Abstract
Cell-cell contacts play a vital role in intracellular signaling, although the molecular mechanisms of these signaling pathways are not fully understood. E-cadherin, an important mediator of cell-cell adhesions, has been shown to be cleaved by γ-secretase. This cleavage releases a fragment of E-cadherin, E-cadherin C-terminal fragment 2 (E-cad/CTF2), into the cytosol. Here, we study the fate and function of this fragment. First, we show that coexpression of the cadherin-binding protein, p120 catenin (p120), enhances the nuclear translocation of E-cad/CTF2. By knocking down p120 with short interfering RNA, we also demonstrate that p120 is necessary for the nuclear localization of E-cad/CTF2. Furthermore, p120 enhances and is required for the specific binding of E-cad/CTF2 to DNA. Finally, we show that E-cad/CTF2 can regulate the p120-Kaiso-mediated signaling pathway in the nucleus. These data indicate a novel role for cleaved E-cadherin in the nucleus.
In multicellular organisms, individual cells are often connected with each other via cell-cell adhesions to form three-dimensionally structured tissues or organs. Formation of tight and compact cell-cell adhesions suppresses free cell movement and provides cells with a positional cue for the establishment of cell polarity. In addition to the structural roles, it has been long known that cell-cell contacts play an important role in various signal transduction pathways (1, 2). In other words, through cell-cell contacts, cells can exchange a variety of information with their neighbors to behave properly in their community.
Cadherins are a family of transmembrane cell-cell adhesion proteins that can be subdivided into several groups including classical cadherins and protocadherins (3). Classical cadherins, of which E-cadherin is the best characterized, play a crucial role in mediating cell-cell contacts at adherens junctions. The extracellular domains of classical cadherins form homophilic ligations. The cytoplasmic domain of cadherin includes two cadherin homology (CH)2 domains: CH2 domain (located at the membrane proximal region) and CH3 domain (located at the distal region). These domains are conserved between classical cadherins. The CH2 and CH3 domains of cadherins interact with p120-catenin (p120) and β-catenin, respectively (4, 5). Furthermore, the CH2 domain of E-cadherin also interacts with Hakai (6, 7).
Cadherins, especially classical cadherins, have been shown to be involved in several signaling pathways regulating cell proliferation, differentiation, and survival (8–11). However, the molecular mechanisms whereby cadherins regulate varied cellular processes are not fully understood. The cytoplasmic domain of cadherins does not possess any intrinsic enzymatic activity that could directly mediate signaling pathways in the cytosol. However, cadherin-binding proteins, especially β-catenin and p120, have been shown to localize in the nucleus and play a key role in signal transduction. The crucial role of β-catenin in the canonical Wnt signaling pathway has been well characterized (12, 13). When cells are stimulated with Wnt, β-catenin is translocated into the nucleus where it binds a transcription factor, T-cell factor/lymphocyte enhancer factor-1 (TCF/LEF-1), and acts as a transcriptional activator (14, 15). More recently, it has been shown that p120 also has a functional role in the nucleus (16–18). A transcriptional repressor Kaiso has been characterized as a p120-binding protein (17). The interaction with p120 prevents Kaiso from binding to DNA and thus suppresses the repressor activity of Kaiso (16, 19). The Kaiso-p120 complex was recently shown to regulate both canonical and non-canonical Wnt signaling pathways (20, 21). Although the functional roles of cadherin-binding proteins in the nucleus have been intensively studied, it is not clear whether the cytoplasmic domain of cadherin itself has a signaling role in the nucleus.
Cadherin-based cell-cell contacts are dynamically regulated. Cadherin at cell-cell contacts can be down-regulated by transcriptional repression and endocytosis (6, 22, 23). Moreover, cadherins are proteolytically cleaved by a number of proteases including extracellular metalloproteases, γ-secretase, and caspase (24–27). γ-Secretase cleavage of several transmembrane molecules following initial cleavage by metalloproteases has emerged as a common mechanism for signaling to the nucleus (28). This is best characterized in Notch signaling where the γ-secretase-cleaved intracellular Notch fragment translocates to the nucleus and regulates gene transcription (29, 30). In recent years, a similar mechanism has also been described for amyloid precursor protein, ErbB4, CD44 as well as N-cadherin and γ-protocadherins (31–38). For all these transmembrane molecules, intracellular domain fragments resulting from γ-secretase cleavage are translocated to the nucleus and have signaling functions. In particular, γ-secretase-mediated cleavage of N-cadherin in neural crest cells, induced by bone morphogenic protein 4, leads to nuclear translocation of the released cytoplasmic domain of N-cadherin (38). The cytoplasmic domain of γ-protocadherins also localizes in the nucleus and activates the promoter of γ-protocadherin locus (37). However, no signaling function for the cleaved cytoplasmic domain of E-cadherin has been described.
In this study, we show that the intracellular domain of E-cadherin released by γ-secretase cleavage can localize in the nucleus and that this localization is specifically enhanced by p120. p120 also promotes the interaction of E-cadherin with DNA. We demonstrate that the cytoplasmic domain of E-cadherin in the nucleus modulates the p120-Kaiso-mediated signaling pathway. Finally, we present data suggesting a possible role of nuclear E-cadherin in the regulation of apoptosis.
EXPERIMENTAL PROCEDURES
Antibodies, Plasmids, and Materials—Antibodies against E-cadherin, p120, N-cadherin, GM130, BiP/GRP78, and Lamp-1 were purchased from BD Biosciences. Anti-HA antibody was obtained from Roche. Anti-FLAG antibody and the peroxidase-conjugated anti-FLAG antibody were purchased from Sigma. Anti-myc antibody was from Upstate (Temecula, CA). Anti-CREB-binding protein (CBP) and anti-α-tubulin antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and Abcam (Cambridge, UK), respectively. Anti-cleaved caspase-3 antibody was from Cell Signaling Technology (Danvers, MA). All antibodies were used at a dilution of 1:1000 for Western blotting and 1:100 for immunofluorescence, except for anti-E-cadherin antibody, which was used at 1:3000 for Western blotting, and anti-GM130, anti-BiP/GRP78, and anti-Lamp-1 antibodies that were used at 1:250 for Western blotting.
