Abstract
The Ras effector and ubiquitin-protein isopeptide ligase family member IMP acts as a steady-state resistor within the Raf-MEK-ERK kinase module. IMP concentrations are regulated by Ras through induction of autodegradation and can modulate signal/response thresholds by directly limiting the assembly of functional KSR1-dependent Raf·MEK complexes. Here, we show that the capacity of IMP to inhibit signal propagation through Raf to MEK is a consequence of disrupting KSR1 homooligomerization and B-Raf/c-Raf hetero-oligomerization. This impairs both the recruitment of MEK to activated Raf family members and the contribution of Raf oligomers to c-Raf kinase activation. Our observations indicate that human KSR1 proteins promote assembly of multivalent Raf·MEK complexes that are required for c-Raf kinase activation and functional coupling of active kinases to downstream substrates. This property is engaged by IMP for modulation of signal amplitude.
The Raf-MEK2-ERK kinase cascade is one of the most extensively studied signal transduction modules on account of its core participation in both normal and pathological cell regulatory networks. A great deal of information has been gathered about the acute participation of the Raf-MEK-ERK signal transduction circuitry in the modulation and/or propagation of a broad range of dynamic cell regulatory events ranging from cellular proliferation and tumorigenesis to differentiation and cell specialization (1–3). Attention is now necessarily becoming focused on understanding the molecular architecture that allows the core enzymatic components of this cascade to be shared among multiple regulatory pathways yet produce distinct biological outcomes in response to discrete inductive signals. There is accumulating evidence that control of the amplitude, duration, and subcellular compartmentalization of ERK activation are all critical determinants of the biological response (4–7). The discovery and characterization of non-catalytic accessory components, or scaffolds, that generate higher order molecular organization to modulate the assembly, activation, and compartmentalization of MAPK cascades are beginning to reveal mechanisms that generate specificity in the coupling of MAPK activation to appropriate biological responses (8–11).
We have recently described IMP (impedes mitogenic signal propagation) as a specifier of ERK activation amplitude through inhibition of functional coupling between Raf and MEK kinases (12). The mode of action of IMP is dependent upon the presence of the Raf-MEK-ERK scaffolding protein KSR1 (kinase suppressor of Ras 1) and is a consequence of impairing the capacity of this scaffold to promote signal propagation through Raf to MEK (13). Here, we have examined how IMP association with KSR1 can limit Raf·MEK·ERK complex formation and function. We find that KSR1 homo-oligomerization is required to couple distinct KSR·MEK and KSR·B-Raf complexes to allow MEK activation. Furthermore, KSR1 promotes formation of B-Raf/c-Raf hetero-oligomers that contribute to c-Raf kinase activation. IMP association blocks both KSR1 homo-oligomerization and B-Raf/c-Raf hetero-oligomerization to impair both MEK activation and the contribution of Raf oligomers to c-Raf kinase activation. These observations reveal the participation of KSR1 both upstream and downstream of Raf kinase activation in human cells and highlight Raf family oligomerization as a basic control point for modulation of signal amplitude by IMP.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection—HEK293 cells were cultured in Dulbecco's modified Eagle's medium without sodium pyruvate (Invitrogen) with 10% fetal bovine serum and transfected with Lipofectamine and Plus Reagent (Invitrogen). HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, and RNA interference was performed with DharmaFECT 1 (Dharmacon).
siRNAs, Plasmids, and Antibodies—The following siRNA sequences were used: IMP-FW (GGACACAGCAGAGGAAAUUUU) and IMP-RV (AAUUUCCUCUGCUGUGUCCUU), c-Raf-FW (GACGUUCCUGAAGCUUGCCUU) and c-Raf-RV (GGCAAGCUUCAGGAACGUCUU), and Control-FW (AUGAACGUGAAUUGCUCAAUU) and Control-RV (UUGAGCAAUUCACGUUCAUUU). pCMV5-Myc-IMP, pcDNA3-HA-KSR1, pCMV5-FLAG-KSR1, pDCR-HA-RasG12V, pLNCX-FLAG-c-Raf, and pSRα-c-Raf-BXB (12); pCMV5-FLAG-KSR1-C′ (amino acids 540–873) (14); and pcDNA3-Pyo-KSR1-C′ (amino acids 542–873) (15) have been described previously. pEFm-B-Raf and pEFm-B-RafG596R (16) and pEFHA-c-Raf-BXB (17) were gifts from Richard Marais. pLNCX-FLAG-B-Raf and pLNCX-FLAG-c-Raf-N′ (amino acids 1–330) (18) were gifts from Jeffrey Frost. Antibodies against c-Myc (A-14, 9E10), HA (Y-11, F-7), B-Raf (H-145, F-7), and c-Raf (C-12) were from Santa Cruz Biotechnology. FLAG antibody was from Sigma. Pyo (Glu-Glu) antibody was from Delta Biolabs. Phospho-MEK and MEK antibodies were from Cell Signaling Technology. Monoclonal antibodies against c-Raf and MEK were from BD Biosciences.
