Abstract
TAR DNA-binding protein 43 (TDP-43) is the disease protein in frontotemporal lobar degeneration with ubiquitin-positive inclusions (FTLD-U) and amyotrophic lateral sclerosis (ALS). Although normal TDP-43 is a nuclear protein, pathological TDP-43 is redistributed and sequestered as insoluble aggregates in neuronal nuclei, perikarya, and neurites. Here we recapitulate these pathological phenotypes in cultured cells by altering endogenous TDP-43 nuclear trafficking and by expressing mutants with defective nuclear localization (TDP-43-ΔNLS) or nuclear export signals (TDP-43-ΔNES). Restricting endogenous cytoplasmic TDP-43 from entering the nucleus or preventing its exit out of the nucleus resulted in TDP-43 aggregate formation. TDP-43-ΔNLS accumulates as insoluble cytoplasmic aggregates and sequesters endogenous TDP-43, thereby depleting normal nuclear TDP-43, whereas TDP-43-ΔNES forms insoluble nuclear aggregates with endogenous TDP-43. Mutant forms of TDP-43 also replicate the biochemical profile of pathological TDP-43 in FTLD-U/ALS. Thus, FTLD-U/ALS pathogenesis may be linked mechanistically to deleterious perturbations of nuclear trafficking and solubility of TDP-43.
TAR DNA-binding protein 43 (TDP-43), encoded by the TARDBP gene on chromosome 1, is a highly conserved, ubiquitously expressed nuclear protein implicated in repression of gene transcription, inhibition of exon splicing, and interactions with splicing factors and nuclear bodies (1, 2). Recently, we identified TDP-43 as the disease protein forming insoluble aggregates in the central nervous system of patients with frontotemporal lobar degeneration (FTLD)2 and amyotrophic lateral sclerosis (ALS). Since FTLD patients often develop motor neuron disease consistent with ALS and since ALS patients can also develop cognitive impairment and FTLD, the presence of TDP-43 neuropathology in both disorders provides a molecular link connecting FTLD and ALS as a clinicopathological spectrum of the same neurodegenerative disorder (i.e. TDP-43 proteinopathy) (3-6).
FTLD includes a group of clinically, genetically and neuro-pathologically heterogeneous neurodegenerative disorders that account for ∼20% of presenile dementia (7-9). Although neurodegenerative tauopathies account for about 40% of familial and sporadic FTLD cases, TDP-43 is the major disease protein found within the ubiquitin-positive, tau- and α-synculein-negative inclusions that account for the majority of the FTLD cases (designated as FTLD-U) (4, 10). TDP-43 inclusions are also present in the spinal cord and brain of sporadic and familial ALS cases with the notable exception of familial ALS due to SOD-1 mutations (3-6).
TDP-43 neuropathology in FTLD-U and ALS is characterized by cytoplasmic, neuritic, and nuclear inclusions in neurons and glia (4, 11-13). We showed previously that the presence of cytoplasmic TDP-43 aggregates in disease neurons is accompanied by a dramatic clearance of normal TDP-43 staining, suggesting a redistribution of TDP-43 from the entire nucleus to a focal point adjacent to the nucleus (4, 13-15). Moreover, normal TDP-43 is found to be condensed as intranuclear inclusions mainly in familial FTLD with granulin (GRN) mutations and a rare disease linked to valosin-containing protein mutations (4, 14). Here we model TDP-43 cytoplasmic, neuritic, and nuclear inclusions in cultured cells and demonstrate that perturbation of endogenous TDP-43 trafficking between the nucleus and the cytoplasm leads to aggregate formation. Furthermore, the expression of mutant TDP-43 with defective nuclear localization (ΔNLS) or nuclear export signals (ΔNES) perturbs endogenous TDP-43 trafficking and recapitulates the unique TDP-43 pathologies that are signatures of the FTLD-U and ALS spectrum of disease. Our data implicate altered TDP-43 trafficking as a pathogenic mechanism underlying FTLD-U and ALS.
EXPERIMENTAL PROCEDURES
Constructs—cDNA encoding human TDP-43 (accession number NM 007375) in the plasmid pENTR-221 was obtained from Invitrogen. The addition of a Myc epitope tag to the 5′-end of TDP-43 was achieved by PCR, using the primers 5′-AAGCTTGATGGAACAAAAACTCATCTCGGAAGAGGATCTGTCTGAATATATTCGGGTAACC-3′ and 5′-TCTAGAGCTACATTCCCCAGCCAGAAGACTTAGA-3′. The PCR product was cloned into the pGEM-T vector (Promega, Madison, WI). Following sequence analysis, the PCR product was then subcloned into pcDNA 3.1 plasmid (Invitrogen) using restriction sites HindIII and XbaI, creating pcDNA 3.1/Myc-TDP-43. N-terminal FLAG-tagged cDNA encoding the longest mouse isoform of TDP-43 (pEFFLAG-TDP-43-L; accession number AY 145556) was kindly provided by Dr. C. K. Shen (Academia Sinica, Taipei, Taiwan). Site-directed mutations (see below) were generated using the plasmids pcDNA 3.1/Myc-TDP-43 (for human TDP-43) and pEFFLAG-TDP-43-L (for mouse TDP-43) as templates. N-terminal green fluorescent protein-tagged TDP-43 (GFP-TDP-43-WT) was generated by subcloning into the enhanced GFP (EGFP)-C1 vector (Clontech, Mountain View, CA).
