Abstract
Ubc7p is a ubiquitin-conjugating enzyme (E2) that functions with endoplasmic reticulum (ER)-resident ubiquitin ligases (E3s) to promote endoplasmic reticulum-associated degradation (ERAD). Ubc7p only functions in ERAD if bound to the ER surface by Cue1p, a membrane-anchored ER protein. The role of Cue1p was thought to involve passive concentration of Ubc7p at the surface of the ER. However, our biochemical studies of Ubc7p suggested that Cue1p may, in addition, stimulate Ubc7p E2 activity. We have tested this idea and found it to be true both in vitro and in vivo. Ubc7p bound to the soluble domain of Cue1p showed strongly enhanced in vitro ubiquitination activity, both in the presence and absence of E3. Cue1p also enhanced Ubc7p function in vivo, and this activation was separable from the established ER-anchoring role of Cue1p. Finally, we tested in vivo activation of Ubc7p by Cue1p in an assay independent of the ER membrane and ERAD. A chimeric E2 linking Ubc7p to the Cdc34p/Ubc3p localization domain complemented the cdc34-2 TS phenotype, and co-expression of the soluble Cue1p domain enhanced complementation by this chimeric Ubc7p E2. These studies reveal a previously unobserved stimulation of Ubc7p E2 activity by Cue1p that is critical for full ERAD and that functions independently of the well known Cue1p anchoring function. Moreover, it suggests a previously unappreciated mode for regulation of E2s by Cue1p-like interacting partners.
A significant component of protein degradation in eukaryotes occurs at the surface of the ER3 (1–4). In this process of ER-associated degradation (ERAD), integral membrane and luminal ER proteins destined for degradation are targeted to the proteasome by the covalent addition of ubiquitin. Attachment of ubiquitin to target proteins occurs by a cascade of enzymes, beginning with a ubiquitin-activating enzyme (E1) hydrolyzing ATP to form a thioester-linked ubiquitin-E1 adduct. The E1 next passes its ubiquitin to a ubiquitin-conjugating enzyme (E2), also as a thioester-linked intermediate. Finally, ubiquitination of the target protein is promoted by a ubiquitin ligase (E3) that facilitates transfer of ubiquitin from the E2 to a lysine on the target protein (or a previously added ubiquitin), thus promoting the polyubiquitination of proteins targeted for degradation.
In the baker's yeast Saccharomyces cerevisiae, Ubc7p is an E2 required for ERAD mediated by two ER-localized E3s, Hrd1p and Doa10p (5–7). Ubc7p is able to engage these ER-localized E3s because Ubc7p is anchored to the ER through interaction with the integral ER membrane protein Cue1p. In fact, Cue1p is required for the Ubc7p-dependent ubiquitination and degradation of substrates in the ER (8, 9). Because proximity and interaction between E2 and E3 are critical for ubiquitination function (10), it has been suggested that the main function of Cue1p is to concentrate Ubc7p at the ER membrane surface, thus allowing fruitful engagement of Ubc7p with ER-localized E3s (8).
Although genetic experiments have established a requirement for Cue1p in ERAD and biochemical studies have confirmed that Cue1p and Ubc7p interact, the effects of Cue1p on the enzymatic activity of Ubc7p have not been explored. In our previous study of Hrd1p specificity with Ubc7p, the presence of Cue1p enhanced Ubc7p activity in biochemical assays of Ubc7p function, suggesting that Ubc7p may be activated by Cue1p (11).
Here we directly test the idea that Cue1p stimulates E2 activity of Ubc7p in vitro and in vivo. We examined the nature of the polyubiquitin chains formed in these E3-independent in vitro reactions. We discovered that in vitro, Ubc7p produced lysine 48-linked polyubiquitin chains, and a soluble portion of Cue1p strongly stimulated this Ubc7p activity in the presence or absence of E3. We then designed chimeric proteins to express in vivo that would separate the established anchoring function of Cue1p from its putative activation function and found that both anchoring and Cue1p-based activation were important for Hrd1p-dependent ERAD. We also developed means to assay Ubc7p activity in a context independent of ERAD or the ER membrane and found that Cue1p activated Ubc7p in a manner entirely independent of ER anchoring. Taken together, these results reveal a previously unknown role for Cue1p as an activator of Ubc7p E2 activity and suggest that other E2s may have similar stimulating cofactors.
EXPERIMENTAL PROCEDURES
Recombinant DNA—All DNA segments synthesized by PCR were verified by sequencing. The Ubc7p-chitin-binding domain/intein fusion vector pRH1946 was previously described (11). Ubc7p-2HA coding region was amplified by PCR and subcloned into pTYB2 (New England Biolabs) to produce the Ubc7p-2HA-chitin-binding domain/intein fusion vector pRH1947. Cue1pΔ™, which lacks amino acids 2–22 of Cue1p (and thus the included transmembrane span) was amplified by PCR from pTX129 (8) and cloned into a pET bacterial expression vector. Then the ribosomal binding site and Cue1pΔ™ were amplified by PCR and subcloned into both pRH1946 and pRH1947 behind the Ubc7p-CBD/intein to produce pRH2061 and pRH2064, whose polycistronic message encoded both Cue1pΔ™ and either untagged or HA-tagged Ubc7p-CBD/intein in one inducible operon. GST was expressed from the pET42b(+) bacterial expression plasmid (Novagen). GST-E3 was the previously described fusion to Hrd1p expressed from pRH1726 (11). His6-tagged mouse UBA1 (E1) and HUBC4 were purified from bacterial lysates as described previously (11–14). Ubc7p with two HA epitope tags was expressed in yeast from the strong TDH3 promoter using the previously described vector pRH373 (9). To express Ubc7p-2HA from the native UBC7 promoter, the identical coding sequence for Ubc7p-2HA was amplified by PCR and subcloned into a yeast expression vector containing the native UBC7 promoter (pRH2193). For expression of Cue1p in yeast, sequence encoding full-length Cue1p was amplified by PCR and subcloned between the TDH3 promoter and three HA epitope tags of an existing yeast expression vector (pRH1334). Membrane-anchored versions of Ubc7p were made by a PCR SOEing method (15, 16). Sequences encoding the N-terminal 22-amino acid transmembrane span of Cue1p and the entire coding region of Ubc7p-2HA were amplified by PCR and joined by PCR SOEing, and this chimeric PCR product was subcloned into a vector allowing expression of membrane-anchored Ubc7p without linker from the strong TDH3 promoter (pRH2190). TM-Ubc7p included amino acids 531–618 of Hmg2p, a portion of the cytosolic linker between the transmembrane domain and conserved cytosolic catalytic domain of Hmg2p. Sequence encoding this 88-amino acid linker was amplified from pRH469 by PCR and joined to sequences encoding the Cue1p transmembrane span and Ubc7p-2HA by PCR SOEing to produce the TM-Ubc7p sequence. This chimeric PCR product was subcloned into a vector, allowing expression of TM-Ubc7p from the strong TDH3 promoter (pRH2191). Similarly, sequence for the linker described above joined to Ubc7p-2HA, without the transmembrane span of Cue1p, was amplified by PCR and subcloned into pRH2191 to produce pRH2457, expressing the N-terminally modified linker-Ubc7p-2HA protein (L-Ubc7p) from the TDH3 promoter. To express Cue1pΔ™ in yeast, the sequence encoding amino acids 23–203 and the adjacent three HA epitope tags was amplified by PCR from pRH1334 and subcloned behind the strong TDH3 promoter in a yeast expression vector (pRH2198). Sequence encoding Cdc34p was amplified from genomic DNA and subcloned into the previously described p416-GPD vector (17) between the TDH3 promoter and CYC1 terminator to produce pRH1939. The native CDC34 promoter was amplified from pRG721 (Richard G. Gardner, University of Washington) and subcloned into pRH1939 to make pRH1971, expressing Cdc34p from the CDC34 promoter. To make Ubc7p-Cdc34, UBC7 sequence was linked by PCR SOEing to sequence encoding amino acids 171–295 of Cdc34p. The resulting DNA was subcloned into pRH1939 to express Ubc7p-Cdc34 from the TDH3 promoter (pRH1968). pRH1969, expressing the function-blocking C89S mutant version of Ubc7p-Cdc34, was made as above, except Ubc7p sequence was amplified from a template with the C89S point mutation. Ubc7p-encoding sequence was also cloned into pRH1939 to express Ubc7p from the same vector as the other E2s. These E2 constructs were then each subcloned into pRH1971 described above to express them from the native CDC34 promoter; pRH1983 expressed Ubc7p-Cdc34, pRH1985 expressed Ubc7p-Cdc34 with C89S, and pRH1987 expressed Ubc7p. Protein molecular weight prediction was performed using the Compute pI/Mw tool on the ExPASy proteomics server (18).
Strains and Media—Yeast strains were cultured as described (19, 20), in minimal media with 2% glucose and amino acid supplements, at 30 °C unless otherwise indicated. Only in the cdc34-2 complementation experiments, strains were grown in synthetic complete media lacking uracil and leucine to maintain plasmid selection. All yeast strains were derived from the same genetic background used in our previous work (19, 20). Strains for evaluating the in vivo degradation of Hmg2p-GFP were derived from RHY853 (21), expressing the catalytic domain of Hmg2p as its sole source of 3-hydroxy-3-methylglutaryl-CoA reductase, and Hmg2p-GFP. To test complementation of Ubc7p and Ubc7p-containing constructs, UBC7 was replaced with the selectable HIS3 marker, producing the ubc7Δ strain RHY1848. Constructs expressing Ubc7p, Cue1p, and membrane-anchored versions of Ubc7p were introduced into this strain to test their restoration of Ubc7p function. To examine restoration of both Ubc7p and Cue1p function, the CUE1 gene was replaced in RHY1848 with the nourseothricin (Clon-Nat) resistance marker natMX (22) to produce the ubc7Δ cue1Δ strain RHY5917. In this strain, Cue1pΔ™ and TM-Ubc7p were expressed (individually and together) to test restoration of ERAD function. The cdc34-2 strains were generated in the following manner. To convert the native CDC34 locus to the cdc34-2 allele encoding the G58R mutation, the pRG721 plasmid encoding cdc34-2 was integrated into the strain RHY2863 (ade2–101 met2 lys2–801 ura3–52 trp1:hisG leu2Δ his3Δ200) at the CDC34 locus, placing the selectable URA3 gene between two copies of CDC34, one wild-type and one mutant. These cells were grown in the presence of uracil to allow spontaneous recombination between the two CDC34 loci and loss of URA3, leaving only one copy of CDC34. Such strains were selected on media with 5-fluoroorotic acid for absence of URA3 and then screened for temperature sensitivity to identify strains that retained only the mutant cdc34-2 allele. This cdc34-2 strain derived from RHY2863 is RHY3802. RHY3802 was transformed with plasmids to express from the TDH3 promoter Cdc34p, Ubc7p-Cdc34, or Ubc7p, to test complementation of the cdc34-2 TS growth phenotype. RHY3802 was also transformed with plasmids to express from the CDC34 promoter Cdc34p, Ubc7p-Cdc34, or Ubc7p, to test complementation of the cdc34-2 TS growth phenotype. In turn, the CDC34 promoter strains were transformed with empty vector or Cue1pΔ™ expression plasmid to assess the activation of Ubc7p by Cue1p. CUE1 was disrupted in RHY3802 by introducing a knock-out cassette containing the natMX gene, conferring resistance to nourseothricin. Loss of the CUE1 gene was confirmed by PCR, yielding the cdc34-2 cue1Δ strain RHY7371. This strain was then transformed with plasmids to express from the CDC34 promoter Cdc34p or Ubc7p-Cdc34 as above. Cue1pΔ™ expression plasmid was also transformed as above to assess activation of Ubc7p-Cdc34 by Cue1p in a cue1Δ strain.