Immunoprecipitation—For co-immunoprecipitation, HEK293 cells were lysed 48 h after transfection in modified radioimmune precipitation assay buffer (20 mm Tris, pH 8.0, 137 mm NaCl, 10% glycerol, 1% Triton X-100, 2 mm EDTA, 20 mm NaF, and protease inhibitors). Lysates were rotated for 20 min at 4 °C, cleared by centrifugation at 17,000 × g for 30 min, and immunoprecipitated overnight with antibody-conjugated beads. Immunoprecipitates were washed four times in lysis buffer. Quantitation of immunoblot signals was performed using the National Institutes of Health ImageJ, following signal capture within the linear range using an Alpha Innotech imaging system.
Kinase Assay—c-Raf-BXB was immunoprecipitated and washed three times in modified radioimmune precipitation assay buffer, two times in the same buffer plus 500 mm NaCl, and two times in 25 mm HEPES, pH 7.5, and 10 mm MgCl2. The kinase reaction was performed as described previously (12).
Reverse Transcription-PCR—Total RNA was prepared with the High Pure RNA isolation kit (Roche Diagnostics). Reverse transcriptase reaction was done with the SuperScript First-Strand Synthesis System (Invitrogen). PCR amplification was carried out using the LightCycler system (Roche Diagnostics). The following primers were used for PCR amplification: IMP-FW (TGCACGGTGTGTCTGGAG) and IMP-RV (GCAAACAGGACACGTGGT). To quantify the transcripts in this study, parallel experiments were done using the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase as the internal control. Values were normalized using glyceraldehyde-3-phosphate dehydrogenase and analyzed using the relative quantification mathematical model (Pfaffl) as described previously (19).
RESULTS
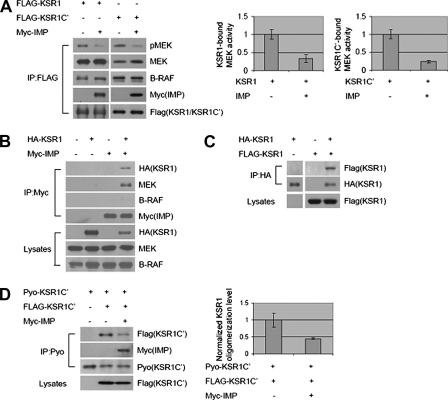
To investigate how IMP limits the capacity of KSR1 to promote productive Raf-MEK interactions and MEK activation, we examined the impact of IMP on KSR1·Raf·MEK complex formation and function. Immunoprecipitation of epitope-tagged KSR1 from serum-starved HEK293 cells resulted in the specific coprecipitation of endogenous MEK and B-Raf (Fig. 1A). A portion of the MEK present in KSR1 complexes was active independently of exogenous stimuli, as indicated by immunoreactivity with antibodies that selectively recognize the activating phosphorylations on MEK1/2. Expression of IMP reduced the population of active MEK in KSR1 complexes without affecting KSR1 association with MEK or B-Raf. IMP was detected in KSR1 complexes; however, direct immunoprecipitation of IMP, under identical conditions, recovered KSR1 and endogenous MEK but not B-Raf (Fig. 1B). In light of our previous finding that IMP can inhibit Raf·MEK complex formation (12), these observations suggested the possibility that IMP impairs the oligomerization of distinct KSR1·MEK and KSR1·B-Raf complexes to limit the accessibility of MEK for activation by B-Raf. Consistent with this, we found that KSR1 proteins can homo-oligomerize through interactions in the carboxyl-terminal half (Fig. 1, C and D) and that IMP expression blocks this interaction (Fig. 1D).
FIGURE 1.
