Abstract
The uncleaved, pro-form of nerve growth factor (proNGF) functions as a pro-apoptotic ligand for the p75 neurotrophin receptor (p75NTR). However, some reports have indicated that proneurotrophins bind and activate Trk receptors. In this study, we have examined proneurotrophin receptor binding and activation properties in an attempt to reconcile these findings. We show that proNGF readily binds p75NTR expressed in HEK293T cells but does not interact with TrkA expressed under similar circumstances. Importantly, proNGF activates TrkA tyrosine phosphorylation, induces Erk and Akt activation, and causes PC12 cell differentiation. We show that inhibiting endocytosis or furin activity reduced TrkA activation induced by proNGF but not that induced by mature NGF and that proNGF123, a mutant form of NGF lacking dibasic cleavage sites in the prodomain, does not induce TrkA phosphorylation in PC12 cells. Therefore, endocytosis and cleavage appear to be prerequisites for proNGF-induced TrkA activity. We also found that proBDNF induces activation of TrkB in cerebellar granule neurons and that proBDNF cleavage by furin and metalloproteases facilitates this effect. Taken together, these data indicate that under physiological conditions, proneurotrophins do not directly bind or activate Trk receptors. However, endocytosis and cleavage of proneurotrophins produce processed forms of neurotrophins that are capable of inducing Trk activation.
The four mammalian neurotrophins comprise a family of related secreted factors required for differentiation, survival, development, and death of specific populations of neurons and non-neuronal cells. The effects of the neurotrophins are mediated by binding to TrkA, TrkB and TrkC receptor tyrosine kinases and to the p75 neurotrophin receptor (p75NTR)4. The Trk receptors play critical roles in mediating neuronal survival and growth and are important modulators of synaptic function (1). p75NTR is a component of distinct cell surface signaling platforms that function to induce apoptosis and mediate neuronal growth inhibition. However, p75NTR also acts as a Trk co-receptor that increases the binding specificity and affinity of Trk receptors for neurotrophins (2, 3).
Neurotrophins are produced as proforms of ∼240 amino acids that are cleaved by furins and proconvertases to yield mature neurotrophins of about 120 amino acids (4). The main functions ascribed to the neurotrophin prodomain include facilitating neurotrophin folding and directing neurotrophins to the regulated secretory pathway (5–7). Several recent studies have indicated that nerve growth factor (NGF) and brain-derived neurotrophic factor (BDNF) are secreted from cells as prodomain-containing forms (proNGF and proBDNF, respectively) (8–10). In some types of primary neurons and in endothelial cells, proneurotrophin binding to the p75NTR-sortilin receptor complex is a potent apoptotic stimulus (8, 9, 11). Additional work demonstrating the predominance of proNGF and proBDNF in vivo (12–14) and its up-regulation following injury (8, 11) adds weight to the hypothesis that proNGF is a potent pro-apoptotic factor mediating p75NTR-dependent apoptosis in vivo.
Nykjaer et al. (15) showed that proNGF has lower affinity than NGF for both p75NTR and TrkA, with Kd shifts from about 1–2 nm to 15–20 nm for each receptor. Sortilin, a member of the VPS10 family, has emerged as a co-receptor that functions in concert with p75NTR to induce apoptosis. Unlike p75NTR and TrkA, sortilin shows considerably higher affinity for proNGF than for NGF (5 nm versus 90 nm, respectively), and proNGF promotes formation of heterotrimeric complexes containing sortilin and p75NTR (15). However, some studies have indicated that proneurotrophins are not selective p75NTR ligands and that they are capable of binding and activating Trk receptors to exert a neurotrophic, rather than a pro-apoptotic effect (16, 17).
In this study, we have compared proneurotrophin binding to p75NTR and TrkA and determined the signaling events activated by these ligands. We report that proNGF readily binds to p75NTR expressed in transfected cells but, under identical conditions, does not bind TrkA. Despite this, exposure to proNGF results in robust TrkA phosphorylation and activation of Akt and Erk signaling cascades in PC12 cells, which express both p75NTR and TrkA. We show that activation of TrkA by proNGF occurs only after proNGF endocytosis and cleavage by furin-like enzymes. We also examined the effect of proBDNF on cerebellar granule neurons, which express p75NTR and TrkB, and show that this ligand activates TrkB and that ligand cleavage by cerebellar granule neuron proteases is a prerequisite for this effect. Therefore, we conclude that proneurotrophins are specific ligands for p75NTR that, once bound and endocytosed, are rapidly converted to mature neurotrophins that activate Trk receptors.
EXPERIMENTAL PROCEDURES
Cell Culture—HEK293T cells were maintained in Dulbecco's modified Eagle's (DME) medium containing 10% bovine calf serum, 2 mm l-glutamine, and 100 μg/ml penicillin and 100 μg/ml streptomycin in a humidified 37 °C incubator with 5% CO2. PC12 and PC12nnr5 cells were maintained in DME containing 6% bovine calf serum, 6% deactivated horse serum, 2 mm l-glutamine, and 100 μg/ml penicillin and streptomycin in a humidified 37 °C incubator with 10% CO2. Cerebellar neurons were prepared from post-natal day 8–10 rat brain, dissociated with trypsin and mechanical trituration, and cultured on poly-l-lysine-coated substrates for 24 h in Sato medium (Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 1% penicillin/streptomycin, 1% N2 supplement, 100 ng/ml l-thyroxine and 80 ng/ml tri-iodothyronine) prior to treatment with BDNF or proBDNF. All cell culture reagents were obtained from Hyclone (Logan, UT).