Protein Purification—All recombinant proteins were expressed in Rosetta(DE3) Escherichia coli (Novagen) grown in LB with appropriate antibiotics. E1, all E2s, and E3 were each purified using the appropriate affinity matrix and previously described methods (11). Single use aliquots of each protein preparation were flash-frozen with liquid nitrogen and stored at –80 °C for later use. E1 and HUBC4 were expressed with the His6 tag and purified using Talon Cell-Thru resin (BD Biosciences). GST and GST-Hrd1p fusion (E3) were purified using glutathione-Sepharose-4B resin (Amersham Biosciences). To purify Ubc7p alone or to co-purify Ubc7p with co-expressed Cue1pΔ™, Ubc7p was expressed as a chitin-binding domain/intein fusion. Each bacterial pellet from 1 liter of culture expressing an intein/CBD fusion was resuspended in 25 ml of intein lysis buffer (ILB; 50 mm Tris, pH 8.0, 500 mm NaCl, 1 mm EDTA, 0.1% Triton X-100) with protease inhibitors (260 μm AEBSF, 105 μm leupeptin, 73 μm pepstatin, 142 μm TPCK) and sonicated as before. Lysate was centrifuged at 20,000 × g for 30 min in an SS34 rotor. Supernatant was filtered through 0.45- and 0.2-μm filters and added to 15 ml of chitin beads (New England Biolabs) equilibrated in ILB and nutated for 90 min at 4 °C. The adsorbed resin was placed in a 2.5-cm column and washed with 350–400 ml of ILB. Next, the resin was nutated in 10 ml of ILB plus 50 mm DTT for 20 h at 4 °C to promote intein cleavage, and chitin beads were washed with ILB to collect intein-cleaved proteins. 40 ml of fluid were collected and concentrated using Amicon Ultra-15 5,000 molecular weight cut-off filters (Millipore). Concentrated protein was dialyzed against 3 × 1 liter of HDBG (25 mm HEPES, 0.7 mm sodium phosphate, 137 mm NaCl, 5 mm KCl, pH 7.4, 10% glycerol) for 24 h in a 0.5-ml 3,000 molecular weight cut-off Slide-a-Lyzer cassette (Pierce). Proteins were ultracentrifuged at 100,000 × g to remove any aggregates, and supernatant was aliquoted and frozen.
For gel filtration analysis, Ubc7p-2HA·Cue1pΔ™ was prepared as above (11) with the following modifications. The protein eluate was collected and concentrated as described and then dialyzed against 3× 1 liter of UbR150 buffer (150 mm NaCl, 50 mm Tris, pH 7.5, 2.5 mm MgCl2, 0.5 mm DTT) for 24 h in a 0.5-ml 3,000 molecular weight cut-off Slide-a-Lyzer cassette (Pierce). This buffer was like ubiquitin reaction buffer with the addition of 150 mm NaCl to reduce nonspecific interactions with the gel filtration resin. The dialyzed protein preparation was centrifuged at 6,000 rpm in an SS34 rotor to precipitate any aggregates prior to gel filtration.
In Vitro Ubiquitination—Ubiquitin was resuspended from lyophilized powder in ubiquitin storage buffer (50 mm Tris, pH 7.5, 50 mm NaCl, 10% glycerol) and frozen. Mutant varieties of ubiquitin were purchased from Boston Biochem, Inc. (Cambridge, MA). Reactions were performed in 1× ubiquitination buffer (50 mm Tris, pH 7.5, 2.5 mm MgCl2, 0.5 mm DTT) with 3 mm ATP, 80 μg/ml ubiquitin, 6 μg/ml E1, 20 μg/ml E2, in a total volume of 15 μl. Protein concentrations were determined by Coomassie staining and comparison with bovine serum albumin standards. In each experiment, proteins common to multiple reactions were added to a reaction mixture and mixed to ensure equal addition of the common components in each reaction. Such partially assembled reactions were aliquoted to individual tubes for the addition of unique reaction components. Reaction mixtures were prepared on ice and then incubated at 30 °C for 2 h and stopped with an equal volume of 2× sample buffer (4% SDS (w/v), 8 m urea, 75 mm MOPS, pH 6.8, 200 mm DTT, 0.2 mg/ml bromphenol blue) and analyzed by SDS-PAGE and immunoblotting or Coomassie staining as indicated.
Immunoprecipitation of Ubc7p with Thioester-linked Ubiquitin—Ubiquitin reactions were prepared as above in 50-μl reactions and then incubated at 30 °C for 2 h. Reactions were stopped by adding 100 μl of SUME (1% (w/v) SDS, 8 m urea, 10 mm MOPS, pH 6.8, 10 mm EDTA) with protease inhibitors (260 μm AEBSF, 105 μm leupeptin, 73 μm pepstatin, 142 μm TPCK) and 5 mm N-ethylmaleimide, followed by the addition of 600 μl of IP buffer (15 mm sodium phosphate, 150 mm NaCl, 10 mm EDTA, 2% Triton X-100, 0.1% SDS, 0.5% deoxycholate) with protease inhibitors above. HA epitope antibody-conjugated resin (Covance) was diluted 6-fold in IP buffer with 0.5 mg/ml bovine serum albumin and incubated for 20 min to block the resin. 120 μl of resin/bovine serum albumin slurry was added to each stopped reaction and incubated at 4 °C for 6 h to precipitate HA-tagged Ubc7p. Beads were washed once with IP buffer and twice with IP wash (50 mm NaCl, 10 mm Tris, pH 7.5), aspirated to dryness, and heated in nonreducing sample buffer (4% SDS (w/v), 8 m urea, 75 mm MOPS, pH 6.8, 0.2 mg/ml bromphenol blue) or reducing sample buffer (same as above plus 200 mm DTT). The immunoprecipitated proteins were resolved by SDS-PAGE and immunoblotted for HA epitope or ubiquitin.
Assay of E2 Charging by E1—Ubiquitin was resuspended from lyophilized powder in ubiquitin storage buffer (50 mm Tris, pH 7.5, 50 mm NaCl, 10% glycerol) and frozen. Reactions were performed in 1× ubiquitination buffer (50 mm Tris, pH 7.5, 2.5 mm MgCl2, 0.5 mm DTT) with 3 mm ATP, 80 μg/ml ubiquitin, 6 μg/ml E1, and the indicated E2 concentration, in a total volume of 15 μl. Reaction mixtures were prepared on ice, incubated at room temperature for 5 min, and then stopped with an equal volume of non-reducing 2x sample buffer (4% SDS (w/v), 8 m urea, 75 mm MOPS, pH 6.8, 0.2 mg/ml bromphenol blue) and analyzed by SDS-PAGE and immunoblotting for ubiquitin or HA-epitope. Pixel quantitation was performed with Adobe Photoshop version 7.0.1.
Protease Protection Assay—Samples of Ubc7p-2HA or Ubc7p-2HA-Cue1pΔ™ were added to 1× ubiquitination buffer at 40 μg/ml on ice, and lyophilized trypsin resuspended in 1× ubiquitination buffer was added at 10 μg/ml. Trypsin digests were then incubated at room temperature, and aliquots were removed at each indicated time, treated with an equal volume of 2× sample buffer with 260 μm AEBSF, and heated to stop proteolysis. Proteolyzed proteins were resolved by SDS-PAGE and immunoblotted for HA epitope.
Gel Filtration—Using an AKTA FPLC system, a 102.5-ml, 51 × 1.6-cm Superose 6 gel filtration column was equilibrated with UbR150 buffer (150 mm NaCl, 50 mm Tris, pH 7.5, 2.5 mm MgCl2, 0.5 mm DTT) and 1 mm AEBSF. 0.6 mg of Ubc7p-2HA·Cue1pΔ™ in 1 ml of UbR150 buffer was loaded onto the column, and 1-ml fractions were collected at a flow rate of 0.4 ml/min UbR150 buffer with AEBSF. A portion of each elution fraction with UV absorbance at 280 nm was resolved by SDS-PAGE and Coomassie-stained or immunoblotted as indicated. The gel filtration column was run in identical conditions with a mixture of protein standards to correlate the fraction distribution with molecular weight. Lyophilized RNase A (13.7 kDa), chymotrypsin (25 kDa), ovalbumin (43 kDa), and albumin (67 kDa) protein standards were reconstituted in UbR150 at 2 mg/ml each, and 1 ml of the protein standard mixture was applied to the column and run as before. A portion of each elution fraction with UV absorbance at 280 nm was resolved by SDS-PAGE and Coomassie-stained.
Flow Cytometry—Log phase cultures (A600 < 0.5) grown in minimal medium at 30 °C were transferred to flow cytometer sample tubes and measured with a BD Biosciences FACScalibur instrument. Flow microfluorimetric data were analyzed, and histograms were generated using CellQuest flow cytometry software. In all cases, histograms represented 10,000 individual cells.
Cycloheximide Chase Assay—Log phase cultures (A600 < 0.1) grown in minimal medium at 30 °C were split into three tubes. One was treated with no drug. The other two were exposed to 50 μg/ml cycloheximide for either 30 min or 2 h. Then each sample was transferred to flow cytometer sample tubes and measured as above.
Microsome Preparation—Five optical density units of log phase cells grown in minimal media were harvested and resuspended in 200 μl of ice cold membrane fractionation buffer (MFB; 20 mm Tris, pH 7.5, 0.1 m NaCl, 0.3 m sorbitol) with protease inhibitors (260 μm AEBSF, 105 μm leupeptin, 73 μm pepstatin, 142 μm TPCK). Glass beads were added to just below the liquid level. Lysis was performed at 4 °C with six cycles of 1 min of vortexing (maximum speed) and 1 min of incubation on ice. Lysate was harvested by removing supernatant from beads and washing beads twice with 200 μl of MFB, pooling the washes and lysate. The resulting pooled lysate was cleared by repeated 10-s microcentrifuge pulses to remove unlysed cells and large debris. The cleared supernatant contains microsome membranes, which were harvested by centrifugation at 21,000 × g for 30 min. The pellet was resuspended in 100 μl of SUME (1% (w/v) SDS, 8 m urea, 10 mm MOPS, pH 6.8, 10 mm EDTA) with protease inhibitors above. After the addition of 100 μl of 2× sample buffer (4% SDS (w/v), 8 m urea, 75 mm MOPS pH 6.8, 200 mm DTT, 0.2 mg/ml bromphenol blue) and heating at 65 °C, the samples were analyzed by SDS-PAGE and anti-HA immunoblotting.
Whole Cell Lysates—Five optical density units of log phase cells grown in minimal medium were harvested and resuspended in 100 μl of SUME (1% (w/v) SDS, 8 m urea, 10 mm MOPS, pH 6.8, 10 mm EDTA) with protease inhibitors above and vortexed with glass beads for 3 min. Then 100 μl of 2× sample buffer (4% SDS (w/v), 8 m urea, 75 mm MOPS, pH 6.8, 200 mm DTT, 0.2 mg/ml bromphenol blue) was added, and samples were heated at 65 °C for 10 min and analyzed by SDS-PAGE and immunoblotting.
Growth Assay for cdc34-2 Temperature-sensitive Phenotype—Log phase cultures (A600 < 0.5) for each strain tested were grown in synthetic complete liquid medial lacking leucine and uracil at 30 °C. These were normalized to equal A600 and then serially diluted 5-fold and deposited with a 48-pin replicator onto plates of synthetic complete medium without uracil and leucine. Plates were incubated at the indicated temperatures for 3 days. Images are representative of three experiments with duplicate plates for each temperature.
RESULTS
Cue1p recruits Ubc7p to the surface of the ER and is necessary for ERAD function. By increasing the local Ubc7p concentration at the ER, Cue1p is thought to promote Ubc7p engagement with the ER-localized ERAD E3s. Our in vitro studies of membrane-anchored Hrd1p reveal that Cue1p reduces the concentration of Ubc7p required to observe ubiquitination (11). Observations that soluble Cue1p lacking a transmembrane span (Cue1pΔ™) binds tightly to Ubc7p in vitro and causes cytosolic localization of GFP-Ubc7p in vivo (23) are also consistent with the model of Cue1p as anchor for Ubc7p. However, our previous in vitro studies of soluble, membrane-free Hrd1p function with Ubc7p suggested increased Ubc7p activity in the presence of Cue1p (11). We considered that Cue1p might be an activator of Ubc7p E2 activity. To explore this possibility, we directly tested Cue1p activation of Ubc7p in a soluble in vitro ubiquitination assay.