IMP blocks KSR1 homo-oligomerization to separate KSR1·B-Raf and KSR1·MEK complexes. A, IMP does not inhibit endogenous B-Raf or MEK binding to KSR1 but inhibits KSR1-bound MEK phosphorylation. FLAG-tagged KSR1 or KSR1-C′ (amino acids 540–873) was immunoprecipitated (IP) from HEK293 cells, and representative immunoblots of phospho-MEK (pMEK), MEK, B-Raf, Myc-IMP, and FLAG-KSR1/KSR1-C′ in FLAG-KSR1/KSR1-C′ immunoprecipitates are shown. MEK activities in immunoprecipitates were analyzed by quantitative immunoblotting. Values are specific activity normalized to MEK activity in the samples with KSR1 or KSR1-C′ expression alone (arbitrarily set at 1). Error bars represent S.D. from three independent experiments. B, both KSR1 and MEK are detected in IMP immunocomplexes, whereas B-Raf is not. Myc-IMP was immunoprecipitated from HEK293 cells. Upper, Western blots for HA-KSR1, MEK, B-Raf, and Myc-IMP in Myc-IMP immunoprecipitates; lower, Western blots for HA-KSR1, MEK, and B-Raf in Triton-soluble cell lysates. C, KSR1 oligomerizes in HEK293 cells. HA-KSR1 was immunoprecipitated from cells coexpressing FLAG-KSR1 or HA-KSR1 or both. Upper, Western blots for FLAG-KSR1 and HA-KSR1 in HA-KSR1 immunoprecipitates; lower, Western blot for FLAG-KSR1 in Triton-soluble cell lysates. D, IMP inhibits KSR1 carboxyl-terminal homo-oligomerization in HEK293 cells. Pyo-KSR1-C′ (amino acids 542–873) was immunoprecipitated from cells coexpressing Pyo-KSR1-C′, FLAG-KSR1-C′, or Myc-IMP. Representative immunoblots of Pyo-KSR1-C′, FLAG-KSR1-C′, or Myc-IMP in Pyo-KSR1-C′ immunoprecipitates and FLAG-KSR1-C′ in Triton-soluble cell lysates are shown. KSR1 oligomerization level was analyzed by quantitative immunoblotting. Values are specific oligomerization level normalized to that in the sample with coexpression of Pyo-KSR1-C′ and FLAG-KSR1-C′ (arbitrarily set at 1). Error bars represent S.D. from three independent experiments.
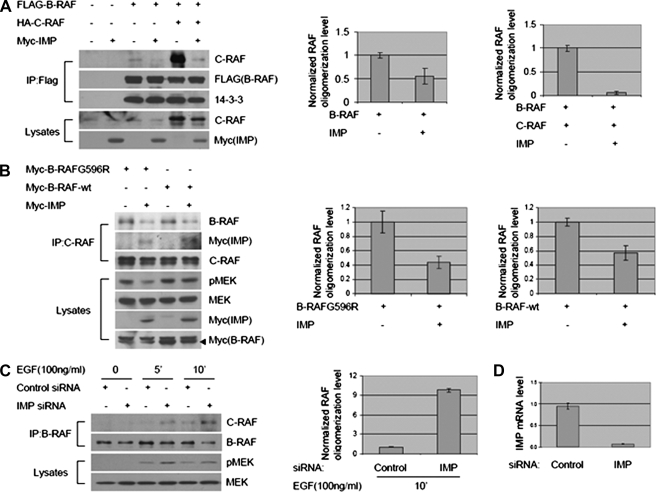
The capacity of KSR homo-oligomerization to participate in Raf-MEK signal propagation was reminiscent of recent observations describing the role of Raf family oligomerization in MEK activation (17). The participation of KSR1 in Raf family oligomerization is unknown; however, B-Raf/c-Raf hetero-oligomer formation correlates with ERK1/2 pathway activation, and recently discovered low activity B-Raf mutations in melanomas require association with c-Raf to induce stimulus-independent ERK1/2 activation (16, 17). Expression of IMP blocked B-Raf·c-Raf (Fig. 2A) complex formation as detected by immunoprecipitation of epitope-tagged B-Raf or by immunoprecipitation of endogenous c-Raf from cells expressing either wild-type B-Raf or the low activity oncogenic variant B-Raf(G596R) (B) (16). Although 14-3-3 association has been implicated as a prerequisite for B-Raf·c-Raf complex formation (16, 17, 20), IMP impaired Raf hetero-oligomerization without affecting the relative amounts of 14-3-3 coprecipitating with B-Raf (Fig. 2A). To examine the contribution of IMP to modulation of native B-Raf·c-Raf complex formation, endogenous IMP was depleted by RNA interference followed by EGF stimulation (Fig. 2D). B-Raf·c-Raf complex formation was induced upon EGF stimulation, and depletion of IMP resulted in an increase in both detectable B-Raf·c-Raf complexes (Fig. 2C). These observations indicate that IMP is both necessary and sufficient to limit generation of active B-Raf·c-Raf protein complexes.
FIGURE 2.
