Abstract
Solubilization of mineralized bone by osteoclasts is largely dependent on the acidification of the extracellular resorption lacuna driven by the vacuolar (H+)-ATPases (V-ATPases) polarized within the ruffled border membranes. V-ATPases consist of two functionally and structurally distinct domains, V1 and V0. The peripheral cytoplasmically oriented V1 domain drives ATP hydrolysis, which necessitates the translocation of protons across the integral membrane bound V0 domain. Here, we demonstrate that an accessory subunit, Ac45, interacts with the V0 domain and contributes to the vacuolar type proton pump-mediated function in osteoclasts. Consistent with its role in intracellular acidification, Ac45 was found to be localized to the ruffled border region of polarized resorbing osteoclasts and enriched in pH-dependent endosomal compartments that polarized to the ruffled border region of actively resorbing osteoclasts. Interestingly, truncation of the 26-amino acid residue cytoplasmic tail of Ac45, which encodes an autonomous internalization signal, was found to impair bone resorption in vitro. Furthermore, biochemical analysis revealed that although both wild type Ac45 and mutant were capable of associating with subunits a3, c, c″, and d, deletion of the cytoplasmic tail altered its binding proximity with a3, c″, and d. In all, our data suggest that the cytoplasmic terminus of Ac45 contains elements necessary for its proper interaction with V0 domain and efficient osteoclastic bone resorption.
Osteoclasts are terminally differentiated multinucleated cells derived from the hematopoietic lineage that specialize in the solubilization of mineralized bone material (1). Upon attachment to the bone surface, the osteoclast undergoes a series of cytoskeletal reorganization and polarization events that culminate in the formation of a unique apical membrane known as the ruffled border. The ruffled border is the “resorptive organelle” of the osteoclast and is formed by the polarized targeting and fusion of acidified intracellular vesicles with the plasma membrane (2–4). These transport vesicles courier acidifying machineries as well as osteolytic enzymes such as, TRACP, cathepsin K, and matrix metalloproteinases (2, 5–7) to the ruffled border membrane domain where each cargo plays a discrete role in the resorptive function. Vacuolar H+-adenosine triphosphatases (V-ATPases)5 that line the ruffled border have long been established to play a vital role in the resorptive process. V-ATPases function to pump H+ into the underlying resorptive lacunae. This elevation in protons results in a highly acidified microenvironment that solubilizes the mineralized component of bone while providing optimal conditions for the degradation of the exposed organic phase by collagenolytic enzymes such cathepsin K (2, 7).
The importance of V-ATPase in osteoclastic bone resorption is exemplified by studies employing specific inhibitors and gene knock-out strategies to impair V-ATPase functions (8–11). Furthermore, mutations in the human TCIRG1 gene, which encodes for a osteoclast specific V-ATPase subunit, a3, results in infantile malignant osteopetrosis, thus substantiating the importance of V-ATPase in osteoclastic bone resorption (12–15). The structural and functional analyses of the V-ATPase complex in osteoclasts might facilitate the development of anti-resorptive agents targeting specifically the osteoclast V-ATPases (16).
V-ATPases belong to a unique class of ATPases responsible for the acidification of intracellular compartments in all eukaryotic cells (17, 18). In phagocytic cells such as macrophages, V-ATPases are known to mediate cytoplasmic pH homeostasis and the acidification of most intracellular organelles. In addition, these macromolecular proton pumps are crucial to a number of fundamental cellular processes, including receptor recycling, protein processing and sorting, and microbial degradation (19, 20). Structurally, V-ATPases has been well modeled and are known to consist of two core functional domains, V1 and V0 (17, 18). The V1 domain is responsible for ATP hydrolysis and is composed of a 570-kDa complex, which is made up of eight different subunits A–H, with a molecular mass of 70–14 kDa, whereas the V0 domain functions as a proton translocation unit located across the limiting membrane.
