Abstract
Eukaryotic proteasomes have been reported to cleave only once within polyglutamine tracts and then only after the N-terminal glutamine (Venkatraman, P., Wetzel, R., Tanaka, M., Nukina, N., and Goldberg, A. L. (2004) Mol. Cell 14, 95–104). We have obtained results that directly conflict with that report. In the presence of the proteasome activator PA28γ(K188E) human red cell proteasomes progressively degraded fluorescein-GGQ10RR or fluorescein-HPHQ10RR into small fragments as shown by size exclusion chromatography and mass spectrometry. MALDI-TOF mass spectrometry revealed that proteolytic products arose from cleavage after every glutamine in fluorescein-HPHQ10RR, and mass accuracy rules out deamidation of glutamine to glutamic acid as an explanation for peptide degradation. Moreover, degradation cannot be attributed to a contaminating protease because peptide hydrolysis was completely blocked by the proteasome-specific inhibitors, lactacystin and epoxomicin. We conclude that proteasomes cleave repetitively anywhere within a stretch of ten glutamine residues. Thus our results cast doubt on the idea that mammalian proteasomes cannot degrade glutamine-expanded regions within pathogenic polyQ-expanded proteins, such as Huntingtin.
Trinucleotide expansions, the cause of a number of human diseases, can occur in nontranscribed regions of the genome in transcribed but nontranslated sequences and in coding regions (1). Expansions that result in altered protein structure involve two amino acids, an alanine encoded by GCG and glutamine (Q) encoded by CAG (2, 3). Both polyalanine and polyglutamine expansions lead to the formation of insoluble inclusions, usually nuclear, that often recruit chaperones, ubiquitin, proteasomes, and a variety of other proteins (4, 5). Glutamine tract expansions occur in a disparate set of proteins with no common biochemical function, and the polyglutamine diseases, such as Huntington, Kennedy, and a number of ataxias, usually occur when the glutamine tract exceeds 35 residues in length (6). Whereas most polyQ-expanded proteins are expressed in numerous tissues, the degenerative process is restricted to a subset of neurons specific for each disease (7).
There are a number of hypotheses concerning the mechanism by which polyglutamine (polyQ) expansions cause disease. The most popular hypothesis is that polyQ-expanded regions interfere with transcription (8–10). A number of findings support this hypothesis including the fact that changes in mRNA transcript levels are among the earliest manifestations of polyQ expression (11). Several articles have presented evidence that polyQ expansions interfere with axonal transport (12–14). Several reports suggest that impairment of the ubiquitin-proteasome system may contribute to polyQ pathology (15–17, see Ref. 18 for review). In this context, we proposed that Huntington and the polyQ ataxias could be analogous to lysosomal storage diseases (19). Our proposal was based on the assumption that proteasomes would become less active upon filling with poorly degradable polyQ fragments. We also hypothesized that the proteasome activator PA28γ, which is highly expressed in neurons, could exacerbate polyQ pathology, because it suppresses the proteasome chymotrypsin-like active site (20). The idea that PA28γ impacts polyQ disease was largely discredited by a recent collaborative study with the laboratory of Gillian Bates that examined proteasome activity in the brains of R6/2 mice, a well-studied model of Huntington disease (21). Disease progression was unaffected by the presence or absence of PA28γ. Moreover, we did not observe any difference in proteasome activity in brain extracts from R6/2 and wild-type mice (21). Similar results were obtained by Bowman et al. (22) and by Diaz-Hernandez et al. (23) who found, respectively, that proteasome activity was unimpaired in the retinas of mice expressing polyQ-expanded ataxin or in brain extracts from the HD94 mouse model of Huntington disease.
On the other hand, several investigators have reported that proteasome activity is inhibited in cells or tissues expressing polyQ-expanded proteins (24, 25). It has been reported that proteasomes do not cleave Gln-Gln bonds beyond the first two glutamines in polyQ tracts (26). This latter finding, if correct, would suggest that polyglutamine fragments might well accumulate within proteasomes. However, as shown by the results presented below, proteasomes are capable of cleaving Q-Q bonds anywhere within a tract of 10 contiguous glutamines. Thus, there is little reason to propose that proteasomes are inactivated by the buildup of polyQ fragments in their central chambers.
EXPERIMENTAL PROCEDURES
Materials—Suc-LeuLeuValTyr-MCA and cbz-LeuLeuGlu-βNA were obtained from Sigma; boc-LeuArgArg-MCA was purchased from Peptides International (Louisville, KY). Lactacystin was purchased from CalBiochem (EMD Chemicals Darmstadt, Germany), and epoxomicin from A. G. Scientific (San Diego, CA). The peptide aldehydes, Ac-PGPH-al and Ac-EPFD-al, were gifts from Jennifer Harris of the Burnham Institute for Medical Research (La Jolla, CA). The peptides, Q5, Q3-R2, GG-Q7-RR, GG-Q10-RR, and HPH-Q10-RR, were synthesized at the University of Utah HSC Core Research Facilities. When employed, carboxyfluorescein was coupled to the N terminus of the peptide while still on the resin. Crude peptides were purified on a C18 column (Grace Vydac, Basel, Switzerland) with an acetonitrile gradient containing 0.1% trifluoroacetic acid. Purity was assessed by re-injection on reversed phase liquid chromatography and by mass spectrometry using either MALDI2 or ESI ionization. Lyophilized samples of the peptides were dissolved by weight in either reticulocyte buffer (40 mm Tris, pH 7.8, 10 mm MgCl2, 20 mm KCl, 2 mm dithiothreitol) or in 10 mm ammonium phosphate, pH 7.8. The concentration of non-fluoresceinated peptides was determined by amino acid analysis, and the concentration of fluoresceinated peptides was assayed by comparison to a fluorescein standard curve.
