Abstract
This report describes the identification and purification of a novel mismatch repair stimulatory factor from HeLa cell extracts. This activity copurifies with a proliferating cell nuclear antigen-dependent 5 ′ → 3 ′ DNA excision activity during several purification steps but is resolved from the excision activity during gel filtration chromatography using Sephacryl S-300. After purification to near homogeneity, the stimulatory factor is associated with three polypeptides with apparent molecular masses of 68, 36, and 30 kDa. Peptide sequencing analysis by tandem mass spectrometry identified the stimulatory factor as the heterotrimeric regulatory factor X (RFX) complex, which regulates transcription of the class II major histocompatibility complex by facilitating histone acetylation and is defective in the human hereditary immunodeficiency syndrome called bare lymphocyte syndrome. This conclusion was confirmed by the facts that purified recombinant RFX stimulates mismatch repair in an in vitro reconstituted mismatch repair system and that depletion of RFX from nuclear extracts or RFX knockdown in cells reduces mismatch repair activity. As expected, RFX knockdown cells display instability in microsatellite sequences. The possible role of RFX in human MMR repair is discussed.
Mismatch repair (MMR)3 is an evolutionarily conserved genome maintenance mechanism that corrects base-base mismatches and insertion-deletion mispairs generated during DNA replication and homologous recombination (reviewed in Refs. 1 and 2). Defects in MMR lead to increased genomic instability and are the genetic basis of certain types of hereditary and sporadic cancers, including hereditary nonpolyposis colorectal cancer (reviewed in Refs. 1–3).
The methyl-directed MMR pathway in Escherichia coli is well characterized and has been reconstituted in vitro (4). In E. coli, 11 proteins carry out the repair reaction in three stages: initiation, excision, and repair DNA synthesis. MutS, MutL, and MutH recognize mismatches and incise the newly synthesized unmethylated DNA strand (initiation). One of four exonucleases (Exo1, Exo VII, Exo X, and RecJ) carries out mismatch removal via 5′ or 3′ excision from the DNA strand break in conjunction with UvrD helicase. Finally, DNA polymerase III holoenzyme, single-strand DNA-binding protein, and DNA ligase together carry out repair DNA synthesis and DNA ligation (for reviews see Refs. 1, 2, and 5).
The eukaryotic MMR pathway resembles E. coli MMR, uses a similar repair mechanism, and is carried out by a similar group of protein factors. For example, both eukaryotic and bacterial MMR are strand-specific, bi-directional, and nick-directed, and eukaryotic MutS- and MutL-like activities are highly homologous to their bacterial counterparts. Human MMR has been successfully reconstituted with purified proteins (6, 7). The reconstituted system includes MutSα or MutSβ, MutLα, RPA, EXO1, HMGB1, PCNA, replication factor C, polymerase δ, and DNA ligase I. Human MMR activities that play roles similar to those of E. coli MutH and helicase II have not yet been identified. It is clear that human MMR is more complex than E. coli MMR. Although redundancy is evident for several components in the eukaryotic MMR pathway, EXO1 is to date the only nuclease known to be involved in eukaryotic MMR. Because Exo1 null mutants in yeast and mice confer only a partial MMR defect in vivo (8, 9), it seems likely that additional nucleases that play a role in MMR in eukaryotic cells remain to be identified.
Our previous studies reported partial purification of a PCNA-dependent 5′ → 3′ DNA excision activity from HeLa nuclear extracts (10). Here, we report that this 5′ → 3′ DNA excision activity resolves into two fractions during purification, one of which is identical to regulatory factor X (RFX), a heterotrimer that regulates transcription of the class II major histocompatibility complex (MHCII) and is defective in the human hereditary immunodeficiency syndrome bare lymphocyte syndrome (BLS) (11–13). The significance of these data for understanding eukaryotic MMR is discussed.
EXPERIMENTAL PROCEDURES
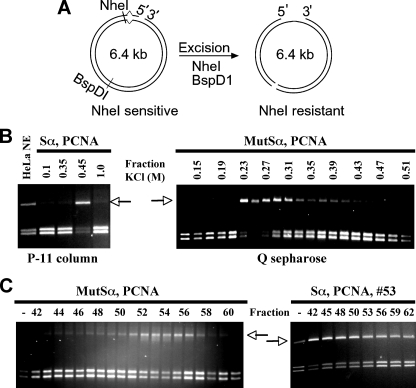
DNA Substrates and Mismatch Excision Assay—DNA heteroduplexes used in this study contained a single G-T mismatch and a strand break 5′ to the mismatch (see Fig. 1). The 5′-nicked substrates were prepared by hybridizing single-stranded f1MR phage to Sau96I-linearized double-stranded DNA of another phage as described previously (14). The mismatch occurs within the overlapping recognition sites for restriction endonucleases HindIII and XhoI, immediately 5′ to the NheI cleavage site (10). The extent of excision was monitored by NheI and BspDI digestion.
