Abstract
Proteins destined for import into the nucleus contain nuclear localization signals (NLSs) that are recognized by import receptors termed karyopherins or importins. Until recently, the only nuclear import sequence that had been well defined and characterized was the classical NLS (cNLS), which is recognized by importin α. However, Chook and coworkers (Lee, B. J., Cansizoglu, A. E., Süel, K. E., Louis, T. H., Zhang, Z., and Chook, Y. M. (2006) Cell 126, 543–558) have provided new insight into nuclear targeting with their identification of a novel NLS, termed the PY-NLS, that is recognized by the human karyopherin β2/transportin (Kapβ2) receptor. Here, we demonstrate that the PY-NLS is conserved in Saccharomyces cerevisiae and show for the first time that the PY-NLS is a functional nuclear targeting sequence in vivo. The apparent ortholog of Kapβ2 in yeast, Kap104, has two known cargos, the mRNA-binding proteins Hrp1 and Nab2, which both contain putative PY-NLS-like sequences. We find that the PY-NLS-like sequence within Hrp1, which closely matches the PY-NLS consensus, is both necessary and sufficient for nuclear import and is also required for receptor binding and protein function. In contrast, the PY-NLS-like sequences in Nab2, which vary from the PY-NLS consensus, are not required for proper import or protein function, suggesting that Kap104 may interact with different cargos using multiple mechanisms. Dissection of the PY-NLS consensus reveals that the minimal PY-NLS in yeast consists of the C-terminal portion of the human consensus, R/H/KX2–5PY, with upstream basic or hydrophobic residues enhancing the targeting function. Finally, we apply this analysis to a bioinformatic search of the yeast proteome as a preliminary search for new potential Kap104 cargos.
In eukaryotic cells, the genetic material is separated from the cellular machinery in the cytoplasm by a nuclear membrane that is studded with regulated channels termed nuclear pore complexes (1–3). Nuclear pores prevent passive diffusion of macromolecules larger than ∼40 kDa (4, 5), so this division can facilitate the precise management of gene expression by controlling access to the genome. However, the presence of the nuclear membrane also necessitates the involvement of nuclear import machinery that can specifically recognize cargo in the cytoplasm, mediate transport through nuclear pores, and effect delivery in the nucleus.
For many import cargos, these import factors are members of the importin/karyopherin β transport receptor superfamily, which has 14 members in Saccharomyces cerevisiae and over 20 members in humans (6). These import receptors recognize protein cargo bearing targeting sequences called nuclear localization signals (NLSs)2 and mediate interactions with the nuclear pore, allowing translocation of the cargo into the nucleus (7). Once inside the nucleus, the import complex encounters RanGTP, a small Ras family GTPase that regulates the directionality of nuclear transport (8). RanGTP binds to the karyopherin β, causing a conformational change and triggering cargo release (9–11). The karyopherin β then returns to the cytoplasm for another round of import.
One main focus of inquiry in the nuclear transport field has been understanding the mechanism by which each karyopherin β receptor recognizes its particular host of cargo proteins. In the most well characterized system of nuclear transport, the classical nuclear import pathway, a specific karyopherin β receptor termed importin β binds to an adaptor protein, importin α, which interacts with cargos carrying classical NLSs (cNLSs). These cNLSs consist of either one or two clusters of basic amino acids and are exemplified by the SV40 large T antigen NLS (126PKKKRRV132) and the nucleoplasmin NLS (155KRPAATKKAGQAKKKK170) (12). A large number of classical substrates have been experimentally verified, and examination of their nuclear targeting signals has led to the development of a useful consensus sequence (12).
In contrast to the classical nuclear import pathway, for most karyopherin β receptors only a handful of substrates have been identified making generalization of binding schemes difficult. Compounding the problem, these cargos often lack sequence or structural homology and, in some cases, even interact with a particular karyopherin β receptor using distinct binding sites and diverse interaction modes (for example, Refs. 13–15). However, a mechanism underlying recognition of a specific class of cargos by a transport receptor that does not mediate cNLS protein import was recently determined. Chook and coworkers (16) reported the structure of a human karyopherin β called karyopherin β2 (Kapβ2) or transportin bound to the NLS of its best characterized cargo, the mRNA-binding protein heterogeneous nuclear ribonucleoprotein A1. Combining the structural information with biochemical studies involving other known Kapβ2 cargos, they then proposed a set of predictive rules that outline the requirements for a putative Kapβ2 NLS: 1) the NLS is structurally disordered, 2) the NLS has a net basic charge, and 3) the NLS has a hydrophobic or basic region upstream of a C-terminal R/H/KX2–5PY motif. The invariant proline and tyrosine residues contained at the end of the consensus led them to term these Kapβ2 recognition sequences “PY-NLSs.” Using these rules, they then searched the human proteome, identified 81 putative Kapβ2 cargos, and showed that a selection of these PY-NLS proteins bound to Kapβ2 in a RanGTP-dependent manner in vitro. These binding experiments suggest that the proteins are substrates of Kapβ2; however, in vivo functional and localization experiments are still necessary to demonstrate that these PY-NLS-containing proteins are imported by Kapβ2 in a PY-NLS-dependent manner.
The apparent ortholog of Kapβ2 in S. cerevisiae is Kap104 (karyopherin 104), which has two known cargos, the mRNA-binding proteins Nab2 (nuclear poly-adenylated RNA-binding protein 2) and Hrp1 (heterogeneous nuclear ribonucleoprotein 1) (17, 18). Previous work has defined the general regions of these cargo proteins that are required for Kap104-mediated nuclear import (18, 19). In Nab2, a region containing an RNA binding motif consisting of a series of arginine-glycine-glycine (RGG) repeats is necessary and sufficient for binding to Kap104 and for nuclear import in vivo (18, 19). Deletion of the CCCH zinc finger region of Nab2 also reduces binding to Kap104 in vitro (18). In Hrp1, the arginine/glycine-rich carboxyl terminus of the protein is required for binding to Kap104 and is sufficient to mediate import in vivo (18). Thus, although the protein domains in Hrp1 and Nab2 that mediate Kap104 binding have been determined, no specific nuclear import sequences have been identified. Much like many Kapβ2 cargos (16), Hrp1 and Nab2 are involved in mRNA biogenesis, with Hrp1 contributing to cleavage and polyadenylation of pre-mRNA 3′-ends (20) and Nab2 aiding in poly(A) tail length regulation and mRNA export (21, 22). Interestingly, the human protein most closely related to Hrp1 is heterogeneous nuclear ribonucleoprotein A1 (PBI-BLAST).