Degradation Assays—The 20 S proteasome was purified from outdated human red blood cells as previously described (27, 28), and recombinant PA28γ and PA28γ(K188E) were purified as outlined in Realini et al. (29). Assays of fluorogenic peptide substrates were carried out in 100-μl reactions in reticulocyte buffer (20 mm Tris, pH 7.8, 5 mm MgCl2, 10 mm KCl, and 1 mm dithiothreitol). Pre-mixes (50 μl) were prepared containing 0.3 μg of proteasome or 0.3 μg of proteasome plus 1 μg of PA28γ(K188E). Reactions were initiated by adding 50 μl of 200 μm fluorogenic peptides in reticulocyte buffer. After incubation, samples were quenched with 200 μl of ETOH, and fluorescence was measured in a PerkinElmer LS5 spectrofluorimeter with excitation/emission at 380/440 nm for MCA substrates and excitation/emission at 335/410 nm for βNA substrates. Assays involving fluoresceinated peptides were equivalent to those just described except that the concentrations of fluorescein-GGQ10RR and fluorescein-HPHQ10RR peptide were 42 μm and 125 μm, respectively, and the proteasome concentration was 1 to 4 μg depending upon the experiment. Also reactions were stopped by freezing on dry ice prior to analysis by gel filtration or mass spectrometry. Sizing chromatography was performed on a 10/30 Superdex Peptide column (GE Healthcare Life Science). Samples (25 μl) were injected onto the column at 0.5 ml per minute in a buffer of 25 mm Tris, pH 7.5 containing 125 mm NaCl. After discarding the initial 10 ml of eluate, 25 1-ml fractions were collected, and their fluorescence was measured at 485 nm excitation and 520 nm emission. Mass spectrometry was carried out in the Proteomics Core Facility of the University of Utah. All of the mass spectral data shown were collected using delayed ion extraction mode on a PerSeptive Biosystems Voyager-DE™ STR MALDI/TOF mass spectrometer. Peptide samples were spotted using the dried-droplet method using a fresh solution of saturated α-cyano-4-hydroxycinnamic acid matrix in 50:50 water/acetonitrile 0.1% trifluoroacetic acid. MALDI spectra were acquired in reflector mode, operating at 10,000 resolution over a mass range from 400 to 5000 Da.
RESULTS
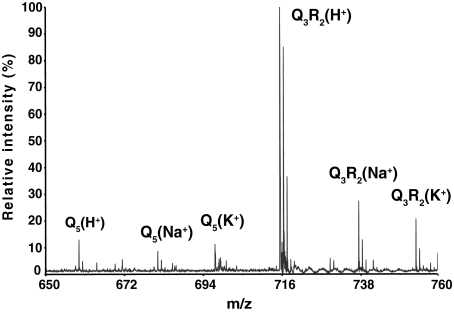
PolyQ Peptides Ionize Poorly and Are Underestimated by Mass Spectrometry—We were surprised by the report from Venkatraman et al. (26) that proteasomes do not cleave beyond the first one or two residues within a polyQ tract, because a previous survey of proteasome specificity showed that glutamines at the P2-P4 positions of fluorogenic tetrapeptides markedly enhanced cleavage after the P1 amino acid (30). Mass spectrometric analysis of the degradation products arising upon proteasomal hydrolysis of the substrates biotin-KKQ10KK and biotin-KKQ20KK constituted some of the evidence, leading to the conclusion that polyQ tracts are resistant to proteasome hydrolysis (26). Reaction mixtures containing rabbit 20 S proteasomes and the proteasome activator PA28αβ were reported to cleave bKKQ10KK at a single site generating bKKQ and Q9KK; with the longer substrate bKKQ20KK ions corresponding to Q17KK and larger peptide products were abundant in the proteasome digest, whereas smaller products were judged to be absent. However, the mass spectrum presented in Fig. 3B of Venkatraman et al. (26) indicated that numerous smaller products might well be present. Furthermore the low ion abundance at masses expected for smaller products could result from proteasomal removal of the readily ionizable lysine residues from each end of the 20-residue glutamine tract in KKQ20KK. If peptides containing only glutamine residues ionize poorly, they would be underestimated by mass spectrometry. We tested this possibility by mass spectrometry of two synthetic peptides, Q5 and Q3R2. When the peptides were mixed at equal molar ratios and subjected to mass spectrometry, the peptide containing arginine residues, Q3R2, was at least 10-fold more abundant in the resulting spectrum (Fig. 1). Moreover, whereas Q3R2 was present mainly as a protonated peptide, the Q5 peptide was almost evenly distributed among sodium, potassium, and protonated ions, thereby making its detection even less efficient. Clearly mass spectrometry does not provide a quantitative measure of products arising upon proteasomal digestion of polyQ peptides. Therefore the low abundance of ions corresponding to small peptides arising from KKQ20KK cannot be taken as evidence for polyQ resistance to proteasome degradation.
FIGURE 1.
Mass spectrum from an equal molar mixture of Q5 and Q3RR. The synthetic peptides Q5 and Q3RR were quantified by amino acid analysis and mixed at a ratio of 1:1 prior to analysis by MALDI-TOF. The spectrum presented above demonstrates that Q3RR ions are present at much higher levels than those from Q5. Note also that protonated Q3RR ions are more prevalent than sodium or potassium Q3RR ions. By contrast Q5 is almost evenly distributed among the three ionic species.