FIGURE 1.
Identification of a novel activity stimulating mismatch-provoked excision. A, schematic diagram of the mismatch-provoked DNA excision assay. The DNA substrate has a G:T mismatch adjacent to an NheI restriction enzyme recognition site and a nick 128 nucleotides 5′ to the mismatch. After mismatch-provoked excision proceeding 5′ to 3′ from the nick toward the mismatch, the excision activity converts the NheI-sensitive double-stranded DNA (left panel) to NheI-resistant ssDNA (right panel). Reaction products are treated with NheI and BspD1 and analyzed by agarose gel electrophoresis (right panel). The BspD1 restriction enzyme recognition site is sensitive to cleavage by BspD1 both before and after mismatch-provoked excision. The assay scores positive for excision activity when the slow migrating DNA species is detected (see arrows in panels B and C), indicating cleavage of the reaction product by BspD1 but not by NheI. The reaction scores negative for excision activity when the more quickly migrating doublet band is seen, indicating cleavage of the substrate by both enzymes. B and C, mismatch-provoked excision assays to detect the excision and the stimulatory activities during column chromatography, as indicated. Unless specified, mismatch-provoked excision assays were performed in the presence of 26 nm MutSα, 67 nm PCNA, and 3 μl of the indicated fractions as described (10). DNA samples were recovered and digested with NheI and BspD1, followed by electrophoresis through 1% agarose gel.
Unless otherwise indicated, mismatch excision assays contained 100 ng (24 fmol) of heteroduplex DNA, 26 nm MutSα,67 nm PCNA, 10 mm Tris-HCl, pH 7.6, 5 mm MgCl2, and 1.5 mm ATP. Samples to be assayed were added in a volume of 2–4 μl. The reactions were incubated at 37 °C for 10 min, and DNA was isolated by phenol extraction followed by ethanol precipitation. The reaction products were digested with NheI and BspDI, analyzed by 1% agarose gel electrophoresis, stained with ethidium bromide, and visualized by UV illumination.
Nuclear Extract Preparation and Protein Purification—HeLa S3 cells were either purchased from the National Cell Culture Center (Minneapolis, MN) or cultured in our own laboratory. The nuclear extracts were prepared as described previously (14). Unless otherwise indicated, fractionation and chromatography were performed at 4 °C. HeLa cell nuclear extract (∼1.0 g of total proteins) was adjusted to 65% saturation with ammonium sulfate, and the precipitate was collected by centrifugation. The precipitate was resuspended in and dialyzed against buffer A (25 mm HEPES, pH 7.5, 0.1 mm EDTA, 2 mm dithiothreitol, 0.1% phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin) containing 0.1 m KCl. The clarified supernatant was loaded onto a phosphocellulose P11 column (7.1 cm2 × 15 cm) equilibrated with buffer A containing 0.1 m KCl. The column was washed with five column volumes of buffer A containing 0.35 m KCl. The 5′ nick-directed excision activity was eluted with 0.45 m KCl in buffer A. The pooled P11 fractions were desalted to 0.1 m KCl in buffer A through five connected 5-ml HiTrap desalting columns (GE Healthcare) and loaded onto a 40 ml (4 cm2 × 10 cm) Q Sepharose column (GE Healthcare) equilibrated in buffer A with 0.1 m KCl. The Q Sepharose column was developed with a linear gradient of 0.1–0.5 m KCl in buffer A. The excision activity eluted at 0.23–0.31 m KCl. Active fractions were pooled, desalted to 0.2 m KCl, and loaded onto a 20-ml (2.5 cm2 × 8 cm) Bio-Rex 70 (Bio-Rad). The column was developed with a linear gradient of 0.2–0.5 m KCl in buffer A. Active fractions eluted from 0.37 to 0.42 m KCl were pooled and concentrated to a volume of 2 ml using an Amicon Ultra-4 filtration device (Millipore). The