Site-directed Mutagenesis of TDP-43—Site-directed mutagenesis (QuikChange kit; Stratagene, La Jolla, CA) was used to create sets of missense mutations for the current study (ΔNLS1, K82A/R83A/K84A; ΔNLS2, K95A/K97A/R98A; ΔNLS1/2, K82A/R83A/K84A/K95A/K97A/R98A; ΔNES1, I239A/L243A; ΔNES2, L248A/I249A/I250A). The sequences of the mutagenized oligonucleotides were as follows: ΔNLS1, 5′-CAACTATCCAAAAGATAACGCAGCAGCAATGGATGAGACAGATGC-3′; ΔNLS2, 5′-GCTTCATCAGCAGTGGCAGTGGCAGCAGCAGTCCAGAAAACATCC-3′; ΔNES1, 5′-GCAGATGATCAGGCTGCGCAGTCTGCTTGTGGAGAGGAC-3′; ΔNES2, 5′-CTTTGTGGAGAGGACGCGGCGGCTAAAGGAATCAGCG-3′. All constructs were subjected to sequence analysis, and the position of each mutation is shown in Fig. 3B.
FIGURE 3.
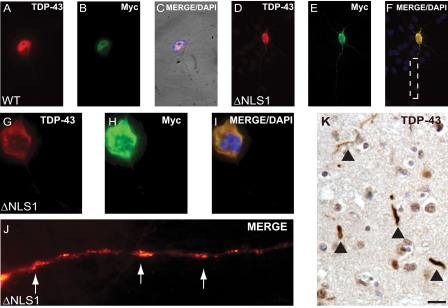
Expression of TDP-43-ΔNLS mutants led to sequestration of endogenous nuclear TDP-43. A, TDP-43 immunohistochemistry in a hippocampal section from a FTLD-U brain. Note clearing of nuclear TDP-43 (arrowheads) in neurons containing cytoplasmic TDP-43 inclusions, as compared with normal neurons (asterics). B, schematic diagram of N-terminal Myc-tagged TDP-43 protein highlighting the location of a bipartite NLS and NES. Both sets of basic aa (red) in NLS were mutated to generate three defective mutants (ΔNLS1, ΔNLS2, and ΔNLS1/2), and both sets of hydrophobic aa (blue) in NES were mutated to generate ΔNES1 and ΔNES2. C-K, double labeling of TDP-43 (red), Myc (green), and counterstaining with DAPI (blue) for nuclei. C-E, QBI-293 cells 72 h after transfection with Myc-TDP-43-WT (WT) and merged image (E) show colocalization of TDP-43 and Myc in nuclei of transfected cells. F-H, QBI-293 cells 24 h after transfection of Myc-TDP-43-ΔNLS1 (ΔNLS1). The merged image (H) shows colocalization of TDP-43 and Myc in the cytoplasm of transfected cells. I-K, QBI-293 cells 72 h after transfection of ΔNLS1. The merged image (K) shows colocalization of TDP-43 and Myc in the cytoplasm of transfected cells. Note the clearing of endogenous nuclear TDP-43 in cells expressing NLS1, as compared with nontransfected cells. Scale bars, 25 μm. *, nontransfected cells.
Antibodies—Commercial antibodies used in this study were as follows: rabbit polyclonal anti-TDP-43 antibody raised to amino acids (aa) 1-260 (Protein Tech Group, Chicago, IL), human specific mouse monoclonal antibody (mAb) raised to the same TDP-43 sequence (2E2-D3) (Abnova, Taipei, Taiwan), anti-MAP2 mAb (AP14) (14), anti-Myc mAb (9E10; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and anti-FLAG and anti-α-tubulin mAb (Sigma). Anti-heterogeneous nuclear ribonucleoprotein A1 mAb (4B10) and anti-heterogeneous nuclear ribonucleoprotein C1/C2 mAb (4F4) were generous gifts from Dr. G. Dreyfuss ((15). Two polyclonal antibodies were produced by immunizing rabbits (Covance Research Products Inc., Denver, PA). The first was an N terminus-specific antibody raised against a synthetic peptide near the N terminus of human TDP-43 corresponding to amino acid residues 6-24. The other is a C terminus-specific TDP-43 antibody raised against an extreme C-terminal synthetic peptide, corresponding to amino acid residues 394-414 of human TDP-43.3
Cell Culture and Transfection—QBI-293 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% l-glutamate. tsBN2 cells, a generous gift from Dr. Mary Dasso (National Institutes of Health, Bethesda, MD), were maintained at 33 °C (permissive temperature) or 39.5 °C (nonpermissive temperature) in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% l-glutamate as previously described (16). Primary cultures of mouse hippocampal neurons were prepared from embryonic day 17 C57/Bl6 mice as previously described (17). Briefly, dissociated cells were plated at a density of 50,000-100,000 cells/cm2 in poly-l-lysine glass bottom culture dishes (MatTak, Ashland, MA) and maintained in Neurobasal medium supplemented with B-27 (Invitrogen) and 0.5 mm l-glutamate. QBI-293 cells were transfected using the Amaxa Nucleofector (Amaxa Inc., Gaithersburg, MD) system and Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Primary neurons were transfected using Lipofectamine 2000 (Invitrogen) at 5-7 days in vitro according to the manufacturer's instructions. In some experiments, naive QBI-293 cells were treated with 50 μm leptomycin B (18) (Sigma) for 16 h.
Immunofluorescence Studies—Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline, permeabilized with 0.02 or 0.2% Triton X-100 (Sigma) in phosphate-buffered saline for 10 min, blocked with 5% powdered milk in phosphate-buffered saline for 2 h, and incubated overnight with primary antibody at 4 °C. Primary antibodies were visualized with secondary antibodies conjugated with Alexa Fluor 488 and Alexa Fluor 594 (Vector Laboratories, Burlingame, CA), and nuclei were detected using DAPI. To demonstrate the presence of TDP-43 aggregates, cells were extracted with 0.2% Triton X-100 prior to fixation to remove detergent-soluble pools of TDP-43. All cells were analyzed using a Nikon TE-2000-E (Nikon, Tokyo, Japan), and images were captured using a CoolSnap-HQ camera (Photometrics, Tuscon, AZ). All micrographs show individual cells representative of the total cell population. Immunohistochemistry on tissue sections from FTLD-U cases using rabbit anti-TDP-43 antibody was conducted as previously described (4).