The V0 domain consists of a 260-kDa integral complex that is made of several subunits with molecular masses of 100–17 kDa (subunit a, d, c/c′, and c″), with six copies of the c/c′ subunits and single copies of a, d and c″ subunits (21). The largest subunit (subunit a) of 100 kDa of the V0 is a transmembrane glycoprotein displaying characteristics of an N-terminal hydrophilic domain and a C-terminal hydrophobic domain with multiple potential transmembrane helices (22). Four isoforms (a1, a2, a3, and a4) of the 100-kDa subunit of the vacuolar proton-translocating ATPase have been identified (23–25). The second largest single subunit (subunit d) of 38 kDa is a hydrophilic protein containing no membrane helices (26) and has been found to be tightly associated with V0 (27). The smallest subunits (subunits c/c′ and c″) of 17–19 kDa are highly hydrophobic proteins with characteristics of proteolipids (18). In addition to the core subunits, a 45-kDa polypeptide, Ac45, has been reported to co-purify with the V0 domain of V-ATPases from bovine adrenal chromaffin granules (28) and is predicted to be oriented toward the luminal side of the complex anchored to the membrane by a single α-helix in the C terminus of the polypeptide. Currently, the interaction of Ac45 with other subunits of V-ATPase complex and its biological function remain to be elucidated.
In the present study, using a subtractive hybridization approach we identified accessory V-ATPase subunit Ac45 in osteoclasts. Ac45 was differentially expressed during osteoclastogenesis and found to localize to the ruffled border and endosomal compartments in polarized osteoclasts. To investigate the potential role of Ac45 in osteoclasts, we expressed a cytoplasmic terminus deletion mutant (Ac45ΔC) that lacks the 26-amino acid internalization signal. Overexpression of Ac45ΔC in osteoclasts showed a dramatic reduction on bone resorption. Furthermore, using immunoprecipitation and bioluminescence resonance energy transfer (BRET) assays, we demonstrate, for the first time, that Ac45 specifically associates with the V0 domain subunits a3, c, and c″. Interestingly, deletion of the C-terminal tail (Ac45ΔC) altered Ac45 association with subunits a3, c″, and d. In all, our studies suggest that the cytoplasmic terminus of Ac45 is required for its proper interaction with V0 domain subunits and plays an important role in osteoclastic bone resorption.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies—Transferrin Alexa Fluor 546, Lysotracker, and rhodamine-conjugated phalloidin were purchased from Invitrogen. Restriction enzymes and reverse transcriptase were purchased from Promega (Sydney, Australia), and DNA polymerase was from GeneWorks (Adelaide, Australia). GST-rRANKL160–318 recombinant proteins were expressed and purified in our laboratory as previously described (29). Anti-GFP rabbit polyclonal antibodies were obtained from Abcam (Sapphire Bioscience Pty. Ltd., Sydney, Australia). Anti-FLAG and anti-c-Myc monoclonal antibodies were purchased from Sigma. A rabbit anti-Ac45 antibody raised against the last 12 amino acids of mouse Ac45 was kindly provided by Dr. E. Jansen (Department of Animal Physiology, University of Nijmegen, The Netherlands). The anti-Ac45 antibody was precleared by absorption with bacterial cell lysates and purified by protein-agarose beads (30). Antibodies against the a3 subunit of the V-type H+-ATPase were raised in guinea pigs and were kindly provided by Dr. Jens C. Fuhrmann (Zentrum fur Molekulare Neurobiologie Hamburg, ZMNH, Universitat Hamburg, Falkenried, Hamburg, Germany) (31). Rabbit polyclonal anti-d2 antibodies were raised against GST-d2 peptide antigen produced in our lab and affinity-purified.