IMP inhibits B-Raf·c-Raf complex formation. A and B, overexpressed IMP suppresses B-Raf·c-Raf complex formation. A, immunoprecipitation (IP) of ectopic B-Raf protein from HEK293 cells coexpressing FLAG-B-Raf, HA-c-Raf, or Myc-IMP. Upper, representative Western blots for c-Raf, FLAG-B-Raf, and 14-3-3 in FLAG-B-Raf immunoprecipitates; lower, representative Western blots for c-Raf and Myc-IMP in Triton-soluble cell lysates. Raf oligomerization level was analyzed by quantitative immunoblotting. Values are specific oligomerization level normalized to that in the samples with B-Raf expression alone or coexpression of B-Raf and c-Raf (arbitrarily set at 1). Error bars represent S.D. from three independent experiments. B, immunoprecipitation of native c-Raf protein from HEK293 cells coexpressing B-Raf(596R), wild-type (wt) B-Raf, or Myc-IMP. Upper, representative Western blots for B-Raf, Myc-IMP, and c-Raf in c-Raf immunoprecipitates; lower, representative Western blots for phospho-MEK (pMEK), MEK, Myc-IMP, and Myc-B-Raf in Triton-soluble cell lysates. Raf oligomerization level was analyzed by quantitative immunoblotting as in A. Values of oligomerization level in the samples with expression of B-Raf(G596R) or wild-type B-Raf alone were arbitrarily set at 1. C and D, inhibiting native IMP enhances EGF-induced B-Raf·c-Raf complex formation and MEK activation. C, immunoprecipitation of endogenous B-Raf from HeLa cells transfected with control or IMP siRNA and stimulated with EGF (100 ng/ml) for 5 or 10 min or left untreatedas indicated. To allow accumulation of the B-Raf·c-Raf complex, cells were pretreated with MEK inhibitor U0126 before EGF stimulation (21). Upper, representative Western blots for c-Raf and B-Raf in B-Raf immunoprecipitates; lower, representative Western blots for phospho-MEK and MEK in Triton-soluble cell lysates. Raf oligomerization level was analyzed by quantitative immunoblotting as in A. The value of oligomerization level in the sample transfected with control siRNA and stimulated with EGF for 10 min was arbitrarily set at 1. D, quantification of IMP mRNA level in cell samples shown in C. mRNA was extracted from duplicated cell samples shown in C, and reverse transcription-PCR assay was performed as described under “Experimental Procedures.” Values are representative of two independent experiments performed in duplicate, and error bars represent S.D. among replicates.
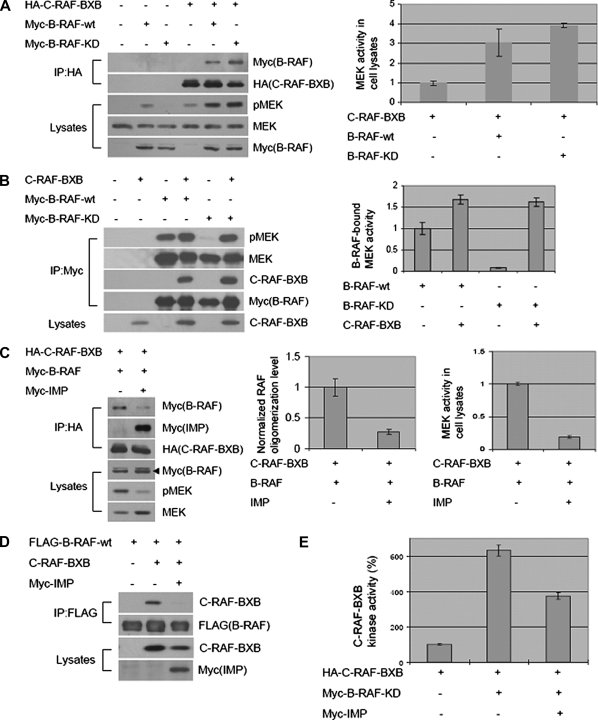
B-Raf/c-Raf heteroduplexes have been reported to have greater specific activity than corresponding B-Raf or c-Raf homoduplexes or monomers (21). The mechanistic basis of this increased activity is unclear, and the mechanism regulating heteroduplex formation is unknown. We employed c-Raf-BXB, an oncogenic c-Raf variant that lacks the amino-terminal regulatory domain and is constitutively active independently of mitogen stimulation, to examine the consequence of IMP-dependent modulation of Raf complex formation on Raf function in serum-starved cells. As shown in Fig. 3A, c-Raf-BXB interacts with both the wild-type and a kinase-dead B-Raf variant with a K483M substitution (B-Raf-KD). Consistent with published observations (21), B-Raf-KD enhanced the activity of c-Raf-BXB in cells as indicated by the increased accumulation of active MEK (Fig. 3A) and by a significant increase in specific activity as measured in vitro (Fig. 3E). Importantly, unlike c-Raf (12), B-Raf associates with MEK independently of stimulation recruiting both c-Raf-BXB and MEK (Fig. 3B). Therefore, Raf family oligomerization may enhance both Raf kinase activity and substrate availability. IMP expression inhibited B-Raf·c-Raf-BXB complex formation together with blocking endogenous MEK activation (Fig. 3, C and D). The capacity of B-Raf-KD to enhance the specific activity of c-Raf-BXB was also impaired by IMP (Fig. 3E).
FIGURE 3.