Solubility and Biochemical Analysis—To examine the solubility profile of TDP-43, sequential extractions were performed. Cells were washed twice with phosphate-buffered saline, lysed in cold RIPA buffer, and sonicated. Cell lysates were cleared by centrifugation at 100,000 × g for 30 min at 4 °C to generate the RIPA-soluble samples. To prevent carry-overs, the resulting pellets were washed twice (i.e. resonicated and recentrifuged). Only the supernatants from the first centrifugation were analyzed. RIPA-insoluble pellets were then extracted with urea buffer (7 m urea, 2 m thiourea, 4% CHAPS, 30 mm Tris, pH 8.5), sonicated, and centrifuged at 100,000 × g for 30 min at 22 °C. Protease inhibitors were added to all buffers prior to use (1 mm PMSF and a mixture of protease inhibitors). Protein concentration was determined by the bicinchoninic acid method (Pierce), and proteins were resolved by 10% SDS-PAGE and transferred to nitrocellulose membranes. Following transfer, nitrocellulose membranes were blocked in 5% powdered milk and incubated in primary antibody overnight at 4 °C. Primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA), and blots were developed with Renaissance Enhanced Luminol Reagents (PerkinElmer Life Sciences). Digital images were acquired using a Fuji Film Intelligent Darkbox II (Fuji Systems, Stamford, CT).
Immunoprecipitation—Transfected and nontransfected cells were lysed in urea buffer as described above and then diluted 10-fold in 50 mm Tris, pH 8.0, containing 1% Triton X-100. Lysates were preabsorbed with Protein A/G-agarose (Santa Cruz Biotechnology) and immunoprecipitated with polyclonal TDP-43 antibody conjugated to Protein A/G-agarose. Immunoprecipitated proteins were eluted with SDS sample buffer (10 mm Tris, pH 6.8, 1 mm EDTA, 40 mm dithiothreitol, 1% SDS, and 10% sucrose), separated by 10% SDS-PAGE, and analyzed by immunoblot as described previously (4).
RESULTS
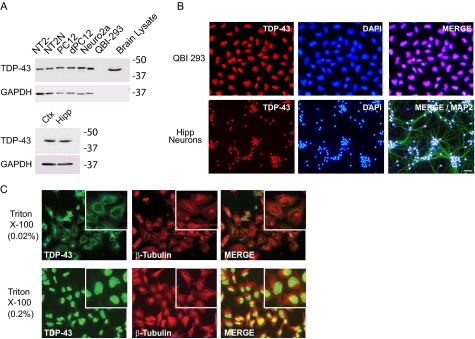
TDP-43 is ubiquitously expressed in neuronal and nonneuronal cells as well as in the brain of multiple species (e.g. rats, mice, and humans) (Fig. 1A). Although TDP-43 predominately localizes to the nucleus of nonneuronal cells (QBI-293) and primary hippocampal neurons (Fig. 1B), low levels of TDP-43 are present in the cytoplasm. Pretreatment of cells with 0.02% Triton X-100, which permeabilizes the plasma membrane but not the nuclear membrane (19), enabled detection of cytoplasmic TDP-43 (Fig. 1C).
FIGURE 1.
Cell biology of TDP-43 expression. A, immunoblot analysis of TDP-43 expression from RIPA extracts of undifferentiated (NT2-) and retinoic acid-treated differentiated neuronal (NT2N) human teratocarcinoma cells; undifferentiated (PC12) and neural growth factor-treated differentiated neural (dPC12) rat pheochromocytoma cells; mouse neuroblastoma (Neuro2a) cells; human embryonic kidney (QBI-293) cells; and Sarkosyl-extracted lysate from human brain (top) and from primary mouse neurons isolated from cortex (Ctx) and hippocampus (Hipp) (bottom). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control (bottom). No glyceraldehyde-3-phosphate dehydrogenase was detected in the Sarkosyl-soluble human brain lysate due to prior extraction with less stringent buffers. B, immunofluorescence detection of endogenous TDP-43 in QBI-293 cells (red), DAPI (blue), and merged images (top) and in primary mouse hippocampal neurons (bottom). In addition, MAP2 (green in the merged image) was used to mark neuronal perikarya and dendrites. Scale bar, 20 μm. C, detection of cytoplasmic (permeabilized with 0.02% Triton X-100; top) or total (permeabilized with 0.2% Triton X-100; bottom) TDP-43 (green), β-tubulin (red), and merged images in QBI-293 cells.
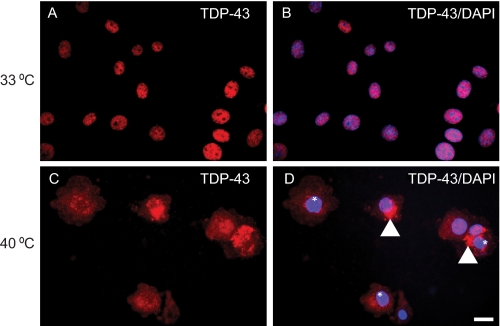
To determine if TDP-43 translocates from the cytoplasm to the nucleus, we used a BHK21-derived cell line (tsBN2) that harbors a temperature-sensitive point mutation in the RCC1 (regulator of chromosome condensation 1) gene (16, 20, 21). At the permissive temperature (33 °C), tsBN2 cells function normally, but at the nonpermissive temperature (39.5 °C), RCC1 rapidly loses its activity, nuclear Ran-GTP redistributes to the cytoplasm, and as a result, nuclear protein import is blocked. At 33 °C, the expression of endogenous TDP-43 localized to the nucleus (Fig. 2, A and B). However, at the nonpermissive temperature of 39.5 °C, endogenous TDP-43 was detected in the cytoplasm in association with the clearance of nuclear TDP-43 and formation of punctuate cytoplasmic aggregates (Fig. 2, C and D). This was specific to tsBN2 cells, since no alteration in the nuclear localization of TDP-43 was observed in QBI-293 cells cultured at 33 or 39.5 °C (data not shown). Thus, inhibition of TDP-43 translocation into nuclei leads to the accumulation and sequestration of TDP-43 as cytoplasmic aggregates.