To construct pcDNA-E-cad/CTF2 and -E-cad/CTF3, the cDNAs of the cytoplasmic domain of E-cadherin were amplified by PCR from pBAT-mouse-E-cadherin using the following primers: pcDNA-E-cad/CTF2, 5′-GGAATTCGCGGCCGCGGTACCATGCGGAGGAGAACGGTGGTC and 3′-GGAATTCGGATCCTAGTCGTCCTCGCCACC; pcDNAE-cad/CTF3, 5′-ATTCGCGGCCGCGGTACCATGAATGTGTATTACTATGATGAAGAAGGAGG and 3′-GGAATTCGGATCCTAGTCGTCCTCGCCACC. Both were cloned into the NotI/BamHI sites of the pcDNA vector. To construct pcDNA-E-cad/CTF2(AAA), the cDNA of the cytoplasmic domain of E-cadherin was amplified by PCR from pLK-E-cadherin(762AAA764) using the same primers as used to construct pcDNA-E-cad/CTF2. To construct pcDNA-E-cad/CTF2(RRRR), all lysine residues were replaced for arginine by site-directed mutagenesis using pcDNA-E-cad/CTF2 as a template. Site-directed mutagenesis was performed using the QuikChange Site-directed mutagenesis kit from Stratagene (La Jolla, CA). To construct pCMV-E-cad/CTF2-NLS-myc and pCMV-E-cad/CTF2(AAA)-NLS-myc, the cytoplasmic domain of E-cadherin was amplified by PCR from pBAT-E-cadherin and pLK-E-cadherin(762AAA764), respectively, using the following primers: 5′-GAATTCGCGGCCGCAATGCGGAGGAGAACGGTGGTC and 3′-GGAATTCGCGGCCGCGTCGTCCTCGCCACCGCCG. These were cloned into a NotI site of the pCMV-myc-nuc pShooter vector (Invitrogen). To construct pcDNA/TO/GFP-E-cad/CTF2-NLS, the cDNA of E-cad/CTF2-NLS was amplified by PCR from pCMV-E-cad/CTF2-NLS-myc using the following primers: 5′-CGGCGCCTCGAGGCGGAGGAGAACGGTGGTC and 3′-CGGCGCCGAATTCCTATGCGGCCCCATTCAGATCC. This was then cloned into the EcoRI and XhoI sites of pcDNA/TO/GFP. To construct pCS2-myc-Kaiso, the cDNA of human Kaiso was amplified by PCR using pCS2-hKaiso as a template, and inserted into the EcoRI site of pCS2-myc. pcDNA-HA-Ecad/CTF2, pcDNA-FLAG-p120, pcDNA-HA-p120, pBSSR-HA-ubiquitin, pcDNA-myc-β-catenin, pcDNA-FLAG-Hakai, pLK-E-cadherin(762AAA764), pcDNA/TO/GFP, pGL3–4x KBS, and pGL3-Basic-2.3kb HMat were described previously (6, 16, 18, 39–41). pME18S-FLAG-ADAM10 was kindly provided by Dr. Eiichiro Nishi (Kyoto University, Kyoto, Japan). pcH110-LacZ was a generous gift from Dr. Walter Birchmeier (Max-Delbrück-Center, Berlin, Germany).
Staurosporine was obtained from Upstate. γ-Secretase inhibitor X was purchased from Calbiochem. Lipofectamine 2000 reagent was obtained from Invitrogen. Hi-PerFect® reagent was purchased from Qiagen (Crawley, UK).
Cell Culture, Immunofluorescence, Immunoprecipitation, DNA Binding Assay, and GST Pull-down Assay—Human embryonic kidney (HEK) 293, Madin-Darby canine kidney (MDCK), MCF-7, COS-1, and L fibroblast cells were cultured as previously described (18, 42). A431 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and penicillin/streptomycin at 37 °C and ambient air supplemented with 5% CO2. MDCK, MCF-7, COS-1, and L fibroblast cells were transfected with Lipofectamine 2000 reagent. HEK293 cells were transfected as previously described (43). MDCK cells stably expressing E-cadherin-myc were produced and cultured as previously described (42). To obtain L fibroblast cell lines stably expressing NLS-tagged E-cad/CTF2, cells were transfected with pcDNA4/TO/GFP-E-cad/CTF2-NLS, and stably transfected cells were selected in medium containing 400 μg ml–1 zeocin (Invitrogen). Immunofluorescence was performed as previously described (40). Images were captured using an Orca camera (Hamamatsu) and Openlab software (Improvision). To obtain epifluorescence images, we used a Zeiss Axipoplan2 microscope using a 40 × 1.3 oil immersion objective at room temperature (Zeiss). Images were captured using a C4742-95 digital camera (Hamamatsu) and Openlab software (Improvision). Confocal images were obtained with a Bio-Rad confocal mounted on a Nikon Optiphot 2 microscope using a 60 × 1.4 oil immersion objective at room temperature, or a Leica SPE confocal microscope with a 63 × 1.3 oil immersion objective at room temperature. Images were acquired using Laser Sharp software (Bio-Rad) or the Leica Application Suite, respectively. Merged images were split using Image J software (National Institutes of Health). Brightness and contrast were adjusted using Photoshop CS (Adobe). The ratio of nucleus/cytoplasm fluorescence intensity in confocal images were analyzed as described (18). Secondary antibodies used were goat anti-mouse Rhodamine Red-X, goat anti-rabbit Cy2, and goat anti-rat Cy2 (Jackson ImmunoResearch Laboratories). To visualize nuclei we used Hoechst 33342 (Invitrogen).
Immunoprecipitation and Western blotting were performed as described (40). To obtain total cell lysates, cells were washed once with phosphate-buffered saline and suspended in Triton X-100 lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 1% Triton X-100) containing 5 μg ml–1 leupeptin, 50 mm phenylmethylsulfonyl fluoride, 7.2 trypsin inhibitor units ml–1 of aprotinin, and 10 mm N-ethylmaleimide. The suspended cells were directly boiled with SDS-PAGE sample buffer. To ensure equal loading, the protein concentration of lysates was quantified using the DC Protein Assay reagent from Bio-Rad (Hercules, CA) and measured on a VERSAmax microplate reader (Molecular Devices, Sunnyvale, CA). Densitometric analyses were performed with the Bio-Rad image acquisition system (Gel Doc 170) and Epson Expression 1680 Pro Scanner (Seiko Epson, Amsterdam, Netherlands), and Quantity One (Bio-Rad). The upsmear bands of E-cad/CTF2 shown in Fig. 4 were not observed in some experiments (e.g. supplemental Fig. S2B). It is highly likely that despite the addition of N-ethylmaleimide to inhibit de-ubiquitination, the de-modification of E-cad/CTF2 occurs very actively after lysing the cells, as the modified bands were frequently lost when the lysates were kept for a longer period.
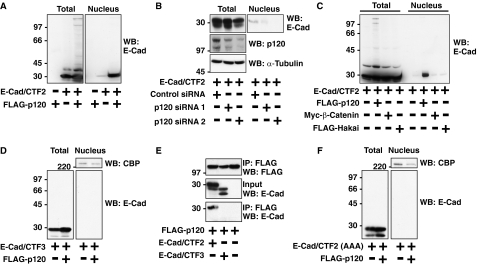
FIGURE 4.
p120 is required for the nuclear localization of E-cad/CTF2. A–D and F, HEK293 cells were transfected as indicated, and total and nuclear lysates were examined by Western blotting (WB) with the indicated antibodies. A, p120 enhances nuclear localization of E-cad/CTF2. B, knockdown of p120 decreases the nuclear localization of E-cad/CTF2. HEK293 cells were transfected with control siRNA, p120 siRNA oligo 1, or p120 siRNA oligo 2 together with E-cad/CTF2. C, E-Cad/CTF2 nuclear localization is increased by p120, but not by other E-cadherin-binding proteins. D, p120 does not induce nuclear localization of E-cad/CTF3. Nuclear fractions were also examined by Western blotting with anti-CREB-binding protein (CBP) antibody. E, E-Cad/CTF3 does not bind to p120. FLAG-p120 was transfected with E-cad/CTF2 or E-cad/CTF3 in HEK293 cells. Immunoprecipitation was performed using anti-FLAG antibody, followed by Western blotting with anti-E-cadherin and anti-FLAG antibodies. F, p120 binding is required for nuclear translocation of E-cad/CTF2. Nuclear fractions were also examined by Western blotting with anti-CBP antibody.
For DNA-cellulose binding assays, cell lysates (100 μg of protein) and/or purified GST or GST-E-cad/CTF2 protein (75 ng) were used in DNA-cellulose assays and competition assays as previously described (18). Purified GST-E-cad/CTF2 and GST proteins were produced, and GST pull-down assays were performed as previously described (40).