concentrated sample was loaded onto a 120-ml (1.2 cm2 × 100 cm) Sephacryl S-300 (S-300) gel filtration column equilibrated in buffer A with 0.1 m KCl. The nuclease activity and excision stimulatory factor did not coelute during this chromatography step. The peak of the excision stimulatory factor was identified by conducting excision assays in the presence of pooled nuclease activity from the S-300 column. Active stimulatory factor fractions were pooled and loaded onto a 10-ml (1.5 cm2 × 6.7 cm) hydroxyapatite (HAP) column equilibrated with a phosphate buffer containing 0.025 m potassium phosphate, pH 7.4, 0.1 m KCl, 2 mm dithiothreitol, 0.1% phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin. The HAP column was washed extensively with the same buffer and eluted with a linear gradient of potassium phosphate (0.025–0.5 m). The stimulatory activity eluted at ∼0.08 M potassium phosphate. Active fractions were pooled, desalted into to buffer A containing 0.05 m KCl, and loaded onto a 1-ml Mono S column (GE Healthcare) equilibrated with buffer A containing 0.05 m KCl. The Mono S column was developed with a linear gradient from 0.05 to 0.5 m KCl in buffer A. The activity eluted from 0.3 to 0.4 m KCl. Active fractions were pooled, desalted into buffer B containing 0.2 m KCl, and loaded onto a 5-ml HiTrap heparin column (GE Healthcare), which was developed with a linear gradient from 0.2 to 1.0 m KCl. Active fractions, which eluted at ∼0.4 m KCl, were pooled, desalted into buffer B containing 0.05 m KCl, and loaded onto a 2-ml ssDNA cellulose column (Sigma). The ssDNA cellulose column was developed with a linear gradient of 0.05–0.5 m KCl in buffer B. Active fractions, which eluted at ∼0.2 m KCl, were pooled and loaded onto a 2-ml HAP column equilibrated with 0.08 m potassium phosphate buffer, pH 7.4, containing 0.1 m KCl. The excision stimulatory factor did not bind to the column under these conditions. Therefore, the unbound fraction was collected and stored for further studies and analysis.
Mass Spectrometry for Protein Identification—An aliquot of the excision stimulatory factor from HAP flow-through was separated by 10% SDS-PAGE (see Fig. 2B). The 68-, 36-, and 30-kDa bands were excised from the gel and analyzed by mass spectrometry as described previously (15). Briefly, each excised gel slice was incubated with trypsin at 37 °C. The tryptic peptides were extracted and analyzed by liquid chromatography/tandem mass spectrometry using a Q-TOF API US mass spectrometer interfaced with a capillary liquid chromatography system (Waters Corp.). The eluate was directed to the electrospray source with a PicoTip emitter (New Objectives). Mass spectra were processed using MassLynx 4.0 software, and proteins were identified using Protein Global Server 1.0/2.0 software. Protein species were identified and confirmed by exporting tandem mass spectrometry peak lists exported from MassLynx to Mascot. Peptide sequences were queried against the NCBI nonredundant protein data base. Protein modifications considered include cysteine carbamidomethylation (fixed), N-terminal acetylation, N-terminal Gln to pyroGlu, methionine oxidation, and serine, threonine, and tyrosine phosphorylation.
FIGURE 2.
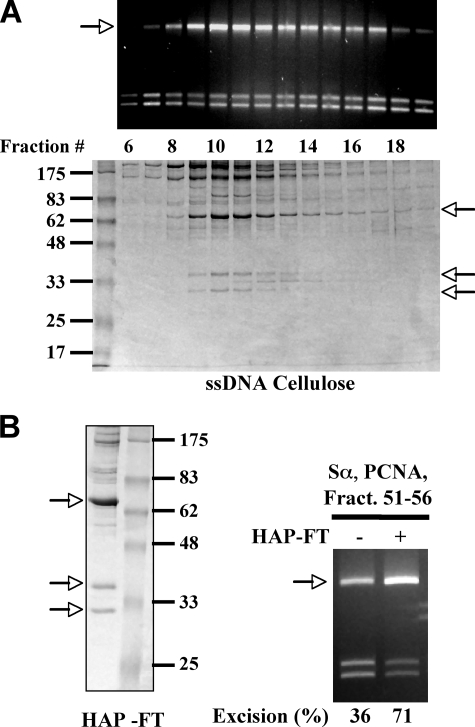
Purification of the stimulatory factor. Protein components and mismatch-provoked excision activity of the individual fractions from the ssDNA cellulose column (A) and HAP (B). Unless mentioned otherwise, mismatch-provoked excision assays were performed in the presence of 26 nm MutSα, 67 nm PCNA, 2 μl of the pool of fractions 51–56, and 3 μl of the indicated fractions. The arrows indicate possible candidates of the stimulatory factor. Protein size markers were 175, 83, 62, 48, 33, 25, and 17 kDa.
