Abstract
Members of the ING (inhibitor of growth) family of chromatin modifying proteins (ING1-ING5) have emerged as critical regulators of gene expression and cellular responses, suggesting that the ING proteins may impinge on specific signal transduction pathways and their mediated effects. Here, we demonstrate a role for the protein ING2 in mediating responses by the transforming growth factor (TGF)-β-Smad signaling pathway. We show that ING2 promotes TGF-β-induced transcription. Both gain-of-function and RNA interference-mediated knockdown of endogenous ING2 reveal that ING2 couples TGF-β signals to the induction of transcription and cell cycle arrest. We also find that the Smad-interacting transcriptional modulator SnoN interacts with ING2 and promotes the assembly of a protein complex containing SnoN, ING2, and Smad2. Knockdown of endogenous SnoN blocks the ability of ING2 to promote TGF-β-dependent transcription, and conversely expression of SnoN augments ING2 enhancement of the TGF-β response. Collectively, our data suggest that ING2 collaborates with SnoN to mediate TGF-β-induced Smad-dependent transcription and cellular responses.
The ING (inhibitor of growth) family of nuclear proteins, consisting of ING1 to ING5, plays important roles in controlling gene expression and cellular functions. A key structural feature of the ING proteins is a conserved plant homeodomain (PHD)6 zinc finger domain that binds to histone H3 in a methylation-sensitive manner (1, 2). ING proteins also specifically bind to phosphatidylinositol monophosphate stress signaling lipids (3-5) via a polybasic basic region located downstream of their PHD region (5). In addition, the ING proteins associate with the histone acetyltransferase and histone deacetylase complexes and thereby control gene expression by remodeling chromatin (6-9). Not surprisingly, the ING proteins strongly influence cellular behaviors including cell proliferation, apoptosis, angiogenesis, and senescence (10). Frequent down-regulation of ING expression in lung, esophageal, and mammary carcinomas suggests that these proteins have tumor-suppressive properties (11-14). Although ING function in transcription and cellular responses has received much attention, how ING function intersects with growth factors and their associated signaling cascades remains poorly understood.
The transforming growth factor (TGF)-β signaling pathways regulate a wide array of biological responses in development and homeostasis (15, 16). Dysfunctions of the TGF-β signaling pathways have been implicated in cancer including mammary, lung, esophageal, colorectal, and pancreatic carcinomas (17-19). The TGF-β proteins act via specific transmembrane serine/threonine kinase receptors, which once activated stimulate the Smad signaling pathway (20, 21). This involves the phosphorylation of the receptor-regulated Smad (R-Smad) proteins. The phosphorylated R-Smad then forms a complex with the copartner Smad4 and accumulates in the nucleus. In the nucleus, the R-Smad-Smad4 complex binds to Smad-binding elements (SBEs) on promoters of TGF-β-responsive genes and regulates target gene expression in collaboration with specific transcriptional coregulators and associated histone acetyltransferases or histone deacetylases (21-23).
The transcriptional protein SnoN has emerged as an important modulator of TGF-β-Smad signaling pathway and responses. SnoN interacts with the R-Smads and co-Smad4 on promoters of TGF-β-responsive genes (24). SnoN exerts a biphasic effect on TGF-β-dependent transcription. At low “physiological” concentrations SnoN acts as a coactivator, and at higher amounts SnoN represses TGF-β-dependent transcription (25). SnoN recruitment of a histone deacetylase complex to TGF-β promoters may account for SnoN repression of TGF-β-dependent transcription (26, 27). However, the mechanisms by which SnoN activates TGF-β-dependent transcription and responses including cell cycle arrest have remained unknown.
In this study, we show that the ING family member ING2 mediates the ability of TGF-β to activate transcription and inhibit cell proliferation. We find that SnoN associates with ING2 to promote the assembly of a protein complex containing ING2, SnoN, and R-Smad2. We also find that SnoN is required for ING2 up-regulation of TGF-β-dependent transcription. Collectively, our data suggest that SnoN collaborates with ING2 to mediate TGF-β-induced Smad2-dependent responses.
EXPERIMENTAL PROCEDURES
Plasmid Construction—pC1 expression vectors encoding human ING1a, 1b, and 2 proteins were generated as described elsewhere (28). Using pCI-ING2 as a template, ING2 was PCR-amplified and subcloned into pCMV5B/FLAG and pCMV5B/Myc mammalian expression vectors to express N-terminal FLAG and Myc epitope-tagged ING2 proteins, respectively. The pCAGIP/FLAG/ING2 vector was generated by subcloning the FLAG/ING2 cDNA insert into a digested pCAGIP/FLAG/FoxH1 vector using convenient endonuclease restriction sites (29). The pCAGIP/FLAG/ING2 plasmid allows the coexpression of FLAG-tagged ING2 and the puromycin resistance marker proteins from an internal ribosomal entry site-containing bicistronic transcript. pCMV5B-based constructs expressing Myc-tagged ΔN, ΔC, and ΔPHD deletion mutant ING2 proteins lacking amino acid residues 1-63, 199-281, and 199-258, respectively, were generated using a PCR-based approach to produce ING2 cDNA missing the corresponding nucleotides. N-terminally FLAG-tagged SnoN, untagged Smad2-, Gal4-, and Gal4-Smad2-expressing vectors have been described before (24, 30, 31). The ING2 RNA interference (RNAi) vector was generated to express ING2 hairpin RNAs and enhanced green fluorescent protein (GFP) under the control of the U6 and CMV promoters, respectively (25, 32). The ING2 RNAi vector was designed to specifically target the region ATGGAGTTACACTCACAGTGT in ING2 mRNA. The SnoN RNAi expressing vector has been described elsewhere (25). 3TP-luciferase, SBE4-luciferase, pG5-E1b-luciferase, and β-galactosidase reporter constructs were described previously (30, 31, 33, 34). All of the constructs were verified by restriction digests and/or DNA sequencing analysis (University of Calgary Core Sequencing Facility).
Cell Lines and Transfections—The mink lung epithelial (Mv1Lu) and human hepatoma (HepG2) cell lines were cultured in minimum essential medium (Invitrogen) containing 1% nonessential amino acids (Invitrogen) and 10% fetal bovine serum (Invitrogen). The human kidney epithelial (293T) cell line was maintained in Dulbecco's modified Eagle's medium (Invitrogen) with high glucose and l-glutamine, supplemented with 10% fetal bovine serum. The HepG2 and Mv1Lu cells were transiently transfected using Lipofectamine (Invitrogen) and FuGENE 6 (Roche Applied Science), respectively, according to the manufacturer's instructions. The 293T cells were transiently transfected using the calcium phosphate method. To generate stable Mv1Lu cell lines expressing a puromycin-resistant marker alone or together with FLAG/ING2, cells were transfected with the pCAGIP-based vectors using Lipofectin (Invitrogen) according to the manufacturer's instructions, followed by selection of stable transfectants using 0.45 μg/ml puromycin (Invitrogen) in the growth medium.
Immunoprecipitation and Immunoblotting—Cell lysates prepared in TNTE buffer containing 0.5% Triton X-100 with protease and phosphatase inhibitors were centrifuged at 14,000 × g for 10 min at 4 °C as described previously (25). Specific protein expression in the cleared lysate was analyzed by Western blotting of the total cell lysate. In protein-protein interaction experiments, 90% of the supernatant was also subjected to immunoprecipitation using mouse anti-FLAG (M2; Sigma), mouse anti-Smad2 (BD Transduction Laboratories), rabbit anti-SnoN (H317; Santa Cruz), rabbit anti-ING2 (Southern Alberta Cancer Research Institute Antibody Facility), or rabbit IgG (Invitrogen) antibodies as described previously (35). The protein composition of total cell lysate and immunoprecipitation samples were resolved by SDS-PAGE followed by immunoblotting using mouse anti-Myc (9E10, Covance), mouse anti-FLAG, rabbit anti-SnoN, mouse anti-Smad2, rabbit ING2, mouse anti-HA (Covance), or rabbit anti-Actin (Sigma) as primary antibodies and horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibody (Amersham Biosciences) as the secondary antibodies as described before (25). ECL (Amersham Biosciences)-generated signals were visualized and quantified using a VersaDoc 5000 Imager together with Quantity One software (Bio-Rad).