PCR-selected Subtractive Hybridization and Expression of Ac45 by RT-PCR—The cDNA-subtracted library was constructed using RAW264.7 (driver) and RAW264.7-derived osteoclasts (tester) according to the procedures of the Clontech PCR-Selected cDNA subtractive hybridization kit (Clontech). The cDNA sequence of the subtracted cDNA library was determined using BigDye termination reaction mix and automated sequencer (PerkinElmer Life Sciences). To determine the tissue distribution of Ac45 mRNA expression in normal mice, total RNA was isolated from various snap-frozen tissues using RNA-zol solution according to the manufacturer's instructions (Ambion Inc., Austin, TX). For RT-PCR, single-stranded cDNA was prepared from 2 μg of total RNA using reverse transcriptase with an oligo(dT) primer. Cycle-dependent PCR was carried out using 2 μl of each cDNA (94 °C, 40 s; 54 °C, 40 s; and 72 °C, 40 s) with mouse Ac45 primers (forward, 5′-AGATCTACCATGATGGCGGCAACAGT-3′; reverse, 5′-AGATCTTCCACAATCTGGGTCAAAGTGA-3′). Primers fora3 (forward, 5′-GGATCCGAATTCATCATGGGCTCTATGTTC-3′; reverse, 5′-GGATCCTCTAGACTAGTCACTGTCCACAGT-3′), c (forward, 5′-GGATCCGAATTCGACATGGCTGACATCAAG-3′; reverse, 5′-GGATCCTCTAGACTACTTTGTGGAGAGGAT-3′), c″ (forward, 5′-AGGATCCGAATTCATGACGGGGCTGGAGTT-3′; reverse, 5′-AGGATCCTCTAGACTAGTCACCCATCTTCA-3′), d1 (forward, 5′-GGATCCGAATTCATGTCGTTCTTCCCGGA-3′; reverse, 5′-GGATCCTCTAGACTAAAAGATGGGGATGTA-3′), d2 (forward, 5′-GGATCCGAATTCATGCTTGAGACTGCAGAG-3′; reverse, 5′-GGTCTAGATTATAAAATTGGAATGTAGCT-3′), and calcitonin receptor (forward, 5′-TGGTTGAGGTTGTGCCCA-3′; reverse, 5′-CTCGTGGGTTTGCCTCATC-3′) were also used to determine their expression during osteoclast formation. As an internal control, the single-stranded cDNA was PCR-amplified for 25 cycles using 36B4 primers (forward, 5′-TCATTGTGGGAGCAGACA-3′; reverse, 5′-TCCTCCGACTCTTCCTTT-3′).
Construction and Expression of GST-Ac45 Fusion Protein— The mouse Ac45 gene was PCR-amplified using mRNA isolated from mouse osteoclasts with the Ac45 primers (forward, 5′-AGATCTACCATGATGGCGGCAACAGT-3′; reverse, 5′-AGATCTTCCACAATCTGGGTCAAAGTGA-3′). The PCR product was cloned into a pCR 2.1 T/A cloning vector to make pCR2.1-Ac45, and its sequence was confirmed. A 606-bp BamHI and EcoRI restriction fragment was then subcloned into the BamHI and EcoRI sites of the pGEX-3X to make pGEX-3X-Ac45. To express glutathione S-transferase (GST) fusion proteins, plasmid pGEX-3X-Ac45 was transformed into the bacterial strain BL-21. Following growth in Luria-Bertani medium containing 100 μg/ml of ampicillin for 3 h at 30 °C, isopropyl β-d-thiogalactopyranoside was added to a final concentration of 0.1 mm, and the bacterial culture was incubated for a further 4 h at 30 °C. Bacteria were harvested and lysed in a standard SDS sampling buffer (Bio-Rad).
Immunofluorescence and Confocal Analysis—For immunofluorescent staining, RAW264.7 cells or osteoclasts cultured on glass coverslips or dentine slices were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature and washed four times with PBS. Fixed cells were treated with 0.1% of Triton X-y100 for 5 min and washed. The anti-Ac45 antibody was added at final dilution of 1:100 and incubated for 1 h at room temperature. A secondary antibody labeled with fluorescence (Sigma) was used at final dilution of 1:300. For the subcellular localization of Ac45, endocytic or lysosome tracers were added to the RAW264.7 cells. Lysotracker was added to cell culture at a final concentration of 1 μm and incubated for 30 min. Transferrin Alexa Fluor 546 was added to a final concentration of 20 μg/ml and incubated for 30 min. For co-localization of Ac45 and a3, the anti-Ac45 antibody was added at final dilution of 1:100, and the anti-a3 was added at a final dilution of 1:200. Secondary antibodies were labeled with rhodomine for the anti-Ac45 antibody and fluorescence for the anti-a3 antibody, respectively. Fluorescent images were collected on a Bio-Rad MRC 1000/1024 UV laser scanning confocal microscope. High numerical aperture Nikon 10×, 40×, and 60× oil immersion objectives were used in this study. In some experiments, optical sectioning of entire cells at 0.1 μm was acquired using a Z step. The serial optical section stacks from each cell were used for reconstruction of three-dimensional images. For F-actin co-staining, 0.3 μm rhodamine-conjugated phalloidin (Invitrogen) was used.