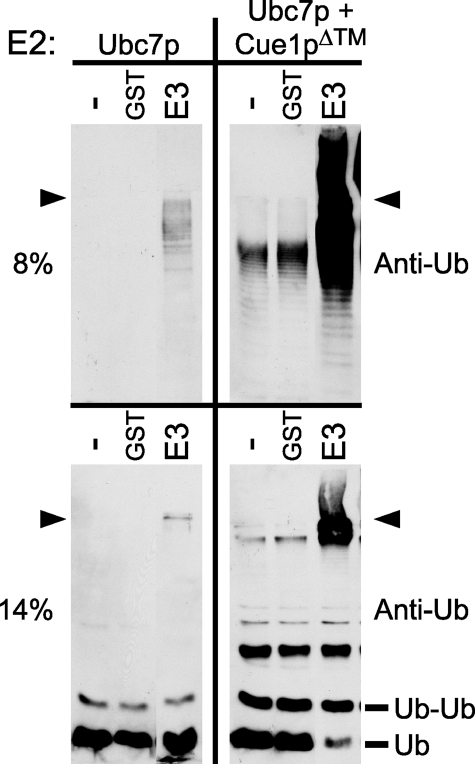
Many RING motif-containing proteins, including Hrd1p, can catalyze the formation of polyubiquitin chains in the presence of E1, E2, ubiquitin, and ATP (7, 11, 12, 24, 25). We adapted this approach to test Ubc7p activity in vitro. Recombinant Ubc7p was expressed from the pTYB2 vector as an inteincleavable fusion to a chitin-binding domain (CBD/intein) and purified using chitin affinity beads. A portion of the Hrd1p cytosolic domain competent for in vitro activity with Ubc7p was fused to GST and affinity-purified using glutathione-Sepharose beads. In vitro reactions were run by combining ATP, ubiquitin, E1, E2, and E3. The formation of polyubiquitin chains in the reactions was evaluated directly by SDS-PAGE and ubiquitin immunoblotting on either 8% (Fig. 1, top) or 14% (Fig. 1, bottom) gels. As can be seen in Fig. 1 (top left), reactions with Ubc7p as E2 formed polyubiquitin chains only in the presence of GST-Hrd1p, and reactions without E3 or GST alone showed no ubiquitin immunoreactivity. To test the effects of Cue1p on Ubc7p activity in vitro, we co-expressed the recombinant Ubc7p described above with Cue1pΔ™, a soluble version of Cue1p lacking its transmembrane span, and purified the Ubc7p·Cue1pΔ™ complex using the same CBD/intein strategy. In contrast to the lone E2, in vitro ubiquitination reactions using Ubc7p·Cue1pΔ™ as the E2 showed strikingly more polyubiquitin formation (Fig. 1, top right). Ubc7p·Cue1pΔ™ catalyzed more polyubiquitination with E3 present but also in the absence of E3. In fact, ubiquitination by Ubc7p·Cue1pΔ™ without E3 was comparable with that of lone Ubc7p with E3, highlighting the strong enhancement of Ubc7p ubiquitination activity by Cue1pΔ™ (Fig. 1, compare top panels). Individual rungs of the polyubiquitin ladder were observed in each of the Ubc7p·Cue1pΔ™ reactions. However, it was puzzling how these ubiquitin-containing chains could have formed without E3 in the reactions. Thus, we wanted to evaluate in more detail the in vitro ubiquitination catalyzed by co-purified Ubc7p and Cue1pΔ™.
FIGURE 1.
Cue1pΔ™ enhanced production of large and small ubiquitin-immunoreactive bands by Ubc7p. In vitro ubiquitination reactions with no E3 (–), GST (GST), or GST-Hrd1p (GST-E3) were prepared with either Ubc7p (left) or Ubc7p·Cue1pΔ™ (right). Identical samples were resolved by SDS-PAGE using 8% gels (top) or 14% gels (bottom), revealing the production of both large and small ubiquitin-immunoreactive bands. Ubiquitination was E3-dependent with Ubc7p alone, whereas Ubc7p·Cue1pΔ™ enhanced ubiquitination both in the presence and absence of E3 (compare left and right panels). The arrowheads indicate the discontinuity between 4% stacking gels and running gels. The molecular weights of monoubiquitin (Ub) and diubiquitin (Ub-Ub) are indicated.
Cdc34p, an E2 associated with the SCF (Skp1/Cul1/F-box) ubiquitin ligase complex, is the E2 most closely related to Ubc7p in yeast (26–28). In the absence of E3, Cdc34p was observed to link two ubiquitin molecules into ubiquitin dimers in vitro, and this activity was strongly enhanced by the addition of purified SCF ubiquitin ligase complex (29). Given the homology between Ubc7p and Cdc34p and that Cue1p has recently been identified as a subunit of larger E3-containing ERAD complexes (30), it seemed possible that the E3-independent, intermediate-sized polyubiquitin chains observed with Ubc7p in vitro might be produced by a mechanism similar to that observed with Cdc34p. To examine this possibility, an identical portion of each reaction above was resolved with 14% SDS-polyacrylamide gels in order to resolve oligoubiquitin structures and immunoblotted for ubiquitin (Fig. 1, bottom). In the presence of Ubc7p·Cue1pΔ™, the in vitro reactions produced multiple ubiquitin-immunoreactive, low molecular weight bands not observed with Ubc7p alone, including a 16 kDa band consistent with a ubiquitin dimer (Fig. 1, bottom, Ub-Ub). Formation of the ubiquitin dimer band with Ubc7p alone was weak compared with Ubc7p·Cue1pΔ™. Although Cue1p strongly enhanced the production of these low molecular weight ubiquitin-immunoreactive bands, there was no further effect of E3 on their production (Fig. 1, bottom right).
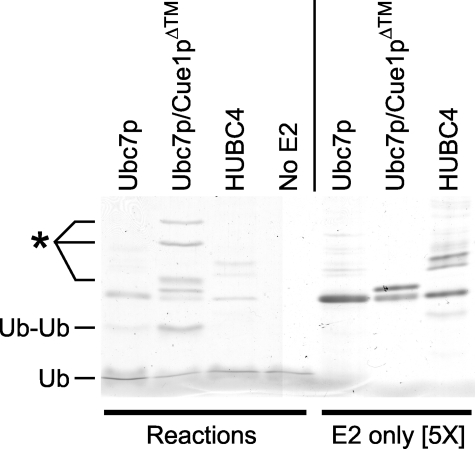
Because these low molecular weight bands were detected by high sensitivity immunoblotting, they could have been low in abundance, representing only a small pool of the protein in the reaction. To examine the extent to which the ubiquitin dimer and other low molecular weight bands were produced, we analyzed the reaction mixes with bulk protein staining. We prepared E3-independent ubiquitination reactions as above, using Ubc7p, Ubc7p·Cue1pΔ™, or human Ubc4 (HUBC4) as E2. These reactions were run as before and then resolved by high percentage SDS-PAGE and stained with Coomassie Brilliant Blue (Fig. 2, left). To distinguish the bands generated in these ubiquitination reactions from bands endemic to the E2 preparations, E2 protein samples were loaded alongside the in vitro reactions (Fig. 2, right) at 5 times their reaction concentration. In the reaction with Ubc7p·Cue1pΔ™ as E2, Coomassie staining easily detected a ubiquitin dimer (Ub-Ub) as well as higher molecular weight bands absent from the other reactions (Fig. 2). In the reaction with lone Ubc7p as E2, a ubiquitin dimer was faintly detectable, but not the higher molecular weight bands seen with Ubc7p·Cue1pΔ™. In the reactions without Cue1p, any faint higher molecular weight bands are similar in size to minor bands present in the E2 preparations (Fig. 2, compare left and right). No bands were produced in the HUBC4 reactions or the reactions without E2. Thus, the E3-independent ubiquitin dimer formation is intrinsic to Ubc7p but not observed in the highly active E2 HUBC4. The E1 in these reactions was not observed, because the high percentage SDS-polyacrylamide gels did not resolve the 115-kDa protein. In the Ubc7p·Cue1pΔ™ reaction, the ubiquitin dimer and larger reaction-product bands were abundant, staining as strongly as the E2 bands. Although immunoblotting indicated that these bands contained ubiquitin, these results did not unambiguously identify the composition of these new bands. Ubiquitin immunoreactivity and the commensurate Coomassie staining band at 16 kDa strongly suggested a dimer of the 8-kDa ubiquitin protein. However, the calculated molecular masses of the E2 test proteins are all similar: HUBC4 is 20.9 kDa, Ubc7p is 19.5 kDa, and Cue1pΔ™ is 20.3 kDa. Thus, it was unclear whether the higher molecular mass bands produced in only the Ubc7p·Cue1pΔ™ reactions were ubiquitinated Ubc7p or Cue1p or ubiquitin multimers.
FIGURE 2.
Coomassie-stained in vitro ubiquitination reactions and E2s. Ubiquitination reactions without E3 and with Ubc7p, Ubc7p·Cue1pΔ™, HUBC4, or no E2, were resolved with 14% SDS-PAGE and Coomassie-stained to observe the size and quantity of small protein products produced by the in vitro ubiquitination reactions (Reactions). Purified E2 preparations were loaded on the same gel at 5 times higher concentration than in the ubiquitination reactions (E2 only [5×]) to identify those bands contributed by the E2 preparations. The molecular weights of monoubiquitin (Ub) and diubiquitin (Ub-Ub) are indicated. *, higher molecular weight bands produced in the Ubc7p·Cue1pΔ™ reaction.
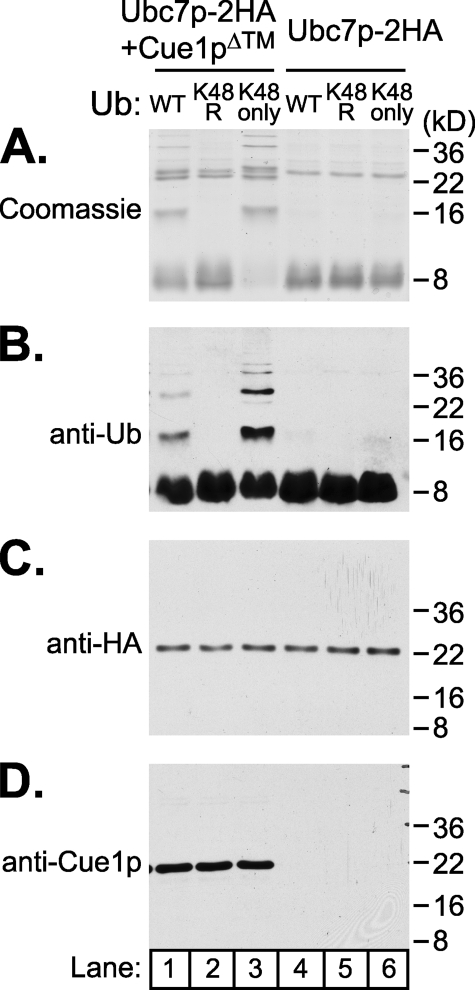
To identify the composition of the products formed by the E3-independent ubiquitination reactions, Cue1p antibodies were obtained (T. Sommer, Max Delbrück Center, Berlin), and the reactions were run with HA-tagged versions of recombinant Ubc7p and Ubc7p·Cue1pΔ™, allowing immunoblotting of each participant. We ran E3-independent in vitro ubiquitination reactions as before, using Ubc7p-2HA·Cue1pΔ™ or Ubc7p-2HA as E2, and resolved the reaction mixes directly by SDS-PAGE. Equal portions of each reaction were loaded and either Coomassie-stained (Fig. 3A) or immunoblotted for ubiquitin, HA epitope, or Cue1p as indicated (Fig. 3, B, C, and D). With Ubc7p-2HA·Cue1pΔ™, Coomassie staining revealed the ubiquitin dimer and higher bands as before, whereas Ubc7p-2HA reactions did not (Fig. 3A, compare lanes 1 and 4). Ubiquitin immunoblotting of the same reaction showed commensurate ubiquitin immunoreactivity (Fig. 3B, lane 1). However, we observed no mobility shift of Ubc7p-2HA or Cue1pΔ™ (Fig. 3, C and D, lane 1), suggesting that the higher molecular weight bands produced in the Ubc7p-2HA·Cue1pΔ™ reactions were composed exclusively of ubiquitin.