FIGURE 2.
TDP-43 expression and accumulation in cytoplasm of temperature-sensitive BN2 cells. A and B, endogenous TDP-43 (red) was detected in the nucleus of tsBN2 cells at 33 °C (permissive temperature). The merge image (B) of TDP-43 (red) and nuclei stain DAPI (blue) confirms that TDP-43 is in the cell nucleus. C and D, at nonpermissive temperature (39.5 °C), TDP-43 (red) was detected as punctate cytoplasmic aggregates (arrowheads) with clearing of nuclear TDP-43 (*) and did not colocalize with DAPI (blue) in tsBN2 cells. D, merged image. Scale bars, 20 μm.
Previous studies in post-mortem brain and spinal cord sections of FTLD-U and ALS cases showed a dramatic clearance of nuclear TDP-43 in neurons with TDP-43 cytoplasmic aggregates (4, 14, 15) (Fig. 3A). To examine if formation of these aggregates leads to the sequestration of nuclear TDP-43 in the cytoplasm, we developed a cell model to study the translocation of nuclear TDP-43 to the cytoplasm. We identified a specific bipartite NLS sequence (i.e. two clusters of basic residues separated by a stretch of 9-12 residues), located at aa residues 82-98 in both human and mouse TDP-43, that is predicted to be required for nuclear targeting (Fig. 3B). To demonstrate that these sequences are required for TDP-43 entry into the nucleus, three defective ΔNLS mutants were generated: 1) ΔNLS1, basic aa Lys-Arg-Lys (residues 82-84) mutated to Ala-Ala-Ala; 2) ΔNLS2, basic aa Lys (residue 95) and Lys-Arg (residues 97 and 98) mutated to Ala; 3) ΔNLS1/2, both ΔNLS1 and ΔNLS2 mutated to Ala (Fig. 3B). WT-TDP-43 and the TDP-43 mutants were N-terminally Myc-tagged (designated as Myc-TDP-43-WT and Myc-TDP-43-NLS) to facilitate the identification of the transgenes independently from the endogenous protein.
Transient transfection of Myc-TDP-43-WT into QBI-293 cells showed complete colocalization with endogenous TDP-43 at 24 and 72 h (Fig. 3, C-E). By contrast, transfection of all Myc-TDP-43-ΔNLS mutants resulted in cytoplasmic expression (Figs. 3, F-H, and S2), confirming that alteration in the basic residues within the specific bipartite NLS sequence impairs nuclear targeting of TDP-43. Similar cytoplasmic localization of all three Myc-TDP-43-ΔNLS mutants, as well as untagged TDP-43-ΔNLS mutants, was observed when expressed in HeLa, Chinese hamster ovary, or Neuro 2a cells (data not shown).
Although endogenous TDP-43 was detectable in the nucleus of QBI-293 cells 24 h post-transfection with TDP-43-ΔNLS mutants (Figs. 3, F-H, and S1, A-C and G-I), it was virtually absent from the nucleus after 72 h (Figs. 3, I-K, and S1, D-F and J-L). To investigate this further, we co-expressed GFP-tagged WT-TDP-43 (GFP-TDP-43-WT) with either Myc-TDP-43-WT or Myc-TDP-43-ΔNLS1. GFP-TDP-43-WT colocalized in the nucleus with Myc-TDP-43-WT (Fig. 4, A-C) and similarly localized to the nucleus when coexpressed with Myc-TDP-43-ΔNLS1 at 24 h (Fig. 4, D-F). However, 72 h post-transfection, GFP-TDP-43-WT shifted to the cytoplasm and colocalized with Myc-TDP-43-ΔNLS1 72 h post-transfection, thereby confirming the abnormal effects of TDP-43-ΔNLS on trafficking of WT endogenous TDP-43 between nucleus and cytoplasm (Fig. 4, G-L).
FIGURE 4.
Cytoplasmic expression of human and mouse TDP-43-ΔNLS results in nuclear clearance of TDP-43. A-C, QBI-293 cells 72 h post-transfection with GFP-TDP-43-WT and Myc-TDP-43-WT. Both GFP and TDP-43 (red) were only detected in the nucleus of transfected cells. D-L, QBI-293 cells 24 h (D-F) or 72 h (G-I) post-transfection with GFP-TDP-43-WT and ΔNLS1. GFP-TDP-43-WT was localized solely to the nucleus (green), and the merged image (F) shows no colocalization of Myc-TDP-43-WT (red) and GFP (green) in cytoplasm 24 h post-transfection. Nuclei were labeled with DAPI (blue). G-L, 72 h post-transfection, GFP-TDP-43-WT was detected in cytoplasm and colocalized with ΔNLS1 (I-L). Note punctate colocalization (yellow) of ΔNLS1 and GFP-TDP-43-WT in the cytoplasm of transfected cells. M-O, QBI-293 cells 72 h after transfection with mouse FLAG-TDP-43-mWT (mWT) and double-labeled with a human-specific mouse anti-TDP-43 (green) and rabbit anti-FLAG antibody (red) with merged image (O) showing expression of endogenous human TDP-43 and FLAG-TDP-43-mWT in nuclei of transfected cells. Note the decreased levels of endogenous human nuclear TDP-43 in cells expressing FLAG-TDP-43-mWT when compared with nontransfected cells (*). P-R, QBI-293 cells 72 h after transfection with mouse FLAG-TDP-43-ΔNLS1/2 (ΔmNLS1/2). The merged image (R) shows the presence of endogenous human TDP-43 (green) and FLAG-TDP-43-ΔNLS1/2 (red) in the cytoplasm of transfected cells. Note that there is clearing of endogenous human nuclear TDP-43 in cells expressing FLAG-TDP-43-ΔNLS1/2 (arrowhead) as compared with nontransfected cells (*). Scale bars, 20 μm.