Western Blot—The proteins were separated on SDS-PAGE gels and then transferred to nitrocellulose by electroblotting as previously described (30). The blots were incubated in 5% dry skim milk in Tris-buffered saline (TBS; 0.05 m Tris, 0.15 m NaCl, pH 7.5) for 1 h and probed with rabbit anti-Ac45 polyclonal antibody, rabbit anti-d2 antibody, mouse anti-α-tubulin antibody, or rabbit anti-GFP antibody at final concentrations of 1:200, 1:1000, 1:2500, or 1:2000 in TBS containing 5% skim milk, respectively. The blots were washed 10 min with TBS three times and incubated with peroxidase conjugated IgG (Sigma) at 1:1000 in TBS containing 5% skim milk. Following three washes with TBS, the antibody reactivity was detected by ECL (Amersham Biosciences) according to the manufacturer's instructions.
Production of Retroviruses—Retroviral vectors pMx-Ac45-IRES-EGFP and pMx-Ac45ΔC-IRES-EGFP were constructed by inserting full-length mouse cDNA of Ac45 and mutated Ac45ΔC, in which the 26-residue cytoplasmic tail (amino acids 437–463) of Ac45 is truncated, into pMx-IRES-EGFP vector (kindly provided by Dr. Kitamura, University of Tokyo). Retrovirus packaging was performed by transfection of the pMx vectors into packaging cell line plat E cells (32). Virus stocks were prepared by collecting the media from cultures 48 h after transfection.
Osteoclastogenesis and Retroviral Transduction—To generate osteoclast precursors, freshly isolated bone marrow cells were cultured for 3 days with 10 ng/ml M-CSF in α-modified essential medium supplemented with 10% fetal calf serum. Adherent cells were used as osteoclast precursors. For retroviral transduction, the osteoclast precursors were transduced with viral supernatants in the presence of polybrene (2 μg/ml). On day 2 of transduction, puromycin (2 μg/ml) was added into the culture to select gene-integrated cells. The cells were then cultured with M-CSF (10 ng/ml) and RANKL (the receptor activator of NF-κB ligand; 100 ng/ml) for 5 or 6 days at 37 °C in an atmosphere of 5% CO2/95% air and were fed every 2 days by replacing half of the spent medium with fresh medium and M-CSF and RANKL. The cells were fixed and stained for TRACP activity to identify osteoclasts. TRACP+ cells containing ≥3 nuclei were scored as osteoclasts or osteoclast-like cells (OCLs). Osteoclastic bone resorption assays were done as previously described (33).
Immunoprecipitation—For co-immunoprecipitation of Ac45 and Ac45ΔC mutant with V0 subunit a3, COS-7 cells were seeded to a density of 2 × 106 in 10-cm Petri dishes and 24 h later were co-transfected with 2 μg each of Ac45-EYFP and Myc-a3 or with Ac45ΔC-EYFP with Myc-a3 using PolyFect transfection reagent (Qiagen). Twenty four hours post-transfection, medium containing transfection complexes were removed and replaced with fresh Dulbecco's modified Eagle's medium and incubated for further 24 h. After 24 h the cells were washed twice in ice-cold 1× PBS prior to lysis with 1 ml of lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, and 1% Nonidet P-40) containing Complete protease inhibitor mixture (Roche Applied Science). The lysates were scraped into 1.5-ml tubes on ice, passed through a 23.5-gauge needle, and cleared by centrifugation at 14000 rpm at 4 °C for 30 min. The lysates were then precleared with incubation with Sepharose G bead slurry (GammaBind G Sepharose; Amersham Biosciences) for 2 h at 4 °C. One milligram of total protein was then incubated at 4 °C overnight with 5 μg of anti-GFP antibody (Abcam; Sapphire Bioscience). Antibody-bound proteins were then captured by incubation with washed (twice with lysis buffer) Sepharose G beads at 4 °C for 2 h. Protein-bound beads were then washed three times with lysis buffer, and proteins were eluted with the addition of 2× SDS sampling buffer containing 2% β-mercaptoethanol. Boiled samples were separated on 10% SDS-polyacrylamide gel followed by overnight transfer to nitrocellulose membrane (Amersham Biosciences). After overnight transfer, the membranes were blocked with 5% (w/v) skim milk powder in TBS-T (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% (v/v) Tween 20) and then probed with anti-Myc (Sigma) antibody diluted 1/1000 in 1% (w/v) skim milk powder in TBS-T. After washing three times with TBS-T, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody diluted 1/5000 in 1% (w/v) skim milk powder in TBS-T. The membranes were then developed using the enhanced chemiluminescence system (Amersham Biosciences).