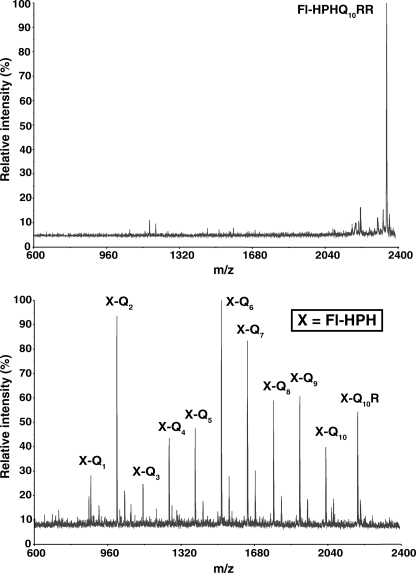
Proteasomes Cleave Anywhere within a Tract of Contiguous Glutamine Residues—Because mass spectrometry is not suitable for quantifying proteolysis, we synthesized a fluorescein (Fl)-labeled peptide, Fl-HPHQ10RR, to assay degradation by size exclusion chromatography. The sequence His-Pro-His was placed next to fluorescein on the assumption that the ionizable His residues would enhance detection of N-terminal products by mass spectrometry, and the Pro residue would suppress proteasome cleavage close to fluorescein. These assumptions were confirmed as shown by the mass spectrum in Fig. 2. When a short proteasomal digest of Fl-HPHQ10RR was assayed by mass spectrometry a number of cleavage products were present, and virtually all contained the Fl-HPH moiety. Peptides resulting from cleavage after each of the ten glutamines were clearly evident in the mass spectrum, and their identification was unequivocal given the accuracy of mass measurement (see Table 1). The absence of products containing only glutamine residues was expected in light of their poor ionization (see Fig. 1). However it was surprising that, with the single exception of Q8R, none of the major products contained arginine. It appears that removal of the two C-terminal basic residues is an early event in the proteolytic digests of Fl-HPHQ10RR.
FIGURE 2.
Mass spectra of Fl-HPHQ10RR and its proteasomal degradation products. The upper panel is the mass spectrum of the substrate Fl-HPHQ10RR; the lower panel shows its degradation products after 30 min of incubation with a 20 S proteasome/PA28γ(K188E) mixture as described under “Experimental Procedures.” Most products are present as H+ and K+ adducts.
TABLE 1.
Mass measurements of Fl-HPHQ10RR degradation products
Masses of the degradation products shown in Fig. 2 are presented in tabular form to emphasize both the high degree of mass accuracy and the presence of multiple ionic species for most products.
| Degradation product | Adduct | Theoretical mass | Observed mass | % Error | Ion intensity |
|---|---|---|---|---|---|
| % | % | ||||
| FL-HPHQ10R | H+ | 2185.18 | 2185.69 | 0.02 | 55 |
| FL-HPHQ10 | K+ | 2067.05 | 2067.77 | 0.03 | 17 |
| H+ | 2029.08 | 2028.85 | 0.01 | 40 | |
| FL-HPHQ9 | K+ | 1938.99 | 1938.76 | 0.01 | 17 |
| H+ | 1901.02 | 1900.80 | 0.01 | 60 | |
| FL-HPHQ8 | K+ | 1810.93 | 1810.71 | 0.01 | 20 |
| H+ | 1772.96 | 1772.75 | 0.01 | 57 | |
| FL-HPHQ7 | K+ | 1682.87 | 1682.66 | 0.01 | 30 |
| H+ | 1644.90 | 1644.70 | 0.01 | 83 | |
| FL-HPHQ6 | K+ | 1554.81 | 1554.51 | 0.02 | 27 |
| H+ | 1516.85 | 1516.64 | 0.01 | 100 | |
| FL-HPHQ5 | K+ | 1426.75 | 1426.46 | 0.02 | 17 |
| H+ | 1388.79 | 1388.57 | 0.01 | 47 | |
| FL-HPHQ4 | K+ | 1298.69 | 1298.41 | 0.02 | 14 |
| H+ | 1260.73 | 1260.53 | 0.02 | 43 | |
| FL-HPHQ3 | H+ | 1132.67 | 1132.45 | 0.02 | 23 |
| FL-HPHQ2 | K+ | 1042.58 | 1042.31 | 0.03 | 22 |
| H+ | 1004.61 | 1004.39 | 0.02 | 94 | |
| FL-HPHQ | H+ | 876.55 | 876.35 | 0.02 | 27 |
| Q8R | H+ | 1199.59 | 1199.64 | 0.00 | 14 |
| Q7R | H+ | 1071.53 | 1071.57 | 0.00 | 15 |
| Q7 | H+ | 915.43 | 915.30 | 0.01 | 15 |
The results in Fig. 2 and Table 1 are in direct conflict with the conclusion that proteasomes cleave only once within polyQ tracts. Conceivably the presence of fluorescein at the N terminus of HPHQ10RR altered substrate degradation by proteasomes such that multiple cleavages in the polyQ tract now occur. To test this possibility, an additional peptide GGQ7RR was synthesized with or without fluorescein at its N terminus. The two peptides were incubated with proteasomes and a mutant proteasome activator PA28γ(K188E), and samples were analyzed by mass spectrometry. Results of this analysis, presented in Table 2, show a clear effect of fluorescein on the distribution of products arising from GGQ7RR. The presence of fluorescein resulted in products lacking C-terminal arginine residues. By contrast, proteasomal degradation of GGQ7RR generated a wider spectrum of peptides some of which contained one or both C-terminal arginine residues. More importantly, the ions listed in Table 2 indicate that hydrolysis occurred after almost every glutamine in the non-fluoresceinated GGQ7R2 substrate. So while an N-terminal fluorescein can affect substrate processing by the proteasome, the enzyme cleaves after multiple glutamines within polyQ tracts even in non-fluoresceinated peptides. Thus, in contrast to the results of Venkatraman et al. (26), we find that human red cell proteasomes cleave anywhere within a stretch of seven or ten contiguous glutamine residues.