Generation of Recombinant ProNGF—PCR-based site directed mutagenesis was used to mutate the furin cleavage site of proNGF from KR to AA (bp 651–657) and add a Myc epitope to the proNGF carboxyl terminus. The PCR product was sequenced to confirm fidelity and then subcloned into pc3.1AP6.str to produce the AP-proNGF fusion construct. An improved Kozak signal and Igκ signal sequence were subsequently added to improve translation initiation and secretion. Secreted AP and AP-proNGF were produced in HEK293T cells and normalized for molar concentrations, as described in Flanagan and Cheng (18). ProNGF123 was prepared and purified as described (19). To produce biotinylated proNGF, cDNA encoding proNGF with the mutated furin cleavage site was subcloned into pEGFPN1-secVSVg-Bgl-BAP, a plasmid encoding a biotin acceptor peptide (BAP) at the amino terminus. BAP-ProNGF was co-transfected with a secreted form of the biotin ligase, BirA, to produce soluble biotinylated ProNGF in HEK293T cells.
Treatments and Reagents—Cells were incubated in serum-free DME supplemented with 0.1% bovine serum albumin (BSA) (DMEB) and 2 mm l-glutamine for 1 h prior to treatment with 25 ng/ml NGF (Cedarlane Laboratories Ltd., Burlington, ON), 50 ng/ml WT-proNGF (Scil Proteins, Saale, Germany), or proNGF123 (19). Final concentrations of inhibitor treatments were 200 nm for K252A, 1 μm for epoxomycin (Calbiochem; 10 μm for BB94 (British Biotech, Oxford, UK), 10 μm for GM6001, 50 μm for decanoyl-RV-KR-chloromethyl ketone (decCMK) (Biomol), and 5 μg/ml brefeldin A (Sigma). In each case, inhibitors were added 1 h prior to ligand or Phorbol 12-myristate 13-acetate (PMA) exposure (Calbiochem).
Immunoprecipitation and Immunoblotting—Samples were lysed in Nonidet P-40 buffer (10 mm Tris, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 10% glycerol) supplemented with a protease inhibitor mixture (Roche, Hertfordshire, UK) and 10 μm sodium orthovanadate. Lysates were immunoprecipitated with anti-TrkA antibody for 2 h at 4°C. The precipitated proteins were then purified with protein A-agarose for 1 h at 4°C. Immunocomplexes were then washed with lysis buffer and eluted with sample buffer and analyzed by SDS-PAGE. For immunoblotting, samples were separated by SDS-PAGE, transferred onto nitrocellulose membrane, which were then blocked in Tris-buffered saline/Tween (10 mmol/liter Tris (pH 8.0), 150 mmol/liter NaCl, 0.2% Tween 20) containing 5% (w/v) dried skim milk powder or 5% bovine serum albumin for phospho-specific antibodies. Membranes were then immunoblotted in blocking solution using anti-NGF (gift from Richard Murphy (Salk Institute, La Jolla, CA)); TrkA203 (gift from David Kaplan (University of Toronto, Ontario, Canada)); RTB, directed against the TrkB extracellular domains (a gift from Louis Reichardt (University of California, San Francisco (UCSF)); anti-p75NTR (20); anti-Myc 9E10 and anti-phospho-tyrosine 4G10 antibodies (Millipore Inc. Lake Placid, NY)); anti-phospho-Erk (Thr-202/Tyr-204), anti-Erk (3A7), anti-phospho-Akt (Ser-473), and anti-Akt from Cell Signaling Technologies (Cambridge, MA). Secondary antibodies were obtained from Jackson Laboratories (West Grove, PA), and incubations with them were performed in blocking solution. Immunoreactive bands were detected using the enhanced chemiluminescence solution kit (Perkin-Elmer Life Sciences, Norwalk, CT).
Alkaline Phosphatase Assays—PC12 cells on 24-well plates were washed twice in Hanks' balanced salt solution (Invitrogen) and then exposed to AP- or APproNGF-conditioned medium for 1 h at either 4 °C or room temperature. Cells were then washed three times with ice-cold HBAH buffer (20 mm Hepes, pH 7.4, 0.5 mg/ml bovine serum albumin, 0.1% sodium azide, Hanks Balanced salt solution) and lysed in 0.1% Triton X-100 in 20 mm Tris, pH 8.0. Lysates were incubated at 65 °C for 30 min to eliminate endogenous AP activity and then centrifuged at 9000 rpm for 5 min to remove insoluble material. AP activity was assayed at room temperature in pNPP reaction buffer (2 m diethanolamine, pH 9.8, 0.5 mm MgCl2, p-nitrophenyl phosphate disodium salt hexahydrate) (Sigma) at room temperature. Data points represent four wells assayed in quadruplicate. Activity was normalized to total protein, assayed using bicinchoninic acid protein Assay (Pierce). Statistical differences were assessed using a one-way analysis of variance (ANOVA) with a Student-Newman-Keuls post-test.
Neurite Outgrowth Assay—PC12 cells were plated in poly-l-lysine-coated 6-well plates at a density of 1.6 × 104 cells/ml in serum-containing medium. After 24 h, cells were switched to DMEB alone or DMEB containing 10 ng/ml NGF, or 50 ng/ml Scil proNGF, and maintained in this media for 3 days. Cells were digitally photographed under phase contrast conditions using an Axiovert 100 microscope.
