Abstract
Intraerythrocytic malaria parasites use host hemoglobin as a major nutrient source. Aspartic proteases (plasmepsins) and cysteine proteases (falcipains) function in the early steps of the hemoglobin degradation pathway. There is extensive functional redundancy within and between these protease families. Plasmepsins are synthesized as integral membrane proenzymes that are activated by cleavage from the membrane. This cleavage is mediated by a maturase activity whose identity has been elusive. We have used a combination of cell biology, chemical biology, and enzymology approaches to analyze this processing event. These studies reveal that plasmepsin processing occurs primarily via the falcipains; however, if falcipain activity is blocked, autoprocessing can take place, serving as an alternate activation system. These results establish a further level of redundancy between the protease families involved in Plasmodium hemoglobin degradation.
Human malaria is a disease of devastating proportions. Approximately 40% of the world population is at risk for acquiring it, and an estimated two million people, mostly children, die of it each year (1). The most deadly form of malaria is caused by infection with the protozoan parasite Plasmodium falciparum. During the disease-causing stage of its life cycle, the parasite lives within human red blood cells and consumes over two-thirds of the hemoglobin of the host cell (2). This massive catabolic process is essential for parasite survival and is facilitated by numerous endo- and exo-peptidases within an acidic organelle termed the food vacuole (3–6).
Two families of proteases are involved in the initial steps of hemoglobin catabolism: three cysteine protease falcipains (FP-2, FP-2′, FP-3) and four aspartic protease plasmepsins (PM I, PM II, HAP, and PM IV). The plasmepsins are synthesized as type II integral membrane proteins that are delivered to the food vacuole through the secretory system via the plasma membrane of the parasite. Once at the plasma membrane, they appear to incorporate into the membranes of the forming cytostomes that deliver host cell hemoglobin to the food vacuole (7, 8). Once at the food vacuole, the plasmepsins are activated and released from the membrane by proteolytic cleavage at a conserved sequence site (see Fig. 1A) (7, 9). Although numerous examples of autocatalytic protease activation exist, PM activation is refractory to inhibitors of aspartic proteases such as pepstatin (a potent inhibitor of plasmepsin activity) as well as to general inhibitors of cysteine, serine, or metalloproteases (7, 9) both in cell culture and cell-free assays. Processing can, however, be inhibited by tripeptide aldehydes such as N-acetyl-l-leucyl-l-leucyl-l-norleucinal (ALLN).3 This has led to a model of plasmepsin processing in which a unique ALLN-sensitive trans-processing activity, or maturase, serves to cleave the proplasmepsins to their mature, active form (9).
FIGURE 1.
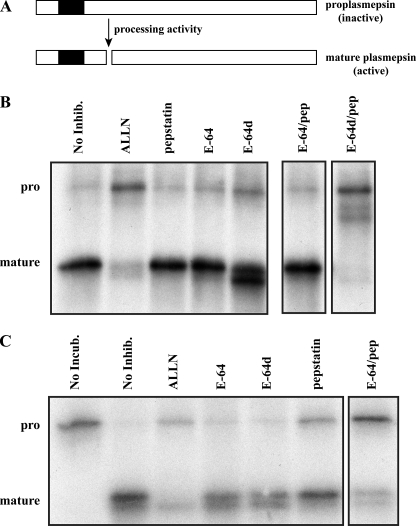
Plasmepsin processing can occur through two redundant activities. A, schematic depicting the activation of the plasmepsins by proteolytic processing. Shaded box, transmembrane domain. B, in vivo processing assay in which metabolically labeled parasites were washed and chased in the presence of various protease inhibitors (10 μm) followed by immunoprecipitation with anti-PM II antibody and visualization by autoradiography. Only treatment with ALLN or with the combination of E-64d/pepstatin (pep) inhibited processing activity. The double band seen in the E-64d lane likely represents alternative autoprocessing activity (see “Discussion”). C, cell-free processing assay confirms in vivo assay. 35S-labeled pro-plasmepsin (pro) was synthesized by metabolic labeling of cells in the presence of brefeldin A followed by production of total parasite lysate. Inhibitors (10 μm) and additional unlabeled parasite lysate were then added, and the reaction was incubated at pH 5.0 at 37 °C for 1 h, followed by immunoprecipitation as above.
We have extended these studies with the identification of the enzymes involved in proplasmepsin processing. These findings significantly revise our current model for this event and have important ramifications for the notion of proplasmepsin processing as a potential drug target.