TABLE 2.
Mass spectrometric identification of proteasome-mediated degradation products from GGQ7RR and Fl-GGQ7RR
Human red cell 20 S proteasomes (4 μg) and 18 μg of PA28γ (K188E) were incubated with 14.5 μg of Fl-GGQ7RR or 10 μg of GGQ7RR in 50 μl of reticulocyte buffer for 3 h at 37 °C prior to identifying degradation products by mass spectrometry. The observed masses for all the products listed below varied less than 0.02% from their theoretical masses.
|
GGQ7RR
|
Fl-GGQ7RR
|
||
|---|---|---|---|
| Product | % Ion intensity | Product | % Ion intensity |
| GGQ7R | 55 | F1-GGQ7R | 4 |
| GGQ7 | 44 | F1-GGQ4 | 20 |
| GGQ6 | 20 | F1-GGQ3 | 100 |
| GGQ5 | 33 | F1-GGQ2 | 22 |
| GGQ4 | 19 | F1-GGQ | 8 |
| Q6RR | 71 | Q6R | 12 |
| Q3RR | 66 | Q3 | 33 |
| Q2RR | 38 | ||
| Q6R | 55 | ||
| Q4R | 55 | ||
| Q3R | 22 | ||
| Q6 | 37 | ||
| Q5 | 41 | ||
| Q4 | 100 | ||
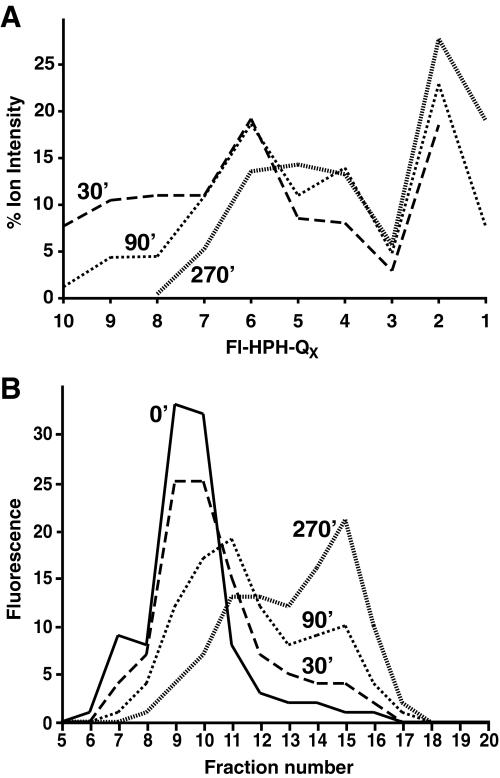
Degradation of Fl-HPHQ10RR and Fl-GGQ10RR Is Markedly Stimulated by the Mutant Proteasome Activator PA28γ(K188E)—To determine whether rates of Fl-HPHQ10RR degradation could be monitored by gel filtration as well as mass spectrometry, the peptide was incubated with 20 S proteasomes and the mutant activator PA28γ(K188E). Samples were taken after 30, 90, or 270 min for analysis by MALDI-TOF or by gel filtration. Relative ion intensities for the proteolytic products, obtained from spectra like that shown in Fig. 1, are plotted in Fig. 3A. Whereas peptides containing 8, 9, or 10 glutamines were still present at 30 min of incubation, they were much less abundant at 90 min and absent by 4.5 h. The disappearance of larger products was accompanied by an increase in smaller peptides, Fl-HPHQ2–4, and the appearance of Fl-HPHQ by 90 min of incubation. Gel filtration profiles of the same reaction mixtures, presented in Fig. 3B, correlate with the mass spectrometric analyses at 90 and 270 min in that there was a substantial and progressive shift to smaller fluoresceinated species eluting in fractions 14–16.
FIGURE 3.
Progressive degradation of Fl-HPHQ10RR by proteasomes. Two additional samples from the reaction that generated the spectrum in Fig. 2 were taken at 90 and 270 min of incubation and analyzed by MALDI-TOF. A, ion frequency of Fl-HPH-peptides containing 1–10 glutamine residues is plotted for samples taken at 30, 90, and 270 min and shows the progressive shortening of peptide products as the reaction proceeds. B, separate timed samples from the same reaction were analyzed by gel filtration on a 10/30 Superdex Peptide column, and the elution profiles show a similar progressive degradation of Fl-HPHQ10RR to small peptides over the 270 min of incubation.
Even though gel filtration underestimated hydrolysis of Fl-HPHQ10RR in the 30-min sample, we employed the column method to analyze the effects of mutant and wild-type proteasome activator PA28γ on degradation of Fl-HPHQ10RR or Fl-GGQ10RR for three reasons. First, gel filtration is more convenient and much less expensive than mass spectrometry. Second, our focus was on proteolysis within the Q tract where peptide bond cleavage generates products small enough to be well separated from substrate on the Superdex peptide column. Third, we were interested in estimating relative activation of proteasomal degradation of polyQ peptides by mutant versus wild-type PA28γ, because faster polyQ degradation by proteasomes might prove therapeutically beneficial (see “Discussion”).