Luciferase Reporter Assays—HepG2 and Mv1Lu cells were seeded in 12-well plates at 1.5 × 105 cells/well and in 24-well plates at 2.5 × 104 cells/well, respectively. One day later cells were cotransfected with the 3TP, SBE4, or pG5-E1b firefly luciferase reporter construct, the pR-TK Renilla luciferase or CMV-β-galactosidase internal control reporter vector, together with a control expression vector or one encoding the TGF-β type I receptor protein (a wild type (WT) version or a constitutively active form in which threonine 204 is converted to aspartate), an ING protein (ING1a, ING1b, wild type, or deletion mutant versions of ING2), SnoN protein, Gal4 (DNA-binding domain, amino acids 1-147) protein, Gal4-Smad2 fusion protein, ING2 RNAi, or SnoN RNAi as outlined in the figure legends. In experiments where activation of the TGF-β signaling pathway was induced by coexpression of TβRI (T204D), the cells were kept in 10% fetal bovine serum until cell lysis. Otherwise, the cells were incubated 1 day post-transfection in 0.2% fetal bovine serum-containing medium in the presence or absence of 100 pm TGF-β (R & D Systems, Minneapolis, MN) for 16-18 h at 37 °C. Two days post-transfection, the cells were lysed and subjected to dual or regular luciferase activity assays as described previously (25, 28). Arbitrary firefly luciferase activity, expressed in relative light units, was normalized to Renilla luciferase or β-galactosidase activity, as indicated in the figure legends, to control for variations in transfection efficiency. Each experimental condition was carried out in triplicate, and the data are presented as the means (±S.D.). The experiments were repeated on independent batches of cells at least two times.
Reverse Transcription-PCR—Mv1Lu cells were lysed using TRIzol lysis reagent (Invitrogen), followed by total RNA extraction and concentration using chloroform and isopropanol/ethanol, respectively. Poly(A)-cDNA was generated by subjecting the mink RNA to reverse transcription using the reverse transcriptase SuperScript II (Invitrogen) and oligo(dT)12-18 (Amersham Biosciences) as the primer. Mink plasminogen activator inhibitor 1 (PAI-1), ING2, and GAPDH cDNA fragments were PCR-amplified using the mink poly(A)-cDNA as template and gene-specific PAI-1 (sense, 5′-GCCTGGCCCTTGTCTTTGGTG-3′, and antisense, 5′-TGAAGTAGAGGGCATTCACCAGC-3′), ING2 (sense, 5′-AGCGGAGCCGGCTGCTCACC-3′, and antisense, 5′-TCCTGCTTGGCCTTGGAGCG-3′), and GAPDH (sense, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′, and antisense, 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′) oligonucleotides as primers. PCR products of approximately 490, 530, and 300 bp were generated for PAI-1, ING2, and GAPDH, respectively. The PCR products were resolved using 1.2% agarose gels and stained with ethidium bromide. The amplified products were visualized using the Versadoc 5000 Imager and quantified using Quantity one software (Bio-Rad). For each sample the density of PAI-1 or ING2 was normalized to the GAPDH value.
Bromodeoxyuridine (BrdUrd) Assay—Transfected or nontransfected Mv1Lu cells plated in 96-well dishes and preincubated for 3 h in 0.2% serum-containing medium were treated for 18-20 h with or without TGF-β at the concentrations specified in Fig. 4. BrdUrd incorporation and detection were carried out using a BrdUrd assay kit I (Roche Applied Science). Briefly, during the last hour in culture, the cells were incubated at 37 °C with 10 mm BrdUrd-containing medium. The cells were fixed at -20 °C using a glycine-ethanol based solution according to the manufacturer's instructions. The cells were then incubated with a mouse anti-BrdUrd antibody alone or together with a rabbit anti-GFP antibody (Molecular Probes) for 1 h followed by several washes with phosphate-buffered saline. The cells were then switched to a solution containing Hoechst 33342 (Invitrogen) together with fluorescein-conjugated anti-mouse secondary antibody (Roche Applied Science) or cy5-anti-mouse and cy2-anti-rabbit antibodies (Jackson Laboratories). A minimum of 200 cells/well were scanned, and the data were generated using the Kinetic Scan Reader (Cellomics, Inc., Pittsburgh, PA). The data were analyzed using the accompanying Target Activation Bio-Application software (36). Each experimental condition was carried out in three to five replicates. Each experiment was repeated at least three independent times. The number of Hoechst-positive cells that were BrdUrd- and/or GFP-positive was determined. The number of BrdUrd-positive cells expressed as a percentage of GFP- or Hoechst-labeled cells scanned per well was then reported by the Bio-Application software. The average percentage of BrdUrd-positive cells of the replicates was calculated and used to determine other parameters as described in legend of Fig. 5. The data presented in Fig. 5 represent the means (±S.E.) of the parameters obtained from three to four independent experiments.
FIGURE 4.
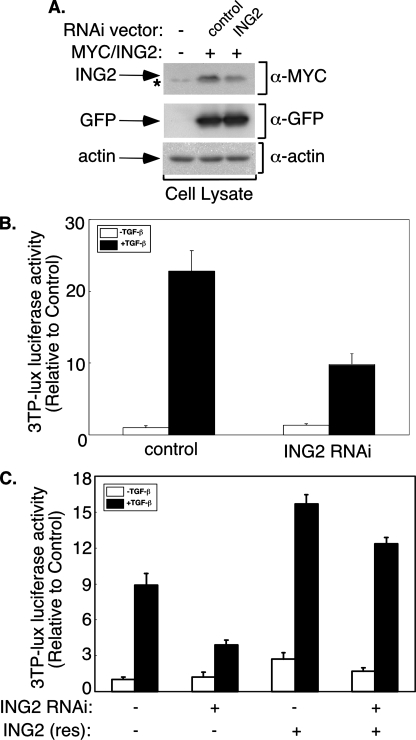
ING2 RNAi suppresses TGF-β-induced transcription. A, ING2, GFP, and actin immunoblotting of lysates of 293T cells coexpressing Myc-tagged ING2 together with a control or an ING2 RNAi plasmid. Quantitative analysis of GFP or actin-normalized ING2 protein density indicates that ING2 RNAi led to a 60-70% reduction in ING2 protein level. The asterisk indicates nonspecific Myc-immunoreactive band. B, effect of ING2 RNAi on TGF-β-induced transcription in Mv1Lu cells. Cells transfected with 3TP-luciferase and CMV-β-galactosidase reporter plasmids together with the control or the ING2 RNAi vector were incubated in the absence or presence of TGF-β. C, the ING2 rescue construct reverses ING2 RNAi-mediated reduction of TGF-β-dependent transcription. The cells were transfected and incubated in the absence or presence of TGF-β as described for B, except for the inclusion of a control mammalian expression vector (-) or one encoding an ING2 RNAi-resistant ING2 protein as indicated. Luciferase activity in the lysates of cells described for B and C were determined as described for Fig. 1D. The data shown in each of B and C represent the means (±S.D.) of triplicate determinations from a representative experiment that was repeated three times.
FIGURE 5.