EXPERIMENTAL PROCEDURES
Materials—Unless otherwise noted, all of the reagents were from Sigma-Aldrich. [35S]Cysteine/methionine (1175 Ci/mmol) and [3H]methionine (1000 Ci/mmol) (PerkinElmer Life Sciences), RPMI and Albumax (Invitrogen), brefeldin A (Calbiochem), pepstatin (Roche Applied Science), HHDG peptide (dabcyl-GABA-NYLGSSND-edans; Anaspec), Z-Leu-Arg-AMC peptide (R & D Systems), and tetanolysin (List Biological Laboratories) were used. E-64-bio was synthesized by the method of Greenbaum et al. (10).
Parasite Culture and Purification—P. falciparum clone 3D7 was grown at 37 °C under 3% O2, 3%CO2, 94% N2 in RPMI 1640 medium supplemented with 25 mm HEPES, 30 mg/l hypoxanthine, 0.225% NaHCO3, and 0.5% Albumax I (Invitrogen). Parasites were grown in 2% human red blood cells and were synchronized using successive rounds of 5% sorbitol treatment. The parasites were purified from host red blood cells by incubation with 50 hemolytic units/ml tetanolysin in PBS for 10 min on ice, followed by two cold PBS washes.
Immunoprecipitation—Rabbit polyclonal antiserum against plasmepsin II (antibody 737) was made using the peptide HSHSSNDNIELNDFQNIMFYGDAEV derived from the mature N terminus of native plasmepsin II prepared as described (7).
Labeled trophozoite stage parasites (routinely 5 × 108) were immunoprecipitated using denaturing conditions. Free parasites were lysed by the addition of 1% SDS (100 μl) at 100 °C for 10 min. The solution was adjusted to 150 mm NaCl, 40 mm Tris, pH 7.5, 6 mm EDTA, 0.5% deoxycholate, 1% Triton X-100, and 0.1% SDS; insoluble material was removed by centrifugation. Antibody was added at a 1:250 dilution (determined to be saturating) along with 70 μl of protein A-Sepharose (1:1 bead slurry to immunoprecipitation buffer). The samples were mixed by nutation at 4 °C for 16–20 h, and immunocomplexes were collected, added to an equal volume of 2× nonreducing sample buffer, boiled 5 min, and analyzed by 12% SDS-PAGE with fluorography.
In Vivo Processing Assay—Inhibitor experiments were performed with synchronized cultures of trophozoite stage parasites. The cultures were washed one time in RPMI prepared without methionine and cysteine, resuspended in the same medium at a density of 2.0 × 108/ml with [35S]methionine/cysteine (350 μCi/ml), and incubated at 37 °C for 30 min (3% O2, 3%CO2). The cultures were chased for the indicated times by removal of labeling media and addition of complete RPMI. The parasites were harvested by tetanolysin purification and processed immediately for immunoprecipitation. Kinetics of processing was calculated by densitometry and calculation of the fraction of plasmepsin II (PM II) processed at each time point. Band intensities were normalized to account for change in specific activity caused by the loss of 35S-labeled cysteine and methionine residues upon processing.
Cell-free Processing Assay—The assay was by the method of Francis et al. (7). Trophozoite stage parasites were metabolically labeled as above except that brefeldin A (5 μg/ml) and [35S]methionine/cysteine were added at the same time. The cultures were incubated for 4–5 h and then harvested and lysed in PBS by gentle sonication. The radiolabeled lysates were incubated in the presence of an equal amount of unlabeled parasite lysate plus inhibitors in 100 mm sodium acetate buffer, pH 5.5, for 1 h at 37 °C. The reactions were stopped by addition of one volume of 2% SDS and boiling. PM II was immunoprecipitated as described above.
In Vitro Transcription/Translation and Radiosequencing—Capped PM II RNA was prepared using linearized plasmid DNA containing the entire open reading frame of plasmepsin II and T7 polymerase Message Machine kit (Ambion) following the manufacturer's instructions. Transcript was added to nuclease-free rabbit reticulocyte lysate (Promega) in the presence of 0.5 μl of [35S]methionine, 10 μg of brewer's yeast tRNA, and 1.8 μl of canine pancreatic microsomes/25 μl of translation reaction. 2 μl of in vitro translated protein (above) was incubated with parasite lysate treated with either 10 μm E-64 or pepstatin for 3 h at 37 °C. Denaturing immunoprecipitations were performed as above, fractionated by SDS-PAGE, and blotted onto polyvinylidene difluoride membrane. Plasmepsin bands were excised and subjected to Edman degradation. Each cycle from the sequencer was assayed for radioactivity in a scintillation counter. Nonspecific background increases with successive cycles as is typical with Edman degradation.
