Abstract
Hepatic steatosis is often associated with insulin resistance and obesity and can lead to steatohepatitis and cirrhosis. In this study, we have demonstrated that hormone-sensitive lipase (HSL) and adipose triglyceride lipase (ATGL), two enzymes critical for lipolysis in adipose tissues, also contribute to lipolysis in the liver and can mobilize hepatic triglycerides in vivo and in vitro. Adenoviral overexpression of HSL and/or ATGL reduced liver triglycerides by 40–60% in both ob/ob mice and mice with high fat diet-induced obesity. However, these enzymes did not affect fasting plasma triglyceride and free fatty acid levels or triglyceride and apolipoprotein B secretion rates. Plasma 3-β-hydroxybutyrate levels were increased 3–5 days after infection in both HSL- and ATGL-overexpressing male mice, suggesting an increase in β-oxidation. Expression of genes involved in fatty acid transport and synthesis, lipid storage, and mitochondrial bioenergetics was unchanged. Mechanistic studies in oleate-supplemented McA-RH7777 cells with adenoviral overexpression of HSL or ATGL showed that reduced cellular triglycerides could be attributed to increases in β-oxidation as well as direct release of free fatty acids into the medium. In summary, hepatic overexpression of HSL or ATGL can promote fatty acid oxidation, stimulate direct release of free fatty acid, and ameliorate hepatic steatosis. This study suggests a direct functional role for both HSL and ATGL in hepatic lipid homeostasis and identifies these enzymes as potential therapeutic targets for ameliorating hepatic steatosis associated with insulin resistance and obesity.
Nonalcoholic fatty liver disease (NAFLD)4 is often associated with obesity, insulin resistance, and metabolic syndrome (1, 2). Nonalcoholic steatohepatitis (NASH), the more virulent form of NAFLD, can lead to cirrhosis. Current treatments for subjects with NAFLD are usually directed at alleviating the associated metabolic symptoms of the patients (3). Insulin sensitizers such as thiazolidinediones or metformin improve insulin sensitivity with concomitant reduction of liver fat contents in human and mouse models (3–5). The amelioration of hepatic steatosis by these agents is likely secondary to improved insulin sensitivity. Imbalances between the input, oxidation, synthesis, and output of fatty acids (FA) all could contribute to hepatic steatosis, and dysregulation of each pathway has been documented in animal models (6). For example, leptin-deficient ob/ob mice are insulin-resistant, dyslipidemic, and have fatty livers despite the up-regulation of FA oxidation genes (7, 8) and increases in mitochondrial and peroxisomal β-oxidation (9). Hepatic steatosis in these animals is attributed to the up-regulation of sterol-responsive element-binding protein (SREBP) 1c, a master regulator of lipogenesis (10), and the consequent increase in de novo lipogenesis (11, 12). FA uptake in the liver is also likely increased as genes involved in FA uptake and transport (e.g. CD36) are up-regulated in these animals (13). NAFLD has also been found in animals with specific genetic defects in FA oxidation (e.g. peroxisome proliferator-activated receptor (PPAR) α deficiency), lipoprotein production (e.g. truncated apolipoprotein B (apoB) proteins or deficiency of microsomal triglyceride transfer protein), fatty acid synthesis (e.g. hepatic SREBP1a overexpression), and FA uptake (e.g. CD36 deficiency) (see review in Ref. 14). In this study, we explored whether the two major adipose lipases, hormone-sensitive lipase (HSL) and adipose triglyceride lipase (ATGL), could remove excess TG from fatty livers in mouse models exhibiting insulin resistance and obesity.
HSL and ATGL together account for >90% of cytosolic TG hydrolase activity in mouse adipose tissues (15). HSL-mediated lipolysis in adipose tissues is tightly regulated by hormones (16). Catecholamines stimulate, whereas insulin inhibits, HSL-mediated lipolysis via the cAMP signaling pathway. Hormone-stimulated HSL hydrolytic action is also regulated by its binding to perilipin A, a lipid droplet-associated protein (16). HSL has broad substrate specificity and can hydrolyze TG, diacylglycerol (DG), and monoacylglycerol as well as cholesteryl esters (CE) (17) and retinyl esters (18). Although normally present at very low levels in the liver, HSL transcripts are up-regulated in livers overexpressing PPARγ (19). Previous studies have shown no clear role for HSL in lipolysis in the liver.
Unlike HSL, ATGL does not have CE hydrolase activity and acts specifically on TG (20–22). Its ability to hydrolyze phospholipids (PL) in vitro appears to vary depending upon the type of assay used (20, 21). Regulation of ATGL in adipose tissue is distinct from that of HSL (23). Unlike HSL, which is translocated from cytosol to lipid droplets only upon hormonal stimulation, ATGL is associated with the lipid droplet in the basal and hormone-stimulated states of the cell (20). In the basal condition, comparative gene identification protein 58 (CGI-58, also called abhydrolase domain containing 5), which is a coactivator of ATGL, is associated with the perilipin A-containing lipid droplet. Activation of cAMP-dependent protein kinase A increases colocalization of ATGL to CGI-58 (24). ATGL is also found at low levels in a variety of tissues (20). Humans with an ATGL mutation have cardiomyopathy and exhibit the characteristics of neutral lipid storage disease, including systemic accumulation of TG in cytoplasmic droplets (25). Ablation of ATGL in mice increases TG mass not only in adipose but also in other tissues, particularly in the heart (26). Hepatic TG was also increased in ATGL-deficient mice. This raises the possibility that ATGL may have a direct role in hepatic lipid homeostasis.
In this study, we determined the role of HSL and ATGL in hepatic lipolysis, and we evaluated the potential of adenoviral overexpression of these enzymes as a means to ameliorate hepatic steatosis in ob/ob mice (27) and mice with diet-induced insulin resistance and obesity.
EXPERIMENTAL PROCEDURES
Generation of Recombinant Adenoviruses—Full-length cDNA fragments of mouse HSL (2.3 kb) and ATGL (1.5 kb) were amplified from adipose cDNA isolated from C57BL/6 (B6) mice and cloned into a pCRII vector using a TA cloning kit as described (28). Upon verification of DNA sequences, the cDNA inserts were then cloned into a NotI site of the pAdTrack-CMV shuttle vector as described previously (29). The shuttle vector also contains jellyfish green fluorescent protein (GFP) that is driven by a second cytomegalovirus promoter. The resultant plasmids (containing either HSL, ATGL, or no insert) were linearized by PmeI digestion and subsequently transformed into Escherichia coli BJ5183 cells containing an adenoviral backbone plasmid, pAdEasy-1 (Stratagene), to generate recombinant plasmids. Recombinant plasmids were isolated, and DNA was purified, linearized, and transfected into 293 cells (ATCC) to obtain viral plaques. Recombinant viruses were further amplified in 293 cells and purified by centrifugation on a cesium chloride gradient. Purified viral particles were titered and stored in a 50% glycerol, 10 mm Tris, 100 mm NaCl, 0.1% bovine serum albumin (BSA) solution at -70 °C. Purified viruses were dialyzed against 3% sucrose, 10 mm Tris, 2 mm MgCl2 solution in 10,000 molecular weight cutoff Slide-A-Lyzer dialysis cassettes (Pierce) overnight prior to use for in vivo infection of animals. Recombinant adenoviruses containing HSL or ATGL are designated AdHSL and AdATGL, respectively. The control recombinant adenovirus that expresses GFP only is designated AdGFP.
Cell Culture and Adenoviral Infection—McA-RH7777 cells, obtained from ATCC, were grown in 6-well plates coated with collagen in Dulbecco's modified Eagle's medium (DMEM) containing penicillin (100 units/ml), streptomycin (100 μg/ml), 10% fetal bovine, and 10% horse serum. All reagents for cell culture were purchased from Invitrogen. Recombinant viruses were titered to yield >90% infection efficiency based on the percentage of cells expressing GFP observed under a fluorescence microscope. For all cell experiments, seeded McA-RH7777 cells were infected with recombinant adenoviruses in DMEM for 90 min and then incubated in the growth medium described above. Twenty four hours after viral infection, cells were washed with phosphate-buffered saline (PBS) and either replenished with the growth medium or switched to a conditioned medium composed of DMEM with either 0.4 mm oleate (OA) (Sigma), which was complexed to 1.5% FFA-free BSA (Sigma), or with 1.5% BSA only.
Mice—Animals were maintained on a 12-h light/dark cycle (light cycle: 7 a.m. to 7 p.m.). All mice were fed a chow diet (PicoLab Rodent Chow, number 5001) unless otherwise indicated. Wild-type B6 and FVB/N (FVB) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). HSL-deficient and ATGL-deficient mice were generated by targeted homologous recombination as described (26, 30). These animals (3–4 months of age) were sacrificed between 8 and 12 a.m., and liver tissues were collected for use in TG hydrolase activity assays. Congenic ob/ob mice of the FVB background were derived as described previously (27). Experiments were performed using both male and female mice ranging from 3 to 5 months of age. Liver samples from one set of ob/ob mice (both male and female) and their lean littermate controls (n = 5–7/group) were collected and subjected to RNA isolation and gene expression analysis. For infection experiments, another set of ob/ob mice were used. These mice (n = 4–8/group) were infected with recombinant adenoviruses (1 × 109 plaque-forming units/mouse) via tail vein injection. Female mice were infected with AdGFP (n = 6), AdHSL (n = 4), or AdATGL (n = 8). Eight days after infection, two animals each from the AdGFP- and AdATGL-infected groups were sacrificed for tissue collection and histological analysis, and the remaining animals (n = 4–6 per group) were subjected to in vivo TG and apoB secretion assays. Male ob/ob mice were infected with either AdGFP (n = 4) or AdHSL (n = 5). These mice were subjected to an in vivo TG secretion assay 5 days after infection and sacrificed for tissue collection 10 days after infection. For glucose measurements, blood was obtained by tail bleed from unanesthetized mice and immediately analyzed for glucose concentration using a One-Touch Ultra glucometer (LifeScan). For the measurement of other aspects of plasma biochemistry, blood was sampled before and at various time points after viral infection via the retroorbital plexus. All blood samples were collected from animals after 4 h of fasting (8 a.m. to 12 p.m.) to minimize the effects of variable food consumption on lipid and glucose levels. Plasma samples were isolated by centrifugation at 4 °C, quick-frozen, and stored at -70 °C.
For infection experiments in diet-induced obese (DIO) mice, 6-week-old male B6 mice were fed a Western-type diet (WTD, Harlan Teklad, TD-88137) for 6 weeks and then subjected to infection experiments (AdGFP, n = 8 or AdATGL, n = 9) as described above. The WTD consisted of 21% (w/w) fat (polyunsaturated/saturated = 0.07), 0.15% (w/w) cholesterol, and 19.5% casein.