Nuclear Fractionation—Cells were washed twice with phosphate-buffered saline and trypsinized thoroughly until well separated. Pelleted cells were resuspended in 150 μl of 2× lysis buffer (50 mm Hepes/NaOH, pH 7.4, 10 mm EGTA, 5 mm MgCl2, 20% glycerol, and 2% Nonidet P-40) containing 5 μg ml–1 leupeptin, 50 mm phenylmethylsulfonyl fluoride, 7.2 trypsin inhibitor units ml–1 of aprotinin, and 10 mm N-ethylmaleimide. Cells were then immediately triturated with a 25-gauge needle 12 times. After centrifugation at 110 × g for 5 min at 4 °C, the supernatant was removed and the pelleted nuclei were washed twice in 1× lysis buffer. The nuclear fractions were then boiled for 10 min with SDS-PAGE sample buffer followed by Western blotting, or lysed in 1 ml of Triton X-100 lysis buffer followed by immunoprecipitation. Purity of nuclear fractions was confirmed by Western blotting with antibodies against several compartment-specific markers (supplemental Fig. S3A).
RNA Interference—siRNA oligos for p120 (p120 siRNA1, CAGCAGAACUCCUCUUGGATT; p120 siRNA2, CAGCAGUCAUUCAUAUGAUTT) were transiently transfected into HEK293 cells using Hi-PerFect® reagent. 5 × 104 cells per well were plated in a 24-well dish. 1.5 μg of siRNA was used with 9 μl of Hi-PerFect reagent per well. As a negative control, we used the non-silencing control siRNA (AF 488) from Qiagen. 48 h after siRNA transfection, cells were further transfected with E-cad/CTF2. After a further 24 h, cells were lysed in Triton X-100 lysis buffer or used for nuclear fractionation.
Luciferase Reporter Assays—HEK293 and COS-1 cells were transfected with pGL3–4x KBS together with FLAG-p120, E-cad/CTF2, E-cad/CTF2-NLS-myc, or E-cad/CTF2(AAA)-NLS-myc. For reporter assays using the matrilysin promoter, HEK293 cells were transfected with pGL3-Basic-2.3kb HMat together with FLAG-p120, E-cad/CTF2-NLS-myc, or E-cad/CTF2(AAA)-NLS-myc. LacZ was used as a control for transfection efficiency in all experiments. Luciferase activity was measured using the luciferase assay kit according to the manufacturer's instructions (Promega, Madison, WI) and a Turner Designs TD-20/20 luminometer. β-Galactosidase activity was measured using the β-galactosidase assay kit according to the manufacturer's instructions (Promega) and the VERSAmax microplate reader (Molecular Devices).
Statistical Analysis—Student's t tests assuming equal or unequal variance and a two-tailed distribution were performed for statistical analysis.
RESULTS
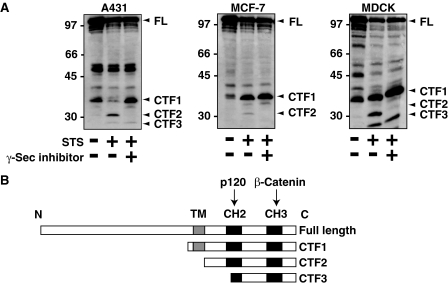
Treatment with Staurosporine Induces γ-Secretase-mediated Cleavage of E-cadherin in Epithelial Cells—First, we examined the proteolytic cleavage of E-cadherin in several epithelial cell lines. As previously reported (25), treatment of the human epithelial cell line A431 with staurosporine lead to the production of cytoplasmic cleavage products, E-cadherin C-terminal fragment 2 (E-cad/CTF2) (33 kDa) and E-cadherin C-terminal fragment 3 (E-cad/CTF3) (29 kDa) (Fig. 1A). Staurosporine is a potent inhibitor of protein kinase C and other protein kinases, which strongly induces apoptosis. E-Cad/CTF2 was also observed in the human breast adenocarcinoma cell line MCF-7 and MDCK cells after treatment with staurosporine (Fig. 1A). A 38-kDa E-cadherin fragment known as E-cadherin C-terminal fragment 1 (E-cad/CTF1) was also present in all three cell lines (Fig. 1A). The identity of a number of other bands between 45 and 100 kDa, particularly in MDCK cells, is not known. The size and domain structure of E-cad/CTF1, E-cad/CTF2, and E-cad/CTF3 are shown in Fig. 1B. E-Cad/CTF1 is the cleavage product induced by extracellular metalloproteases that cleave close to the interface between the extracellular and transmembrane regions of E-cadherin (24–26). E-Cad/CTF3 results from cleavage by caspase-3 after induction of apoptosis (25, 27). E-Cad/CTF2 is the result of γ-secretase cleavage, which occurs following removal of the extracellular region by metalloproteases (25). The resulting E-cad/CTF2 fragment represents the entire intracellular domain of E-cadherin. As expected, we observed that treatment with γ-secretase inhibitor X specifically blocked staurosporine-induced production of E-cad/CTF2 in all three cell lines and lead to an accumulation of its precursor E-cad/CTF1 (Fig. 1A).
FIGURE 1.
γ-Secretase-mediated cleavage of E-cadherin in epithelial cells. A, staurosporine induces γ-secretase-mediated cleavage of E-cadherin in epithelial cell lines. A431, MCF-7, and MDCK cells were treated with staurosporine (STS)(1 μm) in the presence or absence of γ-secretase (γ-sec) inhibitor X (125 nm) for 6 h. Cell lysates were examined by Western blotting with anti-E-cadherin antibody. The positions of full-length E-cadherin (FL) and the C-terminal fragments E-cad/CTF1, E-cad/CTF2, and E-cad/CTF3 are indicated with arrowheads. B, schematics for the domain structure of full-length E-cadherin, E-cad/CTF1, E-cad/CTF2, and E-cad/CTF3. The intracellular region of E-cadherin contains two CH (cadherin homology) domains, CH2 and CH3, which bind to p120 and β-catenin, respectively. TM, transmembrane region.
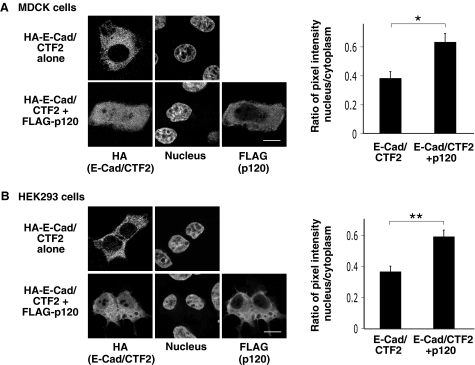
p120 Enhances Nuclear Localization of E-cad/CTF2—To investigate a possible role for E-cad/CTF2 in signaling pathways, we studied whether it can localize in the nucleus, as has been observed for the γ-secretase cleavage products of N-cadherin and γ-protocadherins (35–38). We first expressed HA-tagged E-cad/CTF2 in epithelial cells and examined its subcellular localization. When HA-E-cad/CTF2 alone is expressed in MDCK cells, it was mainly localized in the cytoplasm (Fig. 2A). Interestingly, we found that coexpression of p120 enhanced the nuclear localization of E-cad/CTF2 (Fig. 2A). To quantify this effect of p120, we measured the ratio of pixel intensity within defined regions of the nucleus and cytoplasm. The quantification results showed that coexpression of p120 significantly increased the amount of E-cad/CTF2 in the nucleus (p < 0.002) (Fig. 2A, right). A similar effect of p120 on the nuclear localization of E-cad/CTF2 was observed in HEK293 cells (Fig. 2B) and MCF-7 cells (supplemental Fig. S1).