RESULTS
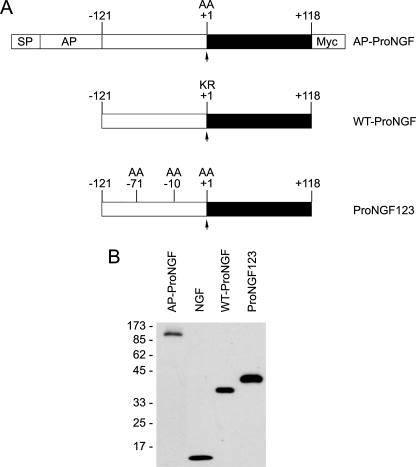
Expression and Secretion of proNGF Fusion Proteins—For our initial studies on proNGF receptor binding and activation, we produced an alkaline phosphatase-proNGF fusion protein (AP-proNGF) in which the main furin cleavage site residues (KR) were mutated to alanine (shown schematically in Fig. 1A). We also used a wild-type untagged form of proNGF (termed WT-proNGF) as well as proNGF123, a cleavage-resistant form of proNGF in which three dibasic cleavage sites within the prodomain were mutated to alanine (19). To confirm that these distinct proNGF proteins were intact and to confirm relative concentrations of these proteins, they were evaluated by immunoblotting using an antibody directed against mature NGF. Fig. 1B shows that recombinant AP-proNGF, WT-proNGF, and proNGF123 are intact and therefore appropriate for use in cell-based assays.
FIGURE 1.
Recombinant proNGF is expressed and secreted in intact form. A, schematic representation of recombinant proNGF proteins used in this study. Position-1 refers to the cleavage site that generates fully processed NGF. AP-proNGF contains an amino-terminal AP cassette, followed by proNGF with a KR to AA mutation at the central dibasic cleavage site followed by a carboxyl-terminal Myc epitope tag. The WT-proNGF is untagged and contains the normal dibasic cleavage site at position-1. In proNGF123, the two amino acids that compose the dibasic cleavage sites at –72, –42, and –1 were mutated to alanine. B, recombinant AP-proNGF produced as described under “Experimental Procedures” was immunoprecipitated using an antibody directed against the Myc epitope tag. The immunoprecipitate was analyzed by immunoblot, together with samples of mature NGF (15 ng), WT-proNGF (25 ng), or proNGF123 (25 ng), using an antibody directed against mature NGF.
Recombinant proNGF Binds to the Surface of 293T Cells Overexpressing p75NTR—Different lines of evidence have shown that proNGF is a selective ligand for p75NTR in vivo, yet some studies have suggested that proNGF can also bind TrkA both in vitro and in vivo. To address this, we examined the binding of AP-proNGF to HEK293T cells that overexpressed either p75NTR or TrkA. These studies revealed that AP-proNGF readily bound to cells that expressed p75NTR, with half-maximal binding observed at ∼5 nm. In contrast, AP-proNGF showed no specific binding to HEK293T cells transfected with a TrkA expression plasmid. (Fig. 2A). To rule out the possibility that TrkA is not expressed or not transported to the cell surface, we assessed the cell surface expression of p75NTR and TrkA by performing a biotinylation assay on transfected HEK293T cells overexpressing these receptors. Fig. 2B shows that both receptors are highly expressed at the cell surface. We therefore conclude that proNGF shows preferential binding to p75NTR and has low affinity for cell surface TrkA expressed at the surface in HEK293T cells.
FIGURE 2.
AP-proNGF binds cell surface p75NTR but not TrkA. A, AP-proNGF binding assays were performed on HEK293T cells transfected with nothing (Mock), with p75NTR, or with TrkA. Values represent four wells assayed in quadruplicate. *, differences between p75NTR and mock transfection with p < 0.001. Error bars represent standard deviation. B, HEK293T cells were transfected with plasmids encoding p75NTR or TrkA, and 24 h later, cells were treated with sulfo-NHS-LC-biotin (0.5 mg/ml) for 30 min. Lysates were immunoprecipitated with streptavidin-conjugated beads and subsequently immunoblotted with p75NTR or TrkA antibodies. SA-PD, streptavidin bead pulldown. Experiments were repeated three times with identical results.
ProNGF Activates TrkA Signaling—Earlier studies have shown that proNGF can activate TrkA phosphorylation but our binding studies indicated that AP-proNGF selectively binds to p75NTR. To address this discrepancy, we compared NGF and proNGF for their ability to induce activation of Erk and Akt. As expected, NGF induced rapid Erk and Akt phosphorylation that was sustained for 60 min. Surprisingly, WT-proNGF also induced robust phosphorylation of Erk and Akt, albeit with slower kinetics than NGF (Fig. 3, A and B). To address whether phosphorylation of ERK and AKT induced by WT-proNGF required TrkA activity, we assessed whether WT-proNGF induced phosphorylation of Erk and Akt in PC12nnr5 cells, which do not express TrkA or in PC12 cells pretreated with the Trk inhibitor, K252A. Fig. 3, C and D shows that WT-proNGF had no effect on Erk and Akt phosphorylation in PC12nnr5 cells or K252A-pretreated cells (Fig. 3E), indicating that TrkA activation is required for the phosphorylation of Akt and Erk induced by WT-proNGF.
FIGURE 3.
WT-ProNGF induces TrkA-dependent signaling cascades. PC12 cells (panels A and B) or PC12nnr5 cells (panels C and D) were incubated in DMEB for 1 h and then treated with 25 ng/ml NGF or 50 ng/ml WT-proNGF for the time points indicated. Cells were lysed in sample buffer, and phosphorylation of ERK (A and C) and AKT (B and D) were examined by immunoblot. E, PC12 cells were exposed to K252A (200 nm) for 1 h prior to addition of NGF (25 ng/ml) or WT-proNGF (50 ng/ml). Experiments were repeated three times, with identical results.