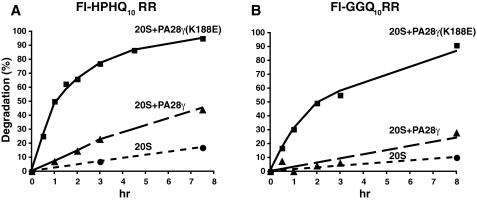
Eukaryotic proteasomes contain seven distinct β-subunits, three of which are catalytically active. Each of the three catalytic subunits exhibits unique substrate specificity, called the chymotrypsin-like (CT), postglutamylpreferring (PGPH), and trypsin-like (T) activities. Whereas PA28αβ increases catalysis by all three subunits, PA28γ only activates the trypsin-like subunit; in fact, wild-type PA28γ suppresses the CT-like active site (29). Previously we isolated a single site mutant of recombinant PA28γ (K188E) that activates all three catalytic subunits (20). Because proteasomal degradation of fluorogenic tetrapeptides with glutamine at the P1 position was markedly stimulated by PA28αβ, but only barely by PA28γ (30), we suspected that PA28γ(K188E) would increase the rate of Fl-HPHQ10RR degradation by proteasomes. As a test of this hypothesis, we incubated the peptide with proteasomes alone or proteasomes plus wild-type or mutant PA28γ, and timed samples were analyzed by gel filtration as described in Fig. 3B. As shown by the results in Fig. 4A, degradation of Fl-HPHQ10RR was increased 10-fold more by PA28γ(K188E) than by wild-type PA28γ; equivalent results were obtained with Fl-GGQ10RR as substrate (Fig. 4B). Although degradation of both substrates was stimulated slightly by PA28γ, this may reflect opening of the “gate” though the proteasome α-ring (31) rather than activation of proteasome β-subunits by PA28γ. In any event, PA28γ(K188E) markedly enhanced cleavage within polyQ tracts, and this finding has implications for the possible treatment of polyglutamine diseases.
FIGURE 4.
The mutant proteasome activator PA28γ(K188E) stimulates degradation of peptides containing polyQ tracts. Fl-HPHQ10RR and Fl-GGQ10RR were incubated with 20 S proteasomes alone (•) or 20 S proteasomes plus PA28γ (▪) or PA28γK188E (▴), and degradation was monitored by the analysis of timed samples on a 10/30 Superdex column as described in the legend to Fig. 3B.
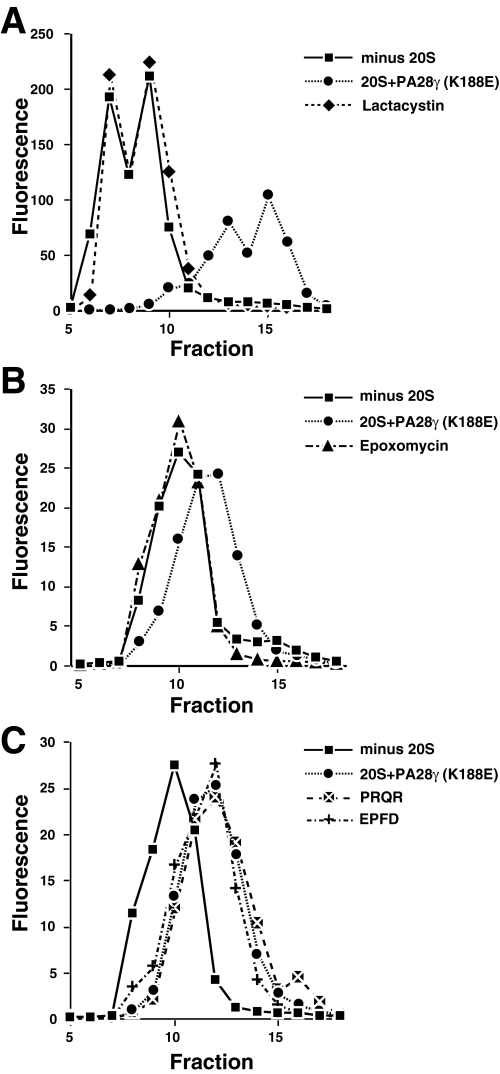
Degradation of Fl-HPHQ10RR and Fl-GGQ7RR Cannot Be Attributed to a Contaminating Protease—The fact that the proteasome activator PA28γ(K188E) markedly increased the rate of polyQ peptide proteolysis (see Fig. 4) is prima facie evidence that the proteasome is the responsible enzyme. But PA28γ(K188E) is a recombinant protein that could be contaminated by an Escherichia coli protease. To test for a possible contaminating protease, we examined the effect of two specific proteasome inhibitors, lactacystin and epoxomicin, on peptide hydrolysis. The gel filtration profile in Fig. 5A shows that degradation of Fl-HPHQ10RR was completely inhibited by lactacystin, and the profile in Fig. 5B demonstrates that Fl-GGQ7RR remained intact in the presence of epoxomicin. Furthermore, mass spectroscopy showed that the unblocked peptide, GGQ10RR, remained intact after 3 h of incubation with lactacystin- or epoxomycin-inhibited proteasomes. Six unblocked fluorogenic di-peptides or amino acid-MCA substrates were not degraded after 90 min of incubation with uninhibited proteasomes. Combined, these results indicate that our proteasome preparation is devoid of contaminating peptidases and provide clear evidence that the fluoresceinated peptides employed in these studies were degraded by the proteasome.
FIGURE 5.
Inhibition of polyQ-peptide degradation by the proteasome inhibitors lactacystin and epoxomicin. Reactions containing 20 S proteasome/PA28γ(K188E) were preincubated with or without proteasome inhibitors for 30 min before adding Fl-polyQ peptide. A, reactions with Fl-HPHQ10RR as substrate were incubated for 6 h before samples were chromatographed on the 10/30 Superdex peptide column. Whereas in the absence of lactacystin Fl-HPHQ7RR was degraded to small fragments (♦), 20 μm of the proteasome inhibitor completely blocked peptide degradation (compare ▪ and •). B, peptide Fl-HPHQ10RR was incubated with 20 S proteasome/PA28γ(K188E) for 3 h in the presence (▴) or absence (♦) of epoxomicin prior to chromatography. Note that epoxomicin completely blocked peptide degradation. C, the peptide Fl-HPHQ7RR was incubated with 20 S proteasome/PA28γ(K188E) in the presence of Ac-PRQR-al (▴) or Ac-EPFD-al (▪) or without any peptide aldehyde (•) for 6 h before chromatography. Note that neither peptide aldehyde inhibited proteolysis of Fl-HPHQ7RR.