FIGURE 2.
p120 enhances the nuclear localization of E-cad/CTF2. MDCK (A) and HEK293 cells (B) were transfected with HA-E-cad/CTF2 and FLAG-p120. Immunostaining was performed with anti-HA and anti-FLAG antibodies. Nuclei were stained with Hoechst dye. Images were acquired using confocal microscopy. Scale bars represent 10 μm. Immunofluorescence intensity with anti-HA antibody was measured within a defined region (diameter 1.97 μm) in the nucleus and cytoplasm. The ratio of nucleus/cytoplasm staining intensity was analyzed in at least 20 cells per transfection. Error bars represent S.E. *, p < 0.002; **, p < 0.00007.
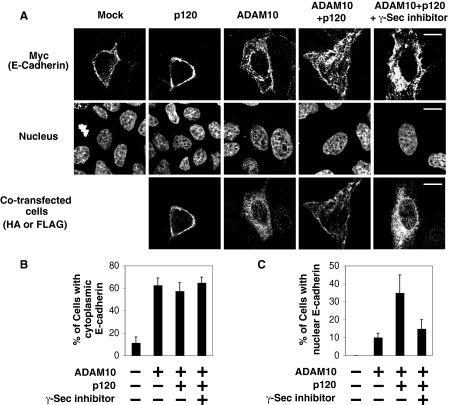
Next we examined whether the cytoplasmic domain is processed from full-length E-cadherin and is translocated to the nucleus. Because the available antibodies against the intracellular region of E-cadherin are not suitable for immunostaining, we used MDCK cells expressing exogenous full-length E-cadherin with a C-terminal myc tag. When E-cadherin-myc alone was expressed, myc immunostaining was mainly observed at cell-cell contacts, which was not affected by coexpression of p120 (Fig. 3A, first and second panels). We next investigated whether the cleaved C terminus of E-cadherin localizes in the nucleus in this system. As staurosporine treatment induced nuclear fragmentation, we transiently expressed a metalloprotease ADAM (a disintegrin and metalloprotease) 10, which cleaves E-cadherin producing E-cad/CTF1, which can be further cleaved to produce E-cad/CTF2 (supplemental Fig. S2) (26). Upon expression of ADAM10, myc staining was decreased at cell-cell contacts, becoming predominantly localized in the cytoplasm, and cells became more spread and flattened (Fig. 3A, third panel). The effect of ADAM10 expression on E-cadherin localization was quantified by counting the percentage of cells showing myc staining in the cytoplasm. Expression of ADAM10 lead to cytoplasmic staining in ∼60% of cells, compared with 10% in mock-transfected cells (Fig. 3B). Neither coexpression of p120 nor treatment with γ-secretase inhibitor X significantly influenced this effect of ADAM10 (Fig. 3B), suggesting that ADAM10 disrupted E-cadherin-based cell-cell contacts independently of p120 and γ-secretase. Furthermore, we analyzed the nuclear myc staining in the cells that lost cell-cell contact staining upon expression of ADAM10. Notably, we found that coexpression of p120 with ADAM10 substantially increased nuclear myc staining (Fig. 3, A, fourth panel, and C). The nuclear myc staining induced by coexpression of ADAM10 and p120 was blocked by treatment with γ-secretase inhibitor X (Fig. 3, A, fifth panel, and C), suggesting that the nuclear localization of the C terminus of E-cadherin is dependent on γ-secretase-mediated processing of E-cad/CTF1. Collectively, data in Figs. 2 and 3 indicate that the γ-secretase cleavage product of E-cadherin, E-cad/CTF2, can localize in the nucleus of epithelial cells and that this localization is enhanced by p120.
FIGURE 3.
Nuclear localization of the cleaved C terminus of E-cadherin. A, p120 induces nuclear localization of the C terminus of E-cadherin in ADAM10-transfected cells. MDCK cells stably expressing C-terminal myc-tagged E-cadherin were transiently transfected with FLAG-ADAM10 and/or HA-p120. γ-Secretase inhibitor X (125 nm) was added for 6 h before fixation where indicated. Immunostaining was performed with anti-myc, anti-HA, or anti-FLAG antibodies. Nuclei were stained with Hoechst dye. Images were acquired using confocal microscopy. Scale bars represent 15 μm. B, quantification of ADAM10-induced cytoplasmic localization of E-cadherin. The percentage of cells showing cytoplasmic myc staining was calculated from 20 transfected cells from two independent transfections. Error bars indicate S.E. C, quantification of nuclear localization of E-cadherin. The percentage of cells showing nuclear myc staining was calculated from 20 transfected cells from two independent transfections. Error bars indicate S.E.
p120 Is Required for Nuclear Localization of E-cad/CTF2 in HEK293 Cells—We used HEK293 cells to further analyze the nuclear localization of E-cad/CTF2, because highly purified nuclei can be obtained by biochemical nuclear fractionation using these cells (supplemental Fig. S3A). First, we confirmed that coexpression of p120 increased the amount of E-cad/CTF2 in the nuclear fraction (Fig. 4A), without affecting the amount of nuclei in this fraction (supplemental Fig. S3B). Interestingly, we found that p120 expression frequently induced higher molecular weight modifications of E-cad/CTF2 that were also sometimes observed in the nuclear fraction (Fig. 4A, see “Experimental Procedures” for details). We next transfected siRNA oligos against p120 to examine the involvement of endogenous p120 proteins in the nuclear localization of E-cad/CTF2. Expression of siRNA oligos efficiently knocked down the expression of endogenous p120 and strongly reduced the amount of E-cad/CTF2 present in the nuclear fraction (Fig. 4B). This reduction of E-cad/CTF2 in the nuclear fraction was observed to a greater extent with siRNA oligo 2, which induced greater knockdown of p120 (Fig. 4B). These data provide further support for an important role of p120 in the nuclear localization of E-cad/CTF2.
Next we investigated the specificity of the effect of p120 on nuclear localization and modification of E-cad/CTF2. Expression of other E-cadherin-binding proteins, β-catenin and Hakai, did not enhance either modification or nuclear localization of E-cad/CTF2 (Fig. 4C). We also analyzed the effect of p120 on another E-cadherin fragment, E-cad/CTF3, that is released following cleavage by caspase-3. When E-cad/CTF3 alone was expressed, it was not detected in the nuclear fraction (Fig. 4D). Coexpression of p120 enhanced neither nuclear localization nor modification of E-cad/CTF3 (Fig. 4D). As caspase-3 cleavage occurs within the CH2 domain that includes the p120-binding site, this cleavage may impair binding of E-cad/CTF3 to p120. To examine this possibility, we analyzed the interaction between p120 and E-cad/CTF2 or E-cad/CTF3 by coimmunoprecipitation. Indeed we found that E-cad/CTF3 was not coimmunoprecipitated with p120, whereas E-cad/CTF2 was coimmunoprecipitated (Fig. 4E). To further investigate the requirement of p120 binding for its enhancement of E-cad/CTF2 nuclear localization, we used a mutant form of E-cad/CTF2 that cannot bind to p120 (E-cad/CTF2(AAA)). This is a triple alanine mutation within the CH2 domain that prevents binding of E-cadherin to p120 (41). We found that E-cad/CTF2(AAA) was not present in the nuclear fraction and that expression of p120 did not induce nuclear localization or modification of E-cad/CTF2(AAA) (Fig. 4F). Collectively, these results suggest that the direct interaction with E-cad/CTF2 is required for p120 to enhance nuclear localization and modification of E-cad/CTF2.
