C., Katsumata, M., Nakamura, T., and Wakabayashi, K. (2002) J. Bone Miner. Res. 17 257-265 [DOI] [PubMed] [Google Scholar]
- 8.Boyce, B. F., Schwarz, E. M., and Xing, L. (2006) Curr. Opin. Rheumatol. 18 427-432 [DOI] [PubMed] [Google Scholar]
- 9.Xing, L., Schwarz, E. M., and Boyce, B. F. (2005) Immunol. Rev. 208 19-29 [DOI] [PubMed] [Google Scholar]
- 10.Zhang, Q., Lu, Y., Proulx, S., Guo, R., Yao, Z., Schwarz, E. M., Boyce, B. F., and Xing, L. (2007) Arthritis Res. Ther. 9 R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tammela, T., Enholm, B., Alitalo, K., and Paavonen, K. (2005) Cardiovasc. Res. 65 550-563 [DOI] [PubMed] [Google Scholar]
- 12.Roy, H., Bhardwaj, S., and Yla-Herttuala, S. (2006) FEBS Lett. 580 2879-2887 [DOI] [PubMed] [Google Scholar]
- 13.Nakagawa, M., Kaneda, T., Arakawa, T., Morita, S., Sato, T., Yomada, T., Hanada, K., Kumegawa, M., and Hakeda, Y. (2000) FEBS Lett. 473 161-164 [DOI] [PubMed] [Google Scholar]
- 14.Tombran-Tink, J., and Barnstable, C. J. (2004) Biochem. Biophys. Res. Commun. 316 573-579 [DOI] [PubMed] [Google Scholar]
- 15.Niida, S., Kaku, M., Amano, H., Yoshida, H., Kataoka, H., Nishikawa, S., Tanne, K., Maeda, N., Nishikawa, S., and Kodama, H. (1999) J. Exp. Med. 190 293-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niida, S., Kondo, T., Hiratsuka, S., Hayashi, S., Amizuka, N., Noda, T., Ikeda, K., and Shibuya, M. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 14016-14021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto, Y., Tanaka, K., Hirata, G., Hanada, M., Matsuda, S., Shuto, T., and Iwamoto, Y. (2002) J. Immunol. 168 5824-5831 [DOI] [PubMed] [Google Scholar]
- 18.Henriksen, K., Karsdal, M., Delaisse, J. M., and Engsig, M. T. (2003) J. Biol. Chem. 278 48745-48753 [DOI] [PubMed] [Google Scholar]
- 19.Deckers, M. M., Karperien, M., van der Bent, C., Yamashita, T., Papapoulos, S. E., and Lowik, C. W. (2000) Endocrinology 141 1667-1674 [DOI] [PubMed] [Google Scholar]
- 20.Su, J. L., Yen, C. J., Chen, P. S., Chuang, S. E., Hong, C. C., Kuo, I. H., Chen, H. Y., Hung, M. C., and Kuo, M. L. (2007) Br. J. Cancer 96 541-545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salven, P., Lymboussaki, A., Heikkila, P., Jaaskela-Saari, H., Enholm, B., Aase, K., von Euler, G., Eriksson, U., Alitalo, K., and Joensuu, H. (1998) Am. J. Pathol. 153 103-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maruyama, K., Asai, J., Ii, M., Thorne, T., Losordo, D. W., and D'Amore, P. A. (2007) Am. J. Pathol. 170 1178-1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao, R., Eriksson, A., Kubo, H., Alitalo, K., Cao, Y., and Thyberg, J. (2004) Circ. Res. 94 664-670 [DOI] [PubMed] [Google Scholar]
- 24.Cha, H. S., Bae, E. K., Koh, J. H., Chai, J. Y., Jeon, C. H., Ahn, K. S., Kim, J., and Koh, E. M. (2007) J. Rheumatol. 34 16-19 [PubMed] [Google Scholar]
- 25.Ristimaki, A., Narko, K., Enholm, B., Joukov, V., and Alitalo, K. (1998) J. Biol. Chem. 273 8413-8418 [DOI] [PubMed] [Google Scholar]
- 26.Sedivy, R., Kalipciyan, M., Mazal, P. R., Wolf, B., Wrba, F., Karner-Hanusch, J., Muhlbacher, F., and Mader, R. M. (2005) Cancer Detect. Prev. 29 8-14 [DOI] [PubMed] [Google Scholar]
- 27.Xing, L., Venegas, A. M., Chen, A., Garrett-Beal, L., Boyce, B. F., Varmus, H. E., and Schwartzberg, P. L. (2001) Genes Dev. 15 241-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing, L., Bushnell, T. P., Carlson, L., Tai, Z., Tondravi, M., Siebenlist, U., Young, F., and Boyce, B. F. (2002) J. Bone Miner. Res. 17 1200-1210 [DOI] [PubMed] [Google Scholar]