IMP blocks B-Raf·c-Raf-BXB complex formation to inhibit B-Raf-dependent c-Raf-BXB activation and c-Raf-BXB-induced MEK activation. A and B, formation of the B-Raf·c-Raf-BXB complex in HEK293 cells coexpressing wild-type (wt) B-Raf, B-Raf-KD, or c-Raf-BXB. A, immunoprecipitation (IP) of c-Raf-BXB. Upper, representative Western blots for Myc-B-Raf and HA-c-Raf-BXB in HA-c-Raf-BXB immunoprecipitates; lower, representative Western blots for phospho-MEK (pMEK), MEK, and Myc-B-Raf in Triton-soluble cell lysates. MEK activities in cell lysates were analyzed by quantitative immunoblotting as in Fig. 1A. The value of MEK activity in the sample with c-Raf-BXB expression alone was arbitrarily set at 1. B, immunoprecipitation of B-Raf. Upper, representative Western blots for phospho-MEK, MEK, c-Raf-BXB, and Myc-B-Raf in Myc-B-Raf immunoprecipitates; lower, representative Western blot for c-Raf-BXB in Triton-soluble cell lysates. MEK activities in B-Raf immunoprecipitates were analyzed by quantitative immunoblotting as in Fig. 1A. The value of MEK activity in the sample with wild-type B-Raf expression alone was arbitrarily set at 1. C and D, IMP inhibits B-Raf·c-Raf-BXB complex formation and c-Raf-BXB-induced MEK activation in HEK293 cells. C, immunoprecipitation of c-Raf-BXB. Upper, representative Western blots for Myc-B-Raf, Myc-IMP, and HA-c-Raf-BXB in HA-c-Raf-BXB immunoprecipitates; lower, representative Western blots for Myc-B-Raf, phospho-MEK, and MEK in Triton-soluble cell lysates. Raf oligomerization level and MEK activities in cell lysates were analyzed by quantitative immunoblotting as in Figs. 1A and 2A. Values of Raf oligomerization level and MEK activity in the sample with coexpression of c-Raf-BXB and B-Raf were arbitrarily set at 1. D, immunoprecipitation of B-Raf. Upper, Western blots for c-Raf-BXB and FLAG-B-Raf in FLAG-B-Raf immunoprecipitates; lower, Western blots for c-Raf-BXB and Myc-IMP in Triton-soluble cell lysates. E, IMP inhibiting B-Raf-dependent c-Raf-BXB activation. c-Raf-BXB kinase activity, from protein immunoprecipitated from HEK293 cells expressing c-Raf-BXB alone or together with kinase-dead B-Raf or Myc-IMP, was measured in vitro using kinase-dead recombinant MEK as the substrate. Values are specific activity normalized to the activity of c-Raf-BXB expressed alone (arbitrarily set at 100%). Error bars represent S.D. from three independent experiments.
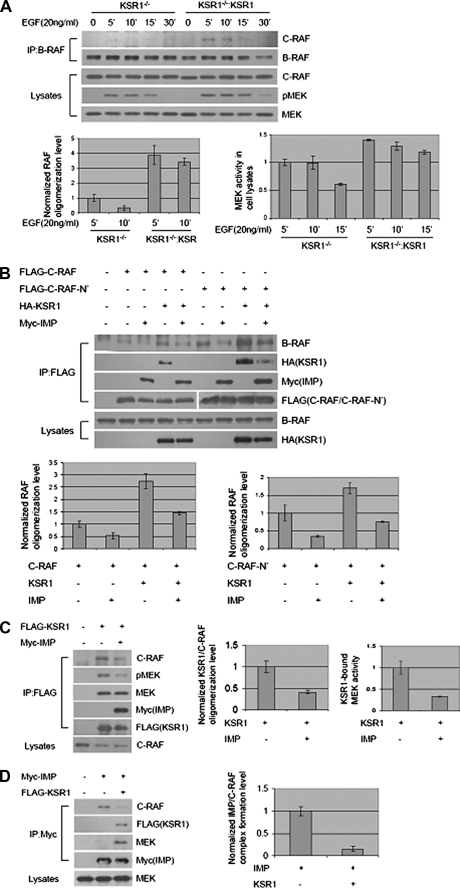
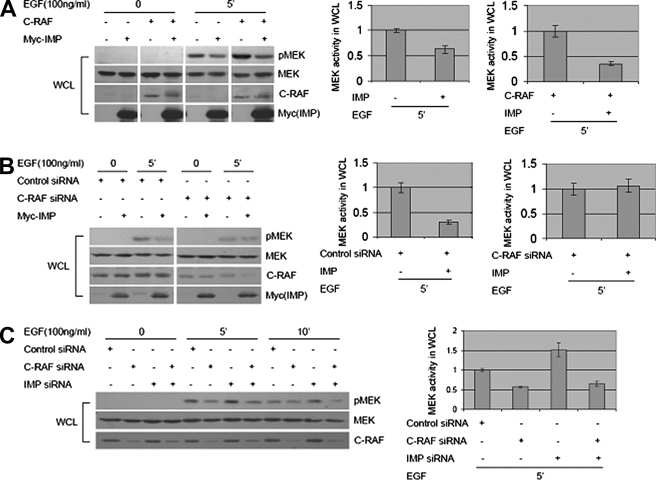
Using KSR1 null mouse embryonic fibroblasts, we have demonstrated previously that the capacity of IMP to limit signaling through the Raf-MEK-ERK cascade is KSR1-dependent (12). This finding together with those above suggest that KSR1 may promote B-Raf/c-Raf heteroduplex formation. Consistent with this, we found that EGF-dependent induction of B-Raf·c-Raf complexes was undetectable in KSR1–/– mouse embryonic fibroblasts but was restored upon reintroduction of KSR1 at near physiological concentrations (Fig. 4A). In addition, KSR1 expression was sufficient to enhance the recovery of endogenous B-Raf in c-Raf immunocomplexes from HEK293 cells, and this was blocked by IMP (Fig. 4B). This phenotype was evident with both full-length c-Raf and the amino-terminal regulatory domain (c-Raf-N′). This observation prompted us to evaluate the impact of IMP on the presence of native c-Raf in KSR1 immunocomplexes. As