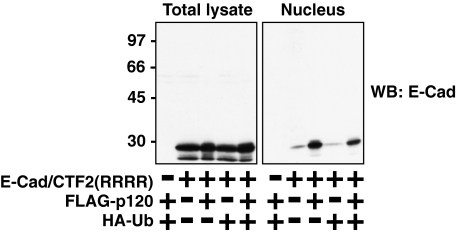
As p120 induces higher molecular weight modifications and nuclear translocation of E-cad/CTF2, we investigated whether these modifications of E-cad/CTF2 play a role in its nuclear translocation. Such upsmear bands are often observed in SDS-PAGE following ubiquitination of proteins. Ubiquitination has a variety of roles such as protein degradation in the proteasome and intracellular protein transport including nuclear import (45, 46). To investigate a possible role of ubiquitination in the nuclear localization of E-cad/CTF2, we mutated all four lysine residues within E-cad/CTF2 to arginine, producing E-cad/CTF2(RRRR), which cannot be ubiquitinated. No higher molecular weight bands of E-cad/CTF2(RRRR) were observed when it was coexpressed with p120 and/or ubiquitin (Fig. 5, left panel), indicating that the modifications indeed occur on lysine residues. Coexpression of ubiquitin did not induce enhanced nuclear localization of E-cad/CTF2(RRRR), however, p120 was still able to enhance its nuclear localization (Fig. 5, right panel). These data suggest that this lysine-based modification is not required for p120-induced nuclear translocation of E-cad/CTF2.
FIGURE 5.
p120 enhances nuclear localization of E-cad/CTF2(RRRR) in which all lysine residues were mutated to arginine. HEK293 cells were transfected as indicated, and total and nuclear lysates were examined by Western blotting (WB) with the anti-E-cadherin antibody.
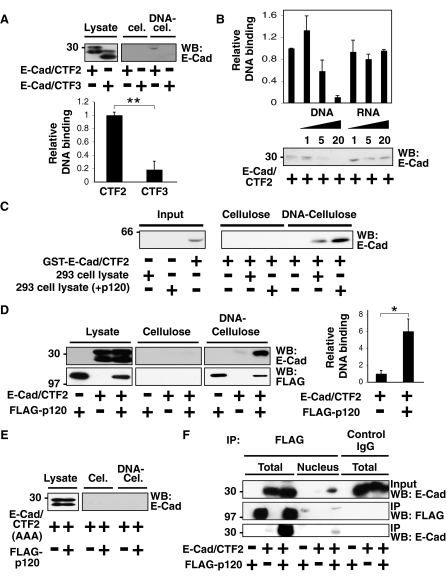
E-Cad/CTF2 Forms a Complex with DNA via p120—To investigate the role of E-cad/CTF2 in the nucleus, we examined its ability to associate with DNA in an in vitro DNA binding assay. This assay uses native calf thymus DNA immobilized on cellulose to precipitate proteins from cell lysates that can bind DNA (18). It is particularly useful when the potential DNA binding sites are not identified. Interestingly, when expressed in HEK293 cells, E-cad/CTF2 in the cell lysate could bind to DNA, whereas no or very little binding of E-cad/CTF3 was detected (Fig. 6A, top). The intensities of Western blot bands were quantified using densitometry, and the binding of E-cad/CTF2 to DNA was significantly higher than that of E-cad/CTF3 (p < 0.004) (Fig. 6A, bottom). We confirmed the specificity of the interaction between E-cad/CTF2 and DNA by performing a competition assay. In this assay, cell lysates were preincubated with increasing amounts of DNA or RNA. Preincubation with DNA suppressed the binding of E-cad/CTF2 to the DNA-cellulose, whereas preincubation with RNA did not (Fig. 6B). This indicates a specific interaction between E-cad/CTF2 and DNA, rather than a nonspecific ionic interaction. Because there are no known DNA binding sites within the E-cadherin cytoplasmic domain, we investigated whether DNA binding was direct by using purified GST-tagged E-cad/CTF2 protein. We found that GST-E-cad/CTF2 alone did not bind to DNA and that it required the HEK293 cell lysate to interact with DNA (Fig. 6C). Lysates from cells overexpressing exogenous p120 further increased the amount of GST-E-cad/CTF2 binding to DNA, suggesting that p120 promotes the interaction between E-cad/CTF2 and DNA (Fig. 6C).
FIGURE 6.
E-Cad/CTF2 forms a complex with DNA in vitro via interaction with p120. A, B and D, E, HEK293 cells were transfected as indicated, and cell lysates were incubated with cellulose or DNA-cellulose beads. The amount of proteins bound to the beads was examined by Western blotting (WB) with the indicated antibodies. A, E-Cad/CTF2 binds to DNA in an in vitro binding assay. B, DNA, but not RNA, inhibits the interaction of E-cad/CTF2 with DNA-cellulose beads. Lysates of HEK293 cells expressing E-cad/CTF2 were preincubated with 1, 5, or 20 μg of DNA or RNA, followed by incubation with DNA-cellulose beads. C, E-Cad/CTF2 indirectly binds to DNA. GST-E-cad/CTF2 was incubated with cellulose or DNA-cellulose beads in the presence or absence of cell lysates of HEK293 cells transfected either with or without p120. For input, 1/50 of the amount of GST-E-cad/CTF2 added to the beads was examined. D, p120 enhances the interaction of E-cad/CTF2 with DNA. Images shown for cellulose and DNA-cellulose are from the same film. E, E-Cad/CTF2(AAA) does not bind DNA. F, E-Cad/CTF2 and p120 interact in the nucleus. Lysates or nuclear fractions of HEK293 cells expressing E-cad/CTF2 and FLAG-p120 were immunoprecipitated with anti-FLAG antibody or control IgG, followed by Western blotting with anti-E-cadherin and anti-FLAG antibodies. A, B, and D, the intensities of Western blot bands were quantified with densitometry from three independent experiments. Error bars indicate S.E. *, p < 0.03; **, p < 0.004.
To further examine the effect of p120 on the DNA binding of E-cad/CTF2, we transfected E-cad/CTF2 with or without exogenous p120 in HEK293 cells. Coexpression of p120 significantly increased the association of E-cad/CTF2 with DNA (p < 0.03) (Fig. 6D). p120 also bound to DNA in this assay (Fig. 6D), suggesting that the association of E-cad/CTF2 with DNA could occur via p120. Indeed E-cad/CTF2(AAA), which is unable to bind to p120, was not detected on the DNA-cellulose, even in the presence of exogenous p120 (Fig. 6E). We then investigated whether E-cad/CTF2 and p120 interact in the nucleus by performing a coimmunoprecipitation using nuclear fractions from HEK293 cells transiently transfected with E-cad/CTF2 and p120 (Fig. 6F). Small amounts of exogenously expressed p120 were detected in the nucleus, and E-cad/CTF2 was coimmunoprecipitated with p120 from the nuclear fraction (Fig. 6F). Collectively, these data indicate that E-cad/CTF2 in the nucleus can specifically associate with DNA via p120.