Our previous survey of proteasome specificity using a combinatorial library of fluorogenic tetrapeptides demonstrated that, in the presence of PA28αβ, proteasomes readily cleave after glutamine residues, but they remain incapable of cleaving XXXGly-MCA or XXXPro-MCA bonds (30). That survey also led to the development of two peptide aldehydes, Ac-EPFD-al and Ac-PRQR-al, which target enzyme PGPH and T active sites, respectively (see Table 3). As shown in Fig. 5C, neither compound inhibited degradation of Fl-GGQ7R2, indicating that the CT-like active site of the proteasome is mainly responsible for cleavage within polyQ tracts.
TABLE 3.
Specificities of Ac-PRQR-al and Ac-EPFD-al as proteasome inhibitors
Reaction mixtures (50 μl) contained 0.32 μg of 20 S proteasome, 1 μg of PA28γ (K188E), and either reticulocyte buffer alone or 2 μm Ac-PRQR-al, 2 μm Ac-EPFD-al, or 20 μm lactacystin. After incubation at 37 °C for 30 min, 50 μl of a 200 μm solution of the indicated fluorogenic peptide was added to initiate degradation. Duplicate samples were quenched with 200 μl of ethanol at 10 and 30 min, and fluorescence was measured at ex/em 380/440 nm for MCA substrates and ex/em 335/410 for the βNA substrate Cbz-LLE. The measured fluorescence was divided by the fluorescence obtained in the absence of inhibitor to produce % inhibition. A negative value indicates stimulation of peptide hydrolysis in the presence of the inhibitor. Note that whereas lactacystin markedly inhibited hydrolysis of sLLVY-MCA (a substrate for the chymotrypsin-like activity of the proteasome) and bLRR-MCA (a substrate for the trypsin-like subunit), the peptide aldehydes are strikingly specific for just one proteasome active site.
|
Fluorogenic peptide
|
% Inhibition
|
||
|---|---|---|---|
| Ac-PRQR-al | Ac-EPFD-al | Lactacystin | |
| Suc-LLVY-MCA | -32 | -5 | 97 |
| Boc-LRR-MCA | 97 | -22 | 63 |
| Cbz-LLE-βNA | 2 | 93 | 10 |
DISCUSSION
The results presented above provide compelling evidence that proteasomes can cleave multiple times within polyQ tracts. This is apparent from the mass spectrometry data presented in Fig. 2 as well as Tables 1 and 2. Proteasomal degradation of Fl-HPHQ10RR and GGQ7RR produced ions corresponding to peptides generated by hydrolysis after each glutamine in Fl-HPHQ10RR and five of the seven glutamines in GGQ7RR. Equally important, both mass spectrometry (Fig. 3A) and gel filtration (Fig. 3B) demonstrated that degradation was progressive, leading eventually to small peptides that contained only one or two glutamines. We conclude that proteasomes can cleave at multiple sites within a polyQ tract, and they can do so repeatedly. The complete inhibition of peptide breakdown by lactacystin or epoxomicin (Fig. 5, A and B) eliminates the possibility that degradation was due to a contaminating protease.
If glutamine residues in our peptide substrates were to deamidate to glutamates, then peptide degradation might be attributed to the PGPH active site of the proteasome. However, the difference between observed and expected masses for each proteolytic product was generally less than 0.22 Da (Table 1). This high degree of mass accuracy rules out glutamine deamidation as a potential explanation for peptide degradation because replacement of NH by O would increase the mass of products by 1 Da. Furthermore, if proteasome cleavage occurred only after glutamic acid residues generated by deamidation, the responsible catalytic subunit of the proteasome would contain the PGPH active site. Accordingly, one would have expected Ac-EPFD-al to inhibit peptide degradation, but it did not (Fig. 5C).
Our results are in direct conflict with those reported by Venkatraman et al. (26). As mentioned, we believe that poor ionization of peptides containing only glutamine (see Fig. 1) led them to the mistaken conclusion that proteasomes do not cleave within polyQ tracts. However, the discrepancy between our results and theirs might reflect differences in proteasome source or reaction components. Because we both used mammalian proteasomes and because PA28γ(K188E) has activation properties very similar to PA28αβ used in their studies, we do not favor the idea that proteasomes cleave polyQ sequences only under our conditions of assay. Rather, we consider it much more likely that proteasomes are generally capable of degrading polyQ sequences provided they remain soluble. In this regard, we have found that the half-life of ataxin-7(Q86)-GFP is the same as ataxin-7(Q10)-GFP in HEK cells and that both are degraded by proteasomes.3
As noted under the “Results,” PA28αβ and PA28γ differ in their activation properties with PA28αβ activating all three proteasome catalytic subunits, while PA28γ only activates the T-like subunit. PA28αβ and PA28γ also differ in their subcellular locations and organ distribution (32). PA28αβ is cytoplasmic, is enriched in immune cells, and is virtually absent in brain. By contrast, PA28γ is nuclear and is highly expressed in neurons. In addition, PA28γ suppresses the proteasome CT-like subunit that is mainly responsible for cleavage within polyQ tracts (see Fig. 5C). These properties led us to speculate that PA28γ might be detrimental to the clearance of polyQ-expanded proteins, and its high level of expression in brain could account for restricting polyglutamine pathologies to that organ (20). Our recent finding that PA28γ status did not affect disease progression in R6/2 mice (21) argues against this hypothesis. Still the ability of proteasomes to cleave within polyQ tracts and PA28γ(K188E) stimulation of polyQ peptide degradation (Fig. 4) raises the possibility of a proteasome-mediated therapy.