The goal of this study was to examine the function of the novel PY-NLS sequence in vivo. Examination of the primary sequences of Hrp1 and Nab2 uncovered putative PY-NLS-like sequences in both proteins. Using a combination of localization, function, and binding studies, we found that the PY-NLS-like sequence within Hrp1 is necessary and sufficient for nuclear import of the protein and is required for protein function. However, the less conserved PY-NLS-like sequences within Nab2 are not required for nuclear accumulation of Nab2. These results demonstrate that the PY-NLS consensus sequence is conserved in S. cerevisiae but also reveal that Kap104 likely utilizes additional mechanisms of cargo recognition. Finally, identification of the functional PY-NLS in yeast allowed us to interrogate the yeast proteome to identify new putative cargos of Kap104. Therefore, we conclude with a preliminary prediction of the likely prevalence of the PY-NLS in S. cerevisiae.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Chemicals—All chemicals were obtained from United States Biological or Sigma unless otherwise noted. All media were prepared and all DNA manipulations were performed according to standard procedures (23, 24). All yeast strains and plasmids used in this study are described in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain/Plasmid | Description | Origin |
|---|---|---|
| FY23 (ACY192) | Wild type, MATa ura3-52 leu2Δ1 trp1 | (30) |
| ESY41-4C (ACY251) | ΔKAP104::HIS3 [KAP104 CEN URA3 AMPR], MATα ade trp leu his | P. A. Silver |
| ACY427 | ΔNAB2::HIS3 [NAB2 CEN URA3 AMPR], MATa leu lys ade his | This study |
| SVL182/PSY1224 (ACY1571) | ΔHRP1::HIS3 [HRP1 CEN URA3 AMPR] | S. R. Valentini |
| pRS315 (pAC3) | CEN LEU2 AMPR | (31) |
| pPS808 (pAC23) | GFP, GAL1-10 promoter, 2μ URA3 AMPR | P. A. Silver |
| pAC717 | NAB2, CEN LEU2 AMPR | (22) |
| pAC753 | NAB2-GFP, CEN LEU2 AMPR | (22) |
| pAC1069 | GFP-GFP, MET25 promoter, CEN URA3 AMPR | (32) |
| pPS1358 (pAC1725) | GFP-HRP1, GAL1-10 promoter, 2μ URA3 AMPR | (20) |
| pAC2023 | NOP1-GFP, CEN URA3 AMPR | This study |
| pAC2325 | HRP1, CEN LEU2 AMPR | This study |
| pAC2329 | GFP-HRP1 R525A/P531A/Y532A, GAL1-10 promoter, 2μ URA3 AMPR | This study |
| pAC2330 | GFP-HRP1 P531A/Y532A, GAL1-10 promoter, 2μ URA3 AMPR | This study |
| pAC2344 | HRP1 R525A/P531A/Y532A, CEN LEU2 AMPR | This study |
| pAC2345 | HRP1 P531A/Y532A, CEN LEU2 AMPR | This study |
| pAC2440 | HRP1 Y532V, CEN LEU2 AMPR | This study |
| pAC2441 | GFP-HRP1 Y532V, GAL1-10 promoter, 2μ URA3 AMPR | This study |
| pAC2442 | NAB2 P332A, CEN LEU2 AMPR | This study |
| pAC2443 | NAB2-GFP PV332A, CEN LEU2 AMPR | This study |
| pAC2444 | NAB2 P407A, CEN LEU2 AMPR | This study |
| pAC2445 | NAB2-GFP P407A, CEN LEU2 AMPR | This study |
| pAC2451 | GFP-GFP-HRP1-(503-534), MET25 promoter, CEN URA3 AMPR | This study |
| pAC2452 | GFP-GFP-HRP1-(522-534), MET25 promoter, CEN URA3 AMPR | This study |
| pAC2472 | GFP-GFP-NAB2-(320-333), MET25 promoter, CEN URA3 AMPR | This study |
| pAC2473 | GFP-GFP-NAB2-(389-408), MET25 promoter, CEN URA3 AMPR | This study |
| pAC2527 | NAB2 P332A/P407A, CEN LEU2 AMPR | This study |
| pAC2528 | GFP-HRP1 P531A, GAL1-10 promoter, 2μ URA3 AMPR | This study |
| pAC2538 | NAB2-GFP P332A/P407A, CEN LEU2 AMPR | This study |
| pAC2539 | HRP1 P531A, CEN LEU2 AMPR | This study |
| pAC2551 | GST-KAP104, pGEX-6P3 AMPR | M. P. Rout |
Microscopy—Green fluorescent protein (GFP) fusion proteins were localized in live cells using direct fluorescence microscopy. The GFP signal was visualized using a GFP-optimized filter (Chroma Technology) on an Olympus BX60 epifluorescence microscope equipped with a Photometrics Quantix digital camera. For most experiments, cells expressing genes expressed from their own promoters were grown overnight in selective medium, diluted in fresh medium, and grown for 3 h prior to localization studies. Cells expressing genes under the control of the GAL1–10 promoter were grown overnight in selective medium with glucose as a sugar source, pelleted, washed, resuspended in fresh medium with galactose as a sugar source, and induced for 6 h prior to localization studies. Cells expressing genes under the control of the MET25 promoter were grown overnight in selective medium, pelleted, washed, resuspended in fresh medium lacking methionine, and induced for 5 h prior to localization studies. Experiments involving the ΔKAP104 deletion strain (ACY251) required an initial growth phase of five nights to reach the appropriate cell density.
In Vitro GST Binding Assay—For binding studies, Kap104 was expressed in Escherichia coli as a GST fusion protein (25), and then GST-Kap104 beads were incubated with yeast lysate from cells expressing wild-type GFP-Hrp1 or R525A/P531A/Y532A GFP-Hrp1. To prepare the GST-Kap104 beads, E. coli cells expressing GST-Kap104 (pAC2551) were resuspended in PBS supplemented with 0.5 mm phenylmethylsulfonyl fluoride, PLAC (3 mg/ml each of pepstatin A, leupeptin, aprotinin, chymostatin), and 2 mm β-mercaptoethanol and lysed by sonication. Lysates were cleared by centrifugation and incubated with 200 μl of prepared glutathione-Sepharose beads (GE Healthcare). Beads were collected and washed three times with PBS. To prepare yeast lysates, wild-type cells (ACY192) containing plasmids encoding wild-type GFP-Hrp1 (pAC1725) or R525A/P531A/Y532A GFP-Hrp1 (pAC2329) were grown overnight in selective medium with glucose as a sugar source, pelleted, washed, and grown overnight in selective medium with galactose as a sugar source to induce expression of the GFP-Hrp1 fusion proteins. Cells were collected and washed twice in dH2O and once in PBSMT (PBS, 5 mm MgCl2, 0.5% Triton X-100). Glass bead lysis was conducted in PBSMT supplemented with protease inhibitors (phenylmethylsulfonyl fluoride and PLAC). Lysates were cleared by centrifugation, and total protein concentration was assessed by Bradford assay. One milligram of total yeast lysate and 50 μl of GST-Kap104 bead slurry were incubated overnight with agitation. The unbound fraction was removed, and the bound fraction was washed three times with PBSMT + 500 mm NaCl and once with PBS. Bound and unbound samples were loaded on a 10% denaturing SDS-PAGE gel. The proteins were transferred to a nitrocellulose membrane and probed with either primary α-GFP (rabbit, 1:3,000 dilution) and secondary α-rabbit HRP-conjugated (1:5,000) antibodies to detect the GFP-Hrp1 fusion proteins or primary α-GST (mouse, 1:5,000) and secondary α-mouse HRP-conjugated (1:5,000) antibodies to detect the GST-Kap104 protein.