To further demonstrate the cytoplasmic sequestration of endogenous nuclear TDP-43 by TDP-43-ΔNLS mutants, we expressed FLAG-tagged mouse TDP-43 with the ΔNLS1/2 mutation (FLAG-TDP-43-ΔmNLS1/2) in QBI-293 cells and demonstrated cytoplasmic sequestration of endogenous human TDP-43 using a human-specific TDP-43 antibody (Fig. 4, M-R). Thus, nuclear depletion of TDP-43 in QBI-293 cells recapitulates the nuclear clearing of this protein in cells harboring TDP-43 cytoplasmic inclusions in ALS and FTLD-U cases (Fig. 3A). This nuclear clearance was specific to TDP-43, since no changes in the distribution of other nuclear proteins, such as heterogeneous nuclear ribonucleoprotein A1 and heterogeneous nuclear ribonucleoprotein C1/C2, were observed (Fig. S2).
The redistribution and sequestration of endogenous TDP-43 were also examined in primary hippocampal neurons transfected with Myc-TDP-43-WT or Myc-TDP-43-ΔNLS1. Although Myc-TDP-43-WT localized in the nucleus (Fig. 5, A-C), transfection with Myc-TDP-43-ΔNLS1 resulted in cytoplasmic and axonal accumulation of Myc-TDP-43 (Fig. 5, D-F). This was associated with the clearance of endogenous nuclear TDP-43 (Fig. 5, G-I) and neuritic aggregates of TDP-43 (Fig. 5J) that showed remarkable verisimilitude to TDP-43 pathology in disease neurons of FTLD-U cases (Figs. 3A and 5K) (22).
FIGURE 5.
Expression of TDP-43-ΔNLS mutants led to aggregate formation in neuronal perikarya and neurites. Mouse hippocampal neurons were transfected with WT (A-C) or ΔNLS1 (D-F) TDP-43 constructs. Note the colocalization of TDP-43 and Myc in the nucleus (A-C) and soma (D-F) of neurons. G-I, higher magnification of a neuron expressing ΔNLS1. The merged image (J) shows colocalization of TDP-43 and Myc in the cytoplasm of a transfected neuron. Note the clearing of endogenous nuclear TDP-43 in I. J, high magnification image of axon in F marked by a dotted box. Neuritic accumulations of TDP-43 present in neurons expressing ΔNLS1 (arrows) are similar to neuritic pathology observed in FTLD-U brains (K). K, immunostaining of neuritic pathology (arrowheads) from frontal cortex of a FTLD-U brain. Scale bars, 20 μm.
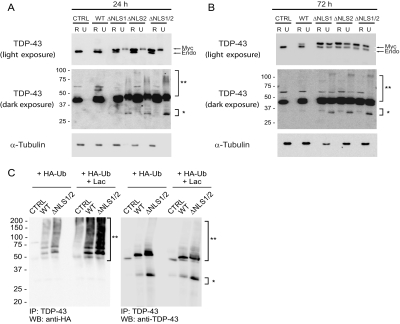
Next, we asked if the expression of Myc-TDP-43-ΔNLS mutants and the sequestration of endogenous TDP-43 lead to the formation of insoluble aggregates composed of both proteins. Immunoblot analysis was conducted on cells transfected with Myc-TDP-43-WT or Myc-TDP-43-ΔNLS mutants harvested at 24 and 72 h post-transfection and sequentially extracted with RIPA and urea buffer. Both endogenous and Myc-TDP-43-WT were recovered exclusively in the RIPA fractions (Fig. 6A). Since Myc-TDP-43 migrated slower than endogenous TDP-43, each was detected separately in immunoblots. Although relatively low levels of nuclear Myc-TDP-43-WT were detected at both 24 and 72 h post-transfection, cytoplasmic expression of all three Myc-TDP-43-ΔNLS mutants were robust at 24 h, resulting in the partial accumulation of the mutant protein in the urea fractions (Fig. 6A). Moreover, darker exposure of the immunoblot revealed the presence of a high Mr smear and C-terminal fragments in the urea fractions that resemble the ubiquitinated TDP-43 high Mr smear as well as the C-terminal fragments seen in FTLD-U and ALS cases (4, 14, 15). N-terminal truncation of TDP-43 in these cells was confirmed with the use of antibodies specific for the N and C termini of TDP-43 (Fig. S3). By 72 h post-transfection, endogenous TDP-43 was also detected in the urea fractions along with the Myc-TDP-43-ΔNLS mutants, consistent with sequestration of endogenous nuclear TDP-43 in the insoluble fraction (Fig. 6B). Again, high Mr smears and C-terminal fragments were also detected 72 h post-transfection.
FIGURE 6.