Quantitative Real Time PCR (qPCR)—Total cellular RNA samples from either McA-RH7777 cells or various mouse tissues (5 μg/sample) were subjected to first strand cDNA synthesis using oligo(dT) primers and a reverse transcriptase from a commercial kit (number 11904-018, Invitrogen). The resulting cDNA samples were then quantified for expression levels of each test gene using gene-specific primers. The primers for mouse SREBP1c used in qPCR are as described previously (31). The primers for all other genes used in the qPCR assays are shown in Table 1. The primer sequences used for HSL and ATGL are shared by mice and rats and thus detect expression of the endogenous transcripts from McA-RH7777 cells. SYBR Green PCR Master Mix (Stratagene) was used for the reactions according to the manufacturer's instructions. The detection of PCR products was performed in duplicate using the Mx3005P Multiplex Quantitative PCR system (Stratagene). Using the standard curve method, the relative amounts of specific PCR products of each target gene were calculated. For normalization, β-actin (or GAPDH) was also amplified from each sample.
TABLE 1.
Primer sequences for quantitative real time PCR
| Gene symbol | Alias used | Primer (strand) | Primer sequence (5′ → 3′) | PCR size | GenBank™ no. |
|---|---|---|---|---|---|
| bp | |||||
| Acox1 | AOXa | 5′ | TCA ACA GCC CAA CTG TGA CTT CCA TTA | 227 | NM_015729 |
| 3′ | TCA GGT AGC CAT TAT CCA TCT CTT CA | ||||
| Actb | β-Actin | 5′ | GTA TCC ATG AAA TAA GTG GTT ACA GG | 143 | NM_007393 |
| 3′ | GCA GTA CAT AAT TTA CAC AGA AGC AAT | ||||
| Actb, rat | β-Actin | 5′ | TTA CTG CCC TGG CTC CTA GC | 170 | NM_031144 |
| 3′ | TTT GCG GTG CAC GAT GGA GG | ||||
| Adfp | ADRP | 5′ | GCA TCG GCT ACG ACG ACA CC | 210 | NM_007408 |
| 3′ | CAG CAT TGC GGA ATA CGG AG | ||||
| Cd36 | CD36 | 5′ | ATT GGT CAA GCC AGC T | 265 | NM_007643 |
| 3′ | TGT AGG CTC ATC CAC TAC | ||||
| Cpt1a | CPT1a | 5′ | CCA GGC TAC AGT GGG ACA TT | 209 | NM_013495 |
| 3′ | GAA CTT GCC CAT GTC CTT GT | ||||
| Fabp1 | L-Fabp | 5′ | TGG ACC CAA AGT GGT CCG CA | 150 | NM_017399 |
| 3′ | AGT TCA GTC ACG GAC TTT AT | ||||
| Fasn | FAS | 5′ | GGT CGT TTC TCC ATT AAA TTC TCA T | 225 | NM_007988 |
| 3′ | CTA GAA ACT TTC CCA GAA ATC TTC C | ||||
| Gapdh | GAPDH | 5′ | ATG ACA TCA AGA AGG TGG TG | 177 | NM_008084 |
| 3′ | CAT ACC AGG AAA TGA GCT TG | ||||
| Lipe | HSLa | 5′ | GCT GGG CTG TCA AGC ACT GT | 160 | NM_010719 |
| 3′ | GTA ACT GGG TAG GCT GCC AT | ||||
| Pnpla2 | ATGLa | 5′ | TGT GGC CTC ATT CCT CCT AC | 158 | NM_025802 |
| 3′ | TCG TGG ATG TTG GTG GAG CT | ||||
| Ppara | PPARαa | 5′ | ATC CAC GAA GCC TAC C | 312 | NM_011144 |
| 3′ | CAC ACC GTA CTT TAG CAA G | ||||
| Scd1 | SCD1 | 5′ | TCG CCC CTA CGA CAA GAA CA | 190 | NM_009127 |
| 3′ | CCG GTC GTA AGC CAG GCC CA | ||||
| Ucp2 | UCP2 | 5′ | CAG AGC ACT GTC GAA GCC TA | 120 | NM_011671 |
| 3′ | GTA TCT TTG ATG AGG TCA TA |
Primer sequences from mouse genes cross-react with rat genes
Western Blot Analysis—Mouse tissues or McA-RH7777 cells were isolated, homogenized in RIPA lysis buffer (sc-24948, Santa Cruz Biotechnology), incubated on ice for 30 min, and centrifuged at 10,000 × g for 10 min. The supernatants were then collected for determination of protein concentration using BCA reagents (Pierce). Total cell or tissue lysates (30–50 μg/sample) were electrophoresed using 10% SDS-PAGE. Proteins were then transferred onto polyvinylidene fluoride membranes (Bio-Rad). Membranes were incubated at room temperature for 1 h with blocking buffer (PBS, pH 7.4, 5% nonfat milk, and 0.1% Tween 20). Membranes were then incubated with specific primary antibody in the blocking buffer. Rabbit polyclonal antibodies used were as follows: anti-HSL (ab45422, Abcam), anti-ATGL (10006409, Cayman), and anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH; ab9485, Abcam). After 2 h of incubation, membranes were washed in 10 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 0.1% Tween 20. Membranes were then incubated with a secondary antibody conjugated to horseradish peroxidase (Bio-Rad) for 1 h. Membranes were washed again and incubated with SuperSignal West Femto Chemiluminescence Substrate (Pierce) for 2 min prior to exposure to x-ray films for 1 s to 10 min at room temperature. The x-ray films were scanned, and the relative signal for each protein band was quantified using the NIH ImageJ program (version 1.38) after calibrating the program to convert pixels to optical density.
Determination of Specific Lipase Activities in Cell and Tissue Lysates—For cell lysates, McA-RH7777 cells were collected 40 h after infection in PBS, pelleted by centrifugation at 200 × g for 2 min, and frozen until ready for assays. To prepare the substrate emulsion, 7.5 mmol of triolein or cholesterol oleate was combined with 2 μCi of [3H]triolein or [14C]cholesterol oleate (PerkinElmer Life Sciences) and 0.6 mg of phosphatidylcholine. This mixture was then dried under nitrogen, resuspended in 1.5 ml of assay buffer (20 mm Tris, 150 mm NaCl, 1 mm EDTA), and sonicated twice for 1 min. 1.2 ml of assay buffer and 0.3 ml of a 20% FFA-free BSA solution were then added, and the mixture was sonicated again four times for 30 s each. The cell pellet was resuspended in 2 ml of homogenization buffer (assay buffer containing 0.2% FFA-free BSA and 1 mm DTT), and cells were lysed using a mechanical homogenizer (Brinkmann Instruments). 100 μl each of the cell homogenate and the substrate emulsion were combined and incubated with shaking at 37 °C for 2 h. Lipolysis was terminated by the addition of 3.25 ml of stop solution (10:9:7 methanol/chloroform/heptane solution containing 12 mm palmitate) and 1 ml of an aqueous pH 10 buffer (SB116-1, Fisher). The solution was centrifuged at 600 × g for 5 min. 1 ml of the supernatant was removed and counted for radioactivity in 10 ml of liquid scintillation mixture. 100 μl of the substrate emulsion was also counted in triplicate to determine the percentage of substrate hydrolyzed. The final substrate concentration was 0.25 mm of triolein or cholesterol oleate per assay. The total amount of triolein or cholesterol oleate hydrolyzed was calculated and adjusted to the protein concentration of the cellular homogenate to yield specific lipase activity in nanomoles of FFA produced per mg of cellular protein/h.
For determination of TG hydrolase activity in tissue lysates, liver tissues of wild-type, HSL-deficient, and ATGL-deficient male mice were homogenized in buffer A (0.25 m sucrose, 1 mm EDTA, 1 mm dithiothreitol, 20 mg/ml leupeptin, 2 mg/ml anti-pain, 1 mg/ml pepstatin, pH 7.0) and then centrifuged at 100,000 × g for 1 h. The lipid-free infranatant (cytosolic fraction) was collected and used for TG hydrolase assays using a previously described procedure (15) similar to assays described above for cell lysates. In brief, liver extracts (200 μg of protein) in a total volume of 100 μl of buffer A were incubated with 100 μl of substrate in a water bath at 37 °C for 60 min in the presence or absence of the HSL inhibitor (NNC0076–0000-0079, Novo Nordisk, Denmark). To increase ATGL-mediated TG hydrolysis, all reactions were carried out in the presence of 400 μg of glutathione S-transferase (GST)-tagged murine CGI-58 (GST-CGI-58), which was purified as described previously (32). The final substrate concentration used per assay was 0.3 mm triolein.
Measurement of Cellular TG Mass—McA-RH7777 cells were washed with PBS 40 h post-infection, and lipids were extracted with hexane/isopropyl alcohol (3/2, v/v) (4 ml/well) at room temperature for 3 h as described (33). The extraction solution was collected, and the remaining cellular proteins were collected in 1 ml of 0.1 n NaOH. The lipid extract was dried under nitrogen gas and resuspended in a buffer from a Trig/GB kit from Roche Diagnostics, and cellular TG mass was determined using this kit. Cellular protein mass was determined using BCA protein assay reagents from Pierce. Cellular TG concentration is expressed as micrograms of TG/mg of cellular protein.
Assessment of Blood Biochemistry—Aspartate aminotransferase (AST) and alanine aminotransferase (ALT) activities in the plasma were assessed using Sigma assay kits 505 and 505-P, respectively. Activity levels are expressed in IU/ml plasma. Plasma TG and total cholesterol (TC) concentrations were measured using Infinity™ colorimetric assay kits from Thermo Electron Corp. FFA and 3-β-hydroxybutyrate (3-HB) levels were measured using the NEFA and Autokit 3-HB colorimetric assay kits, respectively, from Wako Chemicals. Plasma insulin concentrations were measured using an Ultrasensitive Rat Insulin ELISA kit and mouse insulin standards from Crystal Chem.
Liver Lipid Measurements—Total liver lipids were extracted according to a modified method from Folch et al. (34). Briefly, snap-frozen liver tissues (∼100 mg) were homogenized in 5 ml of 1 n NaOH and extracted twice with 5–10 ml of a chloroform/methanol (v/v = 2:1) solution. The organic layer was dried under nitrogen gas and resolubilized in 1 ml of chloroform containing 2% Triton X-100. This extract was dried again and resuspended in 1 ml of water to achieve a final concentration of 2% Triton X-100 (35) and then assayed for TG, FFA, and TC concentration using commercial kits as described above.