FIGURE 3.
E3-indpendent ubiquitination produced multimers of ubiquitin linked through lysine 48. In vitro reactions with either Ubc7p-2HA· Cue1pΔ™ or Ubc7p-2HA were prepared with one of three versions of ubiquitin: wild-type ubiquitin (WT), ubiquitin with lysine 48 changed to arginine (K48R), or ubiquitin with all lysines except lysine 48 changed to arginine (K48only). Reactions were run and then split into four identical portions, resolved by 14% SDS-PAGE and Coomassie-stained or immunoblotted as indicated. E3-independent reaction products enhanced by Cue1p required lysine 48 of ubiquitin, and these higher molecular weight products were composed only of ubiquitin protein (anti-Ub), and not isopeptide conjugates to Ubc7p (anti-HA) or Cue1p (anti-Cue1p) in these reducing conditions.
We wanted to further examine these higher molecular weight ubiquitin-immunoreactive bands to confirm that they were indeed polyubiquitin chains formed by E2 activity. Polyubiquitin chains form by joining the C terminus of one ubiquitin protein with the lysine residue of another ubiquitin in an isopeptide linkage. There are seven lysines on the ubiquitin protein, and each has been observed to receive ubiquitin (31), although the predominant linkage targeting proteasomal degradation is through lysine 48 (32). Mutants of ubiquitin that modify these lysine residues disrupt polyubiquitination while allowing monoubiquitination on substrate lysines (33, 34). To characterize the type of lysine linkages allowed by Ubc7p with Cue1p in this E3-independent process, we prepared reactions using wild type or mutant ubiquitin. K48R-ubiquitin replaces lysine 48 with arginine, and K48only-ubiquitin replaces all lysines with arginine except lysine 48. Reactions using mutant ubiquitin and either Ubc7p-2HA·Cue1pΔ™ or Ubc7p-2HA as E2 were prepared and resolved alongside wild-type ubiquitin reactions (Fig. 3, lanes 2 and 3 and lanes 5 and 6). The bands generated in the E3-independent Ubc7p·Cue1pΔ™ reactions did not form with K48R-ubiquitin but were completely restored by K48only-ubiquitin (Fig. 3, A and B, compare lanes 1–3). The lysine 48 requirement for all of the higher molecular weight ubiquitin-immunoreactive bands strongly suggests that they were composed exclusively of polyubiquitin and that none of these bands resulted from lysine residue ubiquitination of Ubc7p-2HA or Cue1pΔ™. Indeed, there was no change in the mobility of Ubc7p-2HA or Cue1pΔ™ in these reactions (Fig. 3, C and D). It appeared that Cue1pΔ™ enhanced the ability of Ubc7p-2HA to form lysine 48-linked ubiquitin dimers and larger polyubiquitin chains. The high specificity for lysine 48 linkage formation also implies that Cue1p activation of Ubc7p is physiologically relevant, since this is the predominant linkage for proteasomal targeting.
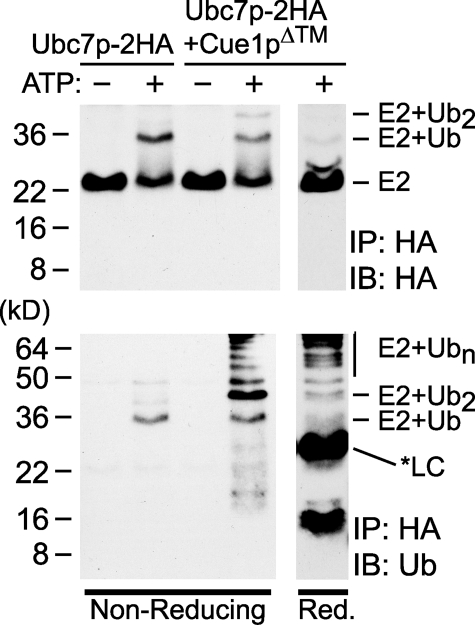
Elegant studies of Ubc7p and its human homolog Ube2g2 reveal that these E2s assemble polyubiquitin chains linked to the E2 through a thioester bond with the conserved catalytic cysteine (23, 35). We wondered if the Cue1p-enhanced polyubiquitin chains were also thioester-linked to Ubc7p. The previous experiments were carried out in reducing conditions that preserve isopeptide bonds formed by ubiquitin conjugation but reverse any thioester bonds that may have been present. In non-reducing conditions, a ubiquitin-Ubc7p thioester bond would be preserved, causing increased mobility of Ubc7p. To determine if polyubiquitin was forming on the catalytic cysteine of Ubc7p, we prepared E3-independent in vitro ubiquitination reactions as before, using either Ubc7p-2HA or Ubc7p-2HA·Cue1pΔ™. Reactions were stopped, the E2 was immunoprecipitated using resin-conjugated antibodies to HA epitope, and the resulting samples were resolved by nonreducing SDS-PAGE and immunoblotted for HA epitope or ubiquitin. In the Ubc7p-2HA reaction, immunoblotting for HA epitope showed an ATP-dependent shift in E2 mobility (Fig. 4, top) and commensurate ubiquitin immunoreactivity that co-precipitated with Ubc7p-2HA (Fig. 4, bottom). This shift was consistent with a thioester conjugate of Ubc7p-2HA to a single ubiquitin and contrasted starkly with the previous reducing condition experiment, where no shift in Ubc7p-2HA was observed (Fig. 3C). In the Ubc7p-2HA·Cue1pΔ™ reactions, HA epitope immunoblots revealed species of Ubc7p-2HA conjugated to both monoubiquitin and diubiquitin (Fig. 4, top). The diubiquitin-Ubc7p was only observed in the presence of Cue1p and not the reactions with Ubc7p alone. These ubiquitin-Ubc7p conjugates were reversed in reducing conditions, suggesting they were thioester-linked to Ubc7p (Fig. 4, right). Ubiquitin immunoblots of these Cue1p-containing reactions showed that large polyubiquitin chains had co-precipitated with Ubc7p-2HA (Fig. 4, bottom), consistent with the Cue1p-dependent activation observed earlier. Importantly, in the nonreducing conditions, ubiquitin immunoreactivity was most prominent at molecular weights higher than E2, with comparatively little immunoreactivity at low molecular weights (Fig. 4, bottom). In contrast, the addition of reducing sample buffer to the Ubc7p immunoprecipitations caused the appearance of ubiquitin dimer and diminution of the monoubiquitinated and diubiquitinated species of Ubc7p (Fig. 4, bottom right). The asterisk (*LC) indicates a light chain antibody band that was detected in the ubiquitin immunoblot, which was released from the HA antibody-conjugated resin only with reducing sample buffer. These nonreducing reactions also contrast with the Cue1p-dependent ubiquitin dimers observed in reducing conditions (Figs. 1, 2, 3). Thus, Cue1p-enhanced Ubc7p could form thioester-conjugated polyubiquitin chains in vitro.
FIGURE 4.
Polyubiquitin conjugates were thioester-linked to Ubc7p in vitro. In vitro reactions with either Ubc7p-2HA or Ubc7p-2HA·Cue1pΔ™ were immunoprecipitated with antibody-conjugated resin to HA epitope. Bound proteins were resolved by nonreducing SDS-PAGE (left) or reducing SDS-PAGE (right), and immunoblotted for HA epitope (top) or ubiquitin (bottom). Ubc7p-2HA showed an ATP-dependent shift in molecular weight in the nonreducing conditions. This shift was improved in the presence of Cue1pΔ™ and was reversed in reducing conditions. Ubiquitin immunoblotting showed high molecular weight ubiquitin chains co-precipitating in these nonreducing conditions. The ubiquitin dimer prevalent in Figs. 1, 2, 3 was not released from the E2 in nonreducing conditions but was liberated in the reducing conditions. The reducing conditions also released an antibody light chain (*LC) from the antibody-conjugated resin that was detected in the ubiquitin immunoblot.
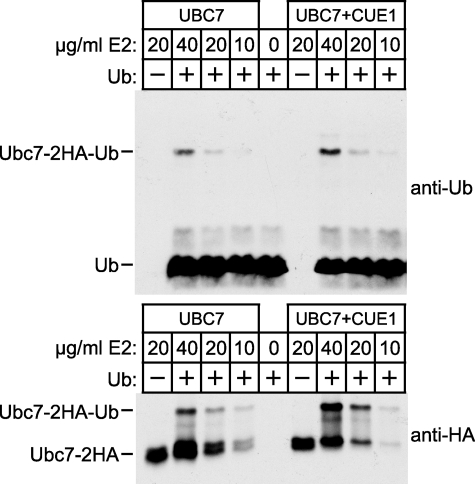
It is well established that Ubc7p (and all E2s) must interact with an E1 to be charged with thioester-linked ubiquitin (36, 37). We considered that Cue1p might enhance Ubc7p activity by promoting the charging of Ubc7p by E1. To examine this, we adapted a previously described method to assay the ubiquitin charging of an E2 by E1 (38). In vitro reactions were run by combining E1, ubiquitin, ATP, and either Ubc7p-2HA or Ubc7p-2HA·Cue1pΔ™. Reactions were incubated for a short time to allow charging of E2 by E1 but not processive polyubiquitin chain formation. Reaction mixes were resolved by nonreducing SDS-PAGE to preserve the E2-ubiquitin thioester linkage and immunoblotted for either HA epitope or ubiquitin. Multiple concentrations of each E2 were tested as indicated. Ubiquitin immunoblotting revealed a band at the molecular weight of a ubiquitin-Ubc7p thioester adduct (Fig. 5, top, Ubc7-2HA-Ub). This band was absent from “no ubiquitin” controls, and its intensity diminished with E2 concentration, suggesting involvement of both ubiquitin and E2 as expected for an E2-ubiqutin adduct. There was only a 1.3-fold difference in the intensity of this band between Ubc7p-2HA and Ubc7p-2HA-Cue1pΔ™ reactions at each E2 concentration tested, whereas a 2-fold reduction in E2 concentration strongly reduced its intensity. HA epitope immunoblotting also revealed a ubiquitin-dependent band of similar size that diminished with E2 concentration (Fig. 5, bottom, Ubc7-2HA-Ub). Although there appeared to be some enhancement of E2 thioester formation in the presence of Cue1pΔ™, this effect diminished at the lower, more physiological concentrations of Ubc7p (11). The presence or absence of Cue1p had no effect on the HA-detectable Ubc7p-Ub band when tested at 10 μg/ml E2, and only a 1.3-fold increase was detected with anti-ubiquitin. Together, these results suggest that Cue1p does not strongly enhance E1 charging of Ubc7p with ubiquitin. Although there may be some small contribution of this mechanism, it seemed that the extremely strong Cue1p-stimulated Ubc7p activity observed earlier was not at the level of ubiquitin transfer from E1 to Ubc7p.
FIGURE 5.
Cue1p did not strongly enhance the ubiquitin charging of Ubc7p by E1. In vitro assays of ubiquitin-Ubc7p thioester formation were performed with the indicated concentrations of Ubc7p-2HA (left lanes) or Ubc7p-2HA·Cue1pΔ™ (right lanes) and resolved with nonreducing SDS-PAGE. Ubiquitin immunoblotting (top) and HA epitope immunoblotting (bottom) both revealed a band the size of ubiquitin-Ubc7p thioester (Ubc7p-2HA-Ub) that diminished with E2 concentration. Ubc7p-2HA and Ubc7p-2HA·Cue1pΔ™ produced ubiquitin-Ubc7p thioester with similar efficiency at the lowest, most physiological E2 concentrations tested.