We next asked whether TrkA was phosphorylated in cells treated with proNGF. PC12 cells were treated with either NGF or WT-proNGF for 15, 30, or 60 min and then TrkA phosphorylation was assessed by immunoprecipitation and immunoblot. NGF causes robust phosphorylation of TrkA that has peaked by 15 min of exposure. TrkA phosphorylation is also induced by WT-proNGF but with this ligand, the time course of TrkA activation lags behind that induced by NGF, with the peak observed only 30 min after WT-proNGF exposure (Fig. 4A).
FIGURE 4.
WT-ProNGF induces robust TrkA phosphorylation and neurite outgrowth. A, PC12 cells were incubated in DMEB for 1 h then treated with NGF (25 ng/ml) or WT-proNGF (50 ng/ml) for the indicated times, then lysed, and TrkA was immunoprecipitated. Immunoblots were performed with anti-phosphotyrosine and anti-TrkA antibodies, as indicated. B, PC12 cells were plated in serum-containing media and 24 h later, were switched to DMEB containing NGF (10 ng/ml) or WT-proNGF (50 ng/ml). Cells were maintained for 3 days and then photographed.
To determine whether proNGF elicits TrkA-dependent biological responses, PC12 cells were maintained in serum-free media containing NGF or WT-proNGF for 48 h and then evaluated for the elaboration of neuritic processes. Both ligands produced a comparable amount of neurite outgrowth in PC12 cells, whereas process extension was not observed in untreated cells (Fig. 4B). Thus, WT-proNGF is functionally equivalent to mature NGF in this assay.
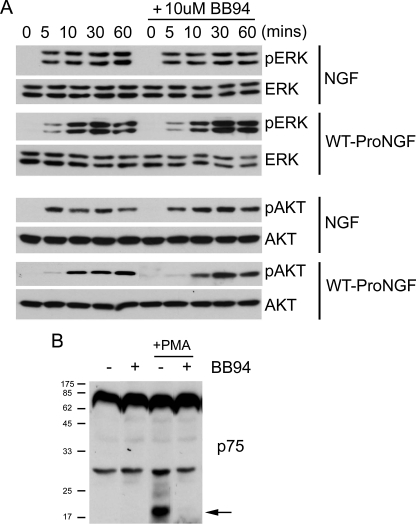
Endocytosis Is Required for proNGF to Activate TrkA Signaling—The experiments above show that AP-proNGF does not directly bind TrkA at the cell surface but proNGF treatment activates TrkA and its downstream signaling cascades. One possible explanation for these findings is that, once bound at the cell surface by p75NTR, proNGF is cleaved, and the newly formed mature NGF goes on to activate TrkA signaling. Previous studies have indicated that proNGF can be cleaved by matrix metalloproteases (MMPs) (9), and we therefore tested whether activity of secreted or transmembrane proteases was required for proNGF-induced TrkA signaling using BB94 and GM6001, chemically distinct metalloproteinase inhibitors. TrkA signaling events were maintained in PC12 cells that were treated with WT-proNGF in the presence of either BB94 (Fig. 5A) or GM6001 (not shown). As a positive control, we asked whether BB94 and GM6001 blocked cleavage of p75NTR induced by phorbol ester and found that these inhibitors almost completely blocked p75NTR cleavage (Fig. 5B), indicating that they are effective metalloprotease inhibitors under our conditions. We conclude that proNGF cleavage by MMPs does not play an essential role in proNGF-induced TrkA activation in PC12 cells.
FIGURE 5.
Inhibiting metalloproteases does not block proNGF-induced activation of TrkA signaling cascades. A, PC12 cells in DMEB were exposed to BB94 (10 μm) for 1 h, then they were treated with NGF (25 ng/ml) or WT-proNGF (50 ng/ml) for the times indicated, and phosphorylation of ERK and AKT was examined by immunoblot. B, PC12 cells were exposed to BB94 (10 μm) for 1 h and then treated with phorbol 12-myristate 13-acetate (PMA) (1 μm) for 90 min. p75NTR cleavage was monitored by immunoblot. Experiment in panel A was repeated three times and in panel B was repeated twice, with identical results.
Recent studies indicate that p75NTR mediates endocytosis of NGF through a slow endocytic route, whereas proNGF is endocytosed rapidly by a p75NTR-sortilin complex (15, 21). We reasoned that proNGF may be endocytosed and then cleaved to mature NGF, which can then go on to activate TrkA. To address this, we inhibited receptor-mediated endocytosis using high sucrose concentrations (22) and examined the effect on proNGF-induced TrkA and Erk activation. We found that sucrose exposure produced a substantial reduction in the phosphorylation of ERK normally induced by either NGF or proNGF (Fig. 6A), consistent with previous studies that have shown that TrkA endocytosis is a prerequisite for NGF-mediated Erk phosphorylation in PC12 cells (23). Interestingly, exposure to high sucrose does not alter TrkA phosphorylation induced by mature NGF but sharply reduces proNGF-induced TrkA activation (92% reduction by densitometry; Fig. 6B). Because AP-proNGF binding assays show that sucrose treatment does not alter cell surface binding to PC12 cells (supplemental Fig. 1), these data suggest that the block in TrkA activation induced by sucrose reflects a decrease in proNGF endocytosis and cleavage.
FIGURE 6.