Histological Analysis—Liver tissue samples were either embedded in Tissue-Tek optimal cutting temperature compound (Sakura Finetek) and sectioned (7 μm) for neutral lipid staining using Oil Red O and a hematoxylin counterstain or fixed in 10% formalin, embedded in paraffin, and sectioned (5 μm) for hematoxylin and eosin staining. Sections were photographed at either ×100 (hematoxylin and eosin) or ×200 (Oil Red O) magnification.
Determination of in Vivo ApoB and TG Secretion Rates—Assessment of apoB and TG secretion rates in animals infected with recombinant adenoviruses (n = 4–6/group) was performed as described previously (36). For the determination of apoB secretion rates, 4-h fasted mice were injected via tail vein with a solution containing 200 μCi of 35S-labeled Promix (SJQ0079, Amersham Biosciences) and 500 mg/kg Triton WR 1339 (Sigma) in 0.9% NaCl. Blood was taken at 0 (just prior to injection), 60, and 120 min after the injection. Plasma samples (10 μl) were subjected to 4% SDS-PAGE followed by fluorography. Both B100 and B48 bands were cut from dried gels and counted in a liquid scintillation counter. B100 and B48 protein counts were normalized to trichloroacetic acid-precipitable counts in each plasma sample and expressed as protein count per 10 μl of plasma (cpm/10 μl) as previously described (36). ApoB secretion rates (cpm/10 μl plasma/hr) were calculated by subtracting normalized protein counts at the 60-min time point from normalized protein counts at the 120-min time point.
The Triton WR1339 method described above was also employed in the absence of 35S-labeled Promix to determine TG secretion rates. Mice were bled at 0 min (before injection) and 60 and 120 min after injection. Plasma samples from the 0-, 60-, and 120-min time points were measured for TG levels. The TG secretion rate was calculated by subtracting the TG level at the 60-min time point from the TG level at the 120-min time point and expressed as mg/dl/h.
Cellular FA Oxidation—McA-RH7777 cells were infected with recombinant adenoviruses for 24 h and then washed with PBS prior to labeling in DMEM (1 ml/well) containing 0.4 mm OA, 1.5% BSA, and [14C]OA (1 μCi/ml) (NEC317, PerkinElmer Life Sciences) for 16 h. Assessment of FA oxidation products was performed as described (37) with modification. Briefly, the labeling medium was collected and centrifuged, and the supernatant was transferred to a 25-ml flask with a center well (882320-0000, Kontes) containing filter paper saturated with 100 μl of 1 m KOH. After the flask was sealed with a stopper, 200 μl of 70% perchloric acid was added to the medium sample to release the [14C]CO2. The flask was then rocked at 37 °C for 1 h. The saturated filter paper containing trapped [14C]CO2 was assessed for radioactivity in a liquid scintillation counter. The acidified medium was centrifuged twice to remove particulate matter, and 200 μl of supernatant was counted to assess the amount of 14C-labeled acid-soluble metabolites (ASM), which include labeled ketone bodies. Cellular lipids were extracted and separated by TLC as described below.
Extraction of Labeled Cellular and Medium Lipids—At the end of cell labeling experiments, medium was collected; cells were washed with PBS, and lipids were extracted with hexane/isopropyl alcohol (3:2, v/v) (4 ml/well) at room temperature for 3 h. The extraction solution was dried under nitrogen gas, and the remaining cellular proteins were collected in 0.1 n NaOH and then measured. Medium lipids were extracted with 20 volumes of chloroform/methanol (2:1, v/v) at room temperature for 2 h. These organic extracts were combined with 5 volumes of water and then centrifuged for 5 min at 2,500 rpm. The lower organic phase was transferred to a new tube and dried under nitrogen gas. All lipids were resuspended in chloroform/methanol to a final volume of 400 μl. Cellular and medium samples were applied to Silica Gel 60 plates (EMD Chemicals) for TLC analysis. Lipids were separated using a hexane/diethyl ether/glacial acetic acid (70:30:1 or 80:20:2 v/v/v for DG and TG separation) mobile phase, and the lipid spots were visualized with iodine vapor and analyzed by liquid scintillation counting.
FA Secretion—McA-RH7777 cells were infected with recombinant adenoviruses for 24 h and then washed with PBS prior to labeling in DMEM (1 ml/well) containing 0.4 mm OA, 1.5% BSA, and 10 μCi of [3H]OA (NET289, PerkinElmer Life Sciences) for 16 h. Cells were then washed three times with PBS and incubated with a chase medium containing either 1.5% BSA only or 0.4 mm OA, 1.5% BSA in the presence of 2 μmol/liter tetrahydrolipstatin (THL) (Roche Diagnostics) dissolved in dimethyl sulfoxide (Sigma). The chase medium was removed either immediately (0 min) after application or after a 120- or 240-min incubation at 37 °C. Lipids were extracted from both collected chase medium and cells and then analyzed by TLC as described above.
Statistical Analysis—Comparisons between two groups for plasma AST or ALT activity levels in infected ob/ob mice were conducted using a nonparametric test, the Mann-Whitney U test, whereas comparisons between two groups for all other experiments were performed using Student's t test (nonpaired and two-tailed). All values are presented as means ± S.D. Statistically significant differences between two groups were defined as those giving a value of p < 0.05.
RESULTS
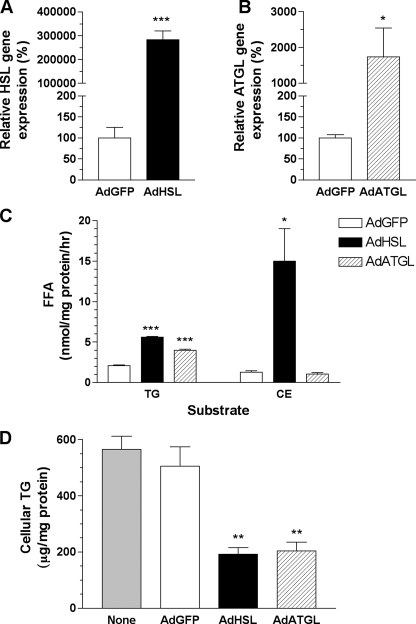
Both HSL and ATGL Reduce Cellular TG Mass in OA-supplemented McA-RH7777 Cells—To determine whether overexpression of HSL or ATGL could remove excess lipids in fatty livers, we generated recombinant adenoviruses containing either HSL or ATGL. Their lipolytic activities were first tested in a rat hepatoma cell line, McA-RH7777, prior to in vivo studies. McA-RH7777 cells were infected with AdHSL, AdATGL, or AdGFP control for 24 h followed by OA supplementation for an additional 16 h prior to harvesting. HSL mRNA and protein levels were increased >2000- and ∼18-fold, respectively, following AdHSL infection in McA-RH7777 cells (Figs. 1A and 2A). TG and CE hydrolase activities in AdHSL-infected cells were increased by 2.6- and 11-fold, respectively, compared with those in AdGFP-infected cells (Fig. 1C). ATGL mRNA and protein levels were increased by 17- and ∼20-fold (over background), respectively, after AdATGL infection (Fig. 1B and Fig. 2A). TG hydrolase activity in AdATGL-infected cells was increased by 1.9-fold compared with that in AdGFP-infected cells (Fig. 1C). Cellular TG mass was reduced by ∼60% in AdHSL-infected (192 ± 23 μg/mg protein) and AdATGL-infected (204 ± 31 μg/mg protein) cells compared with that in AdGFP-infected controls (505 ± 70 μg/mg protein) or uninfected cells (565 ± 47 μg/mg protein) (Fig. 1D). Because these data confirmed that HSL and ATGL were functionally active in cultured liver cells, we proceeded to test whether these enzymes exerted similar effects in vivo.
FIGURE 1.
Adenoviral overexpression of HSL and ATGL and its effect on cellular lipids in McA-RH7777 cells. Cells were infected with AdHSL, AdATGL, or AdGFP for 24 h, followed by OA (0.4 mm, 1.5% BSA) supplementation for an additional 16 h. A and B, total cellular RNA samples (n = 6 wells/group) were isolated and assessed for HSL (A) or ATGL (B) expression by qPCR. HSL or ATGL gene expression levels were normalized to β-actin expression. Expression of either gene in AdHSL- or AdATGL-infected cells is expressed as a percentage of expression (±S.D.) in AdGFP-infected cells. C, cell lysates (n = 3 wells/group) were assessed for TG or CE hydrolase activities using substrate emulsions containing [3H]triolein or [14C]cholesterol oleate, respectively. Measurements were performed in triplicate. Results shown are representative of two experiments. Hydrolase activity is assessed as the average amounts of FFA (±S.D.) produced and expressed in nanomoles/mg of protein/h. D, cellular TG were extracted and assessed by an enzymatic method. Cellular TG are presented as mean ± S.D. microgram/mg protein. Experiments were repeated twice, and each was performed in triplicate. Data from one of the experiments are shown. Comparisons were made against AdGFP-infected cells using Student's t test. p values: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
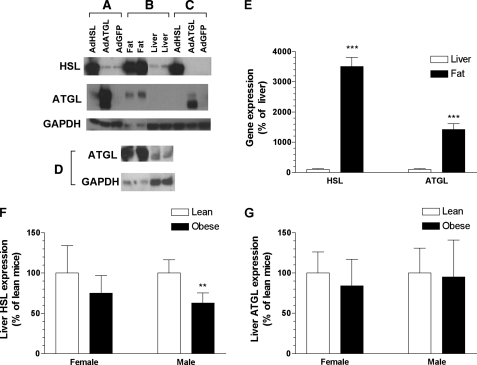
FIGURE 2.
Protein and gene expression of HSL and ATGL in McA-RH7777 cells and mouse tissues. A–D, Western blot analysis of cell lysates from infected McA-RH7777 cells (A), tissues lysates from wild-type mice (B and D), and liver lysates from infected female ob/ob mice (C). For the epididymal fat samples shown in D, 30 μg of protein were used, whereas 50 μg of protein were used for all other samples. Rabbit polyclonal antibodies were used to detect HSL (A–C, top), ATGL (A–C, middle; D, top), and GAPDH (A–D, bottom). GAPDH was used for normalization. Epididymal fat or liver tissues were collected from either B6 mice (B and 2nd and 4th lanes of D) or FVB mice (1st and 3rd lanes of D). The lower molecular weight band detected by anti-ATGL antibody was always present in cells and mice infected with AdATGL and specific to these samples. This band likely represents modified or degraded ATGL. E, gene expression of HSL or ATGL in liver or adipose tissues from male B6 wild-type (n = 5/group) was assessed by qPCR. HSL and ATGL gene expression levels were normalized to GAPDH expression and presented as a percentage of liver gene expression. F and G, gene expression of HSL (F) and ATGL (G) in the liver of male and female FVB-ob/ob mice and their lean littermates (n = 5–7/group) was assessed by qPCR. HSL and ATGL gene expression levels were normalized to β-actin and presented as a percentage of lean levels for each sex. All data are shown as means ± S.D. Comparisons between groups were made using Student's t test. p values: **, p < 0.01; ***, p < 0.001.