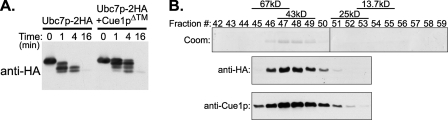
This suggested that Cue1p was acting directly on Ubc7p to promote enhanced activity. We considered that the weak activity of Ubc7p could be due to a lack of structure in solution that is stabilized by the presence of Cue1p. To examine this possibility, we used a limited proteolysis time course and compared the tryptic digests of Ubc7p-2HA with those of Ubc7p-2HA· Cue1pΔ™. Trypsin digestion was performed for the times indicated and stopped with the protease inhibitor AEBSF and sample buffer. Protein samples were analyzed by SDS-PAGE and HA epitope immunoblotting to detect the epitope-containing fragments of Ubc7p. The presence of Cue1p provided Ubc7p-2HA some protection from trypsinolysis, as might be expected from an interacting protein (Fig. 6A). However, the sizes of Ubc7p-2HA fragments produced were similar for Ubc7p-2HA and Ubc7p-2HA·Cue1pΔ™, suggesting that Ubc7p alone in solution without Cue1p was folded and that Cue1p was not causing a gross alteration to the structure of Ubc7p in solution. Although this does not rule out subtle structural changes to Ubc7p that may be induced by contact with Cue1p, Ubc7p activation by Cue1p did not seem to coincide with a transition from unfolded Ubc7p to folded Ubc7p. This is also consistent with the observation that free Ubc7p does have enzymatic activity that is qualitatively identical to its Cue1p-activated form.
FIGURE 6.
Biochemical analysis of Ubc7p and Ubc7p-2HA·Cue1pΔ™ in solution. A, Cue1p did not drastically modify the overall structure of Ubc7p. Limited trypsin proteolysis for the indicated times was performed on Ubc7p-2HA or Ubc7p-2HA·Cue1pΔ™, and then proteolyzed proteins were resolved by SDS-PAGE, and HA epitope immunoblotting revealed proteolysis of Ubc7p-2HA. The presence of Cue1p did not grossly modify the structure of Ubc7p in solution. The presence of the interacting Cue1pΔ™ provided Ubc7p-2HA some protection from proteolysis. However, this did not alter the molecular weights of the discrete bands that formed, suggesting that Ubc7p structure was intact in the absence of Cue1p. B, Cue1p and Ubc7p formed a dimer in solution, not a multimer of dimers. Superose-6 gel filtration chromatography of Ubc7p-2HA·Cue1pΔ™ was performed, and fractions were analyzed by SDS-PAGE. Coomassie staining revealed the peak elution fractions for the Ubc7p-2HA and Cue1pΔ™ proteins. Because these proteins were similar in size (∼20 kDa), we immunoblotted fraction samples to confirm the presence of both proteins. Peak fractions for protein-sizing standards, albumin (67 kDa), ovalbumin (43 kDa), chymotrypsin (25 kDa), and RNase A (13.7 kDa) were determined in identical conditions and are indicated with bars above the fraction numbers.
Another way Cue1p might stimulate the ubiquitination activity of Ubc7p is to facilitate the multimerization of Ubc7p, which could promote the processive building of ubiquitin chains. To examine this possibility, we purified recombinant Ubc7p-2HA co-expressed with Cue1pΔ™ and subjected the proteins to gel filtration chromatography in buffer conditions similar to the in vitro ubiquitination reactions where Cue1p promotes Ubc7p activity. To reduce nonspecific interaction with the gel filtration matrix, we added 150 mm NaCl to the normal 1× ubiquitination reaction buffer (UbR150) and confirmed that Cue1p-activated Ubc7p activity occurred in these conditions (data not shown). All fractions with detectable UV absorbance (280 nm) were resolved by SDS-PAGE and Coomassie-stained. Ubc7p-2HA and Cue1pΔ™ co-migrated through the column in a manner consistent with an ∼40-kDa complex (Fig. 6B, Coom). Because Ubc7p-2HA and Cue1pΔ™ are each ∼20 kDa and their Coomassie-stained bands were nearly superimposable, we also immunoblotted elution fractions to confirm the presence of both Ubc7p-2HA and Cue1pΔ™ (Fig. 6B, anti-HA and anti-Cue1p). The peak fractions occupied by Ubc7p-2HA and Cue1pΔ™ indicated column migration most similar to that of a 43-kDa ovalbumin protein standard. In conditions where Ubc7p activity was enhanced by Cue1pΔ™, these two proteins formed a heterodimer but did not show evidence of further multimerization (Fig. 6B). Thus, we conclude that the mechanism of Ubc7p stimulation does not occur through multimerization of Ubc7p.
Although the E3-independent activation of Ubc7 by Cue1p is surprising and interesting, we wanted to know whether this activity was relevant to the in vivo degradation of ERAD substrates by Ubc7p. It is known that the integral membrane protein Cue1p is required for Ubc7p-dependent ERAD (8, 9, 39–41). Cue1p binds to Ubc7p, localizing this E2 to the ER membrane (8, 9), and overexpression of Ubc7p does not restore ERAD in strains lacking Cue1p (8). Also, deletion of CUE1 reduces cross-linking between Ubc7p and its ERAD substrate Hmg2p (9). Thus, the membrane-tethering function of Cue1p was thought to increase the local concentration of Ubc7p at the ER membrane above a threshold required for ERAD. However, our in vitro studies above suggested an additional role for Cue1p in activating Ubc7p. To examine in vivo the contributions of both anchoring Ubc7p to the ER and activation of Ubc7p by Cue1p, we designed experiments to separate these two functions.
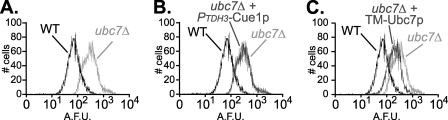
Our in vivo measure of Ubc7p function was degradation of the Hrd1p-dependent ERAD substrate Hmg2p-GFP, which can be assayed both biochemically and by flow cytometry (11, 12, 42–44). Low Hmg2p-GFP levels in a wild-type strain reflect the short half-life of this rapidly degraded protein (19, 44). Hmg2p-GFP cannot be degraded in a ubc7Δ null allele strain, resulting in increased fluorescence compared with a wild-type strain (45). In Fig. 7A, this is shown by superimposing flow cytometry histograms plotting the GFP fluorescence from wild-type and ubc7Δ strains. Expression of Ubc7p-2HA in the ubc7Δ null strain restored degradation of Hmg2p-GFP when expressed from either the native promoter (Fig. 8F) or the strong TDH3 promoter (data not shown). In contrast, expression of Cue1p from the strong TDH3 promoter in the ubc7Δ null strain had no effect on Hmg2p-GFP degradation, since the fluorescence of this strain was equal to that of the ubc7Δ null strain (Fig. 7B).
FIGURE 7.
Partial restoration of ERAD in ubc7Δ strains by ER-anchored Ubc7p. Hmg2p-GFP levels were measured in ubc7Δ strains expressing either empty vector or the indicated ERAD proteins. Histograms plot the number of cells (y axis) having a given arbitrary fluorescence (x axis). A, Hmg2p is degraded in a wild-type strain (WT) and stabilized in a ubc7Δ null strain (ubc7Δ). B, Cue1p overexpression had no effect on ERAD. The histogram of a ubc7Δ null strain expressing Cue1p-3HA from the strong TDH3 promoter is superimposed on wild type and ubc7Δ histograms to test complementation of ubc7Δ. C, the histogram of a ubc7Δ null strain expressing membrane-anchored Ubc7p-2HA (TM-Ubc7p) reveals partial complementation of ubc7Δ.
FIGURE 8.
Soluble Cue1p domain enhanced ERAD by self-anchored TM-Ubc7p in vivo. Hmg2p-GFP levels were measured as above, in cue1Δ ubc7Δ strains with either empty vector or the indicated ERAD proteins. A, Cue1pΔ™ caused no reduction in Hmg2p-GFP levels. B, TM-Ubc7p caused little if any reduction in Hmg2p-GFP. C, when expressed together, TM-Ubc7p and Cue1pΔ™ allowed a reduction in Hmg2p-GFP levels, partially complementing the cue1Δ ubc7Δ double null. D, cycloheximide chase degradation assays were performed to confirm Hmg2p-GFP degradation in wild-type or cue1Δ ubc7Δ strains expressing either empty vector or the indicated ERAD proteins. Hmg2p-GFP levels were measured as above, and reduction of GFP fluorescence was plotted for each strain treated with 50μg/ml cycloheximide for 30 or 120 min. This confirmed that degradation of Hmg2p-GFP was enhanced only in the presence of both TM-Ubc7p and Cue1pΔ™. E, whole cell lysates and microsome fractions of the strains used in Fig. 8, A–D, reveal that TM-Ubc7p is localized to the microsomal membrane fraction, that co-expression of Cue1pΔ™ does not alter TM-Ubc7p levels, and that TM-Ubc7p recruits Cue1pΔ™ to the membrane fraction. F, L-Ubc7p, a version of Ubc7p with an N-terminal flexible linker, weakly complements Ubc7p function in comparison with Ubc7p.
To examine the importance of Ubc7p-anchoring, independent of Cue1p activation of Ubc7p, we wanted to make a chimeric version of Ubc7p with its own membrane anchor. TM-Ubc7p was composed of the single N-terminal transmembrane span of Cue1p, a flexible linker of 88 amino acids, and full-length Ubc7p-2HA. We tested this TM-Ubc7p construct for complementation of the ubc7Δ null allele by expressing it from the strong TDH3 promoter and assaying Hmg2p-GFP levels by flow cytometry as above (Fig. 7C). Hmg2p-GFP levels were lower in the presence of TM-Ubc7p than in the ubc7Δ control. Thus, membrane-anchored Ubc7p partially complemented the ERAD defect in the ubc7Δ null allele strain (Fig. 7C). The presence of the flexible linker was necessary for this partial complementation by TM-Ubc7p; a version of TM-Ubc7p lacking the flexible linker between the transmembrane span and Ubc7p was completely unable to restore any Ubc7p function to a ubc7Δ null allele strain (data not shown). We wanted to confirm that proteolysis of the flexible linker in TM-Ubc7p did not generate a pool of soluble Ubc7p in vivo that restored Ubc7p function. We prepared whole-cell lysates from strains expressing TM-Ubc7p and immunoblotted for HA epitope, confirming the presence of only full-length TM-Ubc7p (Fig. 8E). Thus, we generated a partially functioning version of Ubc7p with its own ER anchor in cis.
If the requirement for Cue1p in ERAD is only to anchor Ubc7p to the ER membrane, then the self-anchored TM-Ubc7p construct should promote ERAD in the absence of both Ubc7p and Cue1p. TM-Ubc7p was expressed from the strong TDH3 promoter in a ubc7Δ cue1Δ strain expressing Hgm2p-GFP, and GFP fluorescence was measured as before. TM-Ubc7p supported little if any degradation of Hmg2p-GFP when expressed in the ubc7Δ cue1Δ strain (Fig. 8B). Interestingly, TM-Ubc7p allowed more degradation of Hmg2p-GFP when tested in a ubc7Δ strain with native CUE1 allele (Fig. 7C). Thus, the partially active TM-Ubc7p was even less effective in the absence of native Cue1p, suggesting that degradation of Hmg2p-GFP in vivo required Cue1p for a function independent of Ubc7p membrane localization.