In Vivo Functional Analyses—The in vivo function of each of the Hrp1 and Nab2 variants was tested using a plasmid shuffle technique (26). Plasmids encoding wild-type or variant Hrp1 or Nab2 proteins were transformed into either ΔHRP1 (ACY1571) or ΔNAB2 (ACY427) cells containing a wild-type HRP1 or NAB2 URA3 maintenance plasmid. Single transformants were grown to saturation in liquid culture, serially diluted (1:10) in dH2O, and spotted on control ura– leu– glu plates or on selective leu– glu plates containing 5-fluoroorotic acid (5-FOA). Plates were incubated at 18, 25, 30, or 37 °C for 3–5 days. For growth curve analysis, cells picked from the 25 °C 5-FOA Hrp1 plasmid shuffle plate that express either wild-type or P531A Hrp1 as the only copy of Hrp1 were grown overnight at 25 °C, normalized to equal starting concentrations, diluted 1:10 in a 96-well plate, and monitored for growth over time using an ultra microplate reader (Bio-Tek Instruments, Inc.). Cells were incubated at 37 °C with shaking, and the optical density was measured at 600 nm every 30 min.
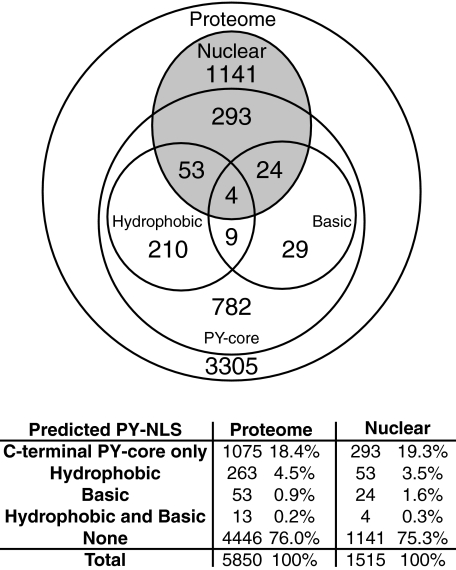
Bioinformatics—A custom library was constructed with different PY-NLS profiles: the consensus sequence for a hydrophobic PY-NLS (16), (LIMHFYVPQ)-(GAS)-(LIMHFYVPRQK)-(LIMHFYVPRQK)-X(7,12)-(RKH)-X(2,5)-PY; the sequence for a basic PY-NLS (16), (KR)-X(0,2)-(KR)-(KR)-X(3,10)-(RKH)-X(1,5)-PY; and the sequence for the C-terminal PY-core of the PY-NLS, (RKH)-X(2,5)-PY. This library was then used with ScanProsite (27) to query two data sets: the yeast proteome represented by the 5,850 proteins in the S. cerevisiae GenBank™ data base (28) and the 1,515 proteins localized to either the nucleus or the nucleolus according to a comprehensive subcellular localization study using the global yeast GFP fusion library (29). The results are summarized in a Venn diagram with corresponding percentages tabulated in a chart. A detailed table of results is presented on the Corbett laboratory Web site.
RESULTS
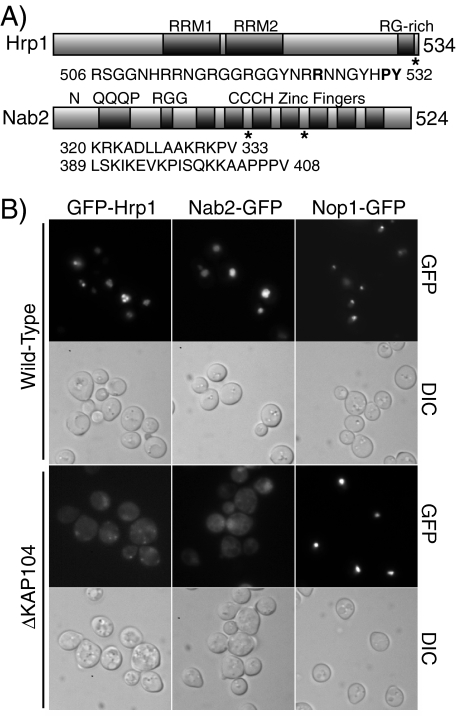
Hrp1 and Nab2 Contain PY-NLS-like Sequences—Scanning the protein sequences of the known Kap104 cargos Hrp1 and Nab2 (17, 18) revealed that Hrp1 contains one sequence and Nab2 contains two sequences that are similar to the established Kapβ2 binding sequence termed the PY-NLS (16) (Fig. 1A). The PY-NLS-like sequence of Hrp1 (residues 506–532) is located at the extreme C terminus of the protein within the region previously shown to be required for import (18). The PY-NLS-like sequences of Nab2 are both contained in the C-terminal CCCH zinc finger domain. PY-NLS-like sequence 1 (residues 320–333) lies between the second and third zinc fingers, and sequence 2 (residues 389–408) is located between the fourth and fifth zinc finger. The sequence in Hrp1 very closely resembles the mammalian PY-NLS, containing the core C-terminal portion of the consensus sequence preceded by a basic stretch of amino acids. In contrast, the putative sequences in Nab2 both contain upstream basic or hydrophobic stretches, but they vary from the PY-NLS consensus in the C-terminal PY-core as both sequences contain a lysine residue followed within two to five residues by a proline and a valine, yielding PV rather than the eponymous PY sequence. This change in the final residue raises the possibility that, although Hrp1 and Nab2 are both established cargos of Kap104 (17, 18), they may interact with the receptor using different sets of interactions or perhaps even in fundamentally different ways.
FIGURE 1.
Hrp1 and Nab2 contain putative PY-NLS-like sequences. A, domain structures of Hrp1 and Nab2. The positions of the PY-NLS-like sequence(s) are indicated by asterisks, and sequences are listed below each protein. B, nuclear localization of Hrp1 and Nab2 is dependent on Kap104. Wild-type and ΔKAP104 cells expressing GFP-Hrp1, Nab2-GFP, or the control protein Nop1-GFP were examined by direct fluorescence microscopy (GFP). Corresponding differential interference contrast (DIC) images are shown.