A number of approaches have been proposed as potential therapies for polyglutamine diseases (33, 34). Glutamine-expanded proteins often misfold and form intracellular aggregates making molecular chaperones attractive candidates for ameliorating the polyQ disease state. A vast literature has shown that overexpression of heat shock proteins increases the survival of cells expressing polyQ-expanded proteins (see Ref. 35 for a review). If polyQ-expanded proteins are inherently toxic, as seems to be the case (36, 37), a reduction in their levels should also prove beneficial. Strikingly, RNAi targeting of mRNAs encoding human polyQ-expanded SCA1 or huntingtin in mouse models of these diseases produced dramatic improvement in motor coordination (38, 39). Increasing the degradation of polyQ-expanded proteins should have the same effect. In this regard, both autophagy-mediated lysosomal destruction of polyQ aggregates (40), and proteasome-mediated degradation of polyQ-expanded proteins have been demonstrated (41). The studies presented above show that a mutant PA28γ speeds the degradation of peptides containing polyQ tracts. If these results extend to intact proteins, then a small molecule able to bind to wild-type PA28γ and convert its proteasome activation properties to those of PA28γ(K188E) might speed the clearance of polyglutamine-expanded proteins in neurons.
Acknowledgments
We thank Vicença Ustrell and Carlos Gorbea for helpful suggestions during the course of these studies and for providing assistance in the preparation of the manuscript. We also thank Robert Schackmann for peptide synthesis and Chad Nelson and Parsawar Krishna for help with the mass spectrometry.
This work was supported, in whole or in part, by National Institutes of Health Grant NS042892 (to M. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MALDI, matrix-assisted laser desorption/ionization; TOF, time-of-flight; ESI, electrospray ionization; PGPH, postglutamyl-preferring; T, trypsin-like; CT, chymotrypsin-like; Fl, fluorescein; MCA, 7-amino-4-methylcoumarin; βNA, β-naphthylamide.
V. Ustrell, G. Goellner, G. Pratt, C. Sloan, and M. Rechsteiner, manuscript in preparation.
References
- 1.Orr, H. T., and Zoghbi, H. Y. (2007) Annu. Rev. Neurosci. 30 575–621 [DOI] [PubMed] [Google Scholar]
- 2.Albrecht, A., and Mundlos, S. (2005) Curr. Opin. Genet. Dev. 15 285–293 [DOI] [PubMed] [Google Scholar]
- 3.Paulson, H. L., Bonini, N. M., and Roth, K. A. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 12957–12958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross, C. A., and Poirier, M. A. (2004) Nat. Med. 10 (suppl.), S10–S17 [DOI] [PubMed] [Google Scholar]
- 5.Weydt, P., and La Spada, A. R. (2006) Expert Opin. Therap. Targets 10 505–513 [DOI] [PubMed] [Google Scholar]
- 6.Klein, F. A., Pastore, A., Masino, L., Zeder-Lutz, G., Nierengarten, H., Oulad-Abdelghani, M., Altschuh, D., Mandel, J. L., and Trottier, Y. (2007) J. Mol. Biol. 371 235–244 [DOI] [PubMed] [Google Scholar]
- 7.Brignull, H. R., Moore, F. E., Tang, S. J., and Morimoto, R. I. (2006) J. Neurosci. 26 7597–7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaffar, G., Breuer, P., Boteva, R., Behrends, C., Tzvetkov, N., Strippel, N., Sakahira, H., Siegers, K., Hayer-Hartl, M., and Hartl, F. U. (2004) Mol. Cell 15 95–105 [DOI] [PubMed] [Google Scholar]
- 9.Riley, B. E., and Orr, H. T. (2006) Genes Dev. 20 2183–2192 [DOI] [PubMed] [Google Scholar]
- 10.Helmlinger, D., Tora, L., and Devys, D. (2006) Trends Genet. 22 562–570 [DOI] [PubMed] [Google Scholar]
- 11.Luthi-Carter, R., Strand, A., Peters, N. L., Solano, S. M., Hollingsworth, Z. R., Menon, A. S., Frey, A. S., Spektor, B. S., Penney, E. B., Schilling, G., Ross, C. A., Borchelt, D. R., Tapscott, S. J., Young, A. B., Cha, J.-H. J., and Olson, J. M. (2000) Hum. Mol. Genet. 9 1259–1271 [DOI] [PubMed] [Google Scholar]
- 12.Gunawardena, S., Her, L. S., Brusch, R. G., Laymon, R. A., Niesman, I. R., Gordesky-Gold, B., Sintasath, L., Bonini, N. M., and Goldstein, L. S. (2003) Neuron 40 25–40 [DOI] [PubMed] [Google Scholar]
- 13.Szebenyi, G., Morfini, G. A., Babcock, A., Gould, M., Selkoe, K., Stenoien, D. L., Young, M., Faber, P. W., MacDonald, M. E., McPhaul, M. J., and Brady, S. T. (2003) Neuron 40 41–52 [DOI] [PubMed] [Google Scholar]