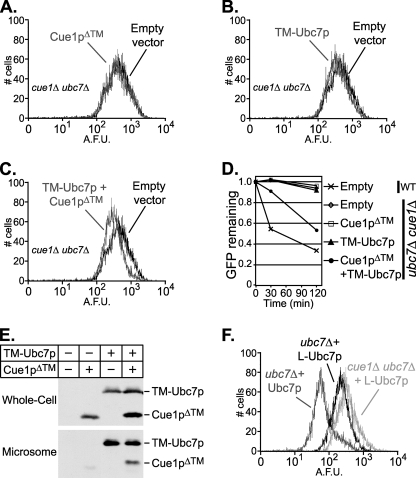
This result implied that Cue1p had a separable activating function in vivo akin to the enhanced Ubc7p ubiquitination observed in vitro. However, the Cue1p that allowed ERAD with TM-Ubc7p in Fig. 7C was native, membrane-anchored Cue1p. To separate the putative Ubc7p-activating function of Cue1p from its established Ubc7p-localizing function, we expressed a soluble Cue1p dissociated from the ER in vivo. A transmembraneless version of Cue1p (similar to the recombinantly expressed Cue1pΔ™ described above but with 3HA-epitope tag) was expressed from the strong TDH3 promoter in strains to test its ability to support ERAD function in vivo. As before, Hmg2p-GFP levels were assayed by flow cytometry. Expression of this Cue1pΔ™ in a wild-type strain did not disrupt Hmg2p-GFP degradation (data not shown). Cue1pΔ™ did not stimulate any Hmg2p-GFP degradation in a cue1Δ null strain carrying a native UBC7 allele, confirming that membrane anchoring is necessary for Cue1p to allow Ubc7p function (data not shown). Not surprisingly, Cue1pΔ™ in a ubc7Δ cue1Δ strain was also unable to promote ERAD (Fig. 8A). However, when Cue1pΔ™ and TM-Ubc7p were expressed together in a ubc7Δ cue1Δ strain, Hmg2p-GFP levels were lowered significantly (Fig. 8C). This change in GFP fluorescence exceeded that caused by either protein expressed alone (Fig. 8, A and B) and was comparable with the shift observed in Fig. 7C, where TM-Ubc7p was shown to partially complement a ubc7Δ allele. Therefore, Cue1pΔ™ enhanced TM-Ubc7p function in a manner similar to native Cue1p.
These results are consistent with a model whereby both ER localization of Ubc7p and stimulation of Ubc7p activity are required in vivo for ERAD function. These results were obtained by comparing Hmg2p-GFP levels between strains that, except for the empty vector or expression plasmid, were isogenic. To confirm that the changes in steady-state GFP levels observed above resulted from degradation of Hmg2p-GFP, cycloheximide chase assays were conducted, in which protein synthesis is inhibited while protein degradation is allowed to proceed. Cycloheximide chase of ubc7Δ cue1Δ strains used in Fig. 8, A–C, confirmed that Cue1pΔ™ co-expressed with TM-Ubc7p allowed improved degradation of Hmg2p-GFP. Log phase cultures of each strain were split and incubated with no drug or with cycloheximide for 30 min or 2 h, after which GFP fluorescence was measured as before. For each strain, mean fluorescence was calculated at each time point. Each strain was normalized to time 0 (no cycloheximide) and plotted in Fig. 8D to reveal the reduction in GFP fluorescence with cycloheximide treatment. There was little effect on GFP fluorescence in the empty vector strain (open diamond) and the strain expressing Cue1pΔ™ alone (open square). The TM-Ubc7p construct revealed little if any reduction in GFP fluorescence upon cycloheximide treatment (Fig. 8D, triangle). The presence of Cue1pΔ™ and TM-Ubc7p together caused significant reduction in Hmg2p-GFP levels, indicating stimulation of ERAD (Fig. 8D, circle). Although it has been reported that levels of cytosolic Ubc7p diminish in the absence of Cue1p (8, 9), we determined by immunoblotting of whole cell lysates that the addition of Cue1pΔ™ had no effect on TM-Ubc7p levels in these strains (Fig. 8E). Importantly, we also confirmed that in these strains, TM-Ubc7p localizes to the microsome membranes and that Cue1pΔ™ is efficiently recruited to the microsome fraction only when co-expressed with TM-Ubc7p (Fig. 8E). The data in Fig. 8D recapitulated the trends observed in Fig. 8, A–C, confirming that membrane-anchored Ubc7p and soluble Cue1p improved degradation of Hmg2p-GFP in vivo. These results are also consistent with the behavior of Ubc7p in our in vitro ubiquitination assays; ubiquitination activity was present with Ubc7p alone but strongly enhanced by the addition of Cue1p.
The in vivo experiments above separated the two roles of Cue1p as activator and localizer of Ubc7p, indicating that each of these roles was necessary for Ubc7p to perform Hrd1p-dependent degradation in the ER. However, it is important to note that we never saw full activity of Ubc7p from the TM-Ubc7p construct. Perhaps Ubc7p was restricted by membrane tethering from achieving its usual orientation with respect to the Hrd1p ERAD complex. To test this idea, we made a version of TM-Ubc7p we call L-Ubc7p, that lacked the transmembrane span of Cue1p but retained the flexible linker appended to the N terminus of Ubc7p-2HA. We then tested this L-Ubc7p for complementation of Ubc7p function. We expressed L-Ubc7p in a cue1Δ ubc7Δ strain and in a ubc7Δ strain with CUE1 intact and measured Hmg2p-GFP levels in these strains as before (Fig. 8F). When CUE1 was present, L-Ubc7p showed increased degradation of Hmg2p-GFP in comparison with the cue1Δ ubc7Δ strain expressing L-Ubc7p (Fig. 8F). The GFP levels in the cue1Δ ubc7Δ strain were superimposable with the cue1Δ ubc7Δ strain containing empty vector (data not shown). However, L-Ubc7p did not fully complement Ubc7p. The reduction in Hmg2p-GFP levels by L-Ubc7p was much less than that of Ubc7p-2HA expressed from the native promoter (Fig. 8F, Ubc7p). The extent of partial complementation by L-Ubc7p was similar to that of TM-Ubc7p with native Cue1p (compare Fig. 8F with Fig. 7C) and to that of TM-Ubc7p with co-expressed Cue1pΔ™ (Fig. 8, compare F with C). From this we conclude that the N-terminal modifications to Ubc7p necessary for in cis membrane anchoring significantly reduced Ubc7p function, explaining why we only observed partial complementation of Ubc7p above.
Importantly, the activation of membrane-anchored TM-Ubc7p by soluble Cue1pΔ™ was almost as good as that seen with membrane-anchored Cue1p and soluble L-Ubc7p. Thus, the presence of the appended TM domain limited the activity of Ubc7p. Nevertheless, the activation of the anchored Ubc7p was very efficient within the confines of this technical limitation.
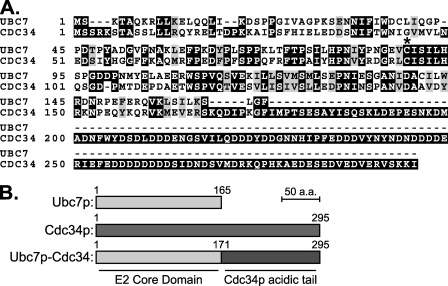
Because of these limitations in the ER-localized experiments, we wanted to examine the in vivo Cue1p enhancement of Ubc7p function in a context removed from the ER and ERAD machinery. Previous studies of Cdc34p suggested an approach. CDC34 is an essential gene with well characterized conditional alleles. Cdc34p associates with the cytosolic SCF ubiquitin ligase complex, which regulates cell cycle progression by targeting key regulatory proteins for ubiquitination and degradation (46). Cdc34p is localized to the SCF ubiquitin ligase complex through an acidic region C-terminal to the conserved E2 domain. It was reported that the yeast E2 Rad6p, when fused to this C-terminal tail domain of Cdc34p, could partially complement a temperature-sensitive (TS) allele of CDC34 (47, 48). Because Cdc34p is the E2 most closely related to Ubc7p in yeast (Fig. 9A), we adapted this idea to study Ubc7p E2 activity.
FIGURE 9.
A strategy to test Ubc7p function, independent of ERAD. A, primary sequence alignment of Ubc7p and Cdc34p. *, the conserved cysteine residue (Cys-89 in Ubc7p, Cys-95 in Cdc34p) where the ubiquitin thioester forms. Cdc34p has an acidic C-terminal tail required for localization to the SCF ubiquitin ligase complex. B, schematic of Ubc7p, Cdc34p, and a construct fusing the E2 domain of Ubc7p with the C-terminal tail of Cdc34p (Ubc7p-Cdc34) to target Ubc7p to the SCF ubiquitin ligase complex.
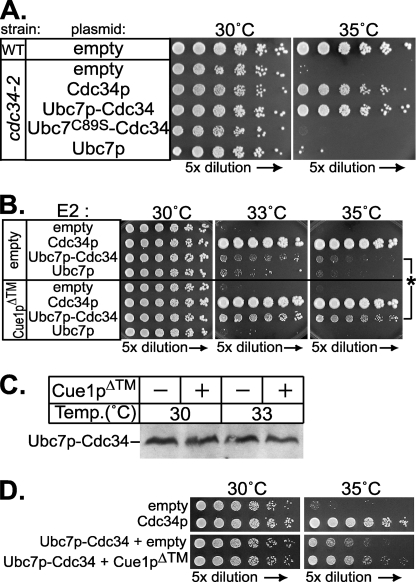
We made a construct appending the C-terminal tail of Cdc34p to full-length Ubc7p (Fig. 9B), thus directing Ubc7p to the soluble SCF complex E3. We expressed this Ubc7p-Cdc34 protein (Ubc7p-Cdc34) in strains whose only copy of CDC34 was the recessive cdc34-2 TS allele. By evaluating rescue of the cdc34-2 TS phenotype caused by Ubc7p-Cdc34, we could assay Ubc7p function independent of Hrd1p, ERAD substrates, or the ER membrane. Ubc7p-Cdc34 expressed from the strong TDH3 promoter complemented the cdc34-2 TS phenotype to a similar extent as full-length Cdc34p (Fig. 10A). This complementation by Ubc7p-Cdc34 required the conserved catalytic cysteine residue of Ubc7p essential for E2 function, since mutation of this residue to serine resulted in no complementation (Fig. 10A, Ubc7C89S-Cdc34). Overexpression of Ubc7p alone (without the Cdc34p tail) failed to complement cdc34-2. Thus, cdc34-2 TS phenotype rescue required both the E2 activity of Ubc7p and localizing tail of Cdc34p. Weaker expression of Rad6p fusions to Cdc34p only partially complemented the cdc34-2 TS phenotype (47). Similarly, expression of Ubc7p-Cdc34 from the weaker CDC34 promoter only weakly complemented the cdc34-2 phenotype (Fig. 10B), allowing some growth at a semirestrictive temperature. This partial complementation by Ubc7p-Cdc34 expressed from the CDC34 promoter allowed us to evaluate enhancement of this Ubc7p activity by Cue1p in a cellular context independent of the ER.
FIGURE 10.