Hrp1 and Nab2 Are Imported by Kap104—To verify that nuclear localization of Hrp1 and Nab2 depends on Kap104 (17), the localization of GFP-tagged Hrp1 or Nab2 was assessed in wild-type or ΔKAP104 cells (Fig. 1B). Hrp1 was tagged with GFP at the N terminus of the protein to preserve the C-terminal location of the PY-NLS-like sequence. Nab2 was tagged at the C terminus with GFP because this fusion protein has previously been shown to function in vivo and to localize to the nucleus (22). In wild-type cells, both GFP-Hrp1 and Nab2-GFP are localized to the nucleus; however, in ΔKAP104 cells, both GFP-Hrp1 and Nab2-GFP are localized throughout the cell, indicating that proper nuclear localization of Hrp1 and Nab2 is dependent on Kap104. This experiment also demonstrates that GFP-tagged versions of the proteins are valid reporters. A non-Kap104-dependent cargo, Nop1-GFP (17, 25), is properly localized within the nucleus in both wild-type and ΔKAP104 cells, showing that not all nuclear import is impaired in ΔKAP104 cells.
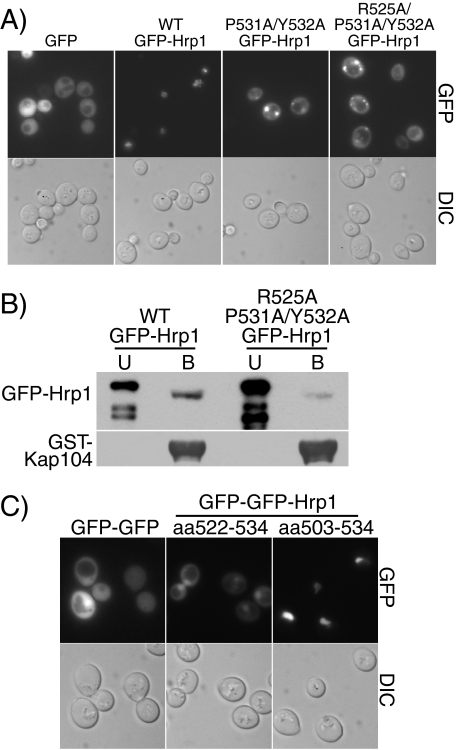
The PY-NLS-like Sequence within Hrp1 Is Necessary and Sufficient for Nuclear Import and Is Necessary for Kap104 Binding—To test whether the putative PY-NLS-like sequence within Hrp1 is necessary for the import of Hrp1 into the nucleus, specific amino acid changes were created in the C-terminal PY-core of the PY-NLS-like sequence of GFP-Hrp1 and the resulting GFP-Hrp1 variants were localized in wild-type cells (Fig. 2A). Wild-type GFP-Hrp1 was localized to the nucleus; however, when either the PY (P531A/Y532A) or both the upstream arginine and the PY (R525A/P531A/Y532A) residues were changed to alanine, GFP-Hrp1 was mislocalized to the cytoplasm, indicating that the PY-NLS-like sequence of Hrp1 is necessary for nuclear accumulation of Hrp1. Both the wild-type and the mutant GFP-Hrp1 partially localized in discrete puncta. These accumulations do not seem to correlate with particular nuclear or cytoplasmic bodies and their cause is unknown. Immunoblotting verified that all of the GFP-Hrp1 proteins were expressed at approximately equal levels (data not shown).
FIGURE 2.
The PY-NLS-like sequence within Hrp1 is necessary and sufficient for Hrp1 import and is necessary for Kap104 binding. A, wild-type cells expressing GFP alone, wild-type GFP-Hrp1, P531A/Y532A GFP-Hrp1, or R525A/P531A/Y532A GFP-Hrp1 were examined by direct fluorescence microscopy (GFP). Corresponding differential interference contrast (DIC) images are shown. B, in vitro binding between GST-Kap104 and either wild-type GFP-Hrp1 or R525A/P531A/Y532A GFP-Hrp1 was examined using glutathione beads as described under “Experimental Procedures.” The unbound (U) and bound (B) fractions were probed with an anti-GFP antibody to detect GFP-Hrp1 fusion proteins or with an anti-GST antibody to detect GST-Kap104. C, wild-type cells expressing a GFP-GFP control, GFP-GFP-Hrp1-(522–534) (containing the Hrp1 PY-core), or GFP-GFP-Hrp1-(503–534) (containing the entire Hrp1 PY-NLS) were examined by direct fluorescence microscopy (GFP). Corresponding differential interference contrast (DIC) images are shown.
To determine whether the PY-NLS-like sequence within Hrp1 is required for the interaction between Hrp1 and Kap104, we tested whether recombinant GST-Kap104 could interact with either wild-type or mutant GFP-Hrp1 in yeast lysate. For this experiment, GST-Kap104 beads were incubated with yeast lysate from cells expressing either wild-type GFP-Hrp1 or R525A/P531A/Y532A GFP-Hrp1 (Fig. 2B). Wild-type GFP-Hrp1 robustly bound to GST-Kap104 whereas R525A/P531A/Y532A GFP-Hrp1 showed greatly reduced binding, demonstrating that the PY-NLS-like sequence within Hrp1 is important for receptor binding.
To test whether the PY-NLS-like sequence within Hrp1 is sufficient to mediate import of a non-nuclear protein, either the C-terminal PY-core (residues 522–534) or the entire PY-NLS-like sequence of Hrp1 (residues 503–534) was fused to GFP-GFP (Fig. 2C). Two GFP molecules were used to create a protein that was too large to efficiently diffuse through the nuclear pore into the nucleus. Accordingly, to accumulate in the nucleus these fusion proteins must utilize an active system of transport. As expected, GFP-GFP alone was mainly localized to the cytoplasm. The C-terminal PY-core fused to GFP-GFP showed significant nuclear accumulation with some remaining cytoplasmic signal. The complete PY-NLS-like sequence fused to GFP-GFP mediated very efficient nuclear targeting with the reporter protein localized exclusively to the nucleus. These results indicate that the PY-NLS-like sequence of Hrp1 is sufficient to mediate import into the nucleus and that even the small C-terminal PY-core lacking the upstream basic or hydrophobic residues can mediate nuclear targeting to some degree. Both Hrp1 GFP-GFP-PY-NLS reporters were mislocalized to the cytoplasm in kap104-16 mutant cells (17) (data not shown), showing that import of these reporter proteins is dependent on Kap104.