Antibody, Antibody Depletion, and Western Blot—An antibody to EXO1 was generated by Genemed Synthesis (San Antonio, TX), using 1.0 mg of recombinant human EXO1 protein expressed and purified from insect cells as described (7). Antibody depletion experiments were performed at 0–4 °C. Nuclear extract (400 μg) was incubated with 5 μl of anti-RFX5 antibody (Bethyl Laboratories, Montgomery, TX) for 30 min on ice. Prewashed protein A-Sepharose (15 μl) (Roche Applied Science) was added, and the reaction was incubated for 1 h and centrifuged at 350 × g for 2 min. The clarified supernatant was used in mismatch excision assays. Control samples were treated as above with a nonspecific antibody. Western blots were performed according to standard procedures using the ECL Plus Western blot kit (Amersham Biosciences).
Expression and Purification of Recombinant RFX—cDNAs encoding RFX5, RFXAP, and RFXANK were kindly provided by Matija Peterlin (University of California, San Francisco). cDNAs were amplified by PCR and subcloned into pFastBac1 transfer vector (Invitrogen). Baculovirus was prepared, and recombinant proteins were expressed using the Bac-to-Bac baculovirus expression system (Invitrogen) as described (7). Briefly, upon expression of the recombinant protein in insect cells, the cells were homogenized using a Dounce homogenizer, and cell extracts were purified using Q Sepharose, HAP, Mono S, and Superdex 200 gel filtration column chromatography. Purified recombinant RFX was stored in buffer B containing 0.15 m KCl in aliquots at –80 °C.
RFX5 Knockdown by shRNA—HeLa cells were transfected with a pmH1P-bsd-based construct (16), which targets position 1785–1803 (AGGAGCATGTGCTTCAAAG) of RFX5. Transfected cells were selected with 40 mg/ml of blasticidin (Invitrogen). Individual blasticidin-resistant clones were selected for analysis of RFX expression by Western blot. Clones with significantly reduced expression of RFX5 were selected, characterized further, and used in this study.
RESULTS
Identification of a Novel Factor That Stimulates Mismatch-provoked Excision—The biochemical mechanism of human MMR is only partly understood at present. Key protein factors involved in this pathway include MutSα, MutSβ, MutLα, PCNA, EXO1, RPA, replication factor C, HMGB1, DNA polymerase δ, and ligase I (1, 6, 7, 17). It has been proposed that additional as yet unidentified protein factors, including one or more exonucleases, play significant roles in human MMR, and additional studies are needed to identify and characterize these protein factors.
Our previous studies identified an activity in HeLa cell nuclear extracts that supports 5′ nick-directed, mismatch-provoked DNA excision (10). In vitro biochemical studies demonstrated that the excision activity is PCNA-dependent, 5′ nick-specific, and heteroduplex-specific (10). Here, a series of chromatographic steps were used to purify this novel MMR excision factor (see “Experimental Procedures”).
The excision activity was monitored during purification by virtue of its ability to conduct 5′-directed mismatch excision in the presence of MutSα and PCNA using a functional assay, which measures the formation of a ssDNA gap during mismatch-provoked excision. The ssDNA gap confers resistance to restriction endonuclease NheI (Fig. 1A), producing a slow moving DNA product (see arrows in Fig. 1, B and C). The excision activity was recovered as a single peak of activity during phosphocellulose P-11, Q-Sepharose (Fig. 1B), and Bio-Rex 70 chromatography (data not shown). However, a dramatic loss in activity was observed after Sepharose S-300 (S-300) gel filtration chromatography. As shown in Fig. 1C (left panel), the mismatch-provoked excision activity was detected in fractions 51–57 of the S-300 column; however, the amount of activity detected in these fractions was reproducibly ≈5-fold lower than the amount of activity detected after Q Sepharose chromatography (Fig. 1, compare B, right panel, with C). This result suggests that the excision activity might include multiple components that partially or completely dissociate during gel filtration.
This possibility was tested by reassaying individual fractions of the S-300 column for mismatch-provoked excision in the presence of the most active (i.e. peak) S-300 fraction (fraction 53). The results showed that fractions 42–48 significantly stimulated product formation (Fig. 1C, right panel), with fraction 42 producing the greatest stimulatory effect (note that fraction 42 is the first fraction after the void volume of the gel filtration column). These results suggest that (i) the 5′ directed mismatch excision activity includes an excision activity (fractions 51–56) and a dissociable stimulatory factor (fractions 42–48) and (ii) the stimulating factor is a high molecular weight protein or protein complex.