Endogenous HSL and ATGL Expression and TG Hydrolase Activities in Mouse Livers—To establish a base line for normal expression levels of ATGL and HSL as well as to determine the degree of overexpression induced by AdATGL and AdHSL infection, we first quantified the endogenous protein and mRNA levels of HSL and ATGL in liver and adipose tissues from wild-type B6 mice. When similar amounts of total protein were subjected to Western blot analysis, the relative abundances of HSL and ATGL proteins were ∼14- and ∼6-fold higher, respectively, in adipose tissues than those in the liver (Fig. 2, B and D). When normalized to GAPDH protein levels, HSL protein levels were ∼48-fold higher, and ATGL levels were ∼24-fold higher in adipose tissues than those in the liver. HSL and ATGL mRNA levels, when normalized to GAPDH gene expression, were 35- and 14-fold higher, respectively, in adipose tissues than those in the liver (Fig. 2E). Although large differences in GAPDH gene expression make it difficult to quantitatively compare expression levels between tissues, our assays show low but detectable levels of HSL and ATGL protein and mRNA in the liver. To determine whether transcription of either gene is altered by leptin deficiency, HSL and ATGL mRNA levels were measured in both male and female ob/ob mice as well as in lean littermate controls. Hepatic HSL mRNA levels were unchanged in female ob/ob mice but were reduced by 37% in male ob/ob mice compared with their lean littermates (Fig. 2F). Hepatic ATGL mRNA levels were similar between ob/ob mice and their lean littermates in both sexes (Fig. 2G).
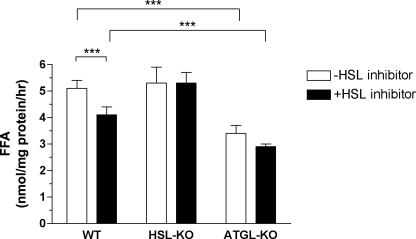
We further assessed the contribution of HSL and ATGL to cytosolic TG lipolysis in the liver by in vitro assays using liver extracts from wild-type, HSL-deficient, and ATGL-deficient male mice (Fig. 3). Compared with that in wild-type mice, TG hydrolase activity was reduced by 33% in ATGL-deficient mice. TG hydrolase activity was not altered in HSL-deficient mice, possibly because of compensatory increases in other TG hydrolases. Addition of HSL inhibitor reduced TG hydrolase activity by 20 and 14.5% in extracts from wild-type and ATGL-deficient mice, respectively. As expected, the HSL inhibitor had no effect on TG hydrolase activity in extracts from HSL-deficient mice. Compared with untreated extracts from wild-type mice, extracts from ATGL-deficient mice treated with HSL inhibitor exhibited a reduction in TG hydrolase activity of 43%. Overall, these data showed that HSL and ATGL together contribute to 43% of the TG hydrolase activity in cytosolic extracts from mouse livers.
FIGURE 3.
Liver TG hydrolase activity of wild-type, HSL-deficient, and ATGL-deficient mice. Cytosolic extracts from liver of wild-type (WT), HSL-deficient (HSL-KO), and ATGL-deficient (ATGL-KO) male mice were used to measure TG hydrolase activity. The assay was performed in the presence of 400 μg of GST-CGI-58 and in the absence or presence of 5 nmol of HSL inhibitor. Measurements were performed in triplicate. Results shown are representative of three experiments (n = 3 mice/group/experiment). Data are presented as means ± S.D. Comparisons between groups were made using Student's t test. p value: ***, p < 0.001.
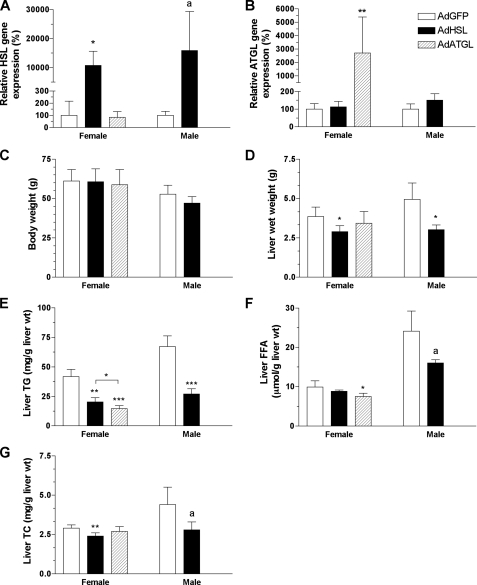
Hepatic Overexpression of HSL or ATGL Reduces Liver TG Mass and Ameliorates Steatosis in ob/ob Mice—Female ob/ob mice, which have been shown to have less severe diabetes than their male counterparts (27), were initially chosen for infection with both AdATGL and AdHSL. As the ablation of genes that moderate FA metabolism often produces more dramatic effects in male mice than in females (38), we also conducted one set of experiments (i.e. AdHSL versus AdGFP) in male mice for contrast. Compared with expression levels in AdGFP-infected ob/ob mice, hepatic HSL expression was increased by >100-fold in both AdHSL-infected female and male ob/ob mice (Fig. 4A) at 8 and 10 days post-infection, respectively, whereas hepatic ATGL expression was increased by ∼27-fold in AdATGL-infected female mice 8 days post-infection (Fig. 4B). HSL and ATGL protein levels in AdHSL- or ATGL-infected livers, respectively, were comparable with those found in the adipose tissue (Fig. 2, B and C).
FIGURE 4.
Body weights, liver weights, and hepatic lipid contents in AdHSL- and AdATGL-infected ob/ob mice. Female and male ob/ob mice were infected with AdGFP, AdHSL, or AdATGL. Animals were sacrificed 8 or 10 days post-infection for females and males, respectively. Hepatic expression of HSL (A) or ATGL (B) was assessed by qPCR and normalized to β-actin expression. Expression of either gene in AdHSL- or AdATGL-infected mice is expressed as a percentage of expression (±S.D.) in AdGFP-infected mice. Body weights (C) and liver weights (D) are expressed in grams. Liver lipids were extracted and assessed for TG (E), FFA (F), and TC (G) by enzymatic methods. All data are shown as means ± S.D. Comparisons were made against AdGFP-infected animals by Student's t test. p values: a, p = 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
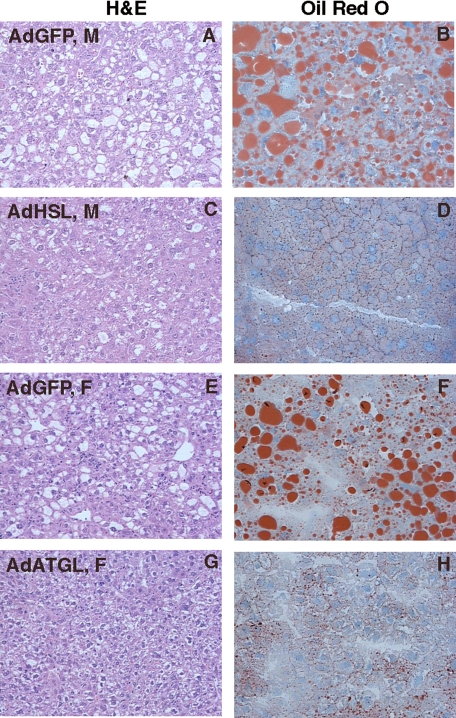
Overexpression of HSL had no effect on body weight in either sex (Fig. 4C). In ob/ob mice infected with AdHSL, liver weights were significantly reduced (25–39%) compared with those in mice infected with AdGFP (female, 2.89 ± 0.39 versus 3.86 ± 0.6 mg/g wet weight, p < 0.05; male, 3.13 ± 0.26 versus 4.95 ± 1.04 mg/g wet weight, p < 0.05; Fig. 4D). Liver TG levels were reduced by 50 and 60% in AdHSL-infected female and male mice, respectively (female, 20.3 ± 3.6 versus 41.9 ± 6.0 mg/g wet weight, p < 0.0001; male, 27.2 ± 4.4 versus 67.2 ± 9.2 mg/g wet weight, p < 0.001; Fig. 4E). Liver FFA levels were not significantly reduced in AdHSL-infected mice (female, 8.8 ± 0.4 versus 9.9 ± 1.6 μmol/g wet weight; male, 16 ± 0.9 versus 24.1 ± 5.1 μmol/g wet weight, p = 0.05; Fig. 4F). Liver total cholesterol (TC) levels were significantly lower in female AdHSL-infected mice (female, 2.4 ± 0.2 versus 2.9 ± 0.2 mg/g wet weight, p < 0.05; male, 2.8 ± 0.5 versus 4.4 ± 1.1 mg/g wet weight, p = 0.05; Fig. 4G). Histological analysis of liver sections (Fig. 5) showed that lipid droplets of various sizes were present in the liver sections from AdGFP-infected male ob/ob mice (Fig. 5, A and B), whereas only very small lipid droplets were present in those from AdHSL-infected male ob/ob mouse livers (Fig. 5, C and D).
FIGURE 5.
Histological analysis of liver sections from AdHSL- and AdATGL-infected ob/ob mice. Male (A–D) and female (E–H) ob/ob mice were infected with AdGFP (A, B, E, and F), AdHSL (C and D), or AdATGL (G and H). Liver sections were either stained with hematoxylin and eosin (H&E) (left panels, ×100 magnification) or stained with Oil Red O and counter-stained with hematoxylin (right panels, ×200 magnification).
As in HSL-infected mice, overexpression of ATGL had no effect on body weight (Fig. 4C). In female ob/ob mice infected with AdATGL, liver weights did not differ from those in AdGFP-infected ob/ob mice (3.44 ± 0.74 versus 3.86 ± 0.6 mg/g wet weight; Fig. 4D). Liver TG levels were reduced by 65% (p < 0.001) compared with AdGFP-infected animals. This reduction was even greater than that resulting from hepatic overexpression of HSL (AdATGL versus AdHSL = 14.6 ± 2.5 versus 20.3 ± 3 mg/g wet weight, p < 0.05; Fig. 4E). Liver FFA levels, but not TC levels, were significantly lower in AdATGL-infected animals compared with those in AdGFP-infected animals (FFA, 7.5 ± 0.8 versus 9.9 ± 1.6 μmol/g wet weight, p < 0.05; Fig. 4F; TC, 2.7 ± 0.3 versus 2.9 ± 0.2 mg/g wet weight; Fig. 4G). As in AdHSL-infected mice (Fig. 5, C and D), and unlike AdGFP-infected male (Fig. 5, A and B) or female ob/ob mice (Fig. 5, E and F), only very small lipid droplets were present in the liver sections from AdATGL-infected female ob/ob mice (Fig. 5, G and H).