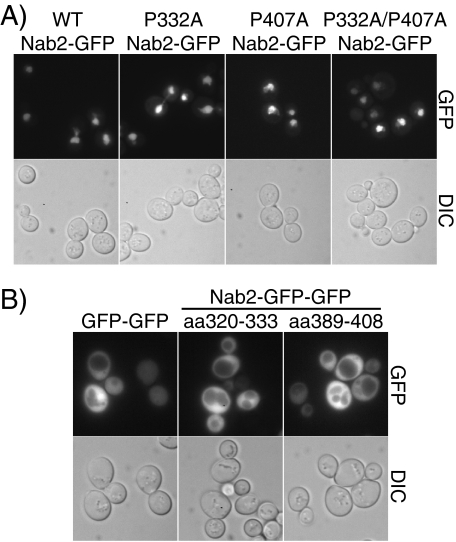
The PY-NLS-like Sequences within Nab2 Are Neither Necessary nor Sufficient for Nab2 Import into the Nucleus—An approach similar to that described for Hrp1 was taken to determine whether the PY-NLS-like sequences within Nab2, which actually contain terminal PV residues rather than PY residues, are required for Nab2 nuclear localization. To test whether either or both of these PY-NLS-like sequences within Nab2 are required for Nab2 nuclear import, wild-type Nab2-GFP, P332A Nab2-GFP, P407A Nab2-GFP, and P332A/P407A Nab2-GFP were localized in wild-type cells. As shown in Fig. 3A, wild-type Nab2-GFP is localized exclusively to the nucleus. Nab2-GFP with amino acid changes in either or both of the two putative PY-NLS-like sequences is also localized exclusively to the nucleus, indicating that neither PY-NLS-like sequence is required for Nab2 import.
FIGURE 3.
The PY-NLS-like sequences within Nab2 are neither necessary nor sufficient for Nab2 nuclear localization. A, wild-type cells expressing GFP alone, wild-type Nab2-GFP, P332A Nab2-GFP, P407A Nab2-GFP, or P332A/P407A Nab2-GFP were examined by direct fluorescence microscopy (GFP). Corresponding differential interference contrast (DIC) images are shown. B, wild-type cells expressing a GFP-GFP control, GFP-GFP-Nab2-(320–333), or GFP-GFP-Nab2-(389–408) were examined by direct fluorescence microscopy (GFP). Corresponding differential interference contrast (DIC) images are shown.
To test whether either of the putative PY-NLS-like sequences within Nab2 is sufficient to mediate nuclear import, either Nab2 PY-NLS-like sequence 1 (residues 320–333) or Nab2 PY-NLS-like sequence 2 (residues 389–408) was fused to GFP-GFP and these reporter proteins were localized in wild-type cells (Fig. 3B). Both Nab2 PY-NLS-GFP-GFP reporters localized throughout the cell, demonstrating that neither Nab2 PY-NLS-like sequence alone is sufficient to mediate import into the nucleus.
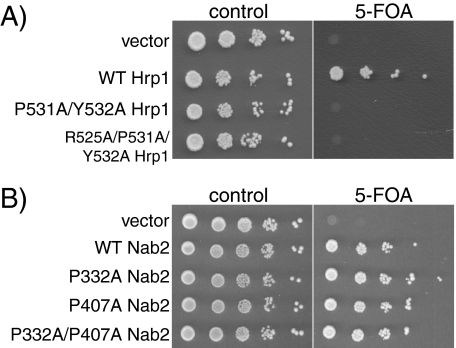
The PY-NLS-like Sequence Is Required for Hrp1 Function but Not for Nab2 Function—Because the PY-NLS-like region of Hrp1 is both necessary and sufficient to mediate Hrp1 nuclear import, this sequence should be required for the function of the essential nuclear Hrp1 protein (20) in vivo. In contrast, because neither of the PY-NLS-like sequences within Nab2 is required for import, we predict that neither of the sequences should be required for Nab2 function. Accordingly, to test whether the PY-NLS-like sequences within Hrp1 or Nab2 are essential for protein function in vivo, plasmid shuffle assays were performed. ΔHRP1 or ΔNAB2 cells containing a wild-type URA3 maintenance plasmid were transformed with plasmids encoding wild-type Hrp1 or Nab2, mutant PY-NLS Hrp1 or Nab2, or vector alone and were plated on control plates or plates containing 5-FOA. 5-FOA is a toxic analog of uracil that selects against URA-containing maintenance plasmids (26). HRP1 and NAB2 are essential genes; therefore, only cells containing a functional copy of HRP1 or NAB2 are able to grow on plates containing 5-FOA. As seen in Fig. 4, cells expressing P531A/Y532A Hrp1 or R525A/P531A/Y532A Hrp1 are not viable, indicating that the PY-NLS-like sequence within Hrp1 is essential for protein function, presumably because import of Hrp1 is required for proper mRNA processing (20). Cells expressing P332A Nab2, P407A Nab2, or P332A/P407A Nab2 as the only copy of Nab2 grow as well as cells expressing wild-type Nab2, indicating that neither PY-NLS-like region within Nab2 is required for cell viability.
FIGURE 4.
The PY-NLS-like sequence within Hrp1 is required for protein function. Protein function in vivo was assessed by a plasmid shuffle assay as described under “Experimental Procedures.” A, ΔHRP1 cells (ACY1571) maintained by a plasmid encoding wild-type Hrp1 and expressing either wild-type or mutant Hrp1 proteins were serially diluted, spotted onto control or 5-FOA plates, and grown at 30 °C for 3 days. B, ΔNAB2 cells (ACY427) maintained by a plasmid encoding wild-type Nab2 and expressing either wild-type or mutant Nab2 proteins were serially diluted, spotted onto control or 5-FOA plates, and grown at 30 °C for 3 days.
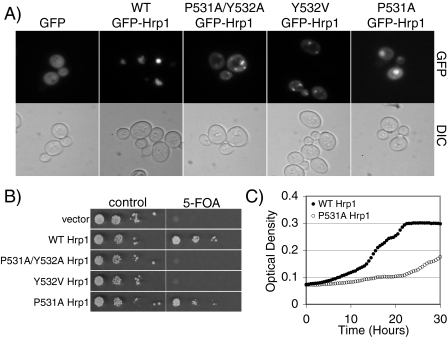
Functional Dissection of the PY-NLS Motif—To assess the contribution of the proline and the tyrosine residues to the function of the PY-NLS motif, we created several variants within the PY-NLS of Hrp1. First, we changed the tyrosine to valine (Y532V Hrp1). In the structure of Kapβ2 bound to heterogeneous nuclear ribonucleoprotein A1, the tyrosine residue of the PY-NLS makes extensive hydrophobic and polar contacts with the receptor (16). This change to a nonpolar residue should disrupt these interactions. This amino acid change also mimics the PY-NLS-like sequences of Nab2, allowing a further test of whether valine can substitute for tyrosine in S. cerevisiae PY-NLS sequences. Second, we changed the proline to alanine (P531A Hrp1). The proline of the heterogeneous nuclear ribonucleoprotein A1 PY-NLS makes hydrophobic contacts with Kapβ2 (16) and may contribute to the unstructured nature of the NLS while bound to Kapβ2. Because alanine is a much smaller, nonpolar side chain, this amino acid change tests whether those interactions and structural changes are required for proper PY-NLS function. Y532V Hrp1 and P531A Hrp1 were tagged at the N terminus with GFP and localized in wild-type cells (Fig. 5A). Much like the other Hrp1 PY-NLS mutants, GFP-Hrp1 Y532V was mislocalized to the cytoplasm, indicating that proper function of the PY-NLS-like sequence was disrupted. Therefore, valine cannot substitute for tyrosine in the Hrp1 PY-NLS-like sequence. GFP-Hrp1 P531A was partially mislocalized to the cytoplasm but also showed nuclear accumulation. This intermediate phenotype suggests that the proline residue may play a smaller, but still important, role in the Hrp1 PY-NLS. Immunoblotting verified that the GFP-Hrp1 variants were expressed at approximately equal levels (data not shown).