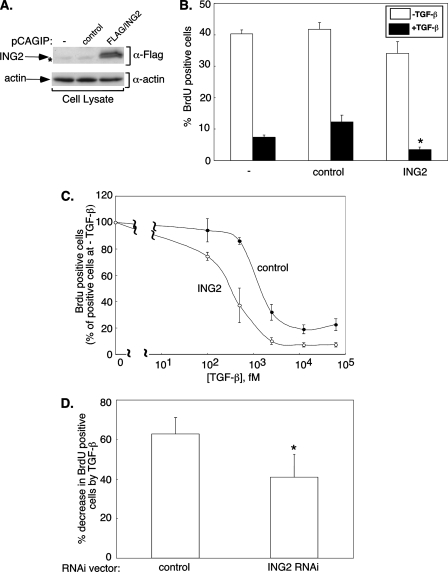
ING2 contributes to TGF-β inhibition of cell proliferation. A, ING2 and actin immunoblotting of lysates of Mv1Lu stable cell lines showing ING2 protein expression. The asterisk indicates nonspecific FLAG-immunoreactive band. B and C, expression of ING2 promotes the ability of TGF-β to inhibit Mv1Lu cell proliferation. B, cells that were untransfected (-), stably expressing the puromycin resistance marker alone (control), or together with a FLAG-tagged ING2 (ING2) were left untreated (-TGF-β) or incubated for 18 h with 62.5 pm TGF-β (+TGF-β). The cells were incubated with BrdUrd-containing medium for 1 h and subjected to indirect immunofluorescence and Hoechst nuclear staining (see “Experimental Procedures”). BrdUrd- and Hoechst-positive cells were imaged and analyzed as outlined under “Experimental Procedures.” The data presented in the bar graph represent the means (±S.E.) of the average of the percent BrdUrd-positive cells from four independent experiments. The asterisk indicates statistically significant difference from the TGF-β-treated untransfected and the resistance marker controls as determined by one-way analysis of variance followed by Student's t test (p ≤ 0.025). C, control or ING2 expressing stable cells were incubated with increasing TGF-β concentrations for 18 h followed by BrdUrd labeling. For each of the control or ING2 expressing cells, the percentage of BrdUrd-positive cells at each TGF-β concentration was expressed relative to that in the absence of TGF-β. The means (±S.E.) of the average values from three independent experiments are plotted on the y axis versus the TGF-β concentration (fM) on the x axis. D, knockdown of endogenous ING2 reduces TGF-β-dependent suppression of cell proliferation. Mv1Lu cells transiently transfected with the control or ING2 RNAi vector were incubated in the absence or presence of 25 pm TGF-β followed by BrdUrd incorporation. BrdUrd and GFP immunolabeling, detection, and analysis were carried out as described under “Experimental Procedures.” For each transfection group, the percentage of decrease in BrdUrd-positive cells by TGF-β treatment was determined by expressing the difference in the percentage of BrdUrd-positive cells in absence and presence of TGF-β relative to the BrdUrd-positive cells in the absence of TGF-β. The data presented in the bar graph represent the means (±S.E.) of the percentage of decrease in BrdUrd-positive cells by TGF-β from four independent experiments. The asterisk indicates a statistically significant decrease from the control as determined by Student's t test (p ≤ 0.025).
RESULTS
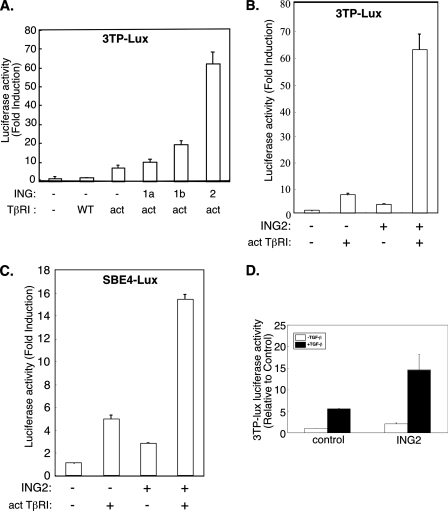
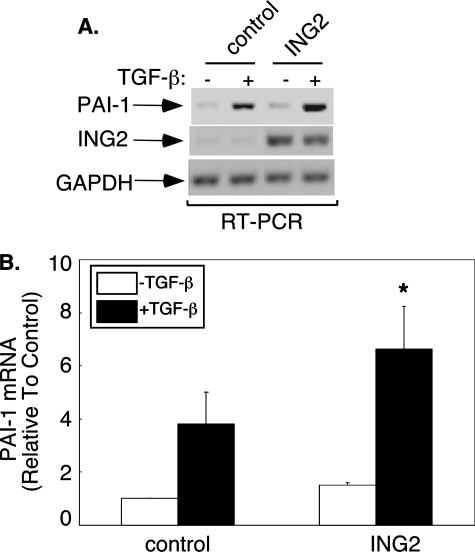
ING2 Acts via the PHD Domain to Stimulate TGF-β-dependent Transcription—The transcriptional and biological functions of the ING proteins suggest that these proteins may regulate specific growth factor-mediated signaling pathways and their responses. The TGF-β family of growth factors has pleiotropic roles that are critical in normal development and homeostasis (15, 16). The Smad signal transducers play a major role in transmitting the TGF-β signal from the cell surface to the nucleus where the Smad proteins regulate gene expression (21). A major focus of research in TGF-β signaling is the elucidation of the nuclear mechanisms by which the Smads mediate TGF-β-dependent gene expression and consequent responses. Both the TGF-β-Smad signaling pathway and the ING transcriptional proteins inhibit cell proliferation and exert tumor-suppressive effects, suggesting the hypothesis that the ING proteins might play a role in TGF-β-dependent responses (10, 13, 17, 18). To test this idea, we first asked whether these proteins modulate TGF-β-induced transcription. We determined the effect of expression of different members of the ING family of proteins on TGF-β induction of a reporter gene that contains TGF-β-responsive promoter elements from the PAI-1 gene driving luciferase enzyme expression (33). In these experiments, the TGF-β pathway was stimulated by coexpressing a constitutively activated form of the TGF-β type I serine/threonine kinase receptor (TβRI) in which threonine 204 is converted to aspartate (Fig. 1A and Ref. 37). Interestingly, ING2 robustly increased TβRI-induced reporter gene expression (Fig. 1A). The effect of ING2 and TβRI on reporter gene expression was synergistic (Fig. 1B). In contrast to ING2, the ING family members ING1a and ING1b, which are the ING family members most closely related to ING2, had less effect on the TβRI-induced response (Fig. 1A). Thus, among the ING proteins, ING2 may be more efficient in inducing TGF-β-dependent transcription. We also found that ING2 increased the ability of the activated type I receptor to induce the SBE4-luciferase reporter gene, which has four tandem repeats of SBEs driving the expression of the luciferase construct (Fig. 1C and Ref. 34). ING2 also promoted the expression of the reporter gene driven by the PAI-1 promoter in HepG2 and the Mv1Lu lung epithelial cell lines upon exposure to the TGF-β ligand (data not shown and Fig. 1D). In other experiments, we found that ING2 expression significantly augmented the ability of TGF-β to induce endogenous PAI-1 gene expression in Mv1Lu cells (Fig. 2), further supporting the idea that ING2 positively controls TGF-β-induced transcription. Collectively, these data indicate that ING2 may function to up-regulate TGF-β-dependent transcription and suggest that ING2 may act via Smad family proteins to mediate the TGF-β response.
FIGURE 1.
ING2 enhances TGF-β-dependent transcription. A, HepG2 cells were transfected with 3TP-luciferase and pRL-TK reporter constructs together with an empty mammalian expression vector (-) or one encoding a WT or a constitutively activated (act) TGF-β type I receptor alone or with a pCI plasmid expressing the ING1a, ING1b, or ING2 proteins. The lysates were subjected to the dual luciferase assay. The firefly luciferase data were normalized to the corresponding Renilla luciferase activity as described under “Experimental Procedures.” The normalized firefly luciferase activity for each transfection is expressed relative to the corresponding values in lysates of cells transfected with empty vector control (fold induction). B, lysates of HepG2 cells, transfected with the reporter constructs described for A together with an empty mammalian expression vector alone, one encoding activated TGF-β type I receptor (act), ING2, alone or together, were processed and analyzed as described for A. C, HepG2 cells were transfected with SBE4-luciferase and pRL-TK reporter constructs together with an empty expression vector, or one encoding activated TGF-β type I receptor, ING2, alone or together. Luciferase assays were assayed and expressed as described for A. D, lysates of Mv1Lu cells transfected with 3TP-luciferase and CMV-β-galactosidase constructs together with an empty expression vector or one encoding Myc-tagged ING2 protein, and either left untreated (-TGF-β) or incubated for 16 h with TGF-β (+TGF-β), were subjected to luciferase and β-galactosidase assays as described under “Experimental Procedures.” For each sample, the luciferase data were normalized to the corresponding β-galactosidase values. The β-galactosidase normalized luciferase data were then expressed relative to the corresponding value obtained from lysates of TGF-β-untreated control cells. The data in each of A, B, and C are the means (±S.D.) of six replicate measurements, and the data in D are the means (±S.D.) of triplicate measurements from a representative experiment that was repeated at least two times.