Target Analysis with E-64-biotin—Synchronized trophozoite stage culture (∼5% parasitemia) was incubated with 10 μm inhibitor for 15 min to block accessible active sites followed by purification of parasites by tetanolysin. The parasites were then washed twice with ice-cold PBS. The parasite pellet was resuspended in 100 μl of lysis/labeling mix (50 mm sodium acetate, pH 5.5, 1.0%Triton X-100, 10 mm dithiothreitol, 25 μm E-64-bio) and incubated at 37 °C for 30 min followed by the addition of 25 μl of 5× SDS sample buffer and boiled for 5 min. 30 μl of sample was resolved by SDS-PAGE and transferred to nitrocellulose. Biotinylated proteins were detected using Vectastain ABC-AmP kit and DuoLuX reagent (Vector Laboratories) as per the manufacturer's instructions.
Fluorogenic Cleavage Assays—Cleavage of HHDG internally quenched fluorogenic peptide was initiated by the addition of 0.5 μl of purified recombinant FP-2 or FP-3 to a final reaction volume of 200 μl (50 mm sodium acetate, pH 5.5, HHDG peptide, 10 mm dithiothreitol) and monitored over time using a Varian Cary Eclipse fluorimeter fitted with a 96-well plate reader. Excitation and emission wavelengths were 355 and 495 nm, respectively. The reaction rates were plotted relative to substrate concentration, and Km and Vmax values were determined by a Michaelis-Menten curve fit using Kaleidagraph (Synergy Software). Enzyme concentration was determined by absorbance at 280 nm using calculated extinction coefficients of 41,870 and 55,280 m–1 cm–1 for mature FP-2 and FP-3, respectively.
RESULTS
Plasmepsin Processing Is Inhibited by Combined Treatment with E-64d and Pepstatin—Pulse-chase experiments on cultured parasites incubated with various protease inhibitors clearly demonstrate that the pro-plasmepsin processing activity is refractory to the general cysteine and aspartic protease inhibitors, E-64 and pepstatin, but sensitive to ALLN as previously reported (7, 9) (Fig. 1B). We tested combinations of inhibitors and surprisingly, the combination of E-64d and pepstatin inhibited pro-PM II processing activity to a similar degree as ALLN. This result indicates the presence of two redundant, yet unique, processing activities. In the case of treatment with pepstatin, a potent plasmepsin inhibitor, processing of pro-PM II appears normal. This is in contrast, however, to E-64d treatment, which resulted in two processed forms of PM II, both of which appear to be the result of autoprocessing (see below). Evidently, both activities (the pepstatin-sensitive and the E-64d-sensitive) can process pro-PM II, albeit with different specificities. Inhibition of pro-PM II processing therefore requires inhibition of both of these activities.
We were concerned that the inhibition of processing seen by the combined treatment with E-64d and pepstatin could be due to a general toxic effect and not through a direct inhibition of processing activity. Therefore, we carried out a complementary experiment using a cell-free processing assay (9) in which pro-PM II was generated in vivo by incubating parasites with brefeldin A during a metabolic labeling with [35S[Met/Cys (Fig. 1C). A sonicate was then prepared from these parasites and incubated with additional unlabeled parasite lysate in the presence of inhibitors. Processing of endogenously synthesized pro-PM II was inhibited by the combination of E-64 and pepstatin as well as by ALLN. This indicates a direct effect of these inhibitors on PM processing activity. The result also revealed, in contrast to the in vivo assay, that the E-64d-sensitive processing activity is accessible and sensitive to E-64 in the cell-free system, indicating that proplasmepsin processing in vivo occurs in a cellular compartment not readily accessible to E-64.
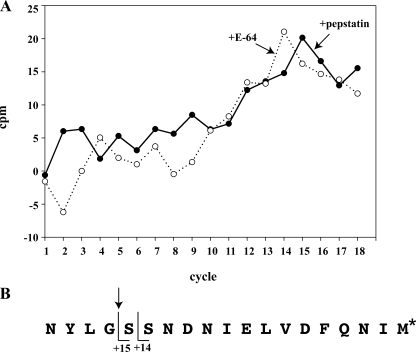
Endogenous Cleavage of PM II Is Inhibited by E-64d—Previous efforts had identified that endogenous processing of pro-PM II occurs at the G124-S125 peptide bond (11). We were therefore interested in the cleavage specificities of each of these redundant processing activities and whether one, or both, contributed to endogenous pro-PM II processing. To address this question we mapped, by radiosequencing, the N terminus of in vitro translated pro-PM II that had been incubated with parasite lysate pretreated with either E-64 or pepstatin (Fig. 2). Radiosequencing of these cleavage products showed that the pepstatin-treated lysate processes pro-PM II at the endogenous Gly124–Ser125 cleavage site (as shown by peak of radiolabeled methionine released 15 amino acids after the N terminus), whereas incubation with E-64-treated lysate resulted in cleavage one amino acid C-terminal to this site at the Ser125–Ser126 peptide bond (peak of radiolabeled methionine released 14 amino acids after the N terminus). Given that endogenous cleavage at Ser125–Ser126 does not occur (9), this experiment indicates that the E-64d-sensitive cysteine protease activity represents the dominant processing activity present in vivo.