Endocytosis is required for WT-proNGF treatment to induce TrkA signaling. A and B, PC12 cells in DMEB were exposed to 0.45 m sucrose for 15 min and then treated with NGF (25 ng/ml) or WT-proNGF (50 ng/ml) for 10 or 30 min, as indicated. A, cells were lysed in sample buffer, and ERK phosphorylation was examined by immunoblot. B, TrkA was immunoprecipitated, and receptor phosphotyrosine levels were examined by immunoblot. C, PC12 cells in DMEB were cooled to 4 °C for 30 min and then treated with NGF (5, 10, or 50 ng/ml) or WT-proNGF (10, 50, 100 ng/ml) for 30 or 60 min, as indicated. TrkA was immunoprecipitated, and phosphotyrosine levels were examined by immunoblot. Experiments in panels A and B were repeated three times, with identical results.
To show that proNGF internalization is a prerequisite for subsequent TrkA activation, we maintained the cells at low temperature to reduce endocytosis. Receptor tyrosine kinases in mammalian cells can undergo ligand-induced dimerization and phosphorylation even when maintained at low temperatures. Endocytosis is completely blocked at 4 °C, and we therefore examined NGF- and WT-proNGF-induced TrkA activation in cells maintained at this temperature. Fig. 6C shows that in PC12 cells maintained at 4 °C, NGF caused robust phosphorylation of TrkA, whereas proNGF was incapable of activating TrkA under these circumstances, even at high proNGF concentrations and extended exposure times. These data indicate that that proNGF endocytosis is a prerequisite for TrkA activation.
proNGF Must be Degraded Intracellularly to Activate TrkA—ProNGF is normally cleaved by furin to produce mature NGF during transit through the secretory pathway. Endocytic vesicles undergo fusion with post-Golgi secretory vesicles and therefore proNGF endocytosed from the cell surface may encounter furin during its endocytic journey. To test this, we first determined whether exposure to the furin inhibitor decCMK altered TrkA phosphorylation or Erk activation induced by WT-proNGF. Fig. 7, A and B show that dec-CMK pre-treatment (30 min) dramatically reduces phosphorylation of TrkA and Erk induced by proNGF but has no effect on TrkA phosphorylation induced by NGF, indicating that proNGF cleavage by furin is required for TrkA activation.
FIGURE 7.
ProNGF cleavage is required for TrkA activation. A, PC12 cells in DMEB were pretreated with dec-CMK (50 μm) for 30 min and then exposed to NGF (25 ng/ml) or WT-proNGF (50 ng/ml). TrkA was immunoprecipitated and phosphotyrosine levels were examined by immunoblot. B, as in A with phosphorylation of ERK within cell lysates was determined by immunoblot. C, PC12 cells in DMEB were pretreated with dec-CMK (50 μm) for 30 min and then exposed to B-proNGF (25 ng/ml) for 60 min. Cells were then washed and lysed, and levels of B-proNGF and phosphorylation of ERK within cell lysates were determined by immunoblot. D, PC12 cells were treated with NGF (25 ng/ml), WT-proNGF (50 ng/ml), or proNGF123 (50 ng/ml) for the indicated times, and TrkA was immunoprecipitated and assessed for phosphotyrosine content by immunoblot. E, PC12 cells were treated with NGF (25 ng/ml) or WT-proNGF (50 ng/ml) in the absence of presence of brefeldin A (10 μg/ml). Cells were lysed in sample buffer, and levels of phospho- and total ERK and AKT as well as levels of p75NTR and TrkA were determined by immunoblot. Experiments in panels A–E were repeated three times, with identical results.
To determine whether furin mediates endosomal cleavage of proNGF, we produced a biotinyated form of proNGF (B-proNGF) and exposed PC12 cells to this ligand in the presence or absence of the furin inhibitor, dec-CMK. Cells were then washed, lysed, and levels of B-proNGF were determined by immunoblot. Fig. 7C shows that B-proNGF can be readily detected in treated cells and reveals that B-proNGF levels are sharply increased, and phospho-Erk levels are decreased, in cells exposed to dec-CMK.
To confirm that cleavage of proNGF is a prerequisite for TrkA activation, we asked whether proNGF123, a triple-mutated form of proNGF that is highly resistant to proteolysis (19) was capable of activating TrkA in PC12 cells. Under the same conditions in which NGF and WT-proNGF caused robust TrkA phosphorylation in PC12 cells, cleavage-resistant proNGF123 did not induce TrkA tyrosine phosphorylation (Fig. 7D). To rule out the possibility that proNGF123 is defective in p75NTR binding, we produced alkaline phosphatase-tagged form of proNGF123 (AP-proNGF123) and using this ligand, found that AP-proNGF123 and AP-proNGF bind p75NTR expressed in HEK293T cells equally well (supplemental Fig. 2). We conclude that endocytosis of proNGF and its subsequent cleavage by furin-like enzymes is a prerequisite for TrkA activation.
Our results indicate that proNGF is processed in endosomes and thereby becomes capable of activating TrkA-dependent Akt and Erk signaling. Because TrkA induces Akt activation mainly at the plasma membrane and TrkA-dependent Erk activation occurs primarily in endosomes (24, 25), we were interested in examining how processed proNGF activates these spatially regulated signaling events. After proNGF cleavage, NGF could activate TrkA that is present within endosomes but alternatively, the ligand could be recycled to the cell surface, released, and then bind TrkA. To distinguish between these possibilities, we treated cells with Brefeldin A to reduce endosome recycling (26) and then assessed proNGF-induced signaling events. Interestingly, Brefeldin A treatment enhanced proNGF-induced Erk activation, and decreased proNGF-induced Akt activation. This result suggests that endosomal TrkA activation does occur but a portion of the NGF, which is liberated from proNGF in endosomes, is recycled to activate cell surface TrkA.