The Excision Stimulatory Factor Is RFX—The stimulatory factor was further purified by chromatography on HAP, MonoS, heparin, and ssDNA cellulose (see “Experimental Procedures”), and column fractions were analyzed for mismatch-provoked excision activity and protein composition by SDS-PAGE. The results from the ssDNA cellulose column are shown in Fig. 2A. During ssDNA cellulose chromatography, the excision-stimulatory activity correlated with multiple major protein bands. However, protein analysis of Mono S and other chromatography steps indicated that the stimulatory factor did not include polypeptides larger than 83 kDa (supplemental Fig. S1). Therefore, we conclude that the stimulatory factor is likely to include three major polypeptides of <83 kDa, which are 68, 36, and 30 kDa in size (Fig. 2, arrows, and supplemental Fig. S1). This conclusion is supported by results of HAP chromatography (Fig. 2B). When the active fractions (fractions 51–56) from the ssDNA cellulose column were pooled and applied to a HAP column, the stimulatory activity, which did not bind to the HAP column, again copurified with polypeptides 68, 36, and 30 kDa in size (Fig. 2B, arrows).
To confirm the identity of these three polypeptides, they were excised from the protein gel, digested with trypsin, and analyzed by mass spectrometry. Peptide sequences produced by tandem mass spectrometry analysis were queried against the nonredundant protein data base using two search algorithms: ProteinLynx and Mascot. The results showed that peptides from all three polypeptides produced high confidence hits in the protein data base (Table 1 and supplemental Table S1). Notably, peptides from all three proteins had Mascot scores of >400, and both methods definitively proved that the three polypeptides that copurify with the mismatch-provoked excision stimulatory factor are the subunits of RFX: RFX5 (68 kDa), RFXAP (36 kDa), and RFXANK (30 kDa). In addition, the electrophoretic mobility of the major bands associated with the stimulatory factor during SDS-PAGE are consistent with the predicted molecular weight of RFX subunits (Fig. 2), and the stimulatory activity copurifies with a polypeptide that cross-reacts with antibody to RFX5 (data not shown and supplemental Fig. S2). A recent study by Garvie et al. (18) has shown that RFX is composed of a 2:1:1 complex of RFX5 ·RFXAP ·RFXANK, which can further associate with a dimmer of RFX5 to form a 4:1:1 large complex. This explains why the stimulatory factor elutes immediately after the void volume of the S300 column and why the 68-kDa (RFX5) band is much abundant than the other two subunits (Figs. 2 and 3A).
TABLE 1.
Identification of RFX subunits by mass spectrometry
FIGURE 3.
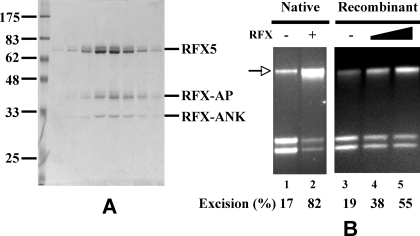
RFX is the stimulatory factor. A, SDS-PAGE of purified RFX. B, RFX stimulates EXO1-catalyzed excision. Mismatch-provoked DNA excision assay was performed in a reconstituted system containing the presence and absence of recombinant RFX. Assays contained EXO1 (3 fmol), MutSα (400 fmol), MutLα (400 fmol), RPA (800 fmol), and HMGB1 (1,000 fmol) with or without native (800 fmol) or recombinant (400 and 800 fmol) RFX.
Both Native and Recombinant RFX Facilitate EXO1-catalyzed Mismatch Excision—EXO1, a 5′ → 3′ exonuclease, is the only nuclease implicated in mammalian MMR (19–21). Therefore, the excision activity was tested for immunoreactivity to antibody to full-length recombinant EXO1 (see “Experimental Procedures”). This experiment suggests that EXO1 is a component of the partially purified mismatch excision activity (data not shown and supplemental Fig. S2). Interestingly, EXO1 and RFX coeluted during early purification steps, and the overlap between the peaks of EXO1 and RFX protein correlated with the peak of excision activity (supplemental Fig. S2). This finding suggests that RFX and EXO1 could be two critical components of a reconstituted/defined in vitro eukaryotic MMR assay, as tested below.
Fig. 3B demonstrates a reconstituted in vitro assay for quantifying mismatch-provoked 5′ → 3′ excision in the presence of purified human MutSα, MutLα, EXO1, RPA, and HMGB1 (7). Note that in these experiments, the amount of EXO1 was somewhat suboptimal, compared with previously published experiments (7). Thus, minimal excision occurred in the absence of the stimulatory factor (Fig. 3B, lane 1) or recombinant RFX (Fig. 3B, lane 3). However, the stimulatory factor (Fig. 3B, lane 2) or recombinant RFX (Fig. 3B, lanes 4 and 5) strongly stimulated the excision reaction. It is interesting to note that native RFX is more active than recombinant RFX in this reaction. This could be due to phosphorylation of native RFX5, because we have identified its phosphorylation at Ser347, Ser185, and Thr614 by mass spectrometry (data not shown). Nevertheless, both proteins stimulate EXO1-catalyzed excision significantly. These data strongly support the conclusion that the excision stimulatory factor is RFX.