Hepatic Overexpression of HSL Improves Liver Function, Has No Effects on Fasting Plasma TG Levels, and Raises Plasma 3-β-Hydroxybutyrate Levels in Male ob/ob Mice— Hepatic overexpression of HSL and ATGL did not worsen liver function in infected ob/ob mice as assessed by activity levels of two liver enzymes, AST and ALT (Table 2). Instead, liver function appeared to be improved in male ob/ob mice infected with AdHSL as plasma activities of AST (42 ± 2 versus 66 ± 9 IU/ml, p < 0.05) and ALT (183 ± 88 versus 813 ± 484 IU/ml, p < 0.05) were significantly reduced compared with those in the control mice.
TABLE 2.
Fasting plasma biochemistry in AdHSL- and AdATGL-infected mice Comparisons between two groups of animals (n = 4–9/group) were made using the Mann-Whitney U test for AST and ALT and Student's t test for all other parameters. p values were derived from comparisons made against AdGFP-infected animals.
| Strain | Diet | Sex | AdGFP | AdHSL | AdATGL | |
|---|---|---|---|---|---|---|
| AST (IU/ml) | FVB-ob/ob | Chow | F | 99 ± 44 | 61 ± 9 | 96 ± 35 |
| FVB-ob/ob | Chow | M | 66 ± 9 | 42 ± 2a | ||
| B6 | WTD | M | 39 ± 23 | 32 ± 16 | ||
| ALT (IU/ml) | FVB-ob/ob | Chow | F | 705 ± 579 | 269 ± 129 | 454 ± 362 |
| FVB-ob/ob | Chow | M | 813 ± 484 | 183 ± 88a | ||
| B6 | WTD | M | 210 ± 86 | 172 ± 73 | ||
| FFA (mmol/liter) | FVB-ob/ob | Chow | F | 1.04 ± 0.11 | 0.88 ± 0.29 | 1.26 ± 0.46 |
| FVB-ob/ob | Chow | M | 1.12 ± 0.24 | 1.00 ± 0.39 | ||
| B6 | WTD | M | 0.49 ± 0.12 | 0.50 ± 0.09 | ||
| TG (mg/dl) | FVB-ob/ob | Chow | F | 128 ± 27 | 151 ± 37 | 158 ± 70 |
| FVB-ob/ob | Chow | M | 148 ± 34 | 166 ± 27 | ||
| B6 | WTD | M | 41 ± 13 | 42 ± 16 | ||
| TC (mg/dl) | FVB-ob/ob | Chow | F | 133 ± 21 | 83 ± 8a | 125 ± 24 |
| FVB-ob/ob | Chow | M | 131 ± 17 | 121 ± 40 | ||
| B6 | WTD | M | 225 ± 28 | 213 ± 35 | ||
| 3-HB (μmol/liter) | FVB-ob/ob | Chow | F | 223 ± 91 | 351 ± 81 | 331 ± 265 |
| FVB-ob/ob | Chow | M | 143 ± 44 | 724 ± 581 | ||
| B6 | WTD | M | 99 ± 54 | 67 ± 64 | ||
| Glucose (mg/dl) | FVB-ob/ob | Chow | F | 144 ± 23 | 313 ± 141 | 203 ± 80 |
| FVB-ob/ob | Chow | M | 439 ± 141 | 401 ± 211 | ||
| B6 | WTD | M | 204 ± 34 | 216 ± 25 | ||
| Insulin (ng/ml) | FVB-ob/ob | Chow | F | 17.6 ± 14.7 | 29.3 ± 13.3 | 14.6 ± 10.4 |
| FVB-ob/ob | Chow | M | 7.1 ± 4.5 | 8.0 ± 3.9 | ||
| B6 | WTD | M | 1.3 ± 0.5 | 1.3 ± 0.5 |
Values shown are p < 0.05
Despite the marked reduction in hepatic TG mass in AdHSL- and AdATGL-infected ob/ob mice, fasting plasma TG levels were not changed compared with those in AdGFP-infected mice (Table 2). Fasting plasma FFA levels were not affected by either AdHSL or AdATGL infection. Fasting plasma TC levels were decreased in female ob/ob mice infected with AdHSL (83 ± 8 mg/dl) compared with AdGFP-infected (133 ± 21 mg/dl) or AdATGL-infected (125 ± 24 mg/dl) mice.
In male ob/ob mice 5 days post-infection, fasting plasma levels of 3-HB, one of the major ketone bodies generated from β-oxidation, were markedly increased by HSL overexpression (AdHSL versus AdGFP = 1076 ± 529 versus 229 ± 156 μmol/liter, p = 0.04). Plasma 3-HB levels for these two groups of animals, however, were not different at the end point of the study (i.e. 10 days post-infection) (Table 2). Because adenoviral overexpression is transient, the lack of effect of HSL overexpression on plasma 3-HB levels likely resulted from a decline in HSL expression over the course of the study. Fasting plasma levels of 3-HB were similar among all groups in female ob/ob mice at the end point (Table 2) as well as at day 3 and day 6 post-infection (not shown). As shown in Table 2, female ob/ob mice have greatly increased insulin concentrations relative to their male counterparts, which was in agreement with previous findings (27). Male ob/ob mice used in this study were severely diabetic with mean fasting glucose levels above 400 mg/dl despite high fasting insulin levels (∼7–8 ng/ml), suggesting a lack of insulin action (Table 2). In contrast, in female ob/ob mice, fasting plasma insulin levels ranged from 5 to 43 ng/ml and fasting plasma glucose levels ranged from 110 to 500 mg/dl, suggesting a varying degree of insulin resistance among these animals (Table 2). Thus, it is likely that the effects of AdHSL (and possibly of AdATGL) on FA oxidation, and hence plasma 3-HB levels, were confounded by the varying degree of insulin action in female ob/ob mice. Nonetheless, these data suggest increased FA β-oxidation as a possible mechanism for the reduction in hepatic TG caused by HSL and ATGL overexpression.
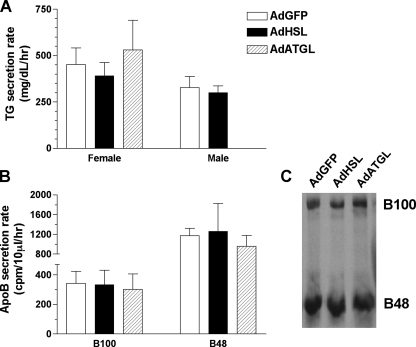
Hepatic Overexpression of HSL or ATGL Does Not Change TG or ApoB Secretion Rates in ob/ob Mice—To determine whether increased lipoprotein secretion contributed to the reduction in hepatic TG mass observed in AdHSL- and AdATGL-infected animals, we assessed in vivo VLDL secretion rates in ob/ob mice using the Triton WR1339 method. The results showed that neither HSL nor ATGL overexpression had an effect on TG (Fig. 6A) or apoB (Fig. 6B) secretion rates in infected ob/ob mice. These data also showed that reduced hepatic TG in these animals did not result from increased TG secretion. Furthermore, these data indicated that although a large portion of the TG storage pools in the liver are hydrolyzed by HSL and ATGL, these do not represent the pools that are destined for VLDL secretion.
FIGURE 6.
In vivo TG and apoB secretion rates in AdHSL- and AdATGL-infected ob/ob mice. A, TG secretion rates were assessed, using the Triton WR1339 method, in female (left panel) and male (right panel) ob/ob mice 8 and 5 days, respectively, after infection with AdGFP, AdHSL, or AdATGL. Animals were bled at 0 min (pre-injection) and at 60 and 120 min post-injection. TG secretion rates are calculated as the increase in TG between 60- and 120-min time points and expressed in mg/dl/h (±S.D.). B, female ob/ob mice were also injected with 35S-labeled Promix in addition to Triton WR1339. Blood samples from the 60-min and 120-min time points were subjected to SDS-PAGE followed by fluorography. B100 (left panel) and B48 (right panel) secretion rates represent differences in protein counts between 60- and 120-min time points and are expressed in cpm/10 μl/h (means ± S.D.). Representative samples from the 60-min time point are shown in the autoradiogram (C).
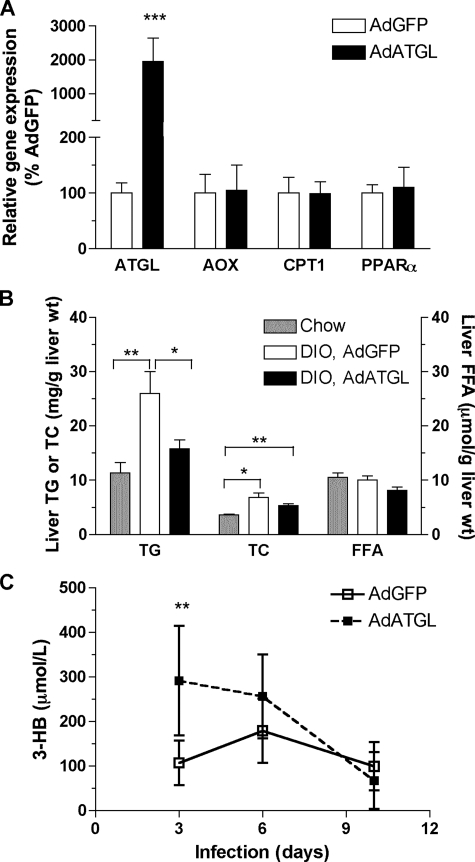
Hepatic Overexpression of HSL Up-regulates Expression of Genes Involved in FA Oxidation but Not in Lipogenesis in Male ob/ob Mice—To assess whether FA oxidation pathways or de novo lipogenesis were affected in AdHSL- or AdATGL-infected ob/ob mice, we assessed expression of genes involved in these pathways. AdHSL-infected male, but not female, ob/ob mice exhibited increased expression of PPARα (41%, p < 0.05) as well as its target gene, acyl-CoA oxidase (AOX, 79%, p < 0.05), a key enzyme in peroxisomal β-oxidation (Table 3). However, the expression of carnitine palmitoyltransferase 1 (a key enzyme in mitochondrial β-oxidation) was not significantly elevated in either group. These data suggested an increase in peroxisomal FA oxidation in AdHSL-infected male ob/ob mice. In addition, we found that expression of mitochondrial UCP2, the up-regulation of which is often associated with fatty livers, was unchanged in both AdHSL- and AdATGL-infected mice (Table 3). The latter corroborated the finding that these animals had improved liver function. Finally, we found no changes in genes involved in FA transport (CD36 and liver fatty acid-binding protein), FA synthesis (fatty-acid synthase, stearoyl-CoA-desaturase 1 (SCD1), and SREBP 1c), or lipid storage (adipose differentiation-related protein (ADRP)) (Table 3). These data suggested a lack of contribution of these pathways toward the reduction of hepatic TG in AdHSL- or AdATGL-infected ob/ob mice.