FIGURE 2.
Cleavage site specificity of E-64 and pepstatin processing activities by radiosequencing. A, plot of [35S]methionine activity released for each sequencing cycle of in vitro translated pro-PM II, which had been incubated with E-64 (dotted line) or pepstatin treated parasite lysate (solid line). B, mapping of the N terminus of processed PM II generated by each treatment. Radiolabeled methionine is indicated by an asterisk, and endogenous cleavage site is indicated by arrowhead.
Alternative Processing Is via Autoprocessing That Proceeds with Slower Kinetics—Alternative processing is only seen in the presence of E-64d (and E-64 in vitro), suggesting to us that it is functionally recessive to the endogenous, E-64d-sensitive processing activity. We hypothesized that this might be the result of a difference in the kinetics of these two activities. To explore this idea, we performed pulse-chase experiments measuring the half-time of pro-PM II processing and determined that there was an ∼60% increase in the t½ of pro-PM II processing in the presence of E-64d (no inhibitor = 15 min versus E-64d = 9.5 min; Fig. 3). This significant rate difference likely explains why pro-PM II processing normally occurs via the E-64d-sensitive activity in vivo.
FIGURE 3.
Alternative processing has slowed kinetics. Pulse-chase experiment demonstrating altered kinetics of pro-PM2 (pro) autoprocessing in the presence of E-64d. The parasites were metabolically labeled for 5 min and then chased in complete medium without (control) or with (+E-64d)25 μm E-64d for indicated times followed by immunoprecipitation with anti-PM II antibody and visualization by autoradiography. t½ values were calculated as 15.0 and 9.5 min for control and E-64d, respectively. E-64d was added to both pulse and chase media.
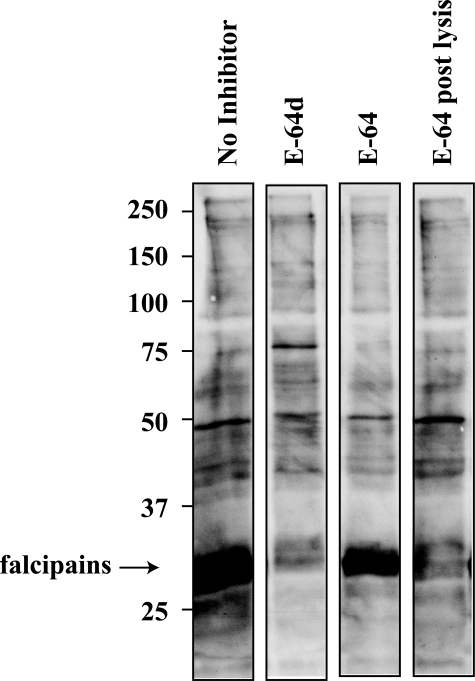
E-64d Efficiently Labels Specific Cysteine Proteases in Vivo—It was clear from the previous experiments that when used in combination with pepstatin, the membrane-permeant E-64d, but not the less permeant E-64, was able to inhibit the endogenous processing activity in vivo. We decided therefore to take advantage of this distinction to aid in identifying the protease target(s) of E-64d. We accomplished this by incubating parasitized red blood cell cultures with either E-64 or E-64d to irreversibly label all accessible, protease active sites (Fig. 4). Following this incubation, the cultures were washed free of inhibitor, and the parasites were purified from the host red blood cells. Total parasite lysate was then prepared and incubated with a biotinylated version of E-64 (E-64-bio) to identify the remaining, unlabeled active sites. These lysates were then resolved by SDS-PAGE and transferred to nitrocellulose, and the biotinylated proteins were detected by streptavidin-horseradish peroxidase. Although numerous bands are labeled in the absence of inhibitor pretreatment, a diffuse protein band that is competed by E-64d but not by E-64 was apparent at ∼30 kDa. When E-64 was added post-lysis, however, it was able to compete. This labeling pattern mimics the sensitivity of processing to these inhibitors (when used in combination with pepstatin; Fig. 1). Previous work by Greenbaum et al. (12) using an identical biotinylated E-64 compound identified this dominant 30-kDa band as comprising falcipains 2 and 3.
FIGURE 4.
Profile of E-64d targets. Competitive labeling in which parasites were incubated in the absence or presence of E-64, or its more membrane-permeant analog E-64d for 15 min is shown. The parasites were then purified, and the lysates were incubated with E-64-bio to label remaining active sites. E-64 (post lysis) represents untreated parasite lysate that was incubated for 5 min at 37 °C with E-64 prior to the addition of E-64-bio.