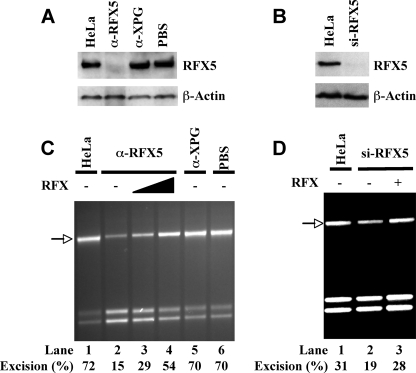
RFX Depletion Inhibits MMR Activity—The role of RFX in mismatch-provoked excision was also examined by depleting or knocking down RFX in HeLa cells. RFX depletion using antibody to RFX5 specifically depleted RFX5 protein by more than 90% (Fig. 4A) and also inhibited mismatch-provoked excision (Fig. 4C, lane 2); similar results were observed with shRNA-mediated RFX knockdown (Fig. 4, B and D); however, control antibody (against nucleotide excision repair enzyme XPG) or phosphate-buffered saline buffer did not deplete RFX protein (Fig. 4A) or inhibit excision activity (Fig. 4C). The addition of recombinant RFX to the RFX5-depleted extract reversed the effect of antibody depletion (Fig. 4C, lanes 3 and 4) as well as shRNA knockdown, supporting a role for RFX in MMR.
FIGURE 4.
Reduced MMR activity in RFX-depleted HeLa nuclear extracts. A, Western blot analysis of RFX5 in immuno-depleted HeLa extracts. HeLa nuclear extracts were treated with the indicated antibodies or phosphate-buffered saline buffer and analyzed by Western blot with antibody to RFX or β-actin (control for protein loading), as indicated. Untreated extract is shown on the left as a negative control. B, Western blot of RFX5 in RFX-knockdown HeLa cells by RNAi. Cell extracts from RFX-RNAi-treated and untreated HeLa cells were blotted with antibody to RFX or β-actin, as indicated. C and D, mismatch-provoked excision assay in RFX-depleted extracts by immunodepletion (C) or RNAi (D). Excision products were indicated by arrows, and excision activities (%) are also shown.
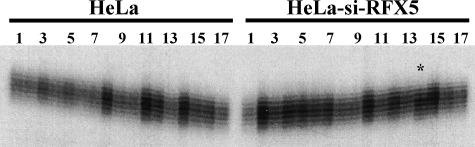
Because defects in MMR causes genome-wide instability at DNA level, including microsatellite instability, subclones were obtained from an RFX5-shRNA knockdown clone and analyzed for microsatellite instability employing three commonly used microsatellite markers, BAT25, BAT26, and D2S123. Although polymorphic changes were not identified in BAT26 and D2S123 (data not shown), instability (one nucleotide contraction) was indeed detected in a subclone of the knockdown cells when BAT25 was analyzed (Fig. 5, clone 14 in the right panel).
FIGURE 5.
Microsatellite instability in RFX-knockdown cells. DNA from single-cell clones of HeLa cells or RFX5-RNAi-treated HeLa cells was used as template to amplify mononucleotide repeat marker, BAT25, in the presence of [α-32P]dCTP as described (26, 27). The PCR products were subjected to electrophoresis and visualized by a phosphorimaging device. Each number represents a subclone. Note that subclone 14 (indicated by a star) derived from the RNAi-treated cells exhibited a different pattern of the BAT25 alleles from that of the rest of clones.
Our results shown here strongly support a role for RFX in MMR. However, it is also noted that depletion of RFX does not completely inhibit MMR (Fig. 4) and only results in a partial mutator phenotype (Fig. 5). These phenomena could be due to the fact that RFX was not completely depleted in the cells/extracts or that RFX is a facilitating factor rather than an essential component in MMR.
DISCUSSION
This study identifies RFX as a novel factor that stimulates 5′ → 3′ mismatch-provoked excision in vitro. Two sources of RFX were used in this study: native RFX purified from HeLa cells and recombinant human RFX, overexpressed and purified from insect cells using a baculovirus expression system. Importantly, both RFX preparations stimulated mismatch-provoked excision in crude HeLa nuclear extracts or in a reconstituted system. These facts validate the results of this study.
RFX plays a well established role in regulating expression of genes encoding the human class II major histocompatibility complex (MHCII) (11–13). Defects in any RFX subunit cause the BLS, a severe immunodeficiency disorder characterized by a lack of MHCII, absence of cellular and humoral T-cell immune response to foreign antigens, and impaired antibody productions, leading to extreme susceptibility to viral, bacterial, and fungal infections. As a result, these patients usually fail to reach puberty (22). The in vitro data presented here suggest a possible role for RFX in MMR. These results pose an obvious and important question: are BLS patients defective in MMR?