- 14.Morfini, G., Pigino, G., and Brady, S. T. (2005) Trends Mol. Med. 11 64–70 [DOI] [PubMed] [Google Scholar]
- 15.Bence, N., Sampat, R., and Kopito, R. (2001) Science 292 1552–1555 [DOI] [PubMed] [Google Scholar]
- 16.Bennett, E. J., Bence, N. F., Jayakumar, R., and Kopito, R. R. (2005) Mol. Cell 17 351–365 [DOI] [PubMed] [Google Scholar]
- 17.Bennett, E. J., Shaler, T. A., Woodman, B., Ryu, K. Y., Zaitseva, T. S., Becker, C. H., Bates, G. P., Schulman, H., and Kopito, R. R. (2007) Nature 448 704–708 [DOI] [PubMed] [Google Scholar]
- 18.Ciechanover, A., and Brundin, P. (2003) Neuron 40 427–446 [DOI] [PubMed] [Google Scholar]
- 19.Goellner, G. M., and Rechsteiner, M. (2003) Int. J. Biochem. Cell Biol. 35 562–571 [DOI] [PubMed] [Google Scholar]
- 20.Li, J., Gao, X., Ortega, J., Nazif, T., Joss, L., Bogyo, M., Steven, A., and Rechsteiner, M. (2001) EMBO J. 20 3359–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bett, J. S., Goellner, G. M., Woodman, B., Pratt, G., Rechsteiner, M., and Bates, G. P. (2006) Hum. Mol. Genet. 15 33–44 [DOI] [PubMed] [Google Scholar]
- 22.Bowman, A. B., Yoo, S. Y., Dantuma, N. P., and Zoghbi, H. Y. (2005) Hum. Mol. Genet. 14 679–691 [DOI] [PubMed] [Google Scholar]
- 23.Diaz-Hernandez, M., Hernandez, F., Martin-Aparicio, E., Gomez-Ramos, P., Moran, M. A., Castano, J. G., Ferrer, I., Avila, J., and Lucas, J. J. (2003) J. Neurosci. 23 11653–11661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seo, H., Sonntag, K. C., and Isacson, O. (2004) Ann. Neurol. 56 319–328 [DOI] [PubMed] [Google Scholar]
- 25.Nishitoh, H., Matsuzawa, A., Tobiume, K., Saegusa, K., Takeda, K., Inoue, K., Hori, S., Kakizuka, A., and Ichijo, H. (2002) Genes Dev. 16 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venkatraman, P., Wetzel, R., Tanaka, M., Nukina, N., and Goldberg, A. L. (2004) Mol. Cell 14 95–104 [DOI] [PubMed] [Google Scholar]
- 27.Hoffman, L., Pratt, G., and Rechsteiner, M. (1992) J. Biol. Chem. 267 22362–22368 [PubMed] [Google Scholar]
- 28.Dubiel, W., Pratt, G., Ferrell, K., and Rechsteiner, M. (1992) J. Biol. Chem. 267 22369–22377 [PubMed] [Google Scholar]
- 29.Realini, C., Jensen, C., Zhang, Z., Johnston, S., Knowlton, R., Hill, C. P., and Rechsteiner, M. (1997) J. Biol. Chem. 272 25483–25492 [DOI] [PubMed] [Google Scholar]
- 30.Harris, J. L., Alper, P. B., Li, J., Rechsteiner, M., and Backes, B. J. (2001) Chem Biol. 8 1131–1141 [DOI] [PubMed] [Google Scholar]
- 31.Whitby, F. G., Masters, E. I., Kramer, L., Knowlton, J. R., Yao, Y., Wang, C. C., and Hill, C. P. (2000) Nature 408 115–120 [DOI] [PubMed] [Google Scholar]
- 32.Rechsteiner, M., Realini, C., and Ustrell, V. (2000) Biochem. J. 345 1–15 [PMC free article] [PubMed] [Google Scholar]
- 33.Di Prospero, N. A., and Fischbeck, K. H. (2005) Nature Rev. 6 756–765 [DOI] [PubMed] [Google Scholar]
- 34.Borrell-Pages, M., Zala, D., Humbert, S., and Saudou, F. (2006) Cell Mol. Life Sci. 63 2642–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muchowski, P. J., and Wacker, J. L. (2005) Nat. Rev. Neurosci. 6 11–22 [DOI] [PubMed] [Google Scholar]
- 36.de Cristofaro, T., Affaitati, A., Feliciello, A., Avvedimento, E. V., and Varrone, S. (2000) Biochem. Biophys. Res. Commun. 272 816–821 [DOI] [PubMed] [Google Scholar]
- 37.Marsh, J. L., Walker, H., Theisen, H., Zhu, Y.-Z., Fielder, T., Purcell, J., and Thompson, L. M. (2000) Hum. Mol. Genet. 9 13025. [DOI] [PubMed] [Google Scholar]
- 38.Xia, H., Mao, Q., Eliason, S. L., Harper, S. Q., Martins, I. H., Orr, H. T., Paulson, H. L., Yang, L., Kotin, R. M., and Davidson, B. L. (2004) Nat. Med. 10 816–820 [DOI] [PubMed] [Google Scholar]
- 39.Harper, S. Q., Staber, P. D., He, X., Eliason, S. L., Martins, I. H., Mao, Q., Yang, L., Kotin, R. M., Paulson, H. L., and Davidson, B. L. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 5820–5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarkar, S., Perlstein, E. O., Imarisio, S., Pineau, S., Cordenier, A., Maglathlin, R. L., Webster, J. A., Lewis, T. A., O'Kane, C. J., Schreiber, S. L., and Rubinsztein, D. C. (2007) Nat. Chem. Biol. 3 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin-Aparicio, E., Yamamoto, A., Hernandez, F., Hen, R., Avila, J., and Lucas, J. J. (2001) J. Neurosci. 21 8772–8781 [DOI] [PMC free article] [PubMed] [Google Scholar]