Falcipains Cleave a PM II Peptide at the Endogenous Processing Site—To directly test the notion that falcipains are responsible for proplasmepsin processing, we assayed the activity of purified, recombinant FP-2 and FP-3 on a synthetic eight-amino acid, fluorogenic peptide comprising the endogenous cleavage site of pro-PM II. Both FP-2 and FP-3 were able to cleave the processing site peptide in a time-dependent fashion with kcat/Km values of 1.4 × 104 and 2.3 × 103 s–1 m–1, respectively (Table 1). Purified, recombinant PM II and PM IV were also tested but were both inactive on this peptide (not shown).
TABLE 1.
Kinetic parameters of FP-2 and FP-3 with internally quenched fluorogenic octapeptide substrate HHDG
The initial rates of peptide cleavage were measured using 21.5 nm FP-2 and 25 nm FP-3. Substrate concentrations were from 0.195 to 12.5 μm.
| Km | kcat | kcat/Km | |
|---|---|---|---|
| m | s-1 | s-1m-1 | |
| FP-2 | 1.9 × 10-6 | 0.027 | 1.4 × 104 |
| FP-3 | 9.8 × 10-6 | 0.023 | 2.3 × 103 |
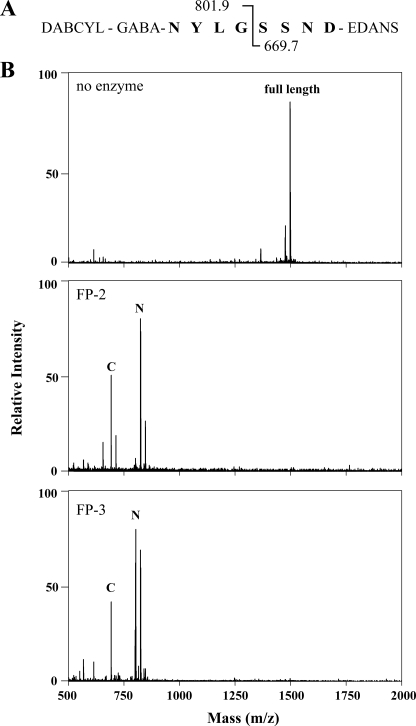
We next used mass spectrometry to map the falcipain cleavage site(s) on this peptide (Fig. 5). Following incubation of peptide with FP-2 or FP-3, the dominant ion species present corresponded to predicted C- and N-terminal fragments of a single cleavage event at the G-S peptide bond. Evidence of alternative cleavage sites was not detected. These results indicate that both enzymes (and also presumably FP-2′ given its extremely high level of identity to FP-2 (13)) recognize and cleave PM II at the endogenous processing site.
FIGURE 5.
Falcipain-2 and 3 both cleave a synthetic peptide at the PM II processing site. A, peptide HHDG comprising the eight amino acids spanning the pro-PM II processing site flanked on the N terminus with dabcyl-GABA and on the C terminus with edans (molecular weight = 1453.6). Predicted masses for both N- and C-terminal fragments after cleavage at the endogenous G/S peptide bond are shown. B, matrix-assisted laser desorption ionization spectra of either uncleaved peptide or cleavage products after 15 min of incubation with FP-2 or FP-3. Predominant ion species of FP-2 cleavage products are 823.8 [M+Na]+, 845.8 [M+2Na]+ (N-terminal fragment, N) and 691.8 [M+Na]+, 713.8 [M+2Na]+ (C-terminal fragment, C). Predominant ion species of FP-3 cleavage products are 801.8 [M+H]+, 823.8 [M+Na]+ (N-terminal fragment, N) and 691.8 [M+Na]+ (C-terminal fragment, C). Ions corresponding to alternative peptide bond cleavage were not detected.
DISCUSSION
Two Redundant Activities Are Capable of Proplasmepsin Processing in Vivo—It has been of great interest to our lab to identify the proplasmepsin processing maturase activity. That general protease inhibitors did not block this activity led us to believe that a unique processing enzyme was likely responsible. The data presented here reveal that processing is not a result of a unique enzymatic activity but rather that proplasmepsin processing can occur through either of two redundant activities. Our in vivo processing experiment demonstrates this clearly because neither E-64d nor pepstatin individually inhibit processing; however, the combination of E-64d and pepstatin does inhibit processing (Fig. 1B). Therefore, blocking of proplasmepsin processing requires inhibition of both an E-64d-sensitive cysteine protease activity as well as inhibition of a pepstatin-sensitive aspartic protease activity.