Although a definitive answer to the question is unknown, our preliminary studies in cell lines seem to support a role for RFX in MMR in vivo. First, HeLa cells with RFX5 knockdown by RNAi exhibited microsatellite instability (Fig. 5), an indicator of MMR deficiency (reviewed in Ref. 1). Second, a B-lymphocyte cell line (a gift from Jeremy Boss, Emory University) derived from an RFX5-deficient BLS patient is defective in strand-specific MMR.4 However, understanding whether or not the BLS patients are defective in MMR requires a direct investigation of BLS patients for MMR capability.
The specificity and mechanism of how RFX stimulates MMR are not yet clear. As a transcription factor, RFX regulates MHCII gene expression by binding to the promoter regions (11, 13). The DNA binding activity of RFX may allow it to interact with MMR components and facilitate mismatch excision; these components may include MutSα, MutLα, PCNA, replication factor C, and EXO1. Recent studies have revealed that RFX, particularly the RFXANK subunit, is required for conquering the repressive influence of chromatin for MHCII gene expression by interacting with nucleosomes and/or by recruiting chromatin-remodeling or -modifying factors (23, 24). The interactions between RFX and chromatin-remodeling or -modifying factors have been shown to induce histone hyperacetylation of a broad chromatin domain at RFX target genes, and it is this change in chromatin structure that allows the recruitment of RNA polymerase II to the target gene loci for transcription (23). Like transcription, chromatin remodeling may be also required for DNA repair. Despite the fact that the human MMR reaction has recently been reconstituted in vitro using a naked mismatched DNA and purified proteins (6, 7), it is anticipated that the protein components adequate for naked DNA heteroduplexes are unlikely sufficient for MMR at the chromatin level. The chromatin remodeling function of RFX may facilitate MMR in vivo, where DNA is wrapped with histone proteins in nucleosomes. Further studies are required to explore this possibility and to determine how RFX stimulates mismatch-provoked excision in vitro.
This study shows that the human mismatch-provoked excision activity separates into two components during Sephacryl S-300 gel filtration chromatography (Fig. 1), one of which is a 5′ → 3′ exonuclease. The identity of the 5′ → 3′ exonuclease was established here using immunological methods. In particular, Western blot analysis showed that the 5′ → 3′ exonuclease copurifies with a polypeptide that is recognized by a highly specific antibody to full-length EXO1 (supplemental Fig. S2 and data not shown). Because mismatch-provoked excision activity is also observed in a reconstituted system using purified EXO1, we conclude that the EXO1 is a critical component of the RFX-stimulated mismatch-provoked excision activity characterized in this study. These findings should stimulate additional molecular and mechanistic studies on the roles of RFX and EXO1 in human MMR.
EXO1 has been shown to be involved in 3′-directed mismatch repair (19). It is worth mentioning that our previous studies failed to detect the presence of EXO1 and 3′-directed mismatch-provoked excision in the RFX-containing partially purified excision activity (10). We realize now that the former was due to the use of a less specific antibody to a fragment of EXO1, and the latter (i.e. no 3′-directed excision) was due to the fact that the partially purified excision activity does not contain MutLα (data not shown), whose endonuclease activity is essential for 3′-directed MMR by EXO1 (25).
Supplementary Material
Acknowledgments
We thank Matija Peterlin for providing RFX cDNAs and Chengtao Her for technical help in RFX-RNAi knockdown.
This work was supported, in whole or in part, by National Institutes of Health Grants CA115942 and GM072756 (to G.-M. L.) and CA104333 (to L. G.). This work was also supported by a grant from the DOE/NASA Low Dose Radiation Research Program (to D. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and supplemental Figs. S1 and S2
Footnotes
The abbreviations used are: MMR, mismatch repair; RFX, regulatory factor X; MHC, major histocompatibility complex class; BLS, bare lymphocyte syndrome; RNAi, RNA interference; PCNA, proliferating cell nuclear antigen; HAP, hydroxyapatite; ssDNA, single-stranded DNA; shRNA, small hairpin RNA; RPA, replication protien A.