Expression of Myc-TDP-43-ΔNLS mutants results in the sequestration of ubiquitinated and insoluble endogenous TDP-43. QBI-293 cells 24 h (A), or 72 h (B) post-transfection with empty vector (CTRL), Myc-TDP-43-WT (WT), Myc-TDP-43-ΔNLS1 (ΔNLS1), Myc-TDP-43-ΔNLS2 (ΔNLS2), or Myc-TDP-43-ΔNLS1/2 (ΔNLS1/2) sequentially extracted with RIPA (R) and urea buffer (U). Immunoblotting was conducted with TDP-43 antibody. Myc-TDP-43 (Myc) migrates slower than endogenous TDP-43 (Endo). Over-exposure of the immunoblot demonstrates the presence of a high Mr smear (**) and C-terminal fragments (*) in the urea fractions of Myc-TDP-43-NLS mutants in transfected cells. α-Tubulin was used as a loading control. C, immunoblots (IB) of immunoprecipitated (IP) QBI-293 cell lysates cotransfected with empty vector, Myc-TDP-43-WT, or Myc-TDP-43-ΔNLS1/2 and HA-tagged ubiquitin (HA-Ub) in the presence (+LAC) or absence of LAC. Note the presence of the LAC-dependent, ubiquitinated TDP-43 positive high-Mr smear and the TDP-43 C-terminal fragments.
To demonstrate unequivocally that pathological TDP-43 in high Mr smears is ubiquitinated and a substrate for proteasomal degradation, Myc-TDP-43-WT or Myc-TDP-43-ΔNLS1/2 was cotransfected with HA-tagged ubiquitin and incubated with or without lactacystin (LAC), a proteasome inhibitor. Immunoprecipitation of the cell lysates with TDP-43 antibodies followed by immunoblotting with anti-HA antibody revealed a ladder and high Mr smear of ubiquitinated TDP-43 species, the abundance of which was enhanced by LAC treatment (Fig. 6C). Endogenous TDP-43 was also ubiquitinated when cells transfected only with HA-tagged ubiquitin construct were treated with LAC, suggesting that normal TDP-43 is degraded by the proteasome. Finally, to visualize the presence of insoluble aggregates in the cytoplasm, cells expressing TDP-43-ΔNLS1 were extracted with 0.2% Triton X-100, and two-color immunofluorescence revealed punctate cytoplasmic aggregates (Fig. S4). Thus, multiple complementary approaches enabled us to demonstrate that increased cytosolic TDP-43, either by direct expression in the cytoplasm or by abrogating nuclear import, leads to the sequestration of nuclear TDP-43 in the cytoplasm as insoluble aggregates that replicate the features of TDP-43 pathology in FLTD-U and ALS.
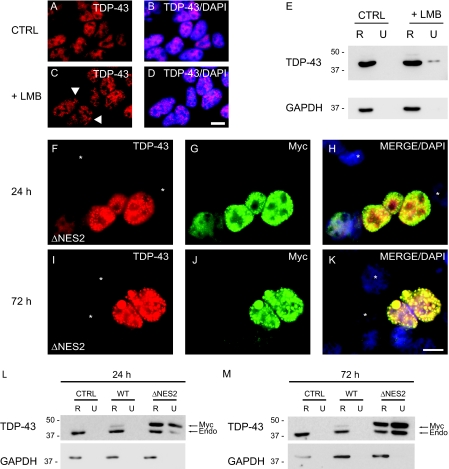
Since TDP-43 also accumulates as intranuclear inclusions in FTLD-U and ALS, we asked if perturbation of nuclear TDP-43 by restricting its export leads to the formation of insoluble TDP-43 nuclear inclusions. Treatment of QBI-293 cells with leptomycin B (LMB), a Streptomyces spp. metabolite that specifically inhibits NES-dependent nuclear export (18), resulted in the rearrangement of endogenous nuclear TDP-43 and induced formation of small punctate TDP-43 nuclear inclusions (Fig. 7, A-D). This was correlated with the presence of urea-soluble but RIPA-insoluble endogenous TDP-43 in LMB-treated but not untreated cells (Fig. 7E).
FIGURE 7.
TDP-43 is sequestered as insoluble nuclear inclusions by restricting nuclear export. A-D, immunofluorescence of endogenous TDP-43 (red) alone (A and C) or merged (B and D) with DAPI (blue) in QBI-293 cells treated with DMSO vehicle (CTRL) (A and B) or LMB (+LMB) (C and D). The arrowheads identify punctuate nuclear inclusions in C. E, QBI-293 cells were treated with DMSO vehicle or LMB and were sequentially extracted with RIPA (R) and urea buffer (U). Immunoblotting was conducted with anti-TDP-43 antibody. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. F-K, double labeling with TDP43 (red), Myc (green) antibodies, and DAPI (blue). QBI-293 cells 24 h (F-H) and 72 h (I-K) after transfection with Myc-TDP-43-ΔNES2 (ΔNES2) and merge images (H and K) show colocalization of TDP-43 (red) and Myc (green) in nuclei of transfected cells. Note the presence of punctate inclusions in the nucleus of transfected cells at both 24 and 72 h. Nontransfected cells are marked withanasterisk.LandM, QBI-293 cells 24 h (L) or 72 h (M)post-transfection with empty vector(CTRL), Myc-TDP-43-WT(WT), or Myc-TDP-43-ΔNES2 (ΔNES2) and sequentially extracted with RIPA (R) and urea buffer (U). Immunoblotting was conducted with TDP-43 antibody. Myc-TDP-43 (Myc) migrates slower than endogenous TDP-43 (Endo), and both Myc-TDP-43-ΔNES2 and endogenous TDP-43 were recovered in the RIPA-insoluble and urea-soluble fraction. Glyceraldehyde-3-phosphate dehydrogenase was used as a loading control. Scale bars, 20μm.