BRET Assays—PCR-based methodology was used to generate the following constructs for BRET assays. These include pcDNA-Rluc-V-ATPase-a3, pcDNA-Rluc-V-ATPase-d, pcDNA-Rluc-V-ATPase-c, pcDNA-Rluc-V-ATPase-c″, pcDNA-Ac45-EYFP, and pcDNA3.1-Ac45 mutant-EYFP. Primers a3 forward (5′-GGATCCGAATTCATCATGGGCTCTATGTTC-3′) and a3 reverse (5′-GGATCCTCTAGACTAGTCACTGTCCACAGT-3′) were used to make pcDNA-Rluc-V-ATPase-a3. Primers c forward (5′-GGATCCGAATTCGACATGGCTGACATCAAG-3′) and c reverse (5′-GGATCCTCTAGACTACTTTGTGGAGAGGAT-3′) were used to make pcDNA3.1-Rluc-V-ATPase c. Primers c″ forward (5′-AGGATCCGAATTCATGACGGGGCTGGAGTT-3′) and c″ reverse (5′-AGGATCCTCTAGACTAGTCACCCATCTTCA-3′) were used to make pcDNA-Rluc-V-ATPase-c″. Primers d1 forward (5′-GGATCCGAATTCATGTCGTTCTTCCCGGA-3′) and d1 reverse (5′-GGATCCTCTAGACTAAAAGATGGGGATGTA-3′) were used to make pcDNA3.1-Rluc-VATPase d. Primers Ac45 forward (5′-AGATCTACCATGATGGCGGCAACAGT-3′) and Ac45 reverse (5′-AGATCTTCCACAATCTGGGTCAAAGTGA-3′) were used to make pcDNA-Ac45-EYFP. Ac45 forward primer (5′-AGATCTACCATGATGGCGGCAACAGT-3′) and Ac45 mutant reverse primer (5′-GGTACCTGCAGACCATAGGTGAATATG-3′) were used to make pcDNA3.1-Ac45ΔC-EYFP, which expressed Ac45 (amino acids 437–463). Sequencing analysis was used to verify the identity of each construct. BRET assays were performed as previously described (34). In brief, COS-7 cells were transiently transfected in 6-well plates using PolyFect (Qiagen), and the cells were assayed 48 h post-transfection. The transfected cells were detached with 0.05% trypsin/PBS and washed twice in PBS. Approximately 20,000 cells/well were assayed in a 96-well plated. The coelenterazine (h form) (Invitrogen) was added to a final concentration of 5 μm, and readings were collected immediately following this addition using the Mithras LB940 BRET plate reader (Berthold Technologies, Inc., Germany). Repeated readings were taken, and the BRET ratios for the co-expression of Rluc and EYFP constructs were normalized against the BRET ratios for the Rluc expression construct alone.