FIGURE 5.
Functional dissection of the PY-NLS motif. A, wild-type cells expressing GFP alone, wild-type GFP-Hrp1, GFP-Hrp1 P531A/Y532A, GFP-Hrp1 Y532V, or GFP-Hrp1 P531A were examined by direct fluorescence microscopy (GFP). Corresponding differential interference contrast (DIC) images are shown. B, ΔHRP1 cells (ACY1571) maintained by a plasmid encoding wild-type Hrp1 and expressing either wild-type or mutant Hrp1 protein were spotted onto control or 5-FOA plates and grown at 30 °C for 3 days. C, wild-type Hrp1 or P531A Hrp1 cells were monitored for growth over time at 37 °C as described under “Experimental Procedures.”
The function of the Y532V and P531A Hrp1 mutants was assessed using the plasmid shuffle assay (Fig. 5B). Cells expressing Y532V Hrp1 as the only form of Hrp1 are non-viable, indicating that the tyrosine residue in the C-terminal PY-core of the Hrp1 PY-NLS-like sequence is required for Hrp1 function. Cells expressing P531A Hrp1 are viable but have a slight growth defect at 30 °C. To determine whether cells expressing P531A Hrp1 as the only copy of Hrp1 are temperature-sensitive for growth, a quantitative growth assay was performed at 37 °C (Fig. 5C). Results of the analysis demonstrate that P531A Hrp1 cells grow much more slowly than wild-type cells at an elevated temperature.
The Prevalence of the PY-NLS in S. cerevisiae—This study has analyzed the PY-NLS-like sequences of the Kap104 cargos Nab2 and Hrp1. The PY-NLS-like sequences of Nab2 are not functional NLSs in vivo; however, it is important to note that Nab2 would not have been predicted to contain a putative PY-NLS. Both of the PY-NLS-like sequences within Nab2 vary from the consensus in the final tyrosine, a residue that we have shown to be essential for proper PY-NLS function in vivo (Fig. 5, A and B). In contrast, we find that Hrp1 does contain a functional PY-NLS, demonstrating that the PY-NLS motif is conserved in yeast. The conservation of the PY-NLS expands our current arsenal of yeast NLS consensus sequences and, combined with a bioinformatics approach, allows us to undertake a preliminary search for new putative Kap104 cargos in S. cerevisiae. Additionally, in Fig. 2C, we showed not only that the entire PY-NLS-like region of Hrp1 could mediate nuclear import but also that a minimal C-terminal PY-core could effect nuclear localization. Therefore, given that both of these sequences can mediate nuclear targeting in vivo, to determine the prevalence of putative PY-NLS-containing proteins in yeast we queried two data sets using the established hydrophobic and basic PY-NLS consensus sequences as well as a shorter sequence consisting of just the core R/H/KX2–5PY motif (Fig. 6). First, we searched the yeast proteome as represented by the 5,850 proteins in the S. cerevisiae GenBank™ data base (28) to reveal the entire complement of potential Kap104 cargos. Second, we scanned the 1,515 proteins localized at steady state to either the nucleus or the nucleolus as determined by a comprehensive localization study utilizing the global yeast GFP fusion library (29). This more focused data set was chosen because all nuclear proteins should contain nuclear targeting sequences, a subset of which should contain PY-NLSs. If one considers the C-terminal PY-core sufficient to mediate import, 1,404 proteins or 24% of the total yeast proteome contain at least one predicted PY-NLS sequence. Additionally, 374 nuclear proteins, ∼25% of the nuclear proteome, contain putative PY-NLSs. A selection of proteins that are likely candidates for Kap104 import are presented in Table 2, which contains a list of yeast proteins that are nuclear or nucleolar, contain a putative hydrophobic or basic PY-NLS, and lack a putative classical NLS (12). A detailed table containing all of the results is available on our Web site.
FIGURE 6.
The prevalence of predicted PY-NLS proteins in S. cerevisiae. Algorithms for hydrophobic and basic PY-NLSs or for the C-terminal PY-NLS core motif (see “Experimental Procedures”) were used to search the 5,850 proteins in the yeast proteome (Proteome) and the 1,515 proteins that are nuclear or nucleolar at steady state (Nuclear). The results of this preliminary analysis are plotted in a Venn diagram and summarized in the chart below. Hydrophobic and basic PY-NLSs, by definition, also contain the C-terminal PY core motif, so proteins denoted as containing “Hydrophobic”or“Basic” PY-NLSs also contain the PY-NLS core.
TABLE 2.