The Pepstatin-sensitive Autoprocessing Activity Is Functionally Recessive—Previous work from our lab has determined that a cleavage motif is conserved for all four food vacuole plasmepsins (9) with the N-terminal residue of endogenous mature plasmepsin II being serine 125 (11). In the studies presented here, we have determined using in vitro translated PM II that the E-64d sensitive activity in parasite lysate correctly processed this substrate, whereas the pepstatin sensitive activity cleaved just one amino acid C-terminal to this site (Fig. 2). These data indicate that the pepstatin-sensitive activity is functionally recessive to the E-64d-sensitive activity. In other words, the pepstatin-sensitive alternative processing is only seen upon treatment with E-64d. Consistent with this is the presence of the alternative processed forms of PM II that are seen upon treatment with E-64d in vivo or in vitro (Fig. 1B). The higher molecular weight band is at the correct size for mature plasmepsin II and likely corresponds to the autoprocessing site we have mapped one amino acid C-terminal from the endogenous site in our in vitro transcription-translation experiments (Fig. 2). The lower molecular weight band is uncharacterized, but presumably is due to a second autoprocessing site given that it is lost upon pepstatin treatment.
We believe the pepstatin-sensitive activity to be the result of alternative plasmepsin autoprocessing. Previous observations have demonstrated that recombinant PM II readily autoprocesses under acidic conditions (14, 15). Kim et al. (16) have extended these studies by demonstrating that recombinant PM II and PM IV undergo autoprocessing in both cis- and trans-catalytic fashions. We also observe in the cell-free assay a minor amount of the lower band present in the absence of inhibitor or in the presence of ALLN (Fig. 1C). This makes sense given that the PM II substrate in this assay is significantly diluted relative to the situation in vivo, conditions that would favor intramolecular autoprocessing. Consistent with this is the finding that ALLN inhibits trans-catalysis more potently than autocatalysis (16).
The Falcipains Are Responsible for the E-64d-sensitive Processing Activity—The plasmepsins traffic through the secretory system to the plasma membrane of the parasite, at which point they appear to incorporate into the membranes of the forming cytostomes that deliver host cell hemoglobin to the food vacuole (7, 8). It is clear that an acidic pH is required for both the E-64d-sensitive processing and the pepstatin-sensitive autoprocessing (9). The neutral pH environment of both the secretory and the cytostomal vesicles (17) limits the location of proplasmepsin processing to the food vacuole. Falcipains 2, 2′, and 3 constitute the predominant food vacuole cysteine protease activity (18) and represent the only characterized cysteine endo-peptidases within this organelle (5, 6). Therefore, the falcipains appear to be the best (only) known candidates for the proplasmepsin maturase.
In our experiments we found that when used in combination with pepstatin, E-64d but not E-64, was able to inhibit PM2 processing (Fig. 1B). This indicates that the cysteine protease processing activity takes place in an intracellular compartment not readily accessible by E-64. Consistent with this, the food vacuole falcipains displayed a decreased sensitivity to E-64 relative to E-64d in our in vivo target analysis experiments (Fig. 4). This result indicates that E-64, as compared with E-64d, does not readily gain access to the food vacuole. This is consistent with the design of E-64d as a membrane-permeant analog of E-64. Studies have shown, however, that E-64 has an inhibitory effect on hemoglobin digestion within the parasite and elicits a “swollen” food vacuole morphology (18). This is probably best explained by the relatively high concentrations (>100 μm) and long incubation times (>10 h) used to produce the effect in these studies compared with our in vivo processing assays.
Through these experiments we propose a model by which the falcipains serve as the endogenous proplasmepsin maturase, and only upon their inhibition, as by E-64d, are alternative autoprocessed forms of PM II generated (Fig. 6). Although both activities are capable of proplasmepsin processing, the falcipains appear to “win out” because of their faster processing kinetics (Fig. 3).
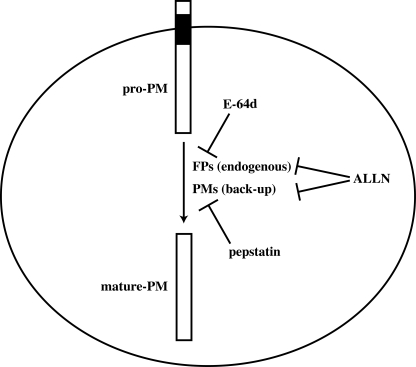
FIGURE 6.
Model of proplasmepsin processing. Upon delivery to the food vacuole, membrane-bound pro-plasmepsin (pro-PM) is processed to its mature, active form (mature-PM) by the E-64d sensitive cysteine protease falcipains (FPs). If, however, falcipain activity is inhibited, then plasmepsins (PMs) can undergo autoprocessing that is sensitive to pepstatin. The previous observation that ALLN (typically referred to as a cysteine protease inhibitor) can block pro-PM processing is explained by its ability to inhibit both the falcipains and the plasmepsins, thus blocking both pathways of pro-PM processing.