To further demonstrate that increased nuclear TDP-43 leads to formation of insoluble nuclear aggregates, we used bioinformatics to identify a leucine-rich NES located at aa 239-250 in human TDP-43 (Fig. 3B) and then determined if this predicted NES sequence is required for TDP-43 export from the nucleus. Two N-terminally Myc-tagged TDP-43 mutants with defective NES sequences were generated: 1) ΔNES1, hydrophobic aa Leu (residue 239) and Ile (residue 243) mutated to Ala; 2) ΔNES2, hydrophobic aa Leu-Ile-Ile (residues 248-250) mutated to Ala-Ala-Ala (Fig. 3B). Although expressed Myc-TDP-43-ΔNES was localized to the nucleus, fine granular intranuclear inclusions were detected in a subset of transfected cells at 24 and 72 h (Fig. 7, F-K). Immunoblot analyses confirmed the presence of insoluble nuclear aggregates composed of both endogenous TDP-43 and Myc-TDP-43-ΔNES, since both Myc-TDP-43-ΔNES and endogenous TDP-43 proteins were recovered from the urea fractions at 24 and 72 h post-transfection (Fig. 7, L and M). Thus, these data suggest that altered TDP-43 nuclear exporting sequences lead to the formation of insoluble nuclear aggregates containing both mutant and endogenous TDP-43.
DISCUSSION
In this study, multiple approaches were used to demonstrate that disruption of TDP-43 nuclear trafficking and localization induced formation of insoluble aggregates that recapitulate features of pathological TDP-43 inclusions in human FTLD-U/ALS cases. First, the disturbance of endogenous TDP-43 distribution resulted in the formation of TDP-43 aggregates in the cytoplasm and nucleus. Second, overexpression of TDP-43 either in the cytoplasm or nucleus also led to the formation of insoluble aggregates in the cytoplasm and nucleus, respectively. Third, insoluble TDP-43 was ubiquitinated and N-terminally truncated. Finally, endogenous TDP-43 was sequestered within cytoplasmic and nuclear aggregates. Thus, these and other data support the hypothesis that perturbation of the delicate balance in trafficking of TDP-43 between nucleus and cytoplasm can lead to the formation of TDP-43 aggregates that recapitulate features of TDP-43 signature lesions of FTLD-U and ALS.
Mutagenesis studies confirmed the presence of functional NES and NLS sequences in TDP-43, since expression of mutant NES and NLS in TDP-43 restricted its distribution to the nucleus and cytoplasm, respectively. We found that both basic motifs within the NLS sequence are independently required for the import of TDP-43 into the nucleus, since alteration of basic residues in either motifs led to its accumulation in the cytoplasm. Similarly, both hydrophobic motifs within the NES sequence are also independently required for TDP-43 export, since modification of either motif prevented its exit to the cytoplasm. Moreover, expression of TDP-43-ΔNES led to the formation of nuclear inclusions composed of endogenous and mutant proteins.
Under physiological conditions, low levels of TDP-43 are found in the cytoplasm, but expression of NLS mutants led to sequestration of endogenous TDP-43 in the cytoplasmic aggregates presumably by preventing newly synthesized TDP-43 being imported into the nucleus and/or by inhibiting the re-entry of existing cytoplasmic TDP-43 into the nucleus.
Moreover, the accumulation of cytoplasmic TDP-43 aggregates also recapitulated the biochemical signature of pathological TDP-43 in FTLD-U and ALS, including a high Mr smear of ubiquitinated TDP-43 and C-terminal TDP-43 fragments. Currently, it is unclear if this cleavage occurs in the nucleus or the cytoplasm, but it is tempting to speculate that TDP-43 C-terminal fragments serve as a nidus for the aggregation and sequestration of TDP-43 into nuclear and/or cytoplasmic inclusions. Further, since most TDP-43 inclusions are cytoplasmic, it is possible that increased trafficking of full-length TDP-43 from the nucleus to the cytoplasm could occur through other pathogenic mechanisms, leading to the accumulation of cytoplasmic TDP-43 aggregates.
Since the cellular functions of TDP-43 are currently not well understood, greater insight into the cellular biology of TDP-43, under both physiological and pathogenic conditions, is needed to further our understanding of this newly identified neurodegenerative proteinopathy and to develop new and more effective therapies for these disorders. To that end, the data reported here are significant in that they implicate aberrant nuclear trafficking and altered solubility of TDP-43 in mechanisms underlying FTLD-U and ALS.
Supplementary Material
Acknowledgments
We thank C. Li for technical assistance.
This work was supportred, in whole or in part, by National Institutes of Health Grants AG17586 and AG10124. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S4.
Footnotes
The abbreviations used are: FTLD, frontotemporal lobar degeneration; FTLD-U, FTLD with ubiquitin-positive inclusions; ALS, amyotrophic lateral sclerosis; LAC, lactacystin; LMB, leptomycin B; NES, nuclear export signal; NLS, nuclear localization signal; GFP, green fluorescent protein; aa, amino acid(s); mAb, monoclonal antibody; DAPI, 4′,6-diamidino-2-phenylindole; RIPA, radioimmune precipitation; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; HA, hemagglutinin.
L. M. Igaz, L. K. Kwong, Y. Xu, A. C. Truax, K. Uryu, M. Neumann, C. M. Clark, L. B. Elman, B. L. Miller, M. Grossman, L. F. McCluskey, J. Q. Trojanowski, and V. M.-Y. Lee, submitted for publication.