FIGURE 2.
ING2 induces the expression of the TGF-β-responsive PAI-1 gene. RNA extracted from untreated or 1 h TGF-β-treated control or ING2-overexpressing Mv1Lu cells was reverse transcribed using oligo(dT) followed by PCR amplification of fragments of PAI-1, ING2, and GAPDH gene products as described under “Reverse Transcription-PCR” under “Experimental Procedures.” The PAI-1, ING2, and GAPDH amplified PCR fragments were resolved, imaged, and quantified (see “Reverse Transcription-PCR” for details). The PAI-1 value for each sample was normalized to its respective GAPDH control and expressed relative to the untreated control. A, a scan of an ethidium bromide-stained agarose gel showing the PAI-1, ING2, and GAPDH reverse transcription-PCR-amplified fragment from a representative experiments that was repeated three times. B, bar graph of the mean (±S.E.) of the PAI-mRNA values from three independent experiments. The asterisk indicates a statistically significant increase from the TGF-β-treated control as determined by Student's t test (p = 0.06).
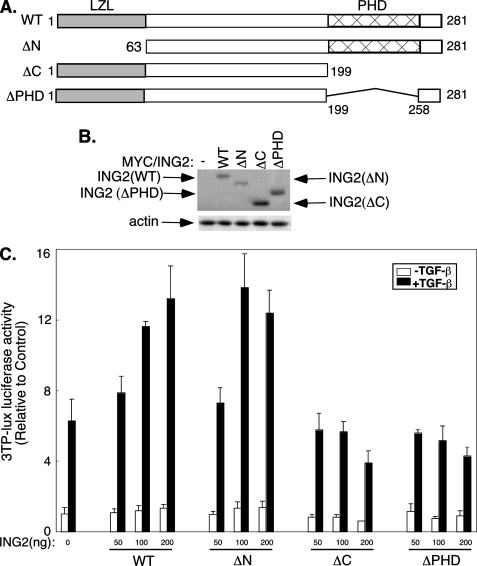
Using structure function analyses, we next determined the domains in ING2 that are required for its enhancement of TGF-β-dependent transcription. We compared the effect of wild type ING2, an N-terminal deletion ING2 mutant (ING2 (ΔN)), a C-terminal deletion ING2 mutant (ING2 (ΔC)), or a PHD deletion ING2 mutant (ING2 (ΔPHD)), on TGF-β-dependent transcription in Mv1Lu cells (Fig. 3). We confirmed that the different ING2 mutants were expressed at equivalent levels by immunoblotting (Fig. 3B). The N-terminal region of ING2 contains a leucine zipper-like protein-protein interaction motif spanning amino acid residues 1-62 that is conserved in other ING proteins (Fig. 3A and Ref. 38). The C-terminal region of the ING2 mutant contains the PHD domain, which plays important roles in chromatin remodeling, and a downstream short C-terminal tail (Fig. 3A) called the polybasic region that binds stress-induced phospholipids (5). ING2 immunoblotting of subcellular fractions and ING2 immunofluorescence of 293T cells showed that wild type and mutant ING2 proteins localized mainly to the nucleus (supplemental Fig. S1). In other experiments, we found that deletion of the N-terminal leucine zipper-like region had little effect on the ability of ING2 to up-regulate TGF-β-induced 3TP-luciferase activity, suggesting that the leucine zipper-like motif is dispensable in ING2 enhancement of TGF-β-dependent transcription (Fig. 3C). In contrast, deletion of the PHD-containing C-terminal region or only the PHD domain blocked the ability of ING2 to increase TGF-β-induced transcription, indicating that the PHD region is required for ING2 function in the TGF-β-induced response (Fig. 3C). In addition, with higher amounts of ING2 mutants lacking the PHD domain, TGF-β-induced reporter gene expression was lower than in the vector control transfected cells, suggesting that ING2 mutants lacking the PHD domain may act in a dominant negative fashion to inhibit endogenous ING2 function (Fig. 3C). Taken together, these data suggest that ING2 acts via its PHD domain to promote TGF-β-dependent transcription.
FIGURE 3.
The PHD domain is required for ING2 enhancement of TGF-β-induced transcription. A, schematic drawing of the WT, N-terminal (ΔN), C-terminal (ΔC), and PHD (ΔPHD) deletion mutants of human ING2 protein encoded by expression vectors used in experiments described for B and C. The amino acids deleted in the respective mutant ING2 relative to the full-length ING2 are indicated. The shaded and the hatched areas depict the leucine zipper-like and PHD domains, respectively. B, immunoblotting of ING2 and actin in extract of Mv1Lu cells transfected with an empty expression vector, wild type ING2 (ING2(WT)), N-terminal-deleted ING2 (ING2(ΔN)), C-terminal-deleted ING2 (IN2(ΔC)), or a PHD-deleted ING2 (ING2(ΔPHD)). The data show that the full-length and deletion mutant ING2 proteins are expressed at equivalent levels. C, lysates of untreated or TGF-β-stimulated Mv1Lu cells transfected with the 3TP-luciferase and CMV-β-galactosidase together with an empty mammalian expression vector (0), or an increasing concentration of plasmid encoding Myc-tagged wild type or one of the deletion mutant ING2 described in A were processed as described for Fig. 1D. The data presented in the graphs in C are the means (±S.D.) of triplicate measurements from a representative experiment that was repeated three independent times.
To assess the function of endogenous ING2 in TGF-β-mediated transcription, we used a plasmid-based method of RNAi to knockdown ING2 in Mv1Lu cells. Expression of hairpin RNAs designed to target ING2 specifically triggered the knockdown of ING2 that was expressed in 293T cells (Fig. 4A). We also found that ING2 RNAi reduced endogenous ING2 mRNA and protein levels in Mv1Lu cells (supplemental Fig. S2). In functional assays, we found that ING2 RNAi reduced TGF-β-dependent expression of the 3TP-luciferase reporter gene in Mv1Lu cells as compared with the vector control transfected cells, suggesting a requirement for ING2 in TGF-β-induced transcription (Fig. 4B). To establish the specificity of the ING2 RNAi effect, we performed a rescue experiment. We constructed an ING2 expression plasmid using a mutated cDNA designed to encode an RNAi-resistant form of ING2. Expression of this RNAi-resistant ING2 reversed the ability of ING2 RNAi to suppress TGF-β induction of 3TP-lucifease reporter gene transcription (Fig. 4C). These data support the conclusion that the effect of ING2 RNAi on TGF-β signaling is the result of specific knockdown of endogenous ING2 rather than off target effects of RNAi. Collectively, the results show that endogenous ING2 promotes TGF-β-dependent transcription.
ING2 Promotes the Ability of TGF-β to Inhibit Cell Proliferation—Having shown that ING2 mediates TGF-β-dependent transcription, we asked whether it contributes to TGF-β-mediated biological responses. One of the most widely studied and important effects of TGF-β is its ability to inhibit cell proliferation. Thus, we investigated the role of ING2 in TGF-β-induced cell cycle arrest in Mv1Lu cells. We first determined the effect of a saturating concentration of TGF-β on BrdUrd incorporation into newly synthesized DNA in Mv1Lu cells that were stably expressing ING2 protein, in cells stably transfected with the vector control, or in cells that were untransfected (Fig. 5A). Untransfected and vector control transfected cells had similar percentages of proliferating cells in the absence of TGF-β (Fig. 5B). Overnight incubation of these cells with TGF-β also led to similar degrees of inhibition of proliferation between both control cells (Fig. 5B). However, ING2 expression significantly enhanced the ability of TGF-β to induce cell cycle arrest as compared with controls (Fig. 5B). In other experiments, we found that in the presence of ING2, a 2.5-3-fold lower concentration of TGF-β induced a 50% reduction in cell proliferation, suggesting that ING2 expression increases cell sensitivity to TGF-β (Fig. 5C). We also determined the role of endogenous ING2 in TGF-β inhibition of cell proliferation. ING2 knockdown in Mv1Lu cells significantly reduced TGF-β inhibition of cell proliferation (Fig. 5D). Together, these data support the conclusion that ING2 promotes the ability of TGF-β to inhibit cell proliferation.