Nuclear or nucleolar yeast proteins that contain putative hydrophobic or basic PY-NLSs, but lack putative classical NLSs
| Gene | Systematic name | Function (adapted from the Saccharomyces genome data base) |
|---|---|---|
| APC4 | YDR118W | Subunit of the anaphase-promoting complex/cyclosome (APC/C) |
| APD1 | YBR151W | Protein of unknown function, required for normal localization of actin patches and for normal tolerance of sodium ions and hydrogen peroxide |
| ENP1 | YBR247C | Protein associated with U3 and U14 small nucleolar RNAs, required for pre-rRNA processing and 40 S ribosomal subunit synthesis |
| FPR1 | YNL135C | Peptidyl-prolyl cis-trans isomerase (PPIase) |
| GAL80 | YML051W | Transcriptional regulator involved in the repression of GAL genes in the absence of galactose |
| HRR25 | YPL204W | Protein kinase involved in regulating diverse events, including vesicular trafficking, DNA repair, and chromosome segregation |
| NRG1 | YDR043C | Transcriptional repressor that recruits the Cyc8p-Tup1p complex to promoters, mediates glucose repression |
| PPX1 | YHR201C | Exopolyphosphatase, hydrolyzes inorganic polyphosphate (poly P) into Pi residues |
| PRP11 | YDL043C | Subunit of the SF3a splicing factor complex, required for spliceosome assembly |
| QNS1 | YHR074W | Glutamine-dependent NAD(+) synthetase |
| RHR2 | YIL053W | Constitutively expressed isoform of dl-glycerol-3-phosphatase, involved in glycerol biosynthesis |
| RPO21 | YDL140C | RNA polymerase II largest subunit B220, part of central core; phosphorylation regulates association with transcription and splicing factors |
| RPS14B | YJL191W | Ribosomal protein 59 of the small subunit, required for ribosome assembly and 20 S pre-rRNA processing |
| RRP7 | YCL031C | Essential protein involved in rRNA processing and ribosome biogenesis |
| SCC2 | YDR180W | Subunit of cohesin-loading factor (Scc2p-Scc4p), a complex required for the loading of cohesin complexes onto chromosomes |
| SGV1 | YPR161C | Cyclin-dependent protein kinase that functions in transcriptional regulation, phosphorylates the C-terminal domain of Rpo21p, which is the largest subunit of RNA polymerase II |
| SIN4 | YNL236W | Subunit of the RNA polymerase II mediator complex; associates with core polymerase subunits to form the RNA polymerase II holoenzyme |
| SOK1 | YDR006C | Protein whose overexpression suppresses the growth defect of mutants lacking protein kinase A activity, involved in cAMP-mediated signaling |
| SPT21 | YMR179W | Protein required for normal transcription at several loci including HTA2-HTB2 and HHF2-HHT2, involved in telomere maintenance |
| TAH11 | YJR046W | DNA replication licensing factor, required for pre-replication complex assembly |
| TFB3 | YDR460W | Subunit of TFIIH and nucleotide excision repair factor 3 complexes, involved in transcription initiation, required for nucleotide excision repair |
| URA5 | YML106W | One of two orotate phosphoribosyltransferase isozymes that catalyze the fifth enzymatic step in de novo biosynthesis of pyrimidines |
| URH1 | YDR400W | Uridine nucleosidase, cleaves N-glycosidic bonds in nucleosides, involved in recycling pyrimidine deoxy- and ribonucleosides via the pyrimidine salvage pathway |
| YSH1 | YLR277C | Putative endoribonuclease, subunit of the mRNA cleavage and polyadenylation specificity complex required for 3′-processing of mRNAs |
DISCUSSION
In this study, we demonstrate that the PY-NLS, a human Kapβ2 binding nuclear targeting sequence (16), is conserved in S. cerevisiae and show for the first time in any organism that the PY-NLS is a functional nuclear import signal in vivo. Through localization, function, and binding studies, we show that the PY-NLS-like sequence within Hrp1, a cargo of the yeast ortholog of Kapβ2, is necessary and sufficient for nuclear import and is required for receptor binding and for protein function. These experiments indicate that the PY-NLS-like sequence within Hrp1 is a true NLS and, significantly, build on previous in vitro binding studies to demonstrate that this PY-NLS can actually mediate import of a protein cargo in vivo.
In contrast, we found that the putative PY-NLS-like sequences within Nab2 are not functional NLS motifs, suggesting that Nab2 must have alternative mechanisms for gaining entry into the nucleus. These mechanisms may involve several different import receptors; however, because Fig. 1B and previous studies (18, 19) show that proper Nab2 localization requires Kap104, these results more directly suggest that Kap104 can interact with different classes of cargo proteins using different mechanisms. In the first mode of interaction, Kap104 recognizes cargo proteins like Hrp1 via PY-NLS motifs, and in the second mode of interaction, Kap104 recognizes cargo proteins like Nab2 via interactions like that mapped within the RGG domain of Nab2 (18, 19).
Our study also assessed the contribution of the signature residues of the PY-NLS, the proline and tyrosine, to the function of the Hrp1 PY-NLS. We found that changing the proline residue to an alanine caused partial mislocalization of Hrp1 accompanied by temperature-sensitive growth, suggesting that, although not essential, the proline is required for proper localization and function of the Hrp1 protein. This moderate effect on Hrp1 localization and function may be due to the position of the PY-NLS only two residues from the C terminus of Hrp1. Typically, proline residues are assumed to contribute to the unstructured nature of NLSs while bound to their receptors. The proline within the PY-NLS of Hrp1 may not be absolutely required for PY-NLS function because the end of the protein may already lack any significant structure. However, a proline residue could make more significant contributions to PY-NLS sequences within other cargos where the motif is located in a more structured region of the protein. Changing the tyrosine in the Hrp1 PY-NLS to a valine completely abrogated PY-NLS function, suggesting that the change in the terminal residue of the PY-NLS-like sequences within Nab2 (PV versus PY) may be responsible for the lack of function of these putative PY-NLSs in vivo.
Interestingly, the PY-NLS sequence of Hrp1 does not exactly match the consensus proposed by Lee et al. (16) for human Kapβ2 binding; the C-terminal PY-core precisely matches, but the pattern of upstream basic residues differs from the established motif. Combined with the results of Fig. 2C where the C-terminal PY-core of Hrp1 alone can mediate import, this variability suggests that the requirements for recognition of yeast PY-NLSs by Kap104 may be slightly less stringent than the requirements for recognition of a PY-NLS by human Kapβ2. It is important to note, however, that the Hrp1 PY-core alone does not mediate proper Hrp1 localization; there is still significant cytoplasmic signal in addition to the nuclear accumulation. For proteins that require strict nuclear localization, the C-terminal PY-core may not be sufficient for proper function even in yeast; however, for many proteins, establishing a nuclear presence to any degree may be sufficient for the protein to accomplish its role in the cell. Therefore, we propose that the minimal PY-NLS in yeast consists of the C-terminal portion of the human PY-NLS, R/H/KX2–5PY, with upstream basic or hydrophobic residues enhancing the ability of the NLS to mediate nuclear import.
Given the variability in the evolving definition of the consensus sequence for the PY-NLS, caution should be used in predicting PY-NLS motifs. However, a preliminary estimate of the likely prevalence of the PY-NLS in yeast using the consensus sequence for the minimal yeast PY-NLS as well as the established consensus sequences for basic and hydrophobic PY-NLSs revealed that 24% of the S. cerevisiae proteome and ∼25% of the proteins that localize to the nucleus or nucleolus at steady state contain a putative PY-NLS. Importantly, this analysis predicts that Hrp1 should contain a putative PY-NLS, and, indeed, our studies show that this PY-NLS is functional in vivo. In contrast, the non-functional PV-containing PY-NLS-like sequences in Nab2 are not identified in this analysis. If we consider only the longer, more stringent hydrophobic or basic PY-NLS consensus sequences, ∼6% of the yeast proteome and ∼5% of the nuclear proteins contain at least one PY-NLS. For comparison, ∼45% of the yeast proteome and ∼57% of nuclear proteins contain a putative classical NLS (12). Obviously, the prevalence of putative PY-NLS-containing proteins is much lower than the prevalence of putative cNLS-containing proteins, but the fraction of putative yeast PY-NLS cargos is still substantial. Examination of the 24 yeast proteins that are nuclear, contain a putative hydrophobic or basic PY-NLS, and lack a cNLS revealed that seven are implicated in RNA biogenesis or trafficking (Table 2). Many Kapβ2 cargos are also involved in RNA processing in higher eukaryotes (16). This similarity brings up the tantalizing possibility that nuclear import via a PY-NLS motif may be a feature common to RNA-related processes in all eukaryotes. However, more functional analyses of predicted PY-NLS cargos are clearly required before we can begin to reliably define the contribution of this import pathway to establishing the nuclear proteome in vivo.