TABLE 3.
Hepatic gene expression in AdHSL- and AdATGL-infected ob/ob mice Hepatic mRNA levels were assessed by qPCR. Data, expressed as means ± S.D., were normalized to β-actin gene expression. Data derived from AdGFP-infected animals are expressed as 100%, and those of the other groups are expressed as a percentage of the AdGFP-infected controls. Comparisons were made against AdGFP-infected animals using Student's t test. L-Fabp is liver fatty acid-binding protein.
|
Pathway/gene
|
Female
|
Male
|
|||
|---|---|---|---|---|---|
| AdGFP | AdHSL | AdATGL | AdGFP | AdHSL | |
| %AdGFP | %AdGFP | %AdGFP | %AdGFP | %AdGFP | |
| FA oxidation | |||||
| AOX | 100 ± 43 | 129 ± 46 | 101 ± 36 | 100 ± 43 | 179 ± 44a |
| CPT1 | 100 ± 45 | 217 ± 202 | 125 ± 43 | 100 ± 42 | 204 ± 80b |
| PPARα | 100 ± 22 | 115 ± 8 | 95 ± 29 | 100 ± 8 | 141 ± 16a |
| Mitochondrial bioenergetics/lipid metabolism | |||||
| UCP2 | 100 ± 36 | 72 ± 14 | 95 ± 22 | 100 ± 54 | 44 ± 14 |
| FA uptake and transport | |||||
| CD36 | 100 ± 31 | 97 ± 25 | 75 ± 34 | 100 ± 21 | 121 ± 27 |
| L-Fabp | 100 ± 38 | 104 ± 36 | 120 ± 50 | 100 ± 3 | 92 ± 36 |
| Lipid synthesis and storage | |||||
| FAS | 100 ± 58 | 135 ± 60 | 146 ± 77 | 100 ± 30 | 89 ± 46 |
| SCD1 | 100 ± 66 | 125 ± 54 | 216 ± 207 | 100 ± 80 | 92 ± 33 |
| SREBP1c | 100 ± 24 | 88 ± 30 | 97 ± 55 | 100 ± 14 | 89 ± 46 |
| ADRP | 100 ± 35 | 87 ± 20 | 103 ± 44 | 100 ± 13 | 108 ± 30 |
p < 0.05
p = 0.06
Hepatic Overexpression of ATGL Yields Similar Phenotypes in DIO and ob/ob Mice—To confirm that the amelioration of steatosis by overexpression of TG hydrolases observed in ob/ob mice also applies to other models of fatty liver, we induced obesity in male B6 mice by 6 weeks of WTD feeding. Compared with age-matched and chow-fed male mice, DIO mice exhibited a 23% increase in body weights (23.1 ± 2.4 versus 28.4 ± 2.4 g, n = 17–19/group; p < 0.0001) and a 38% increase in liver weights (1.04 ± 0.07 versus 1.44 ± 0.23 g, n = 6/group; p = 0.006) but showed no changes in hepatic ATGL mRNA levels (100 ± 20 versus 118 ± 16%, n = 5/group) or HSL mRNA levels (100 ± 26 versus 71 ± 8%, n = 4–5/group, p = 0.07). Male DIO mice were then infected with either AdATGL or AdGFP. The effects of hepatic ATGL expression on female ob/ob mice were, in general, similar to those observed in male DIO mice. At 10 days post-infection, hepatic ATGL mRNA expression was 19-fold higher in AdATGL-infected mice compared with AdGFP-infected mice as assessed by qPCR (Fig. 7A). Compared with chow-fed mice, WTD consumption increased hepatic TG and TC by 129 and 85%, respectively, in AdGFP-infected DIO mice (Fig. 7B). WTD feeding had no effects on hepatic FFA levels. Adenoviral overexpression of ATGL in DIO mice reduced hepatic TG by 40% (26.0 ± 11.4 versus 15.8 ± 4.7 μg/g liver wet weight, p = 0.04) to a level near that found in chow-fed animals (11.3 ± 4.7 μg/g liver wet weight). ATGL overexpression had no effects on hepatic TC levels (Fig. 7B) or body weights (supplemental Fig. 1) compared with AdGTP-infected DIO mice. However, unlike female ob/ob mice, hepatic FFA levels were not changed in AdATGL-infected DIO mice (Fig. 7B), and liver weights in AdATGL-infected DIO mice were slightly higher than those in AdGFP-infected DIO mice (1.7 ± 0.2 versus 1.4 ± 0.2 g, p = 0.01).
FIGURE 7.
Effects of hepatic ATGL overexpression in male DIO mice. Male B6 mice (n = 8–9/group) were fed a WTD for 6 weeks prior to infection with either AdATGL or AdGFP for 10 days. Animals were bled at indicated time points and sacrificed 10 days post-infection. Liver tissues were collected for lipid measurements and RNA isolation. Age-matched and chow-fed male B6 mice (n = 6) were also sacrificed and assessed for hepatic lipid contents. A, hepatic gene expression was assessed by qPCR and normalized to β-actin expression. Expression of each gene in AdATGL-infected mice is expressed as a percentage of expression in AdGFP-infected mice. B, liver TG, TC, and FFA contents. C, fasting plasma 3-HB levels. All data are shown as means ± S.D. Comparisons were made against AdGFP-infected animals or against chow-fed animals by Student's t test. p values: *, p < 0.05; **, p < 0.01, ***, p < 0.001.
At 10 days post-infection, ATGL overexpression had no effects on fasting plasma levels of lipids (FFA, TG, or TC), glucose, or insulin in DIO mice compared with AdGFP-infected mice (Table 2). In addition, ATGL overexpression did not alter VLDL TG or apoB secretion rates in DIO males (supplemental Fig. 2). Although AST and ALT activities did not differ between AdATGL- and AdGFP-infected groups at the end point, activity levels of both enzymes did rise over the course of the infection when compared with day 3 levels, peaking at 6 days post-infection. These data likely reflected the response of these animals to adenoviral infection. ALT activities were higher in AdATGL-infected DIO mice compared with AdGFP-infected mice at day 6 (293 ± 31 versus 218 ± 68 IU/ml, p = 0.01; supplemental Fig. 1).
As in AdATGL-infected ob/ob mice, plasma 3-HB levels in AdATGL-infected DIO mice were not different at the end point (10 days post-infection) of the study (Table 2), but they did show a significant increase at 3 days post-infection compared with AdGFP-infected mice (291 ± 123 versus 107 ± 50 μmol/liter, p = 0.003; Fig. 7C). These data also suggest that adenoviral overexpression of ATGL likely peaked at day 3 and thus exerted a stronger effect on FA oxidation at the earlier time points of the study. As in female AdATGL-infected ob/ob mice (Table 3), expression of genes involved in FA oxidation (AOX, carnitine palmitoyltransferase 1, and PPARα) was not increased in AdATGL-infected DIO mice compared with AdGFP-infected control mice at the end point of the study (Fig. 7A).
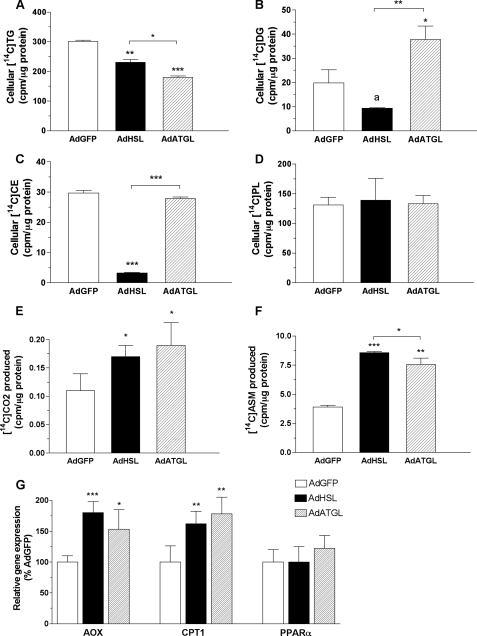
Both HSL and ATGL Promote FA Oxidation in Vitro—To further examine possible mechanisms underlying the reduction of hepatic TG by HSL or ATGL overexpression in ob/ob and DIO mice, we assessed the effects of these enzymes on β-oxidation in OA-supplemented McA-RH7777 cells. Cells were incubated for 16 h in the presence of [14C]OA 24 h post-infection. The net synthesis of 14C-labeled lipids in these cells is shown in Fig. 8 (A–D). Total incorporation of [14C]OA into cellular [14C]TG was decreased 34 and 60% in AdHSL- and AdATGL-infected McA-RH7777 cells, respectively (Fig. 8A). Cellular [14C]DG were reduced by 53% in AdHSL-infected cells but increased by 91% in AdATGL-infected cells (Fig. 8B). Cellular [14C]CE were markedly reduced (90%) in AdHSL-infected, but not in AdATGL-infected, cells (Fig. 8C). Cellular [14C]PL were similar among all groups of cells (Fig. 8D).
FIGURE 8.
Effects of HSL and ATGL overexpression on net TG synthesis and FA oxidation in McA-RH7777 cells. Cells were infected with AdHSL, AdATGL, or AdGFP for 24 h followed by 16 h of incubation in a conditioned medium containing 0.4 mm OA, 1.5% BSA, and 1 μCi of [14C]OA. A–F, cellular TG (A), DG (B), CE (C), and PL (D) were separated by TLC and counted in a scintillation counter. Medium was collected, and oxidation of [14C]OA to [14C]CO2 (E) or [14C]ASM (F) was measured as described under “Experimental Procedures.” Data are presented as means ± S.D. and expressed in counts/min per μg of cellular protein. G, cellular gene expression was assessed by qPCR and normalized to β-actin expression. Levels of mRNA for each gene are expressed as a percentage of those (±S.D.) in AdGFP-infected cells. The experiment was performed in triplicate. Comparisons were made against AdGFP-infected cells or between AdHSL- and AdATGL-infected cells by Student's t test. p values: a, = 0.06; *, p < 0.05; **, p < 0.01, ***, p < 0.001.
Fig. 8 (E and F) also shows that both AdHSL and AdATGL significantly increased oxidation of [14C]OA to [14C]CO2 and production of [14C]ASM (i.e. ketones). Compared with AdGFP-infected cells, the production of [14C]CO2 was increased by 50 and 72% in AdHSL-infected and AdATGL-infected cells, respectively (Fig. 8E). Similarly, incorporation of 14C label into ASM was increased by 120 and 94% in AdHSL- and AdATGL-infected cells, respectively (Fig. 8F). Thus, the almost doubled FA oxidation rates could contribute to the reduction of cellular [14C]TG in AdHSL and AdATGL-infected cells compared with those in AdGFP-infected cells. This increase in FA oxidation was likely because of increases in the expression of AOX and carnitine palmitoyltransferase 1 gene expression in both AdHSL- and AdATGL-infected cells (Fig. 8G). However, PPARα gene expression was not altered in either AdHSL- or AdATGL-infected cells (Fig. 8G). These data confirmed that HSL and ATGL target hydrolytic products from TG storage pools to β-oxidation pathways. We wondered, however, whether there are additional outlets for these products such as direct release of FFA into medium.