The falcipains belong to the papain (C1A) family of cysteine proteases. Published characterization of the substrate specificities for the falcipains reveals differences between FP-2 (and presumably the nearly identical FP-2′) and FP-3 (19), although both enzymes favor substrates with Leu at P2 (20, 21). Consistent with this preference, it is expected that both FP-2 and FP-3 are able to cleave proplasmepsin II at the endogenous cleavage site. That both FP-2 and FP-3 are able to cleave our proplasmepsin peptide, as well as recombinant proplasmepsin II (not shown) is not without precedent because this same analysis did identify peptides that were cleaved with similar kinetics by these enzymes.
Which, if any, of the falcipain enzymes plays a dominant role in proplasmepsin processing in vivo has not been directly addressed in this paper other than our determination that the enzymes cleave the proplasmepsin peptide with an approximate 6-fold difference in catalytic efficiency (kcat/Km = 1.4 × 104 and 2.3 × 103 s–1 m–1 for FP-2 and FP-3, respectively). Also, published data indicate that FP-2′ and FP-3 are expressed later in the erythrocytic cycle of the parasite (21). This would suggest that FP-2 might serve as the dominant proplasmepsin maturase, although the contribution of FP-2′ and FP-3 cannot be ruled out. One possible experiment to test this would be to use the available FP-2 and FP-2′ knock-out cell lines (4, 22) to measure the proplasmepsin processing kinetics of these lines with respect to the parental line. We have attempted this experiment with FP-2 knock-out cells but were unable to detect any measurable difference. It is possible that FP-2′ and/or FP-3 activity was sufficient for processing. These experiments are also challenging given the inherent stage heterogeneity of the parasite cultures.
This work brings into question the idea of a unique proplasmepsin processing activity as a viable drug target. We predict that drugs designed to specifically inhibit the falcipains or the plasmepsins are unlikely to be effective inhibitors of processing activity. Only drugs that inhibit both enzyme families would successfully block proplasmepsin processing. An example of this idea is ALLN, our earlier described maturase inhibitor (7, 9). ALLN can block proplasmepsin processing because it is both a low micromolar inhibitor of the falcipains (not shown) and, surprisingly, a noncompetitive inhibitor of the plasmepsins (our observations and Kim et al. (16)).
We have shown here that pro-PM II can be proteolytically activated by two redundant processing activities and given the conserved processing site, similar processing inhibitor sensitivities, and processing kinetics, we feel that this is likely the case for all food vacuole plasmepsins. Functional redundancy appears to be a common theme in Plasmodium biology. Well documented examples can be found in the multiple invasion pathways of the parasite (23) and the SERA gene family (24). With respect to the plasmepsins, redundancy can also be seen in their contribution, along with the falcipains, to hemoglobin degradation. With the exception of FP-3, single (or multiple) knock-outs of each plasmepsin and falcipain have been successfully generated (22, 25, 26). Furthermore, it has recently been reported that the falcipains appear able to compensate, at least in culture, for the loss of all four food vacuole plasmepsins (27). One interpretation of these results is that these enzymes function by simple mass action to degrade hemoglobin and that gene duplication has served as a mechanism for the parasite to ensure a sufficient level of enzymatic activity. Functional overlap exists among plasmepsin family members and between plasmepsin and falcipain enzyme families. We now provide another level of functional overlap: the ability of the multiple members of the multiple families to activate plasmepsins.
Through the work presented here characterizing proplasmepsin processing, it appears the parasite has struck a balance between specialization and redundancy by maintaining the option to activate the plasmepsins through autoprocessing. To what extent this alternative processing activity is advantageous in the course of natural infection is not understood at this time.
Acknowledgments
We thank Anna Oksman for parasite culturing, Eva Istvan and Jun Liu for helpful discussions, and Ruiguang Ge for assistance with matrix-assisted laser desorption ionization.
This work was supported, in whole or in part, by National Institutes of Health Grants AI-47798 (to D. E. G.) and AI-035800 (to P. J. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ALLN, N-acetyl-l-leucyl-l-leucyl-l-norleucinal; Z, benzyloxycarbonyl; PBS, phosphate-buffered saline; PM, plasmepsin; FP, falcipain.