Cue1p enhanced Ubc7p activity in an ERAD-independent cellular context. A, restoration of Cdc34p function was assayed as complementation of the cdc34-2 temperature-sensitive growth phenotype. Wild-type strains (WT) grew at both 30 °C and 35 °C, but cdc34-2 allele strains do not survive at 35 °C. Cdc34p, Ubc7p, Ubc7p-Cdc34, and Ubc7pC89S-Cdc34 proteins were each expressed from the strong TDH3 promoter in the cdc34-2 strain. Expression of both Cdc34p and Ubc7p-Cdc34 restored the wild-type phenotype. This complementation was abolished when the conserved catalytic cysteine in Ubc7p-Cdc34 was mutated to serine (Ubc7pC89S-Cdc34). Fusion of Ubc7p to the Cdc34p tail domain was required for complementation of the cdc34-2 phenotype, as expression of Ubc7p alone had no effect. B, Cdc34p, Ubc7p, and Ubc7p-Cdc34 were expressed from the weaker CDC34 promoter in cdc34-2 allele strains. These strains also contained either empty vector (top four rows) or a vector expressing Cue1pΔ™ (bottom four rows). These strains were grown at permissive (30 °C) or nonpermissive (33 and 35 °C) temperatures. Here, Ubc7p-Cdc34 only slightly improved complementation of the TS phenotype compared with empty vector and was less effective than Cdc34p at complementing the cdc34-2 TS phenotype. Impressively, the addition of Cue1pΔ™ enhanced complementation only in the presence of Ubc7p-Cdc34, and not Cdc34p or Ubc7p. An asterisk highlights improved complementation by Ubc7p-Cdc34 upon the addition of Cue1pΔ™. C, levels of Ubc7p-Cdc34 were not changed by the addition of Cue1pΔ™. Whole cell lysates of strains grown for 5 h at either permissive (30 °C) or nonpermissive (33 °C) temperature were resolved by SDS-PAGE and immunoblotted for Ubc7p to examine Ubc7p-Cdc34 levels. At both permissive and nonpermissive temperatures, expression of Cue1pΔ™ caused no change in Ubc7p-Cdc34 levels. Thus, enhanced complementation by Cue1pΔ™ with Ubc7p-Cdc34 in Fig. 10B was due to increased activity of Ubc7p-Cdc34. D, using a cdc34-2 strain that was also cue1Δ, we observed complementation of the cdc34-2 TS phenotype by expression of Cdc34p from the CDC34 promoter (top). In this strain, we also observed partial TS rescue by expression of Ubc7p-Cdc34 from the CDC34 promoter, and enhancement of this complementation when Cue1pΔ™ is co-expressed. The similar trends in Fig. 10, B and D, with CUE1 or cue1Δ cdc34-2 strains suggest that the improved complementation of Ubc7p-Cdc34 caused by Cue1pΔ™ is not due to indirect effects, such as titration effects at the membrane surface.
Using cdc34-2 complementation by Ubc7p-Cdc34 as an in vivo, ER-free assay of Ubc7p activity, we tested whether Cue1p enhanced Ubc7p-Cdc34 function. If Cue1p were an enhancer of Ubc7p activity in vivo, then expression of Cue1pΔ™ in these cdc34-2 strains should stimulate the ubiquitination activity of Ubc7p-Cdc34, improving complementation of the cdc34-2 TS phenotype. In a strain with cdc34-2 as the only copy of CDC34, we added empty vector or vectors expressing one of Cdc34p, Ubc7p-Cdc34, or Ubc7p, all from the CDC34 promoter (Fig. 10B). At the permissive temperature (30 °C), all of the strains grew equally well. At the nonpermissive temperatures (33 and 35 °C), the native Cdc34p gene strongly improved growth relative to the empty vector control, indicating complementation of the cdc34-2 TS phenotype. As mentioned above, neither Ubc7p-Cdc34 nor Ubc7p could complement the TS phenotype to the same extent (Fig. 10B). However, expression of Cue1pΔ™ markedly improved complementation of co-expressed Ubc7p-Cdc34 construct but not the strains expressing Cdc34p or normal Ubc7p lacking the Cdc34 tail. Because Cue1p improves the stability of native Ubc7p in vivo (23), we wanted to verify that the Ubc7p-Cdc34 activation by Cue1pΔ™ was not due to a change in cellular levels. We confirmed by immunoblotting that expression of Cue1pΔ™ had no effect on Ubc7p-Cdc34 protein levels at either temperature (Fig. 10C).
The experiments showing Cue1pΔ™-enhanced complementation of cdc34-2 by Ubc7p-Cdc34 were conducted in strains with native Cue1p. It is possible that a fraction of the Ubc7p-Cdc34 expressed in these cells was recruited to the ER by endogenous Cue1p and then displaced by the addition of Cue1pΔ™. To confirm that any such interactions with the ERAD machinery were not responsible for the complementation by Ubc7p-Cdc34, we disrupted the CUE1 locus in the cdc34-2 strain to test complementation of the TS phenotype in the absence of endogenous Cue1p. The cdc34-2 cue1Δ strains expressing Cdc34p from the native promoter fully restored growth at the nonpermissive temperature (Fig. 10D, top). The addition of Ubc7p-Cdc34 partially complemented the TS growth phenotype, and this was enhanced by the addition of Cue1pΔ™ (Fig. 10D, bottom). These results suggest that, even removed from the context of the ER and ERAD, Ubc7p activity was enhanced by Cue1p in a biologically observable manner.
DISCUSSION
In the above studies, we asked whether Cue1p functions as an activator of Ubc7p. Our previous studies of the ERAD ligase Hrd1p first suggested that Cue1p could directly increase Ubc7p activity (11). We began by examining Ubc7p in vitro and found that Cue1p enhanced ubiquitination by Ubc7p, even without an E3 present. Recombinant Ubc7p complexed with soluble Cue1p produced polyubiquitin chains, including dimers of ubiquitin, whereas free Ubc7p did not (Figs. 1 and 2). These polyubiquitin chains formed in vitro exclusively through lysine 48 linkages (Fig. 3). Polyubiquitin could be found conjugated to Ubc7p through its catalytic cysteine (Fig. 4), as has been reported for Ubc7p and its homolog Ube2g2 (23, 35).
We next examined in vivo if Cue1p activation of Ubc7p had a role in the HRD ERAD pathway independent of its known anchoring function. To discern if Cue1p increased in vivo Ubc7p activity, we independently anchored Ubc7p to the ER surface by fusing the ER-localizing transmembrane span of Cue1p to Ubc7p (TM-Ubc7p). This fusion provided minimal E2 function in the absence of Cue1p. The addition of only the soluble cytosolic portion of Cue1p (Cue1pΔ™) enhanced the activity of this self-anchored TM-Ubc7p, showing that Cue1p plays dual roles recruiting Ubc7p to the ER and improving the E2 activity of Ubc7p. Importantly, Cue1pΔ™ had no effect on ERAD with native Ubc7p. Membrane anchoring of Ubc7p and the addition of the Cue1p cytosolic domain were both needed to promote the Ubc7p-dependent ERAD function provided by this self-anchored Ubc7p (Figs. 7 and 8). Importantly, self-anchoring of Ubc7p and soluble Cue1p never attained the levels of ERAD achieved with wild-type Ubc7p and wild-type Cue1p. We determined that this was due to impaired function of Ubc7p when modified at the N terminus, although this may also be due to the role of Cue1p as a participant in ERAD complexes (30). Thus, we turned to a non-ER pathway to further study the ability of Cue1p to independently activate Ubc7p.
In vivo Cue1p enhancement of Ubc7p activity was also observed independent of the ER membrane and ERAD. We demonstrated this by substituting Ubc7p for the E2 homologous region of Cdc34p, which functions with the soluble SCF E3 complex but has no role in ERAD. We assayed the function of the chimeric Ubc7p-Cdc34 E2 by rescue of the TS cdc34-2 growth phenotype (Fig. 10). Ubc7p-Cdc34 expressed from the CDC34 promoter could not rescue the cdc34-2 TS phenotype. However, when soluble Cue1p was co-expressed, identical levels of Ubc7p-Cdc34 were then able to partially rescue the cdc34-2 TS phenotype. The same trend was also observed in cue1Δ cdc34-2 strains. Together, these results indicate that Cue1p has an independent role as a stimulator of Ubc7p enzymatic activity that can be observed both in vivo and in vitro.
We considered several possible mechanisms by which Cue1p might exert its effect on Ubc7p. The enhancement of Ubc7p activity was independent of E3, so it was not a result of improved E2–E3 interaction in the presence of Cue1p. Moreover, Cue1p improved Ubc7p activity in vivo when replacing Cdc34p function at the SCF complex, where Cue1p is unlikely to promote this ectopic E2–E3 pairing. Cue1p did not substantially improve the E1–E2 interaction of Ubc7p, since Cue1p had small effects on the E1-dependent charging of Ubc7p with ubiquitin, particularly at more physiological E2 concentrations. CUE domain proteins are known to bind polyubiquitin, which might explain the in vitro activation observed here. However, Cue1p lacks conserved residues shared by other CUE proteins (49) and has been observed not to bind polyubiquitin (50). Cue1p did not appear to cause gross changes to the folding of Ubc7p in solution, since trypsinolysis of Ubc7p alone preserved similarly sized bands as Ubc7p co-expressed with Cue1pΔ™ (Fig. 6A). Since the effects of Cue1p on Ubc7p were most likely downstream of E2-ubiquitin formation (Fig. 5) and independent of E3, this strongly suggested that Cue1p had a direct effect on Ubc7p to promote E2 activity. Consistent with this, Ubc7p produced lysine 48-linked ubiquitin chains in the presence and absence of Cue1p, suggesting that ubiquitin linkage specificity is intrinsic to Ubc7p. Cue1p did not enhance Ubc7p activity by promoting strong association of multiple Ubc7p molecules; these two ∼20-kDa proteins migrated as an ∼40-kDa complex on a gel filtration column, suggesting that each Cue1p bound only one Ubc7p (Fig. 6B). However, a more transient association between Ubc7p molecules brought about by Cue1p, but not observable by gel filtration, cannot be ruled out. Thus, it seems that Cue1p enhanced the enzymatic activity of Ubc7p directly.
Our observations of Ubc7p activity show parallels with the ubiquitination mechanism of Cdc34p. In detailed kinetic studies, ubiquitin-conjugated Cdc34p released ubiquitin onto unbound ubiquitin from solution only on lysine 48, making a lysine 48-linked dimer (29). This in vitro Cdc34p activity was strongly enhanced by the addition of purified SCF complex. The specific SCF complex component responsible was not addressed, but it was determined that SCF enhanced this reaction by stimulating the Cdc34p-ubiquitin adduct to release ubiquitin (increased Vmax) and not by increasing affinity for ubiquitin in solution (Km did not change). A mutant of Cdc34p was unresponsive to this SCF-dependent activation in vitro and showed reduced E2 function in vivo (51). It is possible that Cue1p similarly activates Ubc7p-mediated production of lysine 48 ubiquitin chains. Cue1p is a ubiquitin ligase complex member, interacting directly with the Ubc7p-utilizing E3 Doa10p and with Hrd1p indirectly through proteins in the Hrd1p E3 complex (30). As proposed for Cdc34p and the SCF complex, Cue1p might act as a “ubiquitin exchange factor” to promote the release of ubiquitin from Ubc7p onto ubiquitin in solution by stabilizing a transition intermediate in the ubiquitin transfer reaction.
Recent studies of the mammalian Ubc7p homolog Ube2g2 have a similar theme of interactor activation. In vitro, Ube2g2 can assemble thioester-linked polyubiquitin chains in the presence of E3, and it appears that these chains can be transferred en mass to a substrate (35). The E3 gp78, a human homolog of Hrd1p, is required for this in vitro action of Ube2g2. gp78 has a CUE-like domain (G2BR) required for association with Ube2g2 and ERAD (24). This domain is specific for Ube2g2 binding, and gp78 without this sequence cannot perform ERAD. This E2 localization domain of gp78 could play a role in Ube2g2 activation, just as Cue1p promotes Ubc7p activity.
The relationship of Ubc7p to Cue1p has similarity to that of the E2 Pex4p and its membrane-anchoring protein Pex22p. Pex4p is required for peroxisome protein import and peripherally associates with peroxisome membranes through Pex22p, which is required for Pex4p function (52). Pex22p and Cue1p have 16% amino acid identity and 30% amino acid similarity, suggesting that Pex22p could influence both the localization and activity of Pex4p, as we have discovered for Cue1p and Ubc7p, and may do so in the same manner.