References
- 1.Ayala, Y. M., Pantano, S., D'Ambrogio, A., Buratti, E., Brindisi, A., Marchetti, C., Romano, M., and Baralle, F. E. (2005) J. Mol. Biol. 348 575-588 [DOI] [PubMed] [Google Scholar]
- 2.Wang, H. Y., Wang, I. F., Bose, J., and Shen, C. K. (2004) Genomics 83 130-139 [DOI] [PubMed] [Google Scholar]
- 3.Davidson, Y., Kelley, T., Mackenzie, I. R., Pickering-Brown, S., Du Plessis, D., Neary, D., Snowden, J. S., and Mann, D. M. (2007) Acta Neuropathol. 113 521-533 [DOI] [PubMed] [Google Scholar]
- 4.Neumann, M., Sampathu, D. M., Kwong, L. K., Truax, A. C., Micsenyi, M. C., Chou, T. T., Bruce, J., Schuck, T., Grossman, M., Clark, C. M., McCluskey, L. F., Miller, B. L., Masliah, E., Mackenzie, I. R., Feldman, H., Feiden, W., Kretzschmar, H. A., Trojanowski, J. Q., and Lee, V. M.-Y. (2006) Science 314 130-133 [DOI] [PubMed] [Google Scholar]
- 5.Arai, T., Hasegawa, M., Akiyama, H., Ikeda, K., Nonaka, T., Mori, H., Mann, D., Tsuchiya, K., Yoshida, M., Hashizume, Y., and Oda, T. (2006) Biochem. Biophys. Res. Commun. 351 602-611 [DOI] [PubMed] [Google Scholar]
- 6.Tan, C. F., Eguchi, H., Tagawa, A., Onodera, O., Iwasaki, T., Tsujino, A., Nishizawa, M., Kakita, A., and Takahashi, H. (2007) Acta Neuropathol. 113 535-542 [DOI] [PubMed] [Google Scholar]
- 7.Grossman, M. (2002) J. Int. Neuropsychol. Soc. 8 566-583 [DOI] [PubMed] [Google Scholar]
- 8.McKhann, G. M., Albert, M. S., Grossman, M., Miller, B., Dickson, D., and Trojanowski, J. Q. (2001) Arch. Neurol. 58 1803-1809 [DOI] [PubMed] [Google Scholar]
- 9.Neary, D., Snowden, J. S., Gustafson, L., Passant, U., Stuss, D., Black, S., Freedman, M., Kertesz, A., Robert, P. H., Albert, M., Boone, K., Miller, B. L., Cummings, J., and Benson, D. F. (1998) Neurology 51 1546-1554 [DOI] [PubMed] [Google Scholar]
- 10.Seelaar, H., Jurgen Schelhaas, H., Azmani, A., Kusters, B., Rosso, S., Majoor-Krakauer, D., de Rijik, M. C., Rizzu, P., Brummelhuis, M. T., van Doorn, P. A., Kamphorst, W., Willemsen, R., and van Swieten, J. C. (2007) Brain 130 1375-1385 [DOI] [PubMed] [Google Scholar]
- 11.Neumann, M., Kwong, L. K., Truax, A. C., Vanmassenhove, B., Kretzschmar, H. A., Van Deerlin, V. M., Clark, C. M., Grossman, M., Miller, B. L., Trojanowski, J. Q., and Lee, V. M.-Y. (2007) J. Neuropathol. Exp. Neurol. 66 177-183 [DOI] [PubMed] [Google Scholar]
- 12.Cairns, N. J., Neumann, M., Bigio, E. H., Holm, I. E., Troost, D., Hatanpaa, K. J., Foong, C., White, C. L., III, Schneider, J. A., Kretzschmar, H. A., Carter, D., Taylor-Reinwald, L., Paulsmeyer, K., Strider, J., Gitcho, M., Goate, A. M., Morris, J. C., Mishra, M., Kwong, L. K., Stieber, A., Xu, Y., Forman, M. S., Trojanowski, J. Q., Lee, V. M.-Y., and Mackenzie, I. R. (2007) Am. J. Pathol. 171 227-240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackenzie, I. R., Bigio, E. H., Ince, P. G., Geser, F., Neumann, M., Cairns, N. J., Kwong, L. K., Forman, M. S., Ravits, J., Stewart, H., Eisen, A., McClusky, L., Kretzschmar, H. A., Monoranu, C. M., Highley, J. R., Kirby, J., Siddique, T., Shaw, P. J., Lee, V. M.-Y., and Trojanowski, J. Q. (2007) Ann. Neurol. 61 427-434 [DOI] [PubMed] [Google Scholar]
- 14.Geisert, E. E., Jr., Johnson, H. G., and Binder, L. I. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 3967-3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi, Y. D., and Dreyfuss, G. (1984) J. Cell Biol. 99 1997-2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohtsubo, M., Kai, R., Furuno, N., Sekiguchi, T., Sekiguchi, M., Hayashida, H., Kuma, K., Miyata, T., Fukushige, S., Murotsu, T., Ohtsubo, M., Kai, R., Furuno, N., Sekiguchi, T., Sekiguchi, M., Hayashida, H., Kuma, K., Miyata, T., Fukushige, S., Murotsu, T., Matsubara, K., and Nishimoto, T. (1987) Genes Dev. 1 585-593 [DOI] [PubMed] [Google Scholar]
- 17.Banker, G., and Goslin, K. (1998) Culturing Nerve Cells, 2nd Ed., pp. 398-425, MIT Press, Cambridge, MA
- 18.Kudo, N., Wolff, B., Sekimoto, T., Schreiner, E. P., Yoneda, Y., Yanagida, M., Horinouchi, S., and Yoshida, M. (1998) Exp. Cell Res. 242 540-547 [DOI] [PubMed] [Google Scholar]
- 19.Peeples, M. E. (1988) Virology 162 255-259 [DOI] [PubMed] [Google Scholar]
- 20.Boche, I., and Fanning, E. (1997) J. Cell Biol. 139 313-325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cornett, J., Cao, F., Wang, C. E., Ross, C. A., Bates, G. P., Li, S. H., and Li, X. J. (2005) Nat. Genet. 37 198-204 [DOI] [PubMed] [Google Scholar]
- 22.Sampathu, D. M., Neumann, M., Kwong, L. K., Chou, T. T., Micsenyi, M., Truax, A., Bruce, J., Grossman, M., Trojanowski, J. Q., and Lee, V. M.-Y. (2006) Am. J. Pathol. 169 1343-1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.