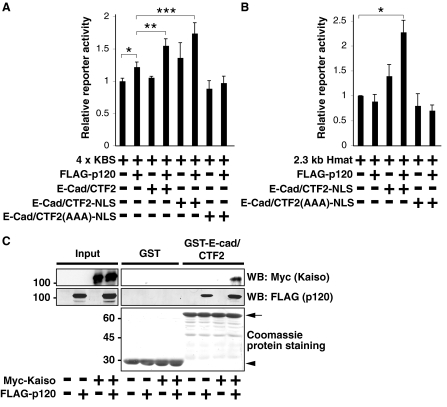
E-Cad/CTF2 in the Nucleus Modulates Gene Transcription from Kaiso-regulated Promoters—To further examine the role of E-cad/CTF2 in the nucleus, we examined its effect on a signaling pathway mediated by p120, which interacts with the transcription factor Kaiso. To analyze the effect of nuclear E-cad/CTF2, we constructed a nuclear-targeted form of E-cad/CTF2, E-cad/CTF2-NLS-myc. In this construct, E-cad/CTF2 was fused to four nuclear localization signals (NLS), and it was shown to predominantly localize in the nucleus (supplemental Fig. S4A). To examine the effect of nuclear E-cad/CTF2 on Kaiso-regulated promoters, we used the pGL3–4x KBS reporter plasmid, which contains four tandem copies of the consensus Kaiso binding site upstream of the luciferase reporter gene. With this construct, the effects of p120, E-cad/CTF2, and E-cad/CTF2-NLS on reporter activity were examined in HEK293 cells. As previously shown (16), we observed a significant increase in reporter activity in the presence of exogenous p120, indicating that p120 relieved the repression mediated by endogenous Kaiso protein (p < 0.038) (Fig. 7A). Interestingly, we found that this p120-mediated increase in reporter activity was significantly enhanced by coexpression of E-cad/CTF2 (p < 0.037) (Fig. 7A). Enhancement of reporter activity was also observed with E-cad/CTF2-NLS (p < 0.025), suggesting that this effect of E-cad/CTF2 can indeed occur in the nucleus (Fig. 7A). CTF2(AAA)-NLS did not enhance reporter activity (Fig. 7A), suggesting that p120 binding is required for E-cad/CTF2 to enhance transcription. A similar effect of E-cad/CTF2 and E-cad/CTF2-NLS on 4x KBS reporter activity was also observed using COS-1 cells (supplemental Fig. S4B). To further examine the ability of nuclear E-cad/CTF2 to relieve Kaiso-mediated repression, we also analyzed the effect of E-cad/CTF2-NLS on the expression of matrilysin using reporter assays. Kaiso has been shown to bind to and repress transcription from the matrilysin promoter (39). The promoter region of matrilysin also contains TCF/LEF-1 binding sites. To exclude the influence of TCF/LEF-1, we used HEK293 cells in which β-catenin is not localized in the nucleus (data not shown). Expression of p120 or E-cad/CTF2-NLS alone had no effect, but coexpression of E-cad/CTF2-NLS and p120 significantly enhanced transcription from the matrilysin promoter (2.3-kb Hmat) in HEK293 cells (p < 0.006) (Fig. 7B). E-Cad/CTF2(AAA)-NLS had no effect on transcription from this promoter (Fig. 7B), suggesting that p120 binding is required for nuclear E-cad/CTF2 to enhance transcription of the matrilysin gene. Because p120 relieves Kaiso-mediated repression through binding to Kaiso, we examined whether E-cad/CTF2 can also bind to Kaiso using a GST pull-down assay. Exogenous Kaiso protein expressed in HEK293 cells specifically bound to GST-E-cad/CTF2, only in the presence of exogenous p120 (Fig. 7C). Collectively, these data suggest that E-cad/CTF2 forms a complex with Kaiso via p120 and that E-cad/CTF2 can enhance the effect of p120 on Kaiso-mediated gene regulation.
FIGURE 7.
Nuclear E-cad/CTF2 modulates Kaiso-regulated gene transcription. HEK293 cells were transfected with the indicated constructs together with LacZ, and luciferase and β-galactosidase reporter activities were measured. Relative reporter activity represents the ratio of luciferase/β-galactosidase activities from five (A) or three (B) independent experiments. A, nuclear E-cad/CTF2 relieves Kaiso-mediated transcriptional repression. *, p < 0.038; **, p < 0.037; ***, p < 0.025. B, nuclear E-cad/CTF2 enhances transcription of matrilysin, *, p < 0.006. C, E-Cad/CTF2 interacts with Kaiso in the presence of p120. Beads coupled to GST or GST-E-cad/CTF2 were incubated with lysates of HEK293 cells expressing myc-Kaiso and/or FLAG-p120. The proteins bound to the beads were analyzed by Coomassie staining and Western blotting (WB) with anti-myc and anti-FLAG antibodies. The arrow and arrowhead indicates the positions of GST-E-cad/CTF2 and GST, respectively.
Expression of Nuclear E-cad/CTF2 Prevents Cells from Undergoing Apoptosis—To further understand the role of E-cad/CTF2 in the nucleus, we established murine L fibroblast cell lines stably expressing GFP-E-cad/CTF2-NLS. L cells were chosen because no classical cadherins are expressed. Therefore p120 is not sequestered at cell-cell junctions and is thus more accessible to bind E-cad/CTF2. First, we confirmed by immunofluorescence that NLS-tagged E-cad/CTF2 is mainly localized in the nucleus in L cells (supplemental Fig. S5). Expression of the nuclear E-cad/CTF2 protein did not induce detectable morphological changes (data not shown), and did not significantly affect the cell proliferation rate (supplemental Fig. S6). As the production of E-cad/CTF2 is enhanced by staurosporine-induced apoptosis (Fig. 1A), we examined whether the expression of E-cad/CTF2-NLS affects apoptosis of cells. After 3–4 h of addition of staurosporine, L cells started to undergo apoptosis, and caspase-3 was proteolytically cleaved and became an active form as observed by immunofluorescence (supplemental Fig. S7, A and B) and Western blotting (supplemental Fig. S7C). Interestingly, we found that this apoptotic process was strongly suppressed in L cells expressing E-cad/CTF2-NLS (supplemental Fig. S7, A–C), suggesting a potential regulatory role of nuclear E-cad/CTF2 in apoptosis.
DISCUSSION
In this study, we describe the crucial role of p120 in the nuclear localization of the γ-secretase cleavage product of E-cadherin, E-cad/CTF2, using several experimental systems. By immunofluorescence, we first observed that coexpression of p120 enhances nuclear localization of E-cad/CTF2 in several cell lines. Second, a similar effect is observed using C-terminal-tagged full-length E-cadherin. Third, using a biochemical nuclear fractionation method, we confirm the effect of p120 and show that this effect is specific for p120 as other cadherin-binding proteins do not induce nuclear translocation of E-cad/CTF2. We also demonstrate the requirement of p120 in E-cad/CTF2 nuclear localization using siRNA oligos that knock down endogenous p120. The direct interaction between p120 and E-cad/CTF2 is also required because E-cad/CTF2(AAA) is not detected in the nuclear fraction. As shown in Fig. 6F, E-cad/CTF2 binds p120 in the nucleus, suggesting that the nuclear translocation of E-cad/CTF2 is mediated by stoichiometric interaction with p120, although the nuclear localization of p120 is not clearly affected by the presence of E-cad/CTF2. However, it is also possible that the effect of p120 is mediated catalytically, for example, through post-translational modification of E-cad/CTF2. p120 is sequestered in multiple cellular locations including adherens junctions and microtubules (5, 47–49), and under conditions where E-cadherin is down-regulated, p120 is released into the cytosol (50, 51). The regulation of p120 subcellular localization could therefore impact on the control of nuclear localization of E-cad/CTF2.