There are other examples of E2 activation by interacting proteins. The yeast E2 Ubc13p mediates assembly of lysine 63-linked polyubiquitin chains but only in the presence of an accessory protein, Mms2p (53). Mms2p is a ubiquitin-conjugating enzyme variant lacking the conserved cysteine residue for ubiquitin thioester formation. Ubc13p or Mms2p alone does not form polyubiquitin chains, but together these proteins form a heterodimer with enhanced E2 activity. A ubiquitin binding site on Mms2p recruits ubiquitin from solution, orienting lysine 63 near the Ubc13p catalytic site to promote polyubiquitin assembly (54, 55). In mammals, there is evidence that different ubiquitin-conjugating enzyme variants partner with the Ubc13p homolog to carry out distinct functions. hUbc13 and hMms2 mediate a DNA damage repair response, whereas hUbc13 and hUev1A mediate NF-κB signaling (56). Thus, E2-interacting proteins can enhance E2 activity, alter the specificity of E2 activity, and localize E2 activity.
We have often wondered why Ubc7p is localized to the ER membrane through a noncovalent interaction with Cue1p rather than a transmembrane span in cis like Ubc6p (57). It is clear that N-terminal fusion to Ubc7p strongly reduces its function. The membrane-embedded E3 complexes that use Ubc7p may have steric or diffusion constraints that cannot accommodate these altered versions of Ubc7p. Alternatively, noncovalent E2-binding partners may simply afford the cell more options for E2 localization than a dedicated, ER-anchored version of Ubc7p.
Although Ubc7p and Cue1p work together to promote ERAD, it is not clear whether other functions, if any, exist for Ubc7p. A recent study shows that in strains without Cue1p, Ubc7p is actively degraded (23). Ubc7p may yet play undiscovered roles in the cell by interacting with other Cue-like proteins that modify the activity or localization of a small pool of Ubc7p. Regulation of E2s by CUE domain proteins or other interaction partners may be a general feature of E2 physiology. These could work to either activate or repress E2 activity in different cellular contexts. In the case of Ubc7p, it is clear that Cue1p not only sequesters this E2 to the surface of the ER but is also required for the enzyme's full activity.
Acknowledgments
We thank Thomas Sommer at the Max Delbrück Center for Molecular Medicine in Berlin for antibodies to Cue1p and Ubc7p and Richard G. Gardner at the University of Washington for cdc34-2 plasmid. We also thank Debra L. Urwin in the laboratory of James T. Kadonaga for assistance with gel filtration chromatography and Michael David for FACScalibur flow cytometry access.
This work was supported, in whole or in part, by National Institutes of Health Grant GM51996-06 from the NIDDK (to R. Y. H.). This work was also supported by an American Heart Association Established Investigator Award (to R. Y. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; E1, ubiquitin-conjugating enzyme; E2, ubiquitin-activating enzyme; E3, ubiquitin ligase; ERAD, endoplasmic reticulum-associated degradation; CBD, chitin-binding domain; HA, hemagglutinin; SOEing, strand overlap extension; GFP, green fluorescent protein; AEBSF, 4-(2-aminoethyl)benzenesulfonyl fluoride; TPCK, 1-chloro-3-tosylamido-4-phenyl-2-butanone; MOPS, 3-(N-morpholino)propanesulfonic acid; DTT, dithiothreitol; GST, glutathione S-transferase; L-Ubc7p, N-terminally modified linker-Ubc7p-2HA protein; ILB, intein lysis buffer; TS, temperature-sensitive.
References
- 1.Sommer, T., and Wolf, D. H. (1997) FASEB J. 11 1227–1233 [DOI] [PubMed] [Google Scholar]
- 2.Hampton, R. Y. (2002) Curr. Opin. Cell Biol. 14 476–482 [DOI] [PubMed] [Google Scholar]
- 3.Kostova, Z., and Wolf, D. H. (2003) EMBO J. 22 2309–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plemper, R. K., and Wolf, D. H. (1999) Trends Biochem. Sci. 24 266–270 [DOI] [PubMed] [Google Scholar]
- 5.Hampton, R. Y., and Bhakta, H. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 12944–12948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiller, M. M., Finger, A., Schweiger, M., and Wolf, D. H. (1996) Science 273 1725–1728 [DOI] [PubMed] [Google Scholar]
- 7.Swanson, R., Locher, M., and Hochstrasser, M. (2001) Genes Dev. 15 2660–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biederer, T., Volkwein, C., and Sommer, T. (1997) Science 278 1806–1809 [DOI] [PubMed] [Google Scholar]
- 9.Gardner, R. G., Shearer, A. G., and Hampton, R. Y. (2001) Mol. Cell Biol. 21 4276–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.VanDemark, A. P., and Hill, C. P. (2002) Curr. Opin. Struct. Biol. 12 822–830 [DOI] [PubMed] [Google Scholar]
- 11.Bazirgan, O. A., Garza, R. M., and Hampton, R. Y. (2006) J. Biol. Chem. 281 38989–39001 [DOI] [PubMed] [Google Scholar]
- 12.Bays, N. W., Gardner, R. G., Seelig, L. P., Joazeiro, C. A., and Hampton, R. Y. (2001) Nat. Cell Biol. 3 24–29 [DOI] [PubMed] [Google Scholar]
- 13.Joazeiro, C. A., Wing, S. S., Huang, H., Leverson, J. D., Hunter, T., and Liu, Y. C. (1999) Science 286 309–312 [DOI] [PubMed] [Google Scholar]
- 14.Mori, S., Tanaka, K., Kanaki, H., Nakao, M., Anan, T., Yokote, K., Tamura, K., and Saito, Y. (1997) Eur. J. Biochem. 247 1190–1196 [DOI] [PubMed] [Google Scholar]
- 15.Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K., and Pease, L. R. (1989) Gene (Amst.) 77 51–59 [DOI] [PubMed] [Google Scholar]
- 16.Horton, R. M., Hunt, H. D., Ho, S. N., Pullen, J. K., and Pease, L. R. (1989) Gene (Amst.) 77 61–68 [DOI] [PubMed] [Google Scholar]
- 17.Mumberg, D., Muller, R., and Funk, M. (1995) Gene (Amst.) 156 119–122 [DOI] [PubMed] [Google Scholar]
- 18.Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003) Nucleic Acids Res. 31 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hampton, R. Y., Gardner, R. G., and Rine, J. (1996) Mol. Biol. Cell 7 2029–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hampton, R. Y., and Rine, J. (1994) J. Cell Biol. 125 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardner, R. G., Swarbrick, G. M., Bays, N. W., Cronin, S. R., Wilhovsky, S., Seelig, L., Kim, C., and Hampton, R. Y. (2000) J. Cell Biol. 151 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldstein, A. L., and McCusker, J. H. (1999) Yeast 15 1541–1553 [DOI] [PubMed] [Google Scholar]
- 23.Ravid, T., and Hochstrasser, M. (2007) Nat. Cell Biol. 9 422–427 [DOI] [PubMed] [Google Scholar]
- 24.Chen, B., Mariano, J., Tsai, Y. C., Chan, A. H., Cohen, M., and Weissman, A. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorick, K. L., Jensen, J. P., Fang, S., Ong, A. M., Hatakeyama, S., and Weissman, A. M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, D., Crowe, E., Stevens, T. A., and Candido, E. P. (2002) Genome Biol. 3 R2.1–R2.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraft, E., Stone, S. L., Ma, L., Su, N., Gao, Y., Lau, O. S., Deng, X. W., and Callis, J. (2005) Plant Physiol. 139 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ptak, C., Gwozd, C., Huzil, J. T., Gwozd, T. J., Garen, G., and Ellison, M. J. (2001) Mol. Cell Biol. 21 6537–6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petroski, M. D., and Deshaies, R. J. (2005) Cell 123 1107–1120 [DOI] [PubMed] [Google Scholar]
- 30.Carvalho, P., Goder, V., and Rapoport, T. A. (2006) Cell 126 361–373 [DOI] [PubMed] [Google Scholar]
- 31.Peng, J., Schwartz, D., Elias, J. E., Thoreen, C. C., Cheng, D., Marsischky, G., Roelofs, J., Finley, D., and Gygi, S. P. (2003) Nat. Biotechnol. 21 921–926 [DOI] [PubMed] [Google Scholar]
- 32.Thrower, J. S., Hoffman, L., Rechsteiner, M., and Pickart, C. M. (2000) EMBO J. 19 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chau, V., Tobias, J. W., Bachmair, A., Marriott, D., Ecker, D. J., Gonda, D. K., and Varshavsky, A. (1989) Science 243 1576–1583 [DOI] [PubMed] [Google Scholar]
- 34.Finley, D., Sadis, S., Monia, B. P., Boucher, P., Ecker, D. J., Crooke, S. T., and Chau, V. (1994) Mol. Cell Biol. 14 5501–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, W., Tu, D., Brunger, A. T., and Ye, Y. (2007) Nature 446 333–337 [DOI] [PubMed] [Google Scholar]
- 36.Glickman, M. H., and Ciechanover, A. (2002) Physiol. Rev. 82 373–428 [DOI] [PubMed] [Google Scholar]
- 37.Hershko, A., and Ciechanover, A. (1998) Annu. Rev. Biochem. 67 425–479 [DOI] [PubMed] [Google Scholar]
- 38.Gosink, M. M., and Vierstra, R. D. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 9117–9121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denic, V., Quan, E. M., and Weissman, J. S. (2006) Cell 126 349–359 [DOI] [PubMed] [Google Scholar]
- 40.Ravid, T., Kreft, S. G., and Hochstrasser, M. (2006) EMBO J. 25 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walter, J., Urban, J., Volkwein, C., and Sommer, T. (2001) EMBO J. 20 3124–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cronin, S. R., and Hampton, R. Y. (1999) Methods Enzymol. 302 58–73 [DOI] [PubMed] [Google Scholar]
- 43.Cronin, S. R., Khoury, A., Ferry, D. K., and Hampton, R. Y. (2000) J. Cell Biol. 148 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gardner, R., Cronin, S., Leader, B., Rine, J., and Hampton, R. (1998) Mol. Biol. Cell 9 2611–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilhovsky, S., Gardner, R., and Hampton, R. (2000) Mol. Biol. Cell 11 1697–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seol, J. H., Feldman, R. M., Zachariae, W., Shevchenko, A., Correll, C. C., Lyapina, S., Chi, Y., Galova, M., Claypool, J., Sandmeyer, S., Nasmyth, K., and Deshaies, R. J. (1999) Genes Dev. 13 1614–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kolman, C. J., Toth, J., and Gonda, D. K. (1992) EMBO J. 11 3081–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silver, E. T., Gwozd, T. J., Ptak, C., Goebl, M., and Ellison, M. J. (1992) EMBO J. 11 3091–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prag, G., Misra, S., Jones, E. A., Ghirlando, R., Davies, B. A., Horazdovsky, B. F., and Hurley, J. H. (2003) Cell 113 609–620 [DOI] [PubMed] [Google Scholar]
- 50.Shih, S. C., Prag, G., Francis, S. A., Sutanto, M. A., Hurley, J. H., and Hicke, L. (2003) EMBO J. 22 1273–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petroski, M. D., Kleiger, G., and Deshaies, R. J. (2006) Mol. Cell 24 523–534 [DOI] [PubMed] [Google Scholar]
- 52.Koller, A., Snyder, W. B., Faber, K. N., Wenzel, T. J., Rangell, L., Keller, G. A., and Subramani, S. (1999) J. Cell Biol. 146 99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hofmann, R. M., and Pickart, C. M. (1999) Cell 96 645–653 [DOI] [PubMed] [Google Scholar]
- 54.Moraes, T. F., Edwards, R. A., McKenna, S., Pastushok, L., Xiao, W., Glover, J. N., and Ellison, M. J. (2001) Nat. Struct. Biol. 8 669–673 [DOI] [PubMed] [Google Scholar]
- 55.VanDemark, A. P., Hofmann, R. M., Tsui, C., Pickart, C. M., and Wolberger, C. (2001) Cell 105 711–720 [DOI] [PubMed] [Google Scholar]
- 56.Andersen, P. L., Zhou, H., Pastushok, L., Moraes, T., McKenna, S., Ziola, B., Ellison, M. J., Dixit, V. M., and Xiao, W. (2005) J. Cell Biol. 170 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sommer, T., and Jentsch, S. (1993) Nature 365 176–179 [DOI] [PubMed] [Google Scholar]