RESULTS
Identification, Expression, and Localization of Ac45 in Osteoclasts—In an effort to identify differentially expressed genes that are important to osteoclast differentiation and function, we employed a subtractive hybridization approach using mRNA derived from RAW264.7 cells treated in the presence or absence of RANKL for 7 days to form multinucleated osteoclasts (supplemental Fig. S1A). From a total of 198 clones analyzed, one clone was found to encode mouse Ac45 (supplemental Fig. S1, B and C). This sequence has been deposited in the GenBank™ (accession number of AY033882) and is identical to an unpublished version of mouse C7–1 sequence (GenBank™ accession number AB031290), with the exception that it encodes an additional 327 bases at its 3′-untranslated region. The homologues of Ac45 gene have previously been cloned from bovine adrenal medulla (28), the Xenopus intermediate pituitary gland (35), and the rat frontal cortex (36), but no counterpart have been identified in yeast V-ATPases based on the genomic sequencing of Saccharomyces cerevisiae (17).
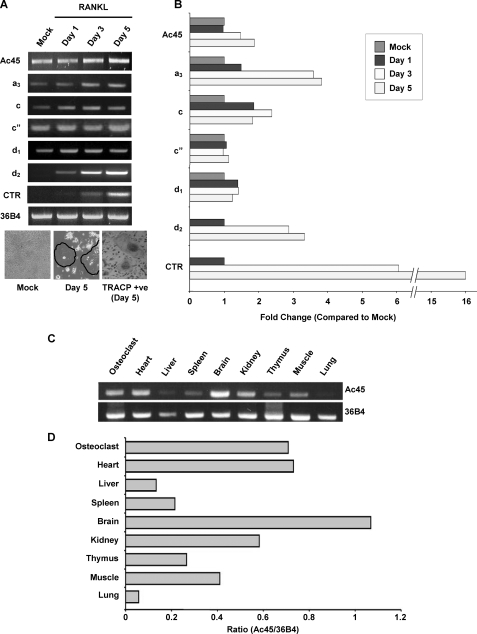
As an initial step to confirm the gene expression of Ac45 in osteoclasts, cycle-dependent PCR was carried out using mouse osteoclast cDNA (supplemental Fig. S1D). To determine the expression of Ac45 during osteoclastogenesis, 30 cycles of PCR amplification within the linear range of amplification was used as compared with other V0 subunits a3, c, c″, d1, and d2 (Fig. 1A). Semi-quantitative RT-PCR analysis using cDNA time course derived from RANKL-stimulated RAW264.7 cells (days 0–5) revealed that Ac45 was up-regulated approximately ∼2-fold (Fig. 1B) during the course of osteoclastogenesis. By comparison, the a3 subunit was found to display ∼4-fold increase, and d2 expression was also highly induced following RANKL stimulation. On the other hand, subunit c″ and d1 appeared to be constitutively expressed in both osteoclasts and their precursor cells (Fig. 1, A and B). In addition, we examined the tissue distribution of Ac45 from selected mouse organs (Fig. 1C). The results revealed that the level of Ac45 mRNA expression was high in the brain, the heart, and osteoclasts followed by kidney, muscle, thymus, spleen, liver, and lung (Fig. 1D). The relative abundance of 36B4 mRNA expression was used as an internal control (Fig. 1, A and C).
FIGURE 1.
A, different V0 subunits expression profiles during osteoclastogenesis. RAW264.7 cells were treated with RANKL (100 ng/ml) for different time periods (0, 1, 3, and 5 days). RT-PCR analysis was carried out using specific primers to Ac45 and V0 subunits including a3, c, c″, d1, and d2. Calcitonin receptor (CTR) and 36B4 primers were used as control for osteoclastogenesis and house keeping gene, respectively. TRACP staining in a parallel experiment was also included with OCLs highlighted with circles. B, the expression of each subunit relative to 36B4 during osteoclastogenesis was expressed as the fold change over mock. C, RT-PCR analysis of Ac45 (upper panel) and 36B4 (lower panel) in various mouse tissues. D, the relative expression of Ac45 in various tissues relative to 36B4.