L. Gu and G.-M. Li, unpublished observations.
References
- 1.Li, G. M. (2008) Cell Res. 18 85–98 [DOI] [PubMed] [Google Scholar]
- 2.Modrich, P., and Lahue, R. (1996) Annu. Rev. Biochem. 65 101–133 [DOI] [PubMed] [Google Scholar]
- 3.Kolodner, R. D., and Marsischky, G. T. (1999) Curr. Opin. Genet. Dev. 9 89–96 [DOI] [PubMed] [Google Scholar]
- 4.Lahue, R. S., Au, K. G., and Modrich, P. (1989) Science 245 160–164 [DOI] [PubMed] [Google Scholar]
- 5.Kunkel, T. A., and Erie, D. A. (2005) Annu. Rev. Biochem. 74 681–710 [DOI] [PubMed] [Google Scholar]
- 6.Constantin, N., Dzantiev, L., Kadyrov, F. A., and Modrich, P. (2005) J. Biol. Chem. 280 39752–39761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang, Y., Yuan, F., Presnell, S. R., Tian, K., Gao, Y., Tomkinson, A. E., Gu, L., and Li, G. M. (2005) Cell 122 693–705 [DOI] [PubMed] [Google Scholar]
- 8.Amin, N. S., Nguyen, M. N., Oh, S., and Kolodner, R. D. (2001) Mol. Cell. Biol. 21 5142–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei, K., Clark, A. B., Wong, E., Kane, M. F., Mazur, D. J., Parris, T., Kolas, N. K., Russell, R., Hou, H., Jr., Kneitz, B., Yang, G., Kunkel, T. A., Kolodner, R. D., Cohen, P. E., and Edelmann, W. (2003) Genes Dev. 17 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo, S., Presnell, S. R., Yuan, F., Zhang, Y., Gu, L., and Li, G. M. (2004) J. Biol. Chem. 279 16912–16917 [DOI] [PubMed] [Google Scholar]
- 11.DeSandro, A., Nagarajan, U. M., and Boss, J. M. (1999) Am. J. Hum. Genet. 65 279–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagarajan, U. M., Louis-Plence, P., DeSandro, A., Nilsen, R., Bushey, A., and Boss, J. M. (1999) Immunity 10 153–162 [DOI] [PubMed] [Google Scholar]
- 13.Reith, W., and Mach, B. (2001) Annu. Rev. Immunol. 19 331–373 [DOI] [PubMed] [Google Scholar]
- 14.Holmes, J., Jr., Clark, S., and Modrich, P. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 5837–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, D., Park, J. S., Chu, J. S., Krakowski, A., Luo, K., Chen, D. J., and Li, S. (2004) J. Biol. Chem. 279 43725–43734 [DOI] [PubMed] [Google Scholar]
- 16.Carmell, M. A., Zhang, L., Conklin, D. S., Hannon, G. J., and Rosenquist, T. A. (2003) Nat. Struct. Biol. 10 91–92 [DOI] [PubMed] [Google Scholar]
- 17.Dzantiev, L., Constantin, N., Genschel, J., Iyer, R. R., Burgers, P. M., and Modrich, P. (2004) Mol. Cell. 15 31–41 [DOI] [PubMed] [Google Scholar]
- 18.Garvie, C. W., Stagno, J. R., Reid, S., Singh, A., Harrington, E., and Boss, J. M. (2007) Biochemistry 46 1597–1611 [DOI] [PubMed] [Google Scholar]
- 19.Genschel, J., Bazemore, L. R., and Modrich, P. (2002) J. Biol. Chem. 277 13302–13311 [DOI] [PubMed] [Google Scholar]
- 20.Tishkoff, D. X., Amin, N. S., Viars, C. S., Arden, K. C., and Kolodner, R. D. (1998) Cancer Res. 58 5027–5031 [PubMed] [Google Scholar]
- 21.Schmutte, C., Marinescu, R. C., Sadoff, M. M., Guerrette, S., Overhauser, J., and Fishel, R. (1998) Cancer Res. 58 4537–4542 [PubMed] [Google Scholar]
- 22.Elhasid, R., and Etzioni, A. (1996) Blood Rev. 10 242–248 [DOI] [PubMed] [Google Scholar]
- 23.Masternak, K., Peyraud, N., Krawczyk, M., Barras, E., and Reith, W. (2003) Nat. Immunol. 4 132–137 [DOI] [PubMed] [Google Scholar]
- 24.Mudhasani, R., and Fontes, J. D. (2005) Mol. Immunol. 42 673–682 [DOI] [PubMed] [Google Scholar]
- 25.Kadyrov, F. A., Dzantiev, L., Constantin, N., and Modrich, P. (2006) Cell 126 297–308 [DOI] [PubMed] [Google Scholar]
- 26.Gu, L., Wu, J., Zhu, B. B., and Li, G. M. (2002) Nucleic Acids Res. 30 2758–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsons, R., Li, G. M., Longley, M. J., Fang, W. H., Papadopoulos, N., Jen, J., de la Chapelle, A., Kinzler, K. W., Vogelstein, B., and Modrich, P. (1993) Cell 75 1227–1236 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.