Previously, it was shown that staurosporine treatment induces the production of E-cad/CTF2 by γ-secretase cleavage in A431 epithelial cells (25). We also observe a similar cleavage product in response to staurosporine in MCF-7 and MDCK epithelial cells. Recently, Bacteroides fragilis toxin was also reported to stimulate γ-secretase-induced cleavage of E-cadherin (52). However, the physiological stimuli that induce the production of E-cad/CTF2 were not clear. Our preliminary results suggest that activation of the hepatocyte growth factor receptor leads to γ-secretase-dependent accumulation of E-cad/CTF2 in MDCK cells.3 However, the amount of E-cad/CTF2 induced by activation of the hepatocyte growth factor receptor is much smaller than that of full-length E-cadherin. Despite extensive efforts, we were unable to isolate pure nuclear fractions from epithelial cells, thus the nuclear localization of endogenous E-cad/CTF2 could not be biochemically determined. Furthermore, the available antibodies against the cytoplasmic domain of E-cadherin are not suitable for immunofluorescent staining. Hence, a novel experimental approach needs to be established to study the fate of the endogenous cleavage product. Our observation that hepatocyte growth factor induces small amounts of E-cad/CTF2 relative to full-length E-cadherin is quite comparable with that of γ-secretase-induced cleavage of Notch. Upon binding to its ligand, Notch is cleaved by γ-secretase to release its intracellular domain, however, the amount is too small to be detected in the nucleus by immunostaining (30, 53). Thus, the main evidence for nuclear localization of Notch has been presented with a reporter assay using a Notch construct C-terminal tagged with GAL4 (30). To demonstrate the physiological significance of E-cad/CTF2 in vivo, a similar reporter system needs to be applied in the future.
Using reporter assays with 4x KBS and the promoter of Kaiso target matrilysin, we have shown that the expression of E-cad/CTF2-NLS potentiates the effect of p120 on the relief of the Kaiso-mediated transcriptional repression. With GST pulldown assays, we further show that E-cad/CTF2 forms a trimeric complex with p120 and Kaiso. These data suggest that the interaction between these three proteins influences gene transcription. Another interesting finding is that E-cad/CTF2 specifically binds to DNA. The association of E-cad/CTF2 and DNA is not direct, as E-cad/CTF2 requires p120 for its DNA binding. Thus, E-cad/CTF2 and p120 can mutually affect their functions; whereas E-cad/CTF2 potentiates the inhibitory role of p120 in Kaiso-mediated transcription, p120 promotes the nuclear localization and DNA binding of E-cad/CTF2. At present, the DNA binding site of the E-cad/CTF2-p120 complex is not clear. It is unlikely that the complex binds to the Kaiso-binding regions because p120 promotes dissociation of Kaiso from DNA (19). Hence, the E-cad/CTF2-p120 complex may be involved in transcriptional regulation or chromatin remodeling of unknown gene(s). Finally, by using L fibroblast cells expressing E-cad/CTF2-NLS, we have also shown that the expression of nuclear E-cad/CTF2 suppresses staurosporine-induced apoptosis. The molecular mechanism of this effect needs to be characterized in future work.
In summary, p120 mediates the nuclear translocation and DNA binding of E-cad/CTF2. In the nucleus, E-cad/CTF2 can modulate the p120-Kaiso-mediated signaling pathway, and can also potentially regulate the apoptotic process. To further understand the nuclear role of E-cad/CTF2, the identification of its DNA binding sites and the clarification of its in vivo functions need to be pursued.
Acknowledgments
We thank Mark Marsh for critical reading of the manuscript. We also thank Walter Birchmeier for providing pcH110-LacZ, Eiichiro Nishi for providing pME18S-FLAG-ADAM10, and Kevin Kelly for constructing pCS2-myc-Kaiso.
Author's Choice—Final version full access.
This work was supported by Medical Research Council funding to the Cell Biology Unit. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
Footnotes
The abbreviations used are: CH, cadherin homology; ADAM10, a disintegrin and metalloprotease 10; CBP, CREB-binding protein; E-cad/CTF, E-cadherin C-terminal fragment; HEK, human embryonic kidney; Hmat, human matrilysin; p120, p120-catenin; siRNA, short interfering RNA; CREB, cAMP-response element-binding protein; TCF/LEF-1, T-cell factor/lymphocyte enhancer factor-1; MDCK, Madin-Darby canine kidney; GST, glutathione S-transferase; HA, hemagglutinin; NLS, nuclear localization signal.
E. C. Ferber and Y. Fujita, unpublished observation.
References
- 1.Jamora, C., and Fuchs, E. (2002) Nat. Cell Biol. 4 E101–108 [DOI] [PubMed] [Google Scholar]
- 2.Fagotto, F., and Gumbiner, B. M. (1996) Dev. Biol. 180 445–454 [DOI] [PubMed] [Google Scholar]
- 3.Yagi, T., and Takeichi, M. (2000) Genes Dev. 14 1169–1180 [PubMed] [Google Scholar]
- 4.McCrea, P. D., Turck, C. W., and Gumbiner, B. (1991) Science 254 1359–1361 [DOI] [PubMed] [Google Scholar]
- 5.Reynolds, A. B., Daniel, J., McCrea, P. D., Wheelock, M. J., Wu, J., and Zhang, Z. (1994) Mol. Cell. Biol. 14 8333–8342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita, Y., Krause, G., Scheffner, M., Zechner, D., Leddy, H. E., Behrens, J., Sommer, T., and Birchmeier, W. (2002) Nat. Cell Biol. 4 222–231 [DOI] [PubMed] [Google Scholar]
- 7.Tricaud, N., Perrin-Tricaud, C., Bruses, J. L., and Rutishauser, U. (2005) J. Neurosci. 25 3259–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larue, L., Antos, C., Butz, S., Huber, O., Delmas, V., Dominis, M., and Kemler, R. (1996) Development 122 3185–3194 [DOI] [PubMed] [Google Scholar]
- 9.Wheelock, M. J., and Johnson, K. R. (2003) Curr. Opin. Cell Biol. 15 509–514 [DOI] [PubMed] [Google Scholar]
- 10.Gumbiner, B. M. (2005) Nat. Rev. Mol. Cell Biol. 6 622–634 [DOI] [PubMed] [Google Scholar]
- 11.Perez-Moreno, M., Jamora, C., and Fuchs, E. (2003) Cell 112 535–548 [DOI] [PubMed] [Google Scholar]
- 12.Brembeck, F. H., Rosario, M., and Birchmeier, W. (2006) Curr. Opin. Genet. Dev. 16 51–59 [DOI] [PubMed] [Google Scholar]
- 13.Willert, K., and Jones, K. A. (2006) Genes Dev. 20 1394–1404 [DOI] [PubMed] [Google Scholar]