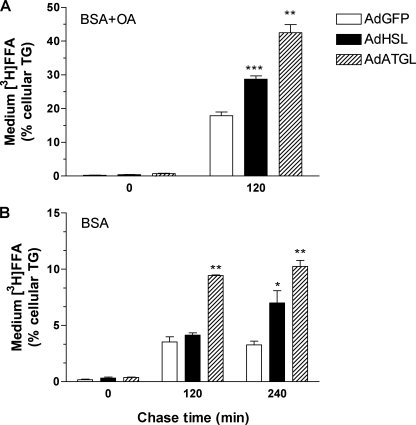
FFA Generated by HSL- and ATGL-mediated Lipolysis in McA-RH7777 Cells Is Removed, in Part, by Direct Release into Medium—To test the hypothesis that some FFA generated by HSL- and ATGL-mediated lipolysis were directly secreted into the medium, we labeled cells with [3H]OA for 16 h and then chased for 120 or 240 min with medium containing either 0.4 mm OA, 1.5% BSA (Fig. 9A), or 1.5% BSA only (Fig. 9B) in the presence of THL, a lipase inhibitor, to block any potential hydrolysis of medium TG. When chased with 0.4 mm OA, 1.5% BSA, medium 3H-labeled FFA counts at the 120-min time point were increased by 20% (p = 0.01) and 41% (p = 0.002) in AdHSL- and AdATGL-infected cells, respectively, compared with AdGFP-infected cells. Expressed as a percentage of cellular [3H]TG, the amount of medium 3H-labeled FFA was increased by 60% (p < 0.001) and 137% (p < 0.01) in AdHSL- and AdATGL-infected cells, respectively, compared with AdGFP-infected cells (Fig. 9A). Significant reuptake of medium [3H]OA was evident in all groups chased with BSA only (Fig. 9B), compared with cells chased with OA (Fig. 9A). Nonetheless, the percentage of medium 3H-labeled FFA was 167% higher in AdATGL-infected cells 120 min after removal of the labeling medium compared with AdGFP-infected cells (Fig. 9B). Although the percentage of medium 3H-labeled FFA in AdHSL-infected cells was unchanged at the 120-min time point, it was increased by 114% at the 240-min time point (Fig. 9B). Similar medium [3H]TG counts were found in all treatment groups. After adjustment to the cellular [3H]TG, the percentage of medium [3H]TG was higher in AdATGL-infected cells 240 min after removal of the labeling medium compared with AdGFP-infected cells (supplemental Fig. 3). However, medium [3H]TG represented less than 1% of cellular [3H]TG and thus was unlikely to be a major contributor to the marked reduction of cellular [3H]TG in AdATGL-infected cells. Overall, these data demonstrated that the reduction of cellular [3H]TG in AdHSL- and AdATGL-infected cells could be attributed, in part, to the direct release of 3H-labeled FFA into the medium.
FIGURE 9.
FFA secretion in AdHSL- and AdATGL-infected McA-RH7777 cells. Cells were infected with AdHSL, AdATGL, or AdGFP for 24 h, followed by 16 h of incubation with a labeling medium containing 0.4 mm OA, 1.5% BSA, and 10 μCi of [3H]OA. The labeling medium was removed, and cells were washed and incubated in the presence of THL with a chase medium containing either 0.4 mm OA, 1.5% BSA (A) or 1.5% BSA only (B) for the indicated amount of time. The experiments were performed in triplicate. All data are presented as means ± S.D. Medium 3H-labeled FFA is expressed as percent of cellular [3H]TG. Cellular [3H]TG counts are shown in supplemental Fig. 3. Comparisons were made against AdGFP-infected cells by Student's t test. p values: *, p < 0.05, **, p < 0.01; ***, p < 0.001.
DISCUSSION
NAFLD can progress into a number of fatal liver diseases, including NASH. To date, no direct treatment is available for NASH. This study explored the potential of two major adipose lipases, HSL and ATGL, as therapeutic targets for fatty liver treatment. Although the role of HSL in hepatic lipid metabolism is unclear in normal physiological conditions, a direct role for ATGL in lipid metabolism is very plausible based on the fatty liver phenotype observed in ATGL-deficient mice (26). In this study, we showed that in male wild-type mice, both HSL and ATGL proteins were detectable in the liver, even though their expression levels were much lower than those in the adipose tissue. In addition, hepatic HSL mRNA levels were lower in male, but not female, ob/ob mice compared with their respective lean littermates. The latter was unexpected as HSL is a known target gene of PPARγ, and adenoviral overexpression of PPARγ in the liver increases HSL expression (19). Up-regulation of PPARγ is often observed in insulin-resistant livers, as is the case with ob/ob mice (39). These results thus suggest transcriptional regulation by factors other than PPARγ for hepatic HSL expression in male ob/ob mice.
A direct role for HSL and ATGL in TG lipolysis in the liver was further supported by their contributions to TG hydrolase activity in cytosolic extracts isolated from mouse livers. Using combinations of genetic and chemical ablation of HSL and/or ATGL activity, we showed that HSL and ATGL, together, contributed ∼43% of liver cytosolic TG hydrolase activity. A 33% reduction of cytosolic TG hydrolase activity in ATGL-deficient mice confirmed a major role for ATGL in hepatic TG lipolysis. Chemical inhibition of HSL in wild-type and ATGL-deficient mice indicates that this enzyme contributes ≥10% of TG hydrolase activity and likely plays a lesser role in hepatic TG lipolysis compared with ATGL. The lack of change in TG hydrolytic rates in the livers of HSL-deficient mice compared with wild-type suggests that expression of other liver lipases may be up-regulated to compensate for the loss of HSL activity in HSL-deficient animals.
We also showed that HSL and ATGL are both active as TG hydrolases in cultured liver cells and mouse livers when introduced by adenoviral infection. Both enzymes reduced cellular TG mass by ∼60% in McA-RH7777 cells supplemented with OA and caused reductions of ∼40–65% in hepatic TG in ob/ob and DIO mice. The reduction of hepatic TG by adenoviral overexpression of HSL and ATGL was not correlated with increased TG and apoB secretion in vivo. The latter likely explains the lack of changes in fasting plasma TG levels. Expression of genes involved in FA uptake/transport and de novo lipogenesis was also unaffected by HSL or ATGL overexpression in ob/ob mice. These data suggested that reduction of steatosis in ob/ob mice was probably not because of decreased FA uptake or de novo lipogenesis. On the other hand, fasting plasma levels of 3-HB were increased at days 3–5 post-infection, but not at the end point, in male mice infected with HSL and ATGL. FA oxidation genes were also increased in HSL-infected, but not in ATGL-infected, male mice at the end point. Taken together, these data suggested that overexpression of these lipase enzymes eliminates excess cellular TG by mobilizing TG storage pools destined for oxidation, but not those destined for lipoprotein secretion.
Studies in McA-RH7777 cells confirmed that HSL and ATGL overexpression increased FA oxidation by up-regulating FA oxidation genes. HSL has been shown to promote FA oxidation in HepG2 cells (40), and transgenic overexpression of HSL in heart prevents starvation-induced lipid accumulation in the heart (41). We have now demonstrated that not only HSL but also ATGL can reduce hepatic steatosis, likely by promoting hepatic FA oxidation in ob/ob or DIO mice. These data also suggest that the accumulation of TG seen in the livers of ATGL-deficient mice (26) is likely caused by decreased FA oxidation in these animals and that ATGL is essential in mobilizing stored TG into FA oxidation pathways under normal conditions. Furthermore, studies of mice deficient in fatty-acid synthase in the liver (FASKOL) suggested that new fats derived from diet or de novo lipogenesis, but not old fats derived from peripheral tissues, are endogenous ligands for PPARα (42). In contrast, our studies showed that HSL-mediated lipolysis in male ob/ob mice provide ligands for activating transcription of PPARα downstream genes, presumably from stored lipid pools.
The apparent increase in FA oxidation did not seem to have adverse effects in ob/ob mice overexpressing HSL or ATGL. Hepatic expression of UCP2, a protein associated with mitochondrial bioenergetics and oxidative stress (43), was not increased in AdHSL- or AdATGL-infected animals. In fact, HSL overexpression seems to improve liver function as these animals had decreased plasma activity levels of AST and ALT. Thus, we wondered whether FFA generated from HSL- or ATGL-mediated lipolysis of hepatic TG have an additional outlet besides β-oxidation pathways. This hypothesis was tested and confirmed in OA-supplemented McA-RH7777 cells labeled with [3H]OA and chased with cold OA. Both HSL and ATGL overexpression markedly increased secretion of 3H-labeled FFA into medium during the chase period, demonstrating that FFA generated from lipolysis of hepatic TG storage pools can be directly released into medium. Although neither AdHSL nor AdATGL increased plasma FFA in infected animals, this is not surprising as FFA are rapidly cleared from the circulation by peripheral tissues in vivo.
Overall, we have demonstrated that both ATGL and HSL contribute to hepatic TG hydrolase activity and likely play a direct role in liver lipolysis under normal physiological conditions. Both enzymes likely maintain hepatic lipid homeostasis by mobilizing TG from storage pools to FA oxidation pathways and possibly also by releasing FFA directly into the circulation. Finally, we have identified both HSL and ATGL as potential therapeutic targets for directly treating fatty liver in human subjects. The fact that overexpression of these enzymes reduces hepatic TG without increasing hepatic apoB or TG secretion may make them especially attractive agents for treating NAFLD patients, many of whom also exhibit profound dyslipidemia.