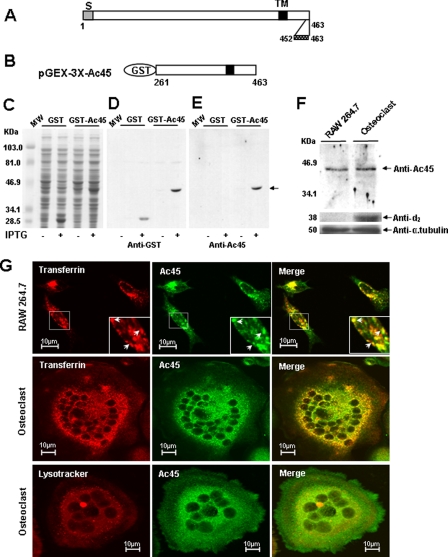
Next, we examined the protein expression and subcellular localization of Ac45 protein using a polyclonal anti-Ac45 antibody that had been raised against a synthetic peptide representing the C terminus (amino acids 452–463) of mouse Ac45. First, to validate the specificity of the anti-Ac45 antibody, a C-terminal region of Ac45 (amino acids 261–463) (Fig. 2, A and B) was expressed as a recombinant GST fusion protein in BL-21 Escherichia coli using the pGEX expression system (Fig. 2C). Western blot analysis showed that the purified anti-Ac45 antibody specifically reacted with the GST-Ac45 (amino acids 261–463) fusion protein but not with GST alone or other bacterial proteins (Fig. 2, D and E). To further confirm the specificity of this antibody to Ac45, RAW264.7 and osteoclast lysates were used to test for endogenous Ac45 expression. A band corresponding to the predicted size (∼45 kDa) was detectable in both RAW264.7 and RAW264.7 cells-derived osteoclast-like cells (Fig. 2F). Furthermore, the protein expression of the d2 subunit was used as a positive control because this subunit is highly up-regulated in osteoclasts as compared with its precursor cells (36).
FIGURE 2.
Expression and localization of Ac45 in OCLs. A and B, expression of GST-Ac45 and Western analysis with an anti-Ac45 antibody. A, molecular structure of the mouse Ac45. S, signal sequence; TM, transmembrane region. C-terminal amino acid residues 452–463 were used for the generation of anti-Ac45 rabbit polyclonal antibody. B, a cDNA fragment encompassing amino acid residues 261–463 of mouse Ac45 was cloned into the pGEX-3X expression vector. C, Coomassie Blue-stained polyacrylamide gel showing the induction and expression of GST-Ac45261–463 fusion proteins in E. coli. MW, molecular mass. D and E, Western blot analysis of GST fusion proteins using a rabbit anti-GST antibody (D) or a rabbit anti-Ac45 antibody (E). F, Western blot analysis of Ac45 in RAW cell-derived osteoclasts and their precursor cells. A band corresponding to a 45-kDa protein was detected using the Ac45 antibody in both in RAW cell-derived osteoclasts and their precursor cells. Protein expression of V-ATPase subunit d2 was used as positive control, and anti-α-tubulin was used as an internal control. G, confocal microscopy analysis of Ac45 protein localization in osteoclastic precursor RAW264.7 cells and in no-resorbing OCLs. RAW264.7 cells or RAW cell-derived osteoclasts were seeded on glass coverslips. Transferrin Alexa Fluor 546 was added to a final concentration of 50 μg/ml and incubated for 30 min. Lysotracker was added to cell culture at 1 μl/ml and incubated for 30 min. The cells were fixed with 4% paraformaldehyde in PBS and stained with purified anti-Ac45 antibody. Fluorescent-labeled secondary antibody was used, and fluorescent images were recorded using a confocal laser scanning microscope (MRC-1000 Bio-Rad). The cells stained with secondary antibody only were used as the negative control and showed no staining signals.