shown in Fig. 4, C and D, IMP significantly inhibited the association of c-Raf with KSR1. This is in stark contrast to the insensitivity of KSR1·B-Raf complexes to IMP expression (Fig. 1A), raising the possibility that c-Raf function may be acutely dependent on scaffolding complexes. To examine this relationship in the context of mitogenic signaling, we tested the relative contributions of IMP and c-Raf to EGF-dependent MEK activation. IMP expression was sufficient to impair EGF-induced MEK activation and to reverse the enhanced EGF-induced MEK activation observed upon c-Raf overexpression (Fig. 5A). Importantly, the reduced levels of EGF-stimulated MEK activation, resulting from RNA interference-mediated c-Raf depletion, were insensitive to IMP expression (Fig. 5B), and the enhanced MEK activation normally observed upon IMP depletion was reversed by codepletion of c-Raf (Fig. 5C).
FIGURE 4.
IMP inhibits KSR1-induced B-Raf·c-Raf complex formation. A, KSR1 is required for B-Raf·c-Raf complex formation. Results are from immunoprecipitation (IP) of endogenous B-Raf from KSR1–/– or KSR1–/–:KSR mouse embryonic fibroblasts (12) stimulated with EGF (20 ng/ml) for 5, 10, 15, or 30 min or left untreated as indicated. Upper, representative Western blots for c-Raf and B-Raf in B-Raf immunoprecipitates; lower, representative Western blots for c-Raf, phospho-MEK (pMEK), and MEK in Triton-soluble cell lysates. Raf oligomerization level and MEK activity in cell lysates were analyzed by quantitative immunoblotting as in Fig. 3C. Values for Raf oligomerization and MEK activity in KSR1–/– cells stimulated with EGF for 5 min were arbitrarily set at 1. B, IMP blocks KSR1-enhanced B-Raf·c-Raf complex formation. FLAG-c-Raf or FLAG-c-Raf-N′ (amino acids 1–330) was immunoprecipitated from HEK293 cells expressing FLAG-c-Raf or c-Raf-N′ alone or together with HA-KSR1 or Myc-IMP as indicated. Upper, representative Western blots for B-Raf, HA-KSR1, Myc-IMP, and FLAG-c-Raf/c-Raf-N′ in FLAG-c-Raf/c-Raf-N′ immunoprecipitates; lower, representative Western blots for B-Raf and HA-KSR1 in Triton-soluble cell lysates. Raf oligomerization level was analyzed by quantitative immunoblotting as in Fig. 2A. Raf oligomerization level in the sample with c-Raf or c-Raf-N′ expression alone was arbitrarily set at 1. C and D, c-Raf and IMP competitively associate with KSR1. C, FLAG-KSR1 was immunoprecipitated from HEK293 cells expressing FLAG-KSR1 alone or together with Myc-IMP. Upper, representative Western blots for c-Raf, phospho-MEK, MEK, Myc-IMP, and FLAG-KSR1 in FLAG-KSR1 immunoprecipitates; lower, representative Western blot for c-Raf in Triton-soluble cell lysates. KSR·c-Raf oligomerization level and KSR1-bound MEK activity were analyzed by quantitative immunoblotting as in Figs. 1A and 2A. Values of KSR1·c-Raf oligomerization level and KSR1-bound MEK activity in the sample with KSR1 expression alone were set at 1. D, Myc-IMP was immunoprecipitated from HEK293 cells expressing Myc-IMP alone or together with FLAG-KSR1.Upper, representative Western blots for c-Raf, FLAG-KSR1, MEK, and Myc-IMP in Myc-IMP immunoprecipitates; lower, representative Western blot for MEK in Triton-soluble cell lysates. IMP·c-Raf complex formation level was analyzed by quantitative immunoblotting as in Fig. 2A. The value of IMP·c-Raf complex formation in the sample with IMP expression alone was set at 1.