Supplementary Material
Acknowledgments
We thank Dr. Andrew Greenberg for the generous gift of anti-ATGL antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants HL62583 (to L. S. H.), DK079221 (to W. S. B.), HL73029, HL45095 (to I. J. G.), and DK063306 (to S. C. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
Footnotes
The abbreviations used are: NAFLD, nonalcoholic fatty liver disease; 3-HB, 3-β-hydroxybutyrate; AdATGL, recombinant adenovirus expressing adipose triglyceride lipase; AdGFP, recombinant adenovirus expressing green fluorescent protein; AdHSL, recombinant adenovirus expressing hormone-sensitive lipase; ALT, alanine aminotransferase; apoB, apolipoprotein B; ASM, acid-soluble metabolites; AST, aspartate aminotransferase; AOX, acyl-CoA oxidase; ATGL, adipose triglyceride lipase; ATGL-KO, ATGL-deficient; B6, C57BL/6; BSA, bovine serum albumin; CE, cholesteryl ester; CGI-58, comparative gene identification protein 58; DG, diacylglycerol; DIO, diet-induced obese; DMEM, Dulbecco's modified Eagle's medium; FA, fatty acid(s); FFA, free fatty acid(s); FVB, FVB/N; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; GST, glutathione S-transferase; HSL, hormone-sensitive lipase; NASH, nonalcoholic steatohepatitis; OA, oleate; PBS, phosphate-buffered saline; PL, phospholipid; PPAR, peroxisome proliferator-activated receptor; SREBP, sterol-responsive element-binding protein; TG, triglyceride(s); THL, tetrahydrolipstatin; WTD, Western-type diet; qPCR, quantitative real time PCR; TC, total cholesterol; VLDL, very low density lipoprotein.
References
- 1.Marchesini, G., Brizi, M., Bianchi, G., Tomassetti, S., Bugianesi, E., Lenzi, M., McCullough, A. J., Natale, S., Forlani, G., and Melchionda, N. (2001) Diabetes 50 1844-1850 [DOI] [PubMed] [Google Scholar]
- 2.Youssef, W., and McCullough, A. J. (2002) Semin. Gastrointest. Dis. 13 17-30 [PubMed] [Google Scholar]
- 3.Portincasa, P., Grattagliano, I., Palmieri, V. O., and Palasciano, G. (2006) Curr. Med. Chem. 13 2889-2900 [DOI] [PubMed] [Google Scholar]
- 4.Lin, H. Z., Yang, S. Q., Chuckaree, C., Kuhajda, F., Ronnet, G., and Diehl, A. M. (2000) Nat. Med. 6 998-1003 [DOI] [PubMed] [Google Scholar]
- 5.Lutchman, G., Promrat, K., Kleiner, D. E., Heller, T., Ghany, M. G., Yanovski, J. A., Liang, T. J., and Hoofnagle, J. H. (2006) Clin. Gastroenterol. Hepatol. 4 1048-1052 [DOI] [PubMed] [Google Scholar]
- 6.Koteish, A., and Diehl, A. M. (2001) Semin. Liver Dis. 21 89-104 [DOI] [PubMed] [Google Scholar]
- 7.Memon, R. A., Tecott, L. H., Nonogaki, K., Beigneux, A., Moser, A. H., Grunfeld, C., and Feingold, K. R. (2000) Endocrinology 141 4021-4031 [DOI] [PubMed] [Google Scholar]
- 8.Ferrante, A. W., Jr., Thearle, M., Liao, T., and Leibel, R. L. (2001) Diabetes 50 2268-2278 [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Ruiz, I., Rodriguez-Juan, C., Diaz-Sanjuan, T., del Hoyo, P., Colina, F., Munoz-Yague, T., and Solis-Herruzo, J. A. (2006) Hepatology 44 581-591 [DOI] [PubMed] [Google Scholar]
- 10.Shimomura, I., Bashmakov, Y., and Horton, J. D. (1999) J. Biol. Chem. 274 30028-30032 [DOI] [PubMed] [Google Scholar]
- 11.Memon, R. A., Grunfeld, C., Moser, A. H., and Feingold, K. R. (1994) Horm. Metab. Res. 26 85-87 [DOI] [PubMed] [Google Scholar]
- 12.Wiegman, C. H., Bandsma, R. H., Ouwens, M., van der Sluijs, F. H., Havinga, R., Boer, T., Reijngoud, D. J., Romijn, J. A., and Kuipers, F. (2003) Diabetes 52 1081-1089 [DOI] [PubMed] [Google Scholar]
- 13.Memon, R. A., Fuller, J., Moser, A. H., Smith, P. J., Grunfeld, C., and Feingold, K. R. (1999) Diabetes 48 121-127 [DOI] [PubMed] [Google Scholar]
- 14.Goldberg, I. J., and Ginsberg, H. N. (2006) Gastroenterology 130 1343-1346 [DOI] [PubMed] [Google Scholar]
- 15.Schweiger, M., Schreiber, R., Haemmerle, G., Lass, A., Fledelius, C., Jacobsen, P., Tornqvist, H., Zechner, R., and Zimmermann, R. (2006) J. Biol. Chem. 281 40236-40241 [DOI] [PubMed] [Google Scholar]
- 16.Holm, C. (2003) Biochem. Soc. Trans. 31 1120-1124 [DOI] [PubMed] [Google Scholar]
- 17.Yeaman, S. J., Smith, G. M., Jepson, C. A., Wood, S. L., and Emmison, N. (1994) Adv. Enzyme Regul. 34 355-370 [DOI] [PubMed] [Google Scholar]
- 18.Wei, S., Lai, K., Patel, S., Piantedosi, R., Shen, H., Colantuoni, V., Kraemer, F. B., and Blaner, W. S. (1997) J. Biol. Chem. 272 14159-14165 [DOI] [PubMed] [Google Scholar]
- 19.Yu, S., Matsusue, K., Kashireddy, P., Cao, W. Q., Yeldandi, V., Yeldandi, A. V., Rao, M. S., Gonzalez, F. J., and Reddy, J. K. (2003) J. Biol. Chem. 278 498-505 [DOI] [PubMed] [Google Scholar]
- 20.Zimmermann, R., Strauss, J. G., Haemmerle, G., Schoiswohl, G., Birner-Gruenberger, R., Riederer, M., Lass, A., Neuberger, G., Eisenhaber, F., Hermetter, A., and Zechner, R. (2004) Science 306 1383-1386 [DOI] [PubMed] [Google Scholar]
- 21.Jenkins, C. M., Mancuso, D. J., Yan, W., Sims, H. F., Gibson, B., and Gross, R. W. (2004) J. Biol. Chem. 279 48968-48975 [DOI] [PubMed] [Google Scholar]
- 22.Villena, J. A., Roy, S., Sarkadi-Nagy, E., Kim, K. H., and Sul, H. S. (2004) J. Biol. Chem. 279 47066-47075 [DOI] [PubMed] [Google Scholar]
- 23.Jaworski, K., Sarkadi-Nagy, E., Duncan, R., Ahmadian, M., and Sul, H. S. (2007) Am. J. Physiol. 293 G1-G4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Granneman, J. G., Moore, H. P., Granneman, R. L., Greenberg, A. S., Obin, M. S., and Zhu, Z. (2007) J. Biol. Chem. 282 5726-5735 [DOI] [PubMed] [Google Scholar]
- 25.Fischer, J., Lefevre, C., Morava, E., Mussini, J. M., Laforet, P., Negre-Salvayre, A., Lathrop, M., and Salvayre, R. (2007) Nat. Genet. 39 28-30 [DOI] [PubMed] [Google Scholar]
- 26.Haemmerle, G., Lass, A., Zimmermann, R., Gorkiewicz, G., Meyer, C., Rozman, J., Heldmaier, G., Maier, R., Theussl, C., Eder, S., Kratky, D., Wagner, E. F., Klingenspor, M., Hoefler, G., and Zechner, R. (2006) Science 312 734-737 [DOI] [PubMed] [Google Scholar]
- 27.Chua, S., Jr., Liu, S. M., Li, Q., Yang, L., Thassanapaff, V. T., and Fisher, P. (2002) Diabetologia 45 976-990 [DOI] [PubMed] [Google Scholar]
- 28.Ko, C., O'Rourke, S. M., and Huang, L. S. (2003) J. Lipid Res. 44 1946-1955 [DOI] [PubMed] [Google Scholar]
- 29.He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W., and Vogelstein, B. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 2509-2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haemmerle, G., Zimmermann, R., Hayn, M., Theussl, C., Waeg, G., Wagner, E., Sattler, W., Magin, T. M., Wagner, E. F., and Zechner, R. (2002) J. Biol. Chem. 277 4806-4815 [DOI] [PubMed] [Google Scholar]
- 31.Shimomura, I., Shimano, H., Horton, J. D., Goldstein, J. L., and Brown, M. S. (1997) J. Clin. Investig. 99 838-845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lass, A., Zimmermann, R., Haemmerle, G., Riederer, M., Schoiswohl, G., Schweiger, M., Kienesberger, P., Strauss, J. G., Gorkiewicz, G., and Zechner, R. (2006) Cell Metab. 3 309-319 [DOI] [PubMed] [Google Scholar]
- 33.Liang, J. J., Oelkers, P., Guo, C., Chu, P. C., Dixon, J. L., Ginsberg, H. N., and Sturley, S. L. (2004) J. Biol. Chem. 279 44938-44944 [DOI] [PubMed] [Google Scholar]
- 34.Folch, J., Lees, M., and Sloane Stanley, G. H. (1957) J. Biol. Chem. 224 497-509 [PubMed] [Google Scholar]
- 35.Carr, T. P., Andresen, C. J., and Rudel, L. L. (1993) Clin. Biochem. 26 39-42 [DOI] [PubMed] [Google Scholar]
- 36.Voyiaziakis, E., Ko, C., O'Rourke, S. M., and Huang, L. S. (1999) J. Lipid Res. 40 2004-2012 [PubMed] [Google Scholar]
- 37.Lewin, T. M., Wang, S., Nagle, C. A., Van Horn, C. G., and Coleman, R. A. (2005) Am. J. Physiol. 288 E835-E844 [DOI] [PubMed] [Google Scholar]
- 38.Augustus, A. S., Buchanan, J., Park, T. S., Hirata, K., Noh, H. L., Sun, J., Homma, S., D'Armiento, J., Abel, E. D., and Goldberg, I. J. (2006) J. Biol. Chem. 281 8716-8723 [DOI] [PubMed] [Google Scholar]
- 39.Boelsterli, U. A., and Bedoucha, M. (2002) Biochem. Pharmacol. 63 1-10 [DOI] [PubMed] [Google Scholar]
- 40.Pease, R. J., Wiggins, D., Saggerson, E. D., Tree, J., and Gibbons, G. F. (1999) Biochem. J. 341 453-460 [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki, J., Shen, W. J., Nelson, B. D., Patel, S., Veerkamp, J. H., Selwood, S. P., Murphy, G. M., Jr., Reaven, E., and Kraemer, F. B. (2001) Am. J. Physiol. 281 E857-E866 [DOI] [PubMed] [Google Scholar]
- 42.Chakravarthy, M. V., Pan, Z., Zhu, Y., Tordjman, K., Schneider, J. G., Coleman, T., Turk, J., and Semenkovich, C. F. (2005) Cell Metab. 1 309-322 [DOI] [PubMed] [Google Scholar]
- 43.Baffy, G. (2005) Front. Biosci. 10 2082-2096 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.