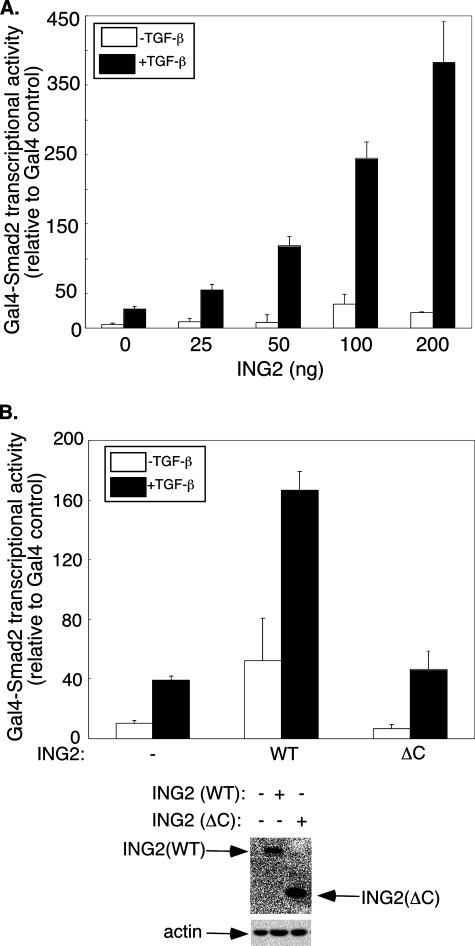
ING2 and SnoN Collaborate to Induce TGF-β-dependent Transcription—The identification of a function for ING2 in TGF-β-dependent transcription and cell cycle arrest led us to explore the mechanisms by which ING2 contributes to TGF-β signaling. We first determined whether ING2 up-regulates the transcriptional activity of TGF-β-regulated Smad2 by using the Gal4-Smad2 fusion heterologous transcription assay as a measure of Smad2 transcriptional activity (30, 31). Expression of ING2 dramatically enhanced the ability of Gal4-Smad2 to mediate TGF-β-induced transcription (Fig. 6A). Deletion of the PHD-containing C-terminal region significantly reduced the ability of ING2 to stimulate Smad2-dependent transcription (Fig. 6B). These data indicate that the ING2 PHD domain plays an important role in TGF-β-dependent transcription by regulating the transcriptional activity of TGF-β-regulated Smad2. In other experiments, we found that ING2 similarly promotes TGF-β-induced Gal4-Smad3 transcriptional activity (supplemental Fig. S3). Collectively, these data indicate that ING2 acts via Smad2 and Smad3 to up-regulate TGF-β-induced transcription.
FIGURE 6.
ING2 increases R-Smad2 transcriptional activity. A, ING2 expression increases TGF-β-induced Gal4-Smad2 transcriptional activity. Lysates of untreated or TGF-β-treated Mv1Lu cells transfected with pG5-E1b-luciferase and CMV-β-galactosidase reporter together with an expression vector encoding Gal4 alone or Gal4-Smad2 fusion and increasing amount of ING2 expressing vector (ng/well) were assayed for luciferase and β-galactosidase activity as described under “Experimental Procedures” and for Fig. 1D. The normalized luciferase activity for Gal4-Smad2 at each condition is expressed relative to the respective Gal4 control. B, the PHD-containing C-terminal region of ING2 is required for ING2 to induce Smad2 transcriptional activity. Lysates of untreated or ligand stimulated Mv1Lu cells transfected with the reporter constructs and Gal4 or Gal4-Smad2 expressing plasmids together with an empty expression vector (-), one encoding WT, or C-terminally deleted ING2 protein (ΔC) were subjected to luciferase and β-galactosidase assays as described for Fig. 1D. The luciferase values are presented as described for A. The data presented in each of A and B represent the means (±S.D.) of a triplicate from a representative experiment that was repeated at least two times.
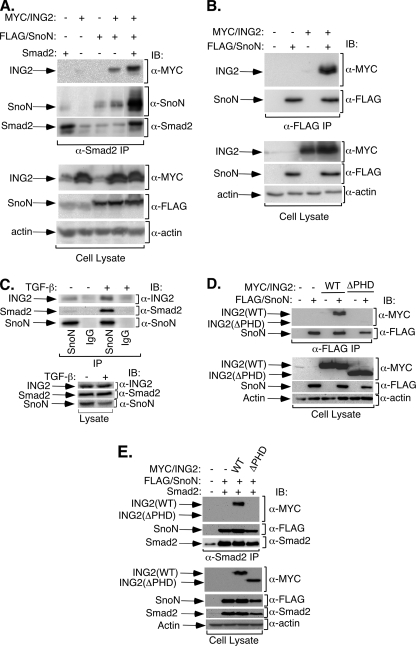
We next investigated the mechanism by which ING2 promotes TGF-β-Smad2 signaling leading to the induction of transcription. In immunoblotting studies, we found that ING2 expression did not appreciably alter the degree of TGF-β-dependent Smad2 phosphorylation (supplemental Fig. S4). Because ING2 is a nuclear protein that operates at the level of chromatin remodeling, we next reasoned that ING2 might form a physical complex with Smad2 and thereby couple the TGF-β signal to the activation of transcription. Because ING2 associated only weakly with Smad2 (data not shown), we considered the possibility that Smad2 might interact with ING2 via a third “adaptor” protein. The transcriptional modulator SnoN robustly interacts with TGF-β-R-Smad2/3 and co-Smad4 (39). In addition, SnoN at its endogenous levels mediates TGF-β-dependent transcription in Mv1Lu cells, thus mimicking the effect of ING2 on TGF-β signaling on transcription (25). We therefore determined whether SnoN might collaborate with ING2 to link Smad2 or Smad3 activity to transcription in these cells. In these studies, we focused on Smad2. We tested the effect of SnoN expression on the ability of endogenous or exogenously expressed Smad2 to associate with ING2. In coimmunoprecipitation analyses, we found that expression of SnoN enhanced the ability of Smad2 to interact with ING2 (Fig. 7A and supplemental Fig. S5). We also found that when overexpressed, SnoN associated efficiently with ING2 (Fig. 7B). Importantly, endogenous SnoN coimmunoprecipitated endogenous Smad2 and ING2 in TGF-β-treated Mv1Lu cells (Fig. 7C). Other studies revealed that the ING2 PHD domain is important for ING2 interaction with SnoN (Fig. 7D). Consistent with these results, SnoN failed to promote an interaction between Smad2 and an ING2 lacking the PHD domain (Fig. 7E). The results of immunofluorescence studies suggested that ING2, SnoN, and Smad2 colocalize in the nucleus (supplemental Fig. S6 and data not shown). Altogether, these data strongly suggest that SnoN promotes formation of a protein complex that contains SnoN, Smad2, and ING2, and that the PHD domain is important for the formation of this complex.
FIGURE 7.