- 14.Molenaar, M., van de Wetering, M., Oosterwegel, M., Peterson-Maduro, J., Godsave, S., Korinek, V., Roose, J., Destree, O., and Clevers, H. (1996) Cell 86 391–399 [DOI] [PubMed] [Google Scholar]
- 15.Behrens, J., von Kries, J. P., Kuhl, M., Bruhn, L., Wedlich, D., Grosschedl, R., and Birchmeier, W. (1996) Nature 382 638–642 [DOI] [PubMed] [Google Scholar]
- 16.Kelly, K. F., Spring, C. M., Otchere, A. A., and Daniel, J. M. (2004) J. Cell Sci. 117 2675–2686 [DOI] [PubMed] [Google Scholar]
- 17.Daniel, J. M., and Reynolds, A. B. (1999) Mol. Cell. Biol. 19 3614–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hosking, C. R., Ulloa, F., Hogan, C., Ferber, E. C., Figueroa, A., Gevaert, K., Birchmeier, W., Briscoe, J., and Fujita, Y. (2007) Mol. Biol. Cell 18 1918–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniel, J. M., Spring, C. M., Crawford, H. C., Reynolds, A. B., and Baig, A. (2002) Nucleic Acids Res. 30 2911–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, S. W., Park, J. I., Spring, C. M., Sater, A. K., Ji, H., Otchere, A. A., Daniel, J. M., and McCrea, P. D. (2004) Nat. Cell Biol. 6 1212–1220 [DOI] [PubMed] [Google Scholar]
- 21.Park, J. I., Kim, S. W., Lyons, J. P., Ji, H., Nguyen, T. T., Cho, K., Barton, M. C., Deroo, T., Vleminckx, K., Moon, R. T., and McCrea, P. D. (2005) Dev. Cell 8 843–854 [DOI] [PubMed] [Google Scholar]
- 22.Batlle, E., Sancho, E., Franci, C., Dominguez, D., Monfar, M., Baulida, J., and Garcia De Herreros, A. (2000) Nat. Cell Biol. 2 84–89 [DOI] [PubMed] [Google Scholar]
- 23.Cano, A., Perez-Moreno, M. A., Rodrigo, I., Locascio, A., Blanco, M. J., del Barrio, M. G., Portillo, F., and Nieto, M. A. (2000) Nat. Cell Biol. 2 76–83 [DOI] [PubMed] [Google Scholar]
- 24.Lochter, A., Galosy, S., Muschler, J., Freedman, N., Werb, Z., and Bissell, M. J. (1997) J. Cell Biol. 139 1861–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marambaud, P., Shioi, J., Serban, G., Georgakopoulos, A., Sarner, S., Nagy, V., Baki, L., Wen, P., Efthimiopoulos, S., Shao, Z., Wisniewski, T., and Robakis, N. K. (2002) EMBO J. 21 1948–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maretzky, T., Reiss, K., Ludwig, A., Buchholz, J., Scholz, F., Proksch, E., de Strooper, B., Hartmann, D., and Saftig, P. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 9182–9187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinhusen, U., Weiske, J., Badock, V., Tauber, R., Bommert, K., and Huber, O. (2001) J. Biol. Chem. 276 4972–4980 [DOI] [PubMed] [Google Scholar]
- 28.Fortini, M. E. (2002) Nat. Rev. Mol. Cell Biol. 3 673–684 [DOI] [PubMed] [Google Scholar]
- 29.Schroeter, E. H., Kisslinger, J. A., and Kopan, R. (1998) Nature 393 382–386 [DOI] [PubMed] [Google Scholar]
- 30.Struhl, G., and Adachi, A. (1998) Cell 93 649–660 [DOI] [PubMed] [Google Scholar]
- 31.Gao, Y., and Pimplikar, S. W. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 14979–14984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marambaud, P., Wen, P. H., Dutt, A., Shioi, J., Takashima, A., Siman, R., and Robakis, N. K. (2003) Cell 114 635–645 [DOI] [PubMed] [Google Scholar]
- 33.Ni, C. Y., Murphy, M. P., Golde, T. E., and Carpenter, G. (2001) Science 294 2179–2181 [DOI] [PubMed] [Google Scholar]
- 34.Okamoto, I., Kawano, Y., Murakami, D., Sasayama, T., Araki, N., Miki, T., Wong, A. J., and Saya, H. (2001) J. Cell Biol. 155 755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uemura, K., Kihara, T., Kuzuya, A., Okawa, K., Nishimoto, T., Bito, H., Ninomiya, H., Sugimoto, H., Kinoshita, A., and Shimohama, S. (2006) Biochem. Biophys. Res. Commun. 345 951–958 [DOI] [PubMed] [Google Scholar]
- 36.Haas, I. G., Frank, M., Veron, N., and Kemler, R. (2005) J. Biol. Chem. 280 9313–9319 [DOI] [PubMed] [Google Scholar]
- 37.Hambsch, B., Grinevich, V., Seeburg, P. H., and Schwarz, M. K. (2005) J. Biol. Chem. 280 15888–15897 [DOI] [PubMed] [Google Scholar]
- 38.Shoval, I., Ludwig, A., and Kalcheim, C. (2007) Development 134 491–501 [DOI] [PubMed] [Google Scholar]
- 39.Spring, C. M., Kelly, K. F., O'Kelly, I., Graham, M., Crawford, H. C., and Daniel, J. M. (2005) Exp. Cell Res. 305 253–265 [DOI] [PubMed] [Google Scholar]
- 40.Hogan, C., Serpente, N., Cogram, P., Hosking, C. R., Bialucha, C. U., Feller, S. M., Braga, V. M., Birchmeier, W., and Fujita, Y. (2004) Mol. Cell. Biol. 24 6690–6700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thoreson, M. A., Anastasiadis, P. Z., Daniel, J. M., Ireton, R. C., Wheelock, M. J., Johnson, K. R., Hummingbird, D. K., and Reynolds, A. B. (2000) J. Cell Biol. 148 189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dupre-Crochet, S., Figueroa, A., Hogan, C., Ferber, E. C., Bialucha, C. U., Adams, J., Richardson, E. C., and Fujita, Y. (2007) Mol. Cell. Biol. 27 3804–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaeper, U., Gehring, N. H., Fuchs, K. P., Sachs, M., Kempkes, B., and Birchmeier, W. (2000) J. Cell Biol. 149 1419–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deleted in proof
- 45.Mukhopadhyay, D., and Riezman, H. (2007) Science 315 201–205 [DOI] [PubMed] [Google Scholar]
- 46.Shcherbik, N., and Haines, D. S. (2004) J. Cell. Biochem. 93 11–19 [DOI] [PubMed] [Google Scholar]
- 47.Chen, X., Kojima, S., Borisy, G. G., and Green, K. J. (2003) J. Cell Biol. 163 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franz, C. M., and Ridley, A. J. (2004) J. Biol. Chem. 279 6588–6594 [DOI] [PubMed] [Google Scholar]
- 49.Yanagisawa, M., Kaverina, I. N., Wang, A., Fujita, Y., Reynolds, A. B., and Anastasiadis, P. Z. (2004) J. Biol. Chem. 279 9512–9521 [DOI] [PubMed] [Google Scholar]
- 50.Roczniak-Ferguson, A., and Reynolds, A. B. (2003) J. Cell Sci. 116 4201–4212 [DOI] [PubMed] [Google Scholar]
- 51.van Hengel, J., Vanhoenacker, P., Staes, K., and van Roy, F. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 7980–7985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, S., Rhee, K. J., Zhang, M., Franco, A., and Sears, C. L. (2007) J. Cell Sci. 120 1944–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Struhl, G., and Greenwald, I. (1999) Nature 398 522–525 [DOI] [PubMed] [Google Scholar]