FIGURE 5.
c-Raf function in mitogenic signaling is sensitive to IMP expression. A, IMP suppresses both EGF-induced and overexpressed c-Raf-enhanced MEK activation. Whole cell lysates (WCL) were collected from HEK293 cells cotransfected with c-Raf or Myc-IMP and stimulated with EGF (100 ng/ml) as indicated. Representative Western blots for phospho-MEK (pMEK), MEK, c-Raf, and Myc-IMP are shown. MEK activities in cells stimulated with EGF for 5 min were analyzed by quantitative immunoblotting as in Fig. 1A. Values of MEK activity in the sample with control or c-Raf expression alone were arbitrarily set at 1. B, overexpressed IMP does not inhibit MEK activation in c-Raf knockdown cells. Whole cell lysates were collected from HEK293 cells transfected with Myc-IMP, control siRNA, or c-Raf siRNA and stimulated with EGF (100 ng/ml) for 5 min or left untreated as indicated. Representative Western blots for phospho-MEK, MEK, c-Raf, and Myc-IMP are shown. MEK activities in cells stimulated with EGF for 5 min were analyzed by quantitative immunoblotting as in A. Values of MEK activity in cells transfected with control siRNA or c-Raf siRNA alone were arbitrarily set at 1. C, inhibition of both IMP and c-Raf expression reverses the increased level of MEK activation caused by depleting IMP protein alone. Whole cell lysates were collected from HeLa cells transfected with control siRNA, c-Raf siRNA, or IMP siRNA and stimulated with EGF (100 ng/ml) for 5 or 10 min or left untreated as indicated. Representative Western blots for phospho-MEK, MEK, and c-Raf are shown. MEK activities in cells stimulated with EGF for 5 min were analyzed by quantitative immunoblotting as in A. The value of MEK activity in cells transfected with control siRNA alone was arbitrarily set at 1.
DISCUSSION
IMP is a Ras effector that controls sensitivity of the ERK1/2 kinase cascade to stimulus-dependent activation by limiting assembly of the core enzymatic components (12). IMP interacts with the MAPK scaffold protein KSR1 and requires KSR1 expression to influence MEK activation. KSR1 has been shown to act as an obligate mediator of functional Raf·MEK complex formation in a variety of cell systems (11, 22). The mechanism by which KSR1 promotes Raf-MEK coupling is unclear but most likely involves direct binding to both Raf and MEK. Our observations suggest that distinct KSR1·MEK and KSR1·B-Raf complexes are present in cells and that pathway activation requires KSR1 homooligomerization to bring Raf together with its substrate. IMP expression inhibits Raf·MEK complex formation in cells, and this is apparently a consequence of blocking KSR1 homooligomerization without affecting the interaction of KSR1 with MEK1 or B-Raf.
Remarkably, IMP has distinct consequences on the mobilization of c-Raf versus B-Raf for mitogenic signaling. In addition to promoting Raf·MEK complex assembly, we found that KSR1 promotes B-Raf/c-Raf hetero-oligomerization, and this is likewise impaired by IMP expression. Recently B-Raf/c-Raf heterooligomerization has been revealed as an essential step for maximal c-Raf kinase activation and is required for biological responses to low activity B-Raf variants in melanomas (16, 17). However, it is still unclear how B-Raf·c-Raf complex formation triggers c-Raf activation and whether this requires B-Raf kinase activity (17, 21). We find that kinase-inactive B-Raf has the capacity to drive activation of the c-Raf catalytic domain, suggesting that B-Raf association either can facilitate autoactivating c-Raf transphosphorylation events or can recruit additional upstream regulators that facilitate c-Raf kinase activation. Importantly, KSR1 promotes B-Raf/c-Raf hetero-oligomerization, and this is blocked by IMP. As a consequence, we find that IMP reverses the capacity of kinase-dead B-Raf to increase the specific activity of the c-Raf catalytic domain, and IMP uncouples c-Raf from KSR1 complexes.
Current paradigms suggest mammalian KSR1 is primarily responsible for coupling active Raf kinases to their substrates, MEK1 and MEK2 (23, 24). In contrast, although Drosophila KSR plays a similar role, it is also clearly required for Raf kinase activation (25, 26). Our observations suggest that, like the Drosophila ortholog, human KSR1 acts both upstream and downstream of Raf kinase activation. However, c-Raf is selectively sensitive to this contribution of KSR1 to signal propagation. A simple role for KSR1 proteins in assembly of multivalent Raf and MEK complexes could easily account for this epistatic relationship. The capacity of IMP to limit the formation of such complexes highlights this regulatory step as a key axis of control for signal modulation.
Acknowledgments
We thank Richard Marais, Jeffrey A. Frost, and Deborah K. Morrison for generously providing some of the reagents used in these studies.
This work was supported, in whole or in part, by National Institutes of Health Grant CA71443 from NCI. This work was also supported by Grant I-1414 from the Robert Welch Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated kinase; siRNA, small interfering RNA; HA, hemagglutinin; EGF, epidermal growth factor; KD, kinase dead.