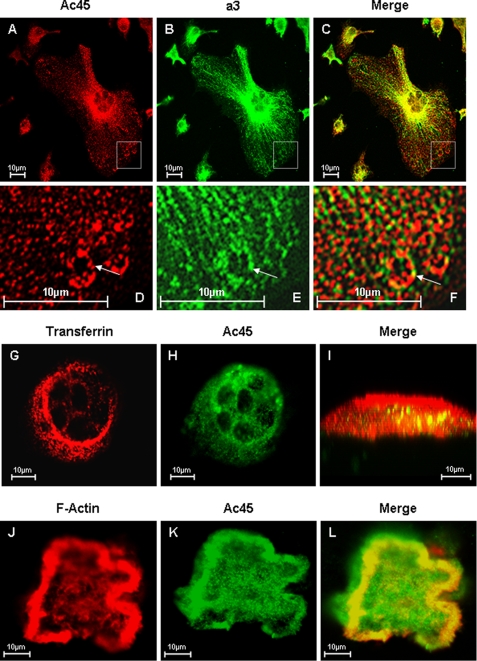
We next examined the subcellular localization of Ac45 in osteoclasts and their mononuclear precursors by immunofluorescent staining. Confocal analyses revealed that Ac45 localizes to punctuate vesicular structures distributed throughout the cytoplasm of both RAW264.7 cells-derived osteoclasts and their precursor cells (Fig. 2G). These structures were often enriched near the perinuclear region of the cell and co-localized with transferrin. Little to no co-localization was observed with Lysotracker, suggesting that Ac45 was preferentially expressed in early rather than late endosomal/lysosomal compartments (Fig. 2G). Importantly, Ac45 showed significant overlap with V-ATPase subunit a3, confirming its association with the V-ATPase complex in osteoclast-like cells (Fig. 3, A–F). In bone resorbing osteoclasts, Ac45 co-localized with transferrin consistent with the localization observed on glass (Fig. 3, G and H). This overlap was largely toward the ruffled border region of the osteoclast (Fig. 3I). In addition, Ac45 showed significant overlap with filamentous actin (F-actin) at the sealing zone, suggesting that Ac45 is targeted toward the ruffled border membrane in polarized osteoclasts (Fig. 3, J–L). Taken together, these findings indicate that Ac45 associates with endocytic compartments within the ruffled border region of osteoclasts during born resorption.
FIGURE 3.
Confocal microscopy analysis of Ac45 co-localization with a3 in OCLs. A–E, osteoclasts cultured on glass coverslips were fixed and permeabilized in 0.1% Triton X-100. The cells were immunostained with primary rabbit Anti-Ac45 and guinea pig anti-a3 for 2 h followed by secondary goat anti-rabbit Alexa Fluor 546 (A) and fluorescein isothiocyanate-conjugated anti-guinea pig (B) for 45 min, respectively. Co-localization was examined following the overlaying of the Ac45 and a3 signals (C). D–F, higher magnification of the boxed area. G–I, osteoclasts seeded onto bone slices for 3 days were incubated with Alexa Fluor 546-conjugated transferrin (G) for 30 min followed by fixation and permeabilization. The cells were immunostained with an anti-Ac45 antibody (H) for 2 h and then with secondary goat anti-rabbit Alexa Fluor 488 for 45 min before confocal analysis. Co-localization between Ac45 and transferrin filled endosomal vesicles were observed in z-x or vertical section imaging (I). J–L, confocal microscopy analysis of Ac45 protein localization in resorbing osteoclasts. Following 3-day culture on bone slices, osteoclasts were fixed and permeabilized in 0.1% Triton X-100. The cells were again immunostained for with an anti-Ac45 antibody (J) and secondary goat anti-rabbit Alexa Fluor 488. F-actin was stained with rhodamine-conjugated phalloidin (K). Ac45 was observed to extensively co-localize with F-actin surrounding the resorption lacunae (L).
Deletion of the Cytoplasmic Tail of Ac45 Impairs Osteoclastic Bone Resorption—It is well accepted that during bone resorption V-ATPases accumulate in the ruffled border membrane of osteoclasts whereby they actively secrete protons to dissolve the underlying mineralized bone. Given the fact that Ac45 also localized to the ruffled border region of osteoclasts, we reasoned that Ac45 might play an important role in the V-ATPase-mediated bone resorption process. To address this possibility we adopted a mutational approach and deleted the 26-amino acid cytoplasmic tail of Ac45 (Ac45ΔC). The 26-residue cytoplasmic tail of Ac45 has been previously shown to contain autonomous targeting information distinct from previously described routing determinants (38). Thus, we hypothesized that the cytoplasmic tail may be important for Ac45 association and function with the V-ATPase complex and therefore osteoclastic bone resorption.