The transcriptional protein SnoN interacts with ING2. A, SnoN enhances ING2 association with Smad2. Lysates of 293T cells coexpressing Myc-tagged ING2 (Myc/ING2), FLAG-tagged SnoN (FLAG/SnoN), and untagged Smad2 (Smad2) were subjected to anti-Smad2 immunoprecipitation (IP) followed by sequential anti-Myc (α-Myc), anti-SnoN (α-SnoN), and anti-Smad2 (α-Smad2) immunoblotting (IB). Expression of ING2 and SnoN were confirmed by immunoblotting the cell lysates with the indicated antibodies. Equal protein loading was confirmed by anti-actin (α-actin) immunoblotting of the lysates. B, overexpressed SnoN and ING2 form a complex. Lysates of 293T cells coexpressing FLAG-tagged SnoN and Myc-tagged ING2 were immunoprecipitated using an anti-FLAG antibody followed by sequential anti-Myc and anti-SnoN antibodies immunoblotting. C, endogenous SnoN interacts with endogenous ING2 and Smad2 in TGF-β-treated Mv1Lu cells. Lysates of Mv1Lu cells incubated in the absence or presence of 100 pm TGF-β for 10 min at 37 °C were immunoprecipitated using a rabbit anti-SnoN (SnoN) or a rabbit-IgG (IgG) antibody followed by sequential immunoblotting with anti-ING2, anti-Smad2, and anti-SnoN antibodies. D, the PHD domain is important for ING2 association with SnoN. Lysates of 293T cells coexpressing FLAG-tagged SnoN and Myc-tagged ING2 were immunoprecipitated using an anti-FLAG antibody followed by sequential anti-Myc and anti-SnoN antibodies immunoblotting. E, deletion of the PHD domain blocks ING2 interaction with Smad2. Lysates of 293T cells coexpressing untagged Smad2, FLAG-tagged SnoN, and Myc-tagged ING2 were immunoprecipitated using an anti-Smad2 antibody followed by sequential anti-Myc, anti-FLAG, and anti-Smad2 antibody immunoblotting. Expression and equal protein loading in B-E were confirmed as described for A.
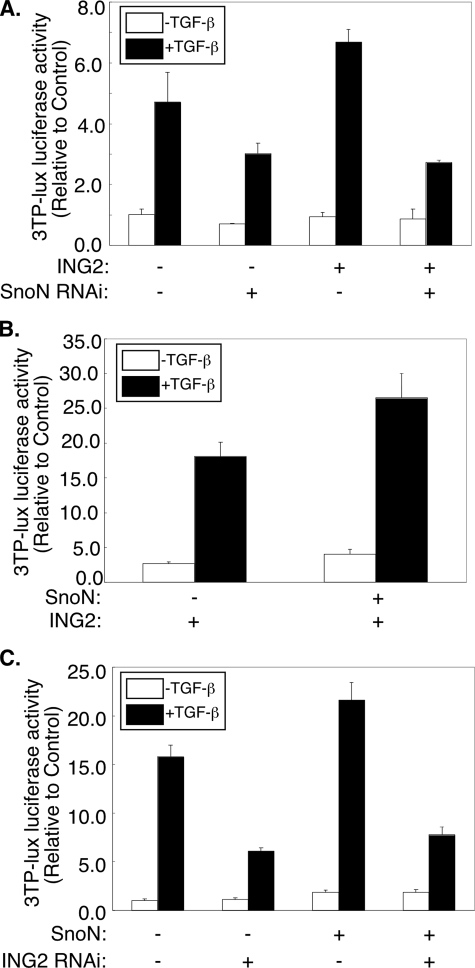
The ability of SnoN to form a protein complex with ING2 and Smad2 suggested that SnoN may contribute to ING2 enhancement of TGF-β signaling in Mv1Lu cells. To test the potential role of SnoN in ING2 upregulation of TGF-β transcription, we determined the effect of SnoN knockdown on ING2 up-regulation of TGF-β-induced 3TP-luciferase activity in Mv1Lu cells. As expected, SnoN knockdown, on its own, inhibited TGF-β-induced expression of the 3TP-luciferase reporter gene (25). Additionally, SnoN knockdown also suppressed ING2 enhancement of TGF-β-induced transcription, suggesting that SnoN contributes to the ability of ING2 to mediate TGF-β-dependent transcription (Fig. 8A). The effect of SnoN RNAi was specific to knockdown of endogenous SnoN and not due to off target effect because we found that expression of a SnoN rescue protein reversed SnoN RNAi reduction of ING2 enhancement of TGF-β-induced transcription (supplemental Fig. S7 and Ref. 25). Consistent with these results, in gain of function experiments, low levels of SnoN expression augmented ING2 enhancement of TGF-β-dependent transcription (Fig. 8B and Ref. 25). Finally, we found ING2 knockdown suppressed the ability of low levels of SnoN to activate transcription of the 3TP-luciferase reporter gene in TGF-β-treated Mv1Lu cells, suggesting that ING2 is required for SnoN to mediate TGF-β-induced transcription (Fig. 8C). Collectively, these data suggest that SnoN and ING2 collaborate to mediate TGF-β-dependent transcription.
FIGURE 8.
SnoN and ING2 collaborate to enhance TGF-β-dependent transcription. A, SnoN knockdown reduces ING2-dependent up-regulation of TGF-β-mediated transcription. Lysates of Mv1Lu cells transfected with the 3TP-luciferase and β-galactosidase reporter constructs together with the control or the SnoN RNAi plasmid and an empty mammalian expression vector or one encoding the ING2 protein and incubated in the absence or presence of TGF-β were subjected to luciferase and β-galactosidase assays as described for Fig. 1D. B, SnoN induces the ability of ING2 to promote TGF-β-dependent transcription. Mv1Lu cells were transfected with the reporter constructs as in Fig. 8A and with an ING2 expressing vector alone or together with a SnoN expressing plasmid. The cells were incubated with or without TGF-β, and the lysates were subjected to luciferase and β-galactosidase assays as described for Fig. 1D. C, ING2 knockdown inhibits SnoN enhancement of TGF-β-induced transcription. Lysates of Mv1Lu cells transfected with the reporter constructs as described for A together with a control or ING2 RNAi vector and an empty expression vector or one encoding SnoN and incubated in the absence or presence of TGF-β were subjected to luciferase and β-galactosidase assays as described for Fig. 1D. In the transfections described in each of B or C, a low amount (1-5 ng/well) of SnoN expressing vector was used (34). The data shown in each of A-C represent the means (±S.D.) of triplicate measurements from a representative experiment that was repeated three times.
In summary, we have identified ING2 as a mediator of TGF-β-dependent transcription and TGF-β inhibition of cell proliferation. We show theSmad2-interacting protein SnoN associates with ING2 and thus assembles a protein complex that includes Smad2, SnoN, and ING2. We also demonstrate that ING2 PHD domain, shown to interact with methylated histones (1, 2), is required for ING2 function in TGF-β-dependent responses and for interaction with SnoN and hence Smad2. Collectively, these data suggest that the Smad2/SnoN/ING2 complex may couple TGF-β-Smad signaling to chromatin remodeling and transcription and consequent biological responses.
DISCUSSION
In this study, we have discovered a novel function for the chromatin remodeling protein ING2 in mediating TGF-β responses. Based on gain-of-function and inhibition of endogenous ING2 studies, we have found that ING2 promotes TGF-β-induced transcription and cell cycle arrest. The PHD domain of ING2 is required for ING2 function in TGF-β signaling. We also identify a novel interaction between ING2 and the transcriptional modulator SnoN that recruits ING2 to the TGF-β-regulated transcription factor Smad2 and thereby induces TGF-β-dependent transcription. We find that PHD domain of ING2 is critical for the ability of ING2 to promote TGF-β-induced transcription and to interact with SnoN and in turn Smad2. In addition to implicating ING2 in TGF-β signaling, this study defines a novel mechanism by which SnoN activates TGF-β-dependent transcription and responses (25). Collectively, our data suggest that SnoN and ING2 collaborate to regulate TGF-β-dependent transcription and consequent biological responses.