- 29.Kaneki, H., Guo, R., Chen, D., Yao, Z., Schwarz, E. M., Zhang, Y. E., Boyce, B. F., and Xing, L. (2006) J. Biol. Chem. 281 4326-4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng, J. Q., Xing, L., Zhang, J. H., Zhao, M., Horn, D., Chan, J., Boyce, B. F., Harris, S. E., Mundy, G. R., and Chen, D. (2003) J. Biol. Chem. 278 29130-29135 [DOI] [PubMed] [Google Scholar]
- 31.Yamashita, T., Yao, Z., Li, F., Zhang, Q., Badell, I. R., Schwarz, E. M., Takeshita, S., Wagner, E. F., Noda, M., Matsuo, K., Xing, L., and Boyce, B. F. (2007) J. Biol. Chem. 282 18245-18253 [DOI] [PubMed] [Google Scholar]
- 32.Chilov, D., Kukk, E., Taira, S., Jeltsch, M., Kaukonen, J., Palotie, A., Joukov, V., and Alitalo, K. (1997) J. Biol. Chem. 272 25176-25183 [DOI] [PubMed] [Google Scholar]
- 33.Zhang, Q., Badell, I. R., Schwarz, E. M., Boulukos, K. E., Yao, Z., Boyce, B. F., and Xing, L. (2005) Arthritis Rheum. 52 2708-2718 [DOI] [PubMed] [Google Scholar]
- 34.Takeshita, S., Kaji, K., and Kudo, A. (2000) J. Bone Miner. Res. 15 1477-1488 [DOI] [PubMed] [Google Scholar]
- 35.Lee, S. K., Gardner, A. E., Kalinowski, J. F., Jastrzebski, S. L., and Lorenzo, J. A. (2006) Bone 38 678-685 [DOI] [PubMed] [Google Scholar]
- 36.Tobe, M., Isobe, Y., Tomizawa, H., Nagasaki, T., Takahashi, H., and Hayashi, H. (2003) Bioorg. Med. Chem. 11 3869-3878 [DOI] [PubMed] [Google Scholar]
- 37.Hamrah, P., Chen, L., Zhang, Q., and Dana, M. R. (2003) Am. J. Pathol. 163 57-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ali, N., Yoshizumi, M., Fujita, Y., Izawa, Y., Kanematsu, Y., Ishizawa, K., Tsuchiya, K., Yano, S., Sone, S., and Tamaki, T. (2005) J. Pharmacol. Sci. 98 130-141 [DOI] [PubMed] [Google Scholar]
- 39.Byrne, A. M., Bouchier-Hayes, D. J., and Harmey, J. H. (2005) J. Cell Mol. Med. 9 777-794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyce, B. F., Yoneda, T., Lowe, C., Soriano, P., and Mundy, G. R. (1992) J. Clin. Invest. 90 1622-1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartzberg, P. L., Xing, L., Hoffmann, O., Lowell, C. A., Garrett, L., Boyce, B. F., and Varmus, H. E. (1997) Genes Dev. 11 2835-2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyce, B. F., Xing, L., Yao, Z., Yamashita, T., Shakespeare, W. C., Wang, Y., Metcalf, C. A., 3rd, Sundaramoorthi, R., Dalgarno, D. C., Iuliucci, J. D., and Sawyer, T. K. (2006) Clin Cancer Res. 12 6291s-6295s [DOI] [PubMed] [Google Scholar]
- 43.Shakespeare, W., Wang, Y., Bohacek, R., Keenan, T., Sunaramoorthi, R., Metcalf III, C., Dilauro, A., Roeloffzen, S., Lui, S., Saltmarsh, J., Paramanathan, G., Dalgarno, D., Narula, S., Pradeepan, S., Schravendilk, M., Keats, J., Ram, M., Liou, S., Adams, S., Wardwell, S., Bogus, J., Iuliucci, J. D., Manfred, W., Xing, L., Boyce, B., and Sawyer, T. (2008) Chem. Biol. Drug Des. 71 97-105 [DOI] [PubMed] [Google Scholar]
- 44.Pettit, A. R., Walsh, N. C., Manning, C., Goldring, S. R., and Gravallese, E. M. (2006) Rheumatology (Oxford) 45 1068-1076 [DOI] [PubMed] [Google Scholar]
- 45.Wei, S., Kitaura, H., Zhou, P., Ross, F. P., and Teitelbaum, S. L. (2005) J. Clin. Invest. 115 282-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao, Z., Xing, L., Qin, C., Schwarz, E. M., and Boyce, B. F. (February 4, 2008) J. Biol. Chem. 10.1074/jbc.M706415200 [DOI] [PMC free article] [PubMed]
- 47.Takayanagi, H., Kim, S., Matsuo, K., Suzuki, H., Suzuki, T., Sato, K., Yokochi, T., Oda, H., Nakamura, K., Ida, N., Wagner, E. F., and Taniguchi, T. (2002) Nature 416 744-749 [DOI] [PubMed] [Google Scholar]
- 48.Karkkainen, M. J., Haiko, P., Sainio, K., Partanen, J., Taipale, J., Petrova, T. V., Jeltsch, M., Jackson, D. G., Talikka, M., Rauvala, H., Betsholtz, C., and Alitalo, K. (2004) Nat. Immunol. 5 74-80 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.