References
- 1.Snow, R. W., Guerra, C. A., Noor, A. M., Myint, H. Y., and Hay, S. I. (2005) Nature 434 214–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldberg, D. E. (2005) Curr. Top. Microbiol. Immunol. 295 275–291 [DOI] [PubMed] [Google Scholar]
- 3.Klemba, M., and Goldberg, D. E. (2002) Annu. Rev. Biochem. 71 275–305 [DOI] [PubMed] [Google Scholar]
- 4.Liu, J., Istvan, E. S., Gluzman, I. Y., Gross, J., and Goldberg, D. E. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8840–8845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahl, E. L., and Rosenthal, P. J. (2005) Mol. Biochem. Parasitol. 139 205–212 [DOI] [PubMed] [Google Scholar]
- 6.Subramanian, S., Sijwali, P. S., and Rosenthal, P. J. (2007) J. Biol. Chem. 282 24961–24969 [DOI] [PubMed] [Google Scholar]
- 7.Francis, S. E., Banerjee, R., and Goldberg, D. E. (1997) J. Biol. Chem. 272 14961–14968 [DOI] [PubMed] [Google Scholar]
- 8.Klemba, M., Beatty, W., Gluzman, I., and Goldberg, D. E. (2004) J. Cell Biol. 164 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banerjee, R., Francis, S. E., and Goldberg, D. E. (2003) Mol. Biochem. Parasitol. 129 157–165 [DOI] [PubMed] [Google Scholar]
- 10.Greenbaum, D., Medzihradszky, K. F., Burlingame, A., and Bogyo, M. (2000) Chem. Biol. 7 569–581 [DOI] [PubMed] [Google Scholar]
- 11.Gluzman, I. Y., Francis, S. E., Oksman, A., Smith, C. E., Duffin, K. L., and Goldberg, D. E. (1994) J. Clin. Investig. 93 1602–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenbaum, D. C., Baruch, A., Grainger, M., Bozdech, Z., Medzihradszky, K. F., Engel, J., DeRisi, J., Holder, A. A., and Bogyo, M. (2002) Science 298 2002–2006 [DOI] [PubMed] [Google Scholar]
- 13.Singh, N., Sijwali, P. S., Pandey, K. C., and Rosenthal, P. J. (2006) Exp. Parasitol. 112 187–192 [DOI] [PubMed] [Google Scholar]
- 14.Hill, J., Tyas, L., Phylip, L. H., Kay, J., Dunn, B. M., and Berry, C. (1994) FEBS Lett. 352 155–158 [DOI] [PubMed] [Google Scholar]
- 15.Luker, K. E., Francis, S. E., Gluzman, I. Y., and Goldberg, D. E. (1996) Mol. Biochem. Parasitol. 79 71–78 [DOI] [PubMed] [Google Scholar]
- 16.Kim, Y. M., Lee, M. H., Piao, T. G., Lee, J. W., Kim, J. H., Lee, S., Choi, K. M., Jiang, J. H., Kim, T. U., and Park, H. (2006) J. Biochem. (Tokyo) 139 189–195 [DOI] [PubMed] [Google Scholar]
- 17.Klonis, N., Tan, O., Jackson, K., Goldberg, D., Klemba, M., and Tilley, L. (2007) Biochem. J. 407 343–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenthal, P. J., McKerrow, J. H., Aikawa, M., Nagasawa, H., and Leech, J. H. (1988) J. Clin. Investig. 82 1560–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramjee, M. K., Flinn, N. S., Pemberton, T. P., Quibell, M., Wang, Y., and Watts, J. P. (2006) Biochem. J. 399 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shenai, B. R., Sijwali, P. S., Singh, A., and Rosenthal, P. J. (2000) J. Biol. Chem. 275 29000–29010 [DOI] [PubMed] [Google Scholar]
- 21.Sijwali, P. S., Shenai, B. R., Gut, J., Singh, A., and Rosenthal, P. J. (2001) Biochem. J. 360 481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sijwali, P. S., Koo, J., Singh, N., and Rosenthal, P. J. (2006) Mol. Biochem. Parasitol. 150 96–106 [DOI] [PubMed] [Google Scholar]
- 23.Cowman, A. F., and Crabb, B. S. (2006) Cell 124 755–766 [DOI] [PubMed] [Google Scholar]
- 24.McCoubrie, J. E., Miller, S. K., Sargeant, T., Good, R. T., Hodder, A. N., Speed, T. P., de Koning-Ward, T. F., and Crabb, B. S. (2007) Infect. Immun. 75 5565–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, J., Gluzman, I. Y., Drew, M. E., and Goldberg, D. E. (2005) J. Biol. Chem. 280 1432–1437 [DOI] [PubMed] [Google Scholar]
- 26.Bonilla, J. A., Moura, P. A., Bonilla, T. D., Yowell, C. A., Fidock, D. A., and Dame, J. B. (2007) Int. J. Parasitol. 37 317–327 [DOI] [PubMed] [Google Scholar]
- 27.Bonilla, J. A., Bonilla, T. D., Yowell, C. A., Fujioka, H., and Dame, J. B. (2007) Mol. Micro. 65 64–75 [DOI] [PubMed] [Google Scholar]