ProBDNF Activates TrkB in Cerebellar Granule Neurons through a p75NTR- and Furin-dependent Mechanism—Many central neurons express TrkB, and proBDNF is widespread in the brain (27). We were interested in determining if proBDNF treatment results in activation of TrkB and downstream signaling cascades in central neurons and if so, whether proBDNF cleavage is a prerequisite for this effect. We chose cerebellar granule neurons for these experiments because they co-expressed TrkB and p75NTR. As expected, mature BDNF induced robust TrkB and Erk phosphorylation in cerebellar granule neurons, and these phosphorylation events were not altered by treatment with dec-CMK, a furin inhibitor, or BB94, a matrix metalloprotease inhibitor (Fig. 8A). Cerebellar granule neurons (CGNs) treated with proBDNF also showed phosphorylation of TrkB and Erk, and dec-CMK and BB94 treatment partially reduced proBDNF-induced TrkB phosphorylation and Erk phosphorylation (Fig. 8B). Therefore, proBDNF treatment of cerebellar granule cells results in TrkB activation and Erk phosphorylation, and full activation of these events requires proBDNF processing by both MMPs and furin.
FIGURE 8.
Processing of proBDNF by metalloproteases and furin facilitates TrkB activation in cerebellar granule neurons. Rat cerebellar granule neurons were pre-incubated with dec-CMK (50 μm) or BB94 (10 μm) for 30 min, then exposed to BDNF (5 ng/ml) or WT-proBDNF (50 ng/ml) for 30 min. TrkB was immunoprecipitated with RTB, and phosphotyrosine content of TrkB in immunoprecipitates (A) and phospho-ERK levels within cell lysates (B) were determined by immunoblot. Experiments in panels A–B were repeated three times, with identical results.
DISCUSSION
The discovery that secreted proneurotrophins activate signaling pathways that are distinct from those of the mature neurotrophins has had a major impact on the neurotrophin field. There is substantial evidence that proneurotrophins act to selectively activate p75NTR-dependent pathways that result in apoptosis or cell migration in vitro, and emerging data indicates that proneurotrophins can activate p75NTR-dependent signaling pathways in physiological settings (8, 11, 28, 29). However, there have been several studies that challenge the hypothesis that proneurotrophins exclusively activate p75NTR signaling paths. Some of these have reported that proneurotrophins bind TrkA at the cell surface and activate its downstream signaling pathways (16, 17). The purpose of our study was to attempt to reconcile opposing views on the receptor binding and signaling properties of the proneurotrophins.
Here, we show that proNGF binds to p75NTR, but not TrkA. We have not addressed the role of sortilin in the binding of proNGF but Nykjaer et al. (15) have reported that HEK293T do not express detectable levels of sortilin and it therefore seems likely that the binding we detect is due solely to interactions with p75NTR. We cannot rule out the possibility that proNGF shows low affinity binding to TrkA at very high ligand concentrations but we did not observe binding of proNGF to TrkA at concentrations up to 12 nm, well beyond the physiological range. In examining signaling properties of proNGF in PC12 cells, the most robust effect observed was the ligand-induced phosphorylation of TrkA and sustained TrkA-dependent phosphorylation of Erk and Akt. Long term exposure to WT-proNGF resulted in neurite outgrowth and survival, indicating that proNGF treatment results in chronic TrkA activation in PC12 cells. Thus, our data are consistent with studies that indicate that proNGF selectively binds p75NTR expressed at the cell surface (9, 30) but also agrees with findings of others who have shown that proneurotrophin treatment results in Trk activation (16, 17).
Our results indicate that proteolytic processing of proNGF that occurs in the endosome is required for subsequent TrkA activation. We show that inhibition of furin blocks proNGF cleavage and proNGF-induced TrkA activation and that proNGF123, which is protease resistant, is incapable of activating the receptor. Furin is active in several cellular compartments, and although steady levels of furin are highest in the TGN, it is well established that furin is trafficked through the TGN and endosomes to reach the cell surface (31), and endocytic vesicles that contain furin can function as processing compartments (32–34). We used two methods to explore the possibility that endocytosis is a prerequisite for proNGF-induced activation of TrkA. Blocking endocytosis using either high sucrose concentration or reduced temperature sharply reduced proNGF-induced TrkA activation but did not block TrkA activation induced by mature NGF. We conclude, therefore, that endocytosis and cleavage of proNGF by furin-like enzymes are prerequisites required for TrkA activation.
During the course of our studies, Althaus and Klöppner (30) showed that proNGF is processed to mature NGF in pig oligodendrocytes through a furin-dependent pathway, similar to the findings shown here. We have demonstrated that proNGF-induced TrkA activation occurs within minutes, similar to the time frame reported for proNGF cleavage in pig oligodendrocytes (30), and it therefore seems likely that proNGF enters the cell through a rapid endocytic route. Rapid endocytosis of a p75NTR-proNGF complex was previously observed and is in fact a feature distinguishing it from the p75NTR-NGF complex, which enters the cell through a slower endocytic route (15, 21). Sortilin has been implicated in the rapid endocytosis of the p75NTR-proNGF complex as well as in vesicular sorting events, and it will be interesting to determine whether sortilin promotes delivery of proNGF to an intracellular compartment where furin-based cleavage occurs. Retrieval of furin and sortilin from endosomes to the trans-Golgi network both require direct binding to phosphofurin acidic cluster sorting protein-1 (PACS-1) (35, 36), and furin and sortilin may therefore share endosomal trafficking routes. Because p75NTR and sortilin function as proNGF co-receptors that induce apoptosis in a subset of cells, it seems likely that the biological response of cells exposed to proNGF will, in part, depend on the relative ratio of p75NTR, sortilin, furin, and TrkA.