References
- 1.Pearson, G., Robinson, F., Beers Gibson, T., Xu, B. E., Karandikar, M., Berman, K., and Cobb, M. H. (2001) Endocr. Rev. 22 153–183 [DOI] [PubMed] [Google Scholar]
- 2.Wellbrock, C., Karasarides, M., and Marais, R. (2004) Nat. Rev. Mol. Cell Biol. 5 875–885 [DOI] [PubMed] [Google Scholar]
- 3.Dhillon, A. S., Hagan, S., Rath, O., and Kolch, W. (2007) Oncogene 26 3279–3290 [DOI] [PubMed] [Google Scholar]
- 4.Whitehurst, A., Cobb, M. H., and White, M. A. (2004) Mol. Cell. Biol. 24 10145–10150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mackeigan, J. P., Murphy, L. O., Dimitri, C. A., and Blenis, J. (2005) Mol. Cell. Biol. 25 4676–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raman, M., Chen, W., and Cobb, M. H. (2007) Oncogene 26 3100–3112 [DOI] [PubMed] [Google Scholar]
- 7.McKay, M. M., and Morrison, D. K. (2007) Oncogene 26 3113–3121 [DOI] [PubMed] [Google Scholar]
- 8.Bumeister, R., Rosse, C., Anselmo, A., Camonis, J., and White, M. A. (2004) Curr. Biol. 14 439–445 [DOI] [PubMed] [Google Scholar]
- 9.Morrison, D. K., and Davis, R. J. (2003) Annu. Rev. Cell Dev. Biol. 19 91–118 [DOI] [PubMed] [Google Scholar]
- 10.Kolch, W. (2005) Nat. Rev. Mol. Cell Biol. 6 827–837 [DOI] [PubMed] [Google Scholar]
- 11.Claperon, A., and Therrien, M. (2007) Oncogene 26 3143–3158 [DOI] [PubMed] [Google Scholar]
- 12.Matheny, S. A., Chen, C., Kortum, R. L., Razidlo, G. L., Lewis, R. E., and White, M. A. (2004) Nature 427 256–260 [DOI] [PubMed] [Google Scholar]
- 13.Ory, S., and Morrison, D. K. (2004) Curr. Biol. 14 R277–R278 [DOI] [PubMed] [Google Scholar]
- 14.Joneson, T., Fulton, J. A., Volle, D. J., Chaika, O. V., Bar-Sagi, D., and Lewis, R. E. (1998) J. Biol. Chem. 273 7743–7748 [DOI] [PubMed] [Google Scholar]
- 15.Therrien, M., Michaud, N. R., Rubin, G. M., and Morrison, D. K. (1996) Genes Dev. 10 2684–2695 [DOI] [PubMed] [Google Scholar]
- 16.Wan, P. T., Garnett, M. J., Roe, S. M., Lee, S., Niculescu-Duvaz, D., Good, V. M., Jones, C. M., Marshall, C. J., Springer, C. J., Barford, D., and Marais, R. (2004) Cell 116 855–867 [DOI] [PubMed] [Google Scholar]
- 17.Garnett, M. J., Rana, S., Paterson, H., Barford, D., and Marais, R. (2005) Mol. Cell 20 963–969 [DOI] [PubMed] [Google Scholar]
- 18.Tran, N. H., Wu, X., and Frost, J. A. (2005) J. Biol. Chem. 280 16244–16253 [DOI] [PubMed] [Google Scholar]
- 19.Whitehurst, A. W., Bodemann, B. O., Cardenas, J., Ferguson, D., Girard, L., Peyton, M., Minna, J. D., Michnoff, C., Hao, W., Roth, M. G., Xie, X. J., and White, M. A. (2007) Nature 446 815–819 [DOI] [PubMed] [Google Scholar]
- 20.Weber, C. K., Slupsky, J. R., Kalmes, H. A., and Rapp, U. R. (2001) Cancer Res. 61 3595–3598 [PubMed] [Google Scholar]
- 21.Rushworth, L. K., Hindley, A. D., O'Neill, E., and Kolch, W. (2006) Mol. Cell. Biol. 26 2262–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrison, D. K. (2001) J. Cell Sci. 114 1609–1612 [DOI] [PubMed] [Google Scholar]
- 23.Muller, J., Ory, S., Copeland, T., Piwnica-Worms, H., and Morrison, D. K. (2001) Mol. Cell 8 983–993 [DOI] [PubMed] [Google Scholar]
- 24.Roy, F., Laberge, G., Douziech, M., Ferland-McCollough, D., and Therrien, M. (2002) Genes Dev. 16 427–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anselmo, A. N., Bumeister, R., Thomas, J. M., and White, M. A. (2002) J. Biol. Chem. 277 5940–5943 [DOI] [PubMed] [Google Scholar]
- 26.Douziech, M., Sahmi, M., Laberge, G., and Therrien, M. (2006) Genes Dev. 20 807–819 [DOI] [PMC free article] [PubMed] [Google Scholar]