SnoN is commonly thought to act as a transcriptional corepressor (26, 27). However, recent evidence supports the idea that under certain conditions SnoN may also act as a transcriptional coactivator (40). Thus, at “physiological” levels, SnoN mediates TGF-β-induced transcription in Mv1Lu cells (25). We have also found that knockdown of endogenous SnoN inhibited the ability of TGF-β to induce transcription in TGF-β-responsive epithelial Madin-Darby canine kidney cells (supplemental Fig. S8). We have also found that SnoN may positively regulate TGF-β-dependent transcription in primary mammalian neurons (supplemental Fig. S9). TGF-β-Smad2 signaling and SnoN play critical roles in the control of axonal growth in primary neurons (41, 42). TGF-β-Smad2 signaling is constitutively active in neurons (42). Consistent with these results, we found that knockdown of endogenous Smad2 significantly reduces the activity of the TGF-β-responsive 3TP-lucifearse reporter gene in primary rat cerebellar granule neurons (supplemental Fig. S9A). In addition, knockdown of endogenous SnoN suppressed, whereas expression of a constitutively active SnoN enhances, the reporter gene in primary neurons (supplemental Fig. 9). Together these data show that SnoN stimulates TGF-β-dependent transcription in neurons. Drosophila dSno was also shown recently to mediate the Drosophila TGF-β signaling pathway, suggesting that the ability of SnoN to activate transcription is evolutionarily conserved (51). In future studies, it will be important to investigate whether ING2 or other ING family members play a role in SnoN ability to positively regulate transcription in different cell types and systems.
Recently, the mechanisms by which the ING proteins regulate cell proliferation, apoptosis, senescence, angiogenesis, and DNA repair has been the subject of intense investigation. The ING proteins may function in part by regulating the p53 signaling pathway. In particular, INGs associate with and thereby control the acetylation status of p53 (28, 43). ING-dependent acetylation of p53 is proposed to drive specific gene expression leading to apoptosis and senescence (28, 43). However, INGs also function in p53-independent manner. In support of this idea, ING1 has been shown to induce apoptosis or inhibit cell proliferation in wild type and p53 mutant cells types (44, 45). In addition, INGs inhibit the nuclear factor κB signaling pathway by at least two different mechanisms (46, 47). Our study places ING2 in TGF-β signaling and suggests that ING2 function may be more generalized than previously believed.
TGF-β is known to potently inhibit cell proliferation (48). Loss of responsiveness to TGF-β inhibition of cell proliferation is a hallmark of many types of cancer (17, 48). Reduced ING protein expression has also been reported in several forms of tumors including mammary and lung carcinomas (49). Thus, our findings suggest the hypothesis that loss of ING proteins may allow cancer cells to evade the tumor-suppressive effects of TGF-β. It will be important in future studies to investigate this hypothesis by concurrent analyses of ING protein levels and responsiveness to TGF-β effects of cancer cells.
Our finding that ING2 plays a positive role in TGF-β-induced transcription and inhibition of cell proliferation raises the important question of whether ING2 contributes to other TGF-β-regulated responses. If future investigations indicate that ING2 mediates only a subset of TGF-β-regulated responses, then this would support the idea that ING2 may contribute to specifying distinct responses to the TGF-β-Smad signaling pathway.
According to several reports, ING2 can exist as a component of histone acetyltransferase coactivator or histone deacetylase corepressor complexes, suggesting that ING2 may activate or repress gene expression (1, 2, 9, 43). Consistent with this idea, ING2 can activate transcription of specific genes including p21 and HSP70 while suppressing that of other including α-fetoprotein and cyclin D (1, 28, 43, 47). Our findings are consistent with ING2 acting as part of a histone acetyltransferase complex to promote TGF-β-dependent transcription and responses.
The PHD-containing C terminus of ING proteins is believed to play an important role in their function (10). This region has been implicated in several ING2 actions including ING2 regulation of p53-dependent responses (3). However, the N-terminal region of the ING proteins can contribute to other ING proteins actions. For example, ING1b protein acts through its N terminus and not C terminus to induce HSP70 expression and mediate TNF-α-induced apoptosis in fibroblasts (47). In the current study, we find that the PHD-containing region in ING2 is important for the ability of ING2 to interact with SnoN and to up-regulate TGF-β-mediated transcription. Thus, different structural domains of ING proteins appear to endow these proteins with the ability to function in distinct signaling pathways (reviewed in Ref. 50).
In conclusion, the results of this study reveal that the ING proteins can function to mediate TGF-β signaling and responses. In addition, our findings show that the transcriptional modulator SnoN is able to act via ING2 to mediate TGF-β-dependent transcription and responses.
Supplementary Material
Acknowledgments
We thank J. Wrana, I. von Both, and A. Smith for providing plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grant NS051255 (to A. B.) and by National Institutes of Health Training Grant GM077226 (to M. A. H.). This work was also supported by grants from the Alberta Cancer Board and the Canadian Institutes of Health Research (to S. B.) and the Harvard Medical School-Merck Sponsored Research Program. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S9.
Footnotes
The abbreviations used are: PHD, plant homeodomain; TGF, transforming growth factor; RNAi, RNA interference; R-Smad, receptor-regulated Smad; SBE, Smad-binding element; GFP, green fluorescent protein; CMV, cytomegalovirus; WT, wild type; PAI, plasminogen activator inhibitor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; BrdUrd, bromodeoxyuridine.
References
- 1.Pena, P. V., Davrazou, F., Shi, X., Walter, K. L., Verkhusha, V. V., Gozani, O., Zhao, R., and Kutateladze, T. G. (2006) Nature 442 100-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi, X., Hong, T., Walter, K. L., Ewalt, M., Michishita, E., Hung, T., Carney, D., Pena, P., Lan, F., Kaadige, M. R., Lacoste, N., Cayrou, C., Davrazou, F., Saha, A., Cairns, B. R., Ayer, D. E., Kutateladze, T. G., Shi, Y., Cote, J., Chua, K. F., and Gozani, O. (2006) Nature 442 96-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gozani, O., Karuman, P., Jones, D. R., Ivanov, D., Cha, J., Lugovskoy, A. A., Baird, C. L., Zhu, H., Field, S. J., Lessnick, S. L., Villasenor, J., Mehrotra, B., Chen, J., Rao, V. R., Brugge, J. S., Ferguson, C. G., Payrastre, B., Myszka, D. G., Cantley, L. C., Wagner, G., Divecha, N., Prestwich, G. D., and Yuan, J. (2003) Cell 114 99-111 [DOI] [PubMed] [Google Scholar]