Once processed from proNGF to NGF, the ligand could be transported to the cell surface to activate TrkA or it could activate TrkA within endocytic vesicles. There is precedence for ligand cleavage within endocytic vesicles that is followed by subsequent recycling and receptor activation (37), and activation of TrkA within an endosome seems possible, given earlier studies (24, 38). The subcellular location of TrkA activation may have a significant impact on the precise signaling events activated because TrkA induces Akt activation mainly at the plasma membrane, whereas TrkA-dependent Erk activation occurs primarily in endosomes (24, 25). By reducing recycling with brefeldin A, we found that proNGF-induced Erk activation was increased, whereas proNGF-induced Akt activation was decreased. Our interpretation of this result is that after cleavage, some of the NGF produced does activate TrkA in endosomes but that a portion of the liberated NGF is recycled and becomes available to activate TrkA at the cell surface.
Both proNGF and proBDNF are apoptotic ligands that activate p75NTR. ProBDNF may also have other functions, most notably in the central nervous system, where it has been proposed to bind p75NTR to induce long term depression and where activity-dependent cleavage of proBDNF by extracellular MMPs may regulate the availability of mature BDNF to modulate synaptic facilitation (29). We show that in primary cerebellar cells, which express both p75NTR and TrkB, proBDNF exposure results in TrkB activation and induces TrkB-dependent phosphorylation of Erk and Akt. Inhibitors of furin (dec-CMK) reduce this proBDNF-induced activity, suggesting that at least some of the TrkB activation result from intracellular cleavage of the ligand. However, BB94 also reduced TrkB activation induced by proBDNF, suggesting that cleavage of proBDNF by soluble matrix metalloproteases or cell surface ADAMs family members also mediate processing of this ligand.
Taken together, these studies indicate that proneurotrophin exposure results in Trk receptor activation and downstream signaling cascades. In accordance with previous findings, proNGF shows a strong preference for binding p75NTR at the cell surface compared with TrkA. We demonstrate that processing of the proneurotrophins to the mature ligand is a prerequisite for subsequent Trk receptor activation in both PC12 cells and primary CGNs. The precise processing mechanisms employed by PC12 cells and CGNs differ, where proNGF appears to be processed in PC12 cells mainly via furin-based cleavage in the endosome, whereas proBDNF processing in CGNs occurs via both MMPs and furin cleavage. This suggests that the physiological outcome of proneurotrophin exposure will be cell-type-specific and will reflect the levels of cell surface neurotrophin receptors and the availability of extracellular and endosomal proteases.
Supplementary Material
Acknowledgments
The secVSVg-Bgl-BAP and BirA plasmids were kind gifts from Michael Barry (Mayo Clinic, Rochester, NY).
This work was supported, in whole or in part, by National Institutes of Health Grant NS24380. This work was also supported by Grant MOP37850 from the Canadian Institute of Health Research (CIHR) and a grant from the United States Public Health Service. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: p75NTR, p75 neurotrophin receptor; NGF, nerve growth factor; proNGF, pro-form of nerve growth factor; AP-proNGF, alkaline phosphatase-tagged pro-nerve growth factor; MMP, matrix metalloprotease; TGN, trans-golgi network; DMEB, Dulbecco's modified eagle media supplemented with 0.1% bovine serum albumin; extracellular signal-regulated kinase; Erk, extracellular signal-regulated kinase; BDNF, brain-derived neurotrophic factor; BAP, biotin acceptor peptide; wt, wild-type; B-proNGF, biotinyated form of proNGF; CGN, cerebellar granule neurons.