Acknowledgments
We thank Seth Kelly for construction of the Nop1-GFP plasmid and for creation of the laboratory Web site, Dr. Pamela Silver for the gift of the ΔKAP104 mutant, Dr. Michael Rout for the gift of the GST-Kap104 plasmid, and Dr. Sandro Valentini for the gift of the ΔHRP1 mutant.
This work was supported, in whole or in part, by grants from the National Institutes of Health (to A. H. C., S. E. D., and R. E. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: NLS, nuclear localization signal; cNLS, classical NLS; GFP, green fluorescent protein; GST, glutathione S-transferase; PBS, phosphate-buffered saline; HRP, horseradish peroxidase; 5-FOA, 5-fluoroorotic acid.
References
- 1.Fahrenkrog, B., and Aebi, U. (2003) Nat. Rev. Mol. Cell Biol. 4 757–766 [DOI] [PubMed] [Google Scholar]
- 2.Stoffler, D., Fahrenkrog, B., and Aebi, U. (1999) Curr. Opin. Cell Biol. 11 391–401 [DOI] [PubMed] [Google Scholar]
- 3.Allen, T. D., Cronshaw, J. M., Bagley, S., Kiseleva, E., and Goldberg, M. W. (2000) J. Cell Sci. 113 1651–1659 [DOI] [PubMed] [Google Scholar]
- 4.Paine, P. L., Moore, L. C., and Horowitz, S. B. (1975) Nature 254 109–114 [DOI] [PubMed] [Google Scholar]
- 5.Bonner, W. M. (1978) Protein Migration and Accumulation in Nuclei, Academic Press, New York
- 6.Mosammaparast, N., and Pemberton, L. F. (2004) Trends Cell Biol. 14 547–556 [DOI] [PubMed] [Google Scholar]
- 7.Cook, A., Bono, F., Jinek, M., and Conti, E. (2007) Annu. Rev. Biochem. 76 647–671 [DOI] [PubMed] [Google Scholar]
- 8.Quimby, B. B., and Dasso, M. (2003) Curr Opin. Cell Biol. 15 338–344 [DOI] [PubMed] [Google Scholar]
- 9.Lee, S. J., Matsuura, Y., Liu, S. M., and Stewart, M. (2005) Nature 435 693–696 [DOI] [PubMed] [Google Scholar]
- 10.Görlich, D., Panté, N., Kutay, U., Aebi, U., and Bischoff, F. R. (1996) EMBO J. 15 5584–5594 [PMC free article] [PubMed] [Google Scholar]
- 11.Chook, Y. M., and Blobel, G. Nature 399 230–237 [DOI] [PubMed]
- 12.Lange, A., Mills, R. E., Lange, C. J., Stewart, M., Devine, S. E., and Corbett, A. H. (2007) J. Biol. Chem. 282 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, S., Sekimoto, T., Yamashita, E., Nagoshi, E., Nakagawa, A., Imamoto, N., Yoshimura, M., Sakai, H., Chong, K., Tsukihara, T., and Yoneda, Y. (2003) Science 302 1571–1575 [DOI] [PubMed] [Google Scholar]
- 14.Cingolani, G., Petosa, C., Weis, K., and Müller, C. W. (1999) Nature 399 221–229 [DOI] [PubMed] [Google Scholar]
- 15.Cingolani, G., Bednenko, J., Gillespie, M. T., and Gerace, L. (2002) Mol. Cell 10 1345–1353 [DOI] [PubMed] [Google Scholar]
- 16.Lee, B. J., Cansizoglu, A. E., Süel, K. E., Louis, T. H., Zhang, Z., and Chook, Y. M. (2006) Cell 126 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aitchison, J. D., Blobel, G., and Rout, M. P. (1996) Science 274 624–627 [DOI] [PubMed] [Google Scholar]
- 18.Lee, D. C., and Aitchison, J. D. (1999) J. Biol. Chem. 274 29031–29037 [DOI] [PubMed] [Google Scholar]
- 19.Truant, R., Fridell, R. A., Benson, R. E., Bogerd, H., and Cullen, B. R. (1998) Mol. Cell. Biol. 18 1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kessler, M. M., Henry, M. F., Shen, E., Zhao, J., Gross, S., Silver, P. A., and Moore, C. L. (1997) Genes Dev. 11 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hector, R. E., Nykamp, K. R., Dheur, S., Anderson, J. T., Non, P. J., Urbinati, C. R., Wilson, S. M., Minvielle-Sebastia, L., and Swanson, M. S. (2002) EMBO J. 21 1800–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green, D. M., Marfatia, K. A., Crafton, E. B., Zhang, X., Cheng, X., and Corbett, A. H. (2002) J. Biol. Chem. 277 7752–7760 [DOI] [PubMed] [Google Scholar]
- 23.Adams, A., Gottschling, D. E., Kaiser, C. A., and Stearns, T. (1997) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 24.Sambrook, J., and Russell, D. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 25.Leslie, D. M., Zhang, W., Timney, B. L., Chait, B. T., Rout, M. P., Wozniak, R. W., and Aitchison, J. D. (2004) Mol. Cell. Biol. 24 8487–8503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boeke, J. D., Trueheart, J., Natsoulis, G., and Fink, G. R. (1987) Methods Enzymol. 154 164–175 [DOI] [PubMed] [Google Scholar]
- 27.Gattiker, A., Gasteiger, E., and Bairoch, A. (2002) Appl. Bioinformatics 1 107–108 [PubMed] [Google Scholar]
- 28.Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., and Wheeler, D. L. (2007) Nucleic Acids Res. 35 D21–D25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., and O'Shea, E. K. (2003) Nature 425 686–691 [DOI] [PubMed] [Google Scholar]
- 30.Winston, F., Dollard, C., and Ricupero-Hovasse, S. L. (1995) Yeast 11 53–55 [DOI] [PubMed] [Google Scholar]
- 31.Sikorski, R. S., and Hieter, P. (1989) Genetics 122 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodel, A. E., Harreman, M. T., Pulliam, K. F., Harben, M. E., Holmes, J. S., Hodel, M. R., Berland, K. M., and Corbett, A. H. (2006) J. Biol. Chem. 281 23545–23556 [DOI] [PubMed] [Google Scholar]