- 4.Jones, D. R., Bultsma, Y., Keune, W. J., Halstead, J. R., Elouarrat, D., Mohammed, S., Heck, A. J., D'Santos, C. S., and Divecha, N. (2006) Mol. Cell 23 685-695 [DOI] [PubMed] [Google Scholar]
- 5.Kaadige, M. R., and Ayer, D. E. (2006) J. Biol. Chem. 281 28831-28836 [DOI] [PubMed] [Google Scholar]
- 6.Loewith, R., Meijer, M., Lees-Miller, S. P., Riabowol, K., and Young, D. (2000) Mol. Cell. Biol. 20 3807-3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vieyra, D., Loewith, R., Scott, M., Bonnefin, P., Boisvert, F. M., Cheema, P., Pastyryeva, S., Meijer, M., Johnston, R. N., Bazett-Jones, D. P., McMahon, S., Cole, M. D., Young, D., and Riabowol, K. (2002) J. Biol. Chem. 277 29832-29839 [DOI] [PubMed] [Google Scholar]
- 8.Kuzmichev, A., Zhang, Y., Erdjument-Bromage, H., Tempst, P., and Reinberg, D. (2002) Mol. Cell. Biol. 22 835-848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doyon, Y., Cayrou, C., Ullah, M., Landry, A. J., Cote, V., Selleck, W., Lane, W. S., Tan, S., Yang, X. J., and Cote, J. (2006) Mol. Cell 21 51-64 [DOI] [PubMed] [Google Scholar]
- 10.Russell, M., Berardi, P., Gong, W., and Riabowol, K. (2006) Exp. Cell Res. 312 951-961 [DOI] [PubMed] [Google Scholar]
- 11.Toyama, T., Iwase, H., Watson, P., Muzik, H., Saettler, E., Magliocco, A., DiFrancesco, L., Forsyth, P., Garkavtsev, I., Kobayashi, S., and Riabowol, K. (1999) Oncogene 18 5187-5193 [DOI] [PubMed] [Google Scholar]
- 12.Chen, L., Matsubara, N., Yoshino, T., Nagasaka, T., Hoshizima, N., Shirakawa, Y., Naomoto, Y., Isozaki, H., Riabowol, K., and Tanaka, N. (2001) Cancer Res. 61 4345-4349 [PubMed] [Google Scholar]
- 13.Feng, X., Hara, Y., and Riabowol, K. (2002) Trends Cell Biol. 12 532-538 [DOI] [PubMed] [Google Scholar]
- 14.Kameyama, K., Huang, C. L., Liu, D., Masuya, D., Nakashima, T., Sumitomo, S., Takami, Y., Kinoshita, M., and Yokomise, H. (2003) Clin. Cancer Res. 9 4926-4934 [PubMed] [Google Scholar]
- 15.Roberts, A. B., and Sporn, M. B. (1990) in Peptide Growth Factors and Their Receptors (Sporn, M. B., and Roberts, A. B., eds) pp. 419-472, Springer-Verlag, Heidelberg
- 16.Massague, J. (1990) Annu. Rev. Cell Biol. 6 597-641 [DOI] [PubMed] [Google Scholar]
- 17.Massague, J., Blain, S. W., and Lo, R. S. (2000) Cell 103 295-309 [DOI] [PubMed] [Google Scholar]
- 18.Derynck, R., Akhurst, R. J., and Balmain, A. (2001) Nat. Genet. 29 117-129 [DOI] [PubMed] [Google Scholar]
- 19.Jakowlew, S. B. (2006) Cancer Metastasis Rev. 25 435-457 [DOI] [PubMed] [Google Scholar]
- 20.Shi, Y., and Massague, J. (2003) Cell 113 685-700 [DOI] [PubMed] [Google Scholar]
- 21.Massague, J., Seoane, J., and Wotton, D. (2005) Genes Dev. 19 2783-2810 [DOI] [PubMed] [Google Scholar]
- 22.Attisano, L., and Wrana, J. L. (2000) Curr. Op. Cell Biol. 12 235-243 [DOI] [PubMed] [Google Scholar]
- 23.Wotton, D., and Massague, J. (2001) Curr. Top. Microbiol. Immunol. 254 145-164 [PubMed] [Google Scholar]
- 24.Stroschein, S. L., Wang, W., and Luo, K. (1999) J. Biol. Chem. 274 9431-9441 [DOI] [PubMed] [Google Scholar]
- 25.Sarker, K. P., Wilson, S. M., and Bonni, S. (2005) J. Biol. Chem. 280 13037-13046 [DOI] [PubMed] [Google Scholar]
- 26.Liu, X., Sun, Y., Weinberg, R. A., and Lodish, H. F. (2001) Cytokine Growth Factor Rev. 12 1-8 [DOI] [PubMed] [Google Scholar]
- 27.Luo, K. (2004) Curr. Opin. Genet. Dev. 14 65-70 [DOI] [PubMed] [Google Scholar]
- 28.Kataoka, H., Bonnefin, P., Vieyra, D., Feng, X., Hara, Y., Miura, Y., Joh, T., Nakabayashi, H., Vaziri, H., Harris, C. C., and Riabowol, K. (2003) Cancer Res. 63 5785-5792 [PubMed] [Google Scholar]
- 29.von Both, I., Silvestri, C., Erdemir, T., Lickert, H., Walls, J. R., Henkelman, R. M., Rossant, J., Harvey, R. P., Attisano, L., and Wrana, J. L. (2004) Dev. Cell 7 331-345 [DOI] [PubMed] [Google Scholar]
- 30.Hayashi, H., Abdollah, S., Qiu, Y., Cai, J., Xu, Y. Y., Grinnell, B. W., Richardson, M. A., Topper, J. N., Gimbrone, M. A., Jr., Wrana, J. L., and Falb, D. (1997) Cell 89 1165-1173 [DOI] [PubMed] [Google Scholar]
- 31.Abdollah, S., Macías-Silva, M., Tsukazaki, T., Hayashi, H., Attisano, L., and Wrana, J. L. (1997) J. Biol. Chem. 272 27678-27685 [DOI] [PubMed] [Google Scholar]
- 32.Hsu, Y. H., Sarker, K. P., Pot, I., Chan, A., Netherton, S. J., and Bonni, S. (2006) J. Biol. Chem. 281 33008-33018 [DOI] [PubMed] [Google Scholar]
- 33.Wrana, J. L., Attisano, L., Carcamo, J., Zentella, A., Doody, J., Laiho, M., Wang, X.-F., and Massagué, J. (1992) Cell 71 1003-1014 [DOI] [PubMed] [Google Scholar]
- 34.Zawel, L., Dai, J. L., Buckhaults, P., Zhou, S., Kinzler, K. W., Vogelstein, B., and Kern, S. E. (1998) Mol. Cell 1 611-617 [DOI] [PubMed] [Google Scholar]
- 35.Bonni, S., Wang, H. R., Causing, C. G., Kavsak, P., Stroschein, S. L., Luo, K., and Wrana, J. L. (2001) Nat. Cell Biol. 3 587-595 [DOI] [PubMed] [Google Scholar]
- 36.Gasparri, F., Mariani, M., Sola, F., and Galvani, A. (2004) J. Biomol. Screen 9 232-243 [DOI] [PubMed] [Google Scholar]
- 37.Wieser, R., Wrana, J. L., and Massague, J. (1995) EMBO J. 14 2199-2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He, G. H., Helbing, C. C., Wagner, M. J., Sensen, C. W., and Riabowol, K. (2005) Mol. Biol. Evol. 22 104-116 [DOI] [PubMed] [Google Scholar]
- 39.Stroschein, S. L., Bonni, S., Wrana, J. L., and Luo, K. (2001) Genes Dev. 15 [DOI] [PMC free article] [PubMed]
- 40.Pot, I., and Bonni, S. (2008) Curr. Mol. Med., in press [DOI] [PubMed]
- 41.Stegmuller, J., Konishi, Y., Huynh, M. A., Yuan, Z., Dibacco, S., and Bonni, A. (2006) Neuron 50 389-400 [DOI] [PubMed] [Google Scholar]
- 42.Stegmuller, J., Huynh, M. A., Yuan, Z., Konishi, Y., and Bonni, A. (2008) J. Neurosci. 28 1961-1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedeux, R., Sengupta, S., Shen, J. C., Demidov, O. N., Saito, S., Onogi, H., Kumamoto, K., Wincovitch, S., Garfield, S. H., McMenamin, M., Nagashima, M., Grossman, S. R., Appella, E., and Harris, C. C. (2005) Mol. Cell. Biol. 25 6639-6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helbing, C. C., Veillette, C., Riabowol, K., Johnston, R. N., and Garkavtsev, I. (1997) Cancer Res. 57 1255-1258 [PubMed] [Google Scholar]
- 45.Coles, A. H., Liang, H., Zhu, Z., Marfella, C. G., Kang, J., Imbalzano, A. N., and Jones, S. N. (2007) Cancer Res. 67 2054-2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garkavtsev, I., Kozin, S. V., Chernova, O., Xu, L., Winkler, F., Brown, E., Barnett, G. H., and Jain, R. K. (2004) Nature 428 328-332 [DOI] [PubMed] [Google Scholar]
- 47.Feng, X., Bonni, S., and Riabowol, K. (2006) Mol. Cell. Biol. 26 9244-9255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seoane, J. (2006) Carcinogenesis 27 2148-2156 [DOI] [PubMed] [Google Scholar]
- 49.Gong, W., Suzuki, K., Russell, M., and Riabowol, K. (2005) Int. J. Biochem. Cell Biol. 37 1054-1065 [DOI] [PubMed] [Google Scholar]
- 50.Soliman, M. A., and Riabowol, K. (2007) Trends Biochem. Sci. 32 509-519 [DOI] [PubMed] [Google Scholar]
- 51.Takaesu, N. T., Hyman-Walsh, C., Ye, Y., Wisotzkey, R. G., Stinchfield, M. J., O'Connor, M. B., Wotton, D., and Newfeld, S. J. (2006) Genetics 174 1299-1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.