References
- 1.Huang, E. J., and Reichardt, L. F. (2001) Annu. Rev. Neurosci. 24 677–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chao, M. V. (2003) Nat. Rev. Neurosci. 4 299–309 [DOI] [PubMed] [Google Scholar]
- 3.Roux, P. P., and Barker, P. A. (2002) Prog. Neurobiol. (Oxf.) 67 203–233 [DOI] [PubMed] [Google Scholar]
- 4.Seidah, N. G., Benjannet, S., Pareek, S., Chretien, M., and Murphy, R. A. (1996) FEBS Lett. 379 247–250 [DOI] [PubMed] [Google Scholar]
- 5.Paoletti, F., Konarev, P. V., Covaceuszach, S., Schwarz, E., Cattaneo, A., Lamba, D., and Svergun, D. I. (2006) Biochem. Soc. Trans. 34 605–606 [DOI] [PubMed] [Google Scholar]
- 6.Kliemannel, M., Golbik, R., Rudolph, R., Schwarz, E., and Lilie, H. (2007) Protein Sci. 16 411–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hauburger, A., Kliemannel, M., Madsen, P., Rudolph, R., and Schwarz, E. (2007) FEBS Lett. 581 4159–4164 [DOI] [PubMed] [Google Scholar]
- 8.Beattie, M. S., Harrington, A. W., Lee, R., Kim, J. Y., Boyce, S. L., Longo, F. M., Bresnahan, J. C., Hempstead, B. L., and Yoon, S. O. (2002) Neuron 36 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, R., Kermani, P., Teng, K. K., and Hempstead, B. L. (2001) Science 294 1945–1948 [DOI] [PubMed] [Google Scholar]
- 10.Hasan, W., Pedchenko, T., Krizsan-Agbas, D., Baum, L., and Smith, P. G. (2003) J. Neurobiol. 57 38–53 [DOI] [PubMed] [Google Scholar]
- 11.Harrington, A. W., Leiner, B., Blechschmitt, C., Arevalo, J. C., Lee, R., Morl, K., Meyer, M., Hempstead, B. L., Yoon, S. O., and Giehl, K. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 6226–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng, S., Wuu, J., Mufson, E. J., and Fahnestock, M. (2005) J. Neurochem. 93 1412–1421 [DOI] [PubMed] [Google Scholar]
- 13.Fahnestock, M., Michalski, B., Xu, B., and Coughlin, M. D. (2001) Mol. Cell. Neurosci. 18 210–220 [DOI] [PubMed] [Google Scholar]
- 14.Bierl, M. A., Jones, E. E., Crutcher, K. A., and Isaacson, L. G. (2005) Neurosci. Lett. 380 133–137 [DOI] [PubMed] [Google Scholar]
- 15.Nykjaer, A., Lee, R., Teng, K. K., Jansen, P., Madsen, P., Nielsen, M. S., Jacobsen, C., Kliemannel, M., Schwarz, E., Willnow, T. E., Hempstead, B. L., and Petersen, C. M. (2004) Nature 427 843–848 [DOI] [PubMed] [Google Scholar]
- 16.Fahnestock, M., Yu, G., Michalski, B., Mathew, S., Colquhoun, A., Ross, G. M., and Coughlin, M. D. (2004) J. Neurochem. 89 581–592 [DOI] [PubMed] [Google Scholar]
- 17.Fayard, B., Loeffler, S., Weis, J., Vogelin, E., and Kruttgen, A. (2005) J. Neurosci. Res. 80 18–28 [DOI] [PubMed] [Google Scholar]
- 18.Flanagan, J. G., and Cheng, H. J. (2000) Methods Enzymol. 327 198–210 [DOI] [PubMed] [Google Scholar]
- 19.Pagadala, P. C., Dvorak, L. A., and Neet, K. E. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 17939–17943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roux, P. P., Colicos, M. A., Barker, P. A., and Kennedy, T. E. (1999) J. Neurosci. 19 6887–6896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bronfman, F. C., Tcherpakov, M., Jovin, T. M., and Fainzilber, M. (2003) J. Neurosci. 23 3209–3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heuser, J. E., and Anderson, R. G. (1989) J. Cell Biol. 108 389–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howe, C. L., Valletta, J. S., Rusnak, A. S., and Mobley, W. C. (2001) Neuron 32 801–814 [DOI] [PubMed] [Google Scholar]
- 24.Howe, C. L., and Mobley, W. C. (2004) J. Neurobiol. 58 207–216 [DOI] [PubMed] [Google Scholar]
- 25.Zhang, Y., Moheban, D. B., Conway, B. R., Bhattacharyya, A., and Segal, R. A. (2000) J. Neurosci. 20 5671–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iglesias-Bartolome, R., Crespo, P. M., Gomez, G. A., and Daniotti, J. L. (2006) FEBS J. 273 1744–1758 [DOI] [PubMed] [Google Scholar]
- 27.Michalski, B., and Fahnestock, M. (2003) Brain Res. Mol. Brain Res. 111 148–154 [DOI] [PubMed] [Google Scholar]
- 28.Peters, E. M., Hendrix, S., Golz, G., Klapp, B. F., Arck, P. C., and Paus, R. (2006) J. Histochem. Cytochem. 54 275–288 [DOI] [PubMed] [Google Scholar]
- 29.Woo, N. H., Teng, H. K., Siao, C. J., Chiaruttini, C., Pang, P. T., Milner, T. A., Hempstead, B. L., and Lu, B. (2005) Nat. Neurosci. 8 1069–1077 [DOI] [PubMed] [Google Scholar]
- 30.Althaus, H. H., and Klöppner, S. (2006) J. Neurochem. 98 506–517 [DOI] [PubMed] [Google Scholar]
- 31.Thomas, G. (2002) Nat. Rev. Mol. Cell Biol. 3 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen, C. D., Huff, M. E., Matteson, J., Page, L., Phillips, R., Kelly, J. W., and Balch, W. E. (2001) EMBO J. 20 6277–6287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen, Y., Molloy, S. S., Thomas, L., Gambee, J., Bachinger, H. P., Ferguson, B., Zonana, J., Thomas, G., and Morris, N. P. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 7218–7223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Band, A. M., Maatta, J., Kaariainen, L., and Kuismanen, E. (2001) FEBS Lett. 505 118–124 [DOI] [PubMed] [Google Scholar]
- 35.Scott, G. K., Gu, F., Crump, C. M., Thomas, L., Wan, L., Xiang, Y., and Thomas, G. (2003) EMBO J. 22 6234–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott, G. K., Fei, H., Thomas, L., Medigeshi, G. R., and Thomas, G. (2006) EMBO J. 25 4423–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Borgne, R. (2006) Curr. Opin. Cell Biol. 18 213–222 [DOI] [PubMed] [Google Scholar]
- 38.Jeanneteau, F., and Chao, M. V. (2006) Novartis Found. Symp. 276 181–189; Discussion 189–192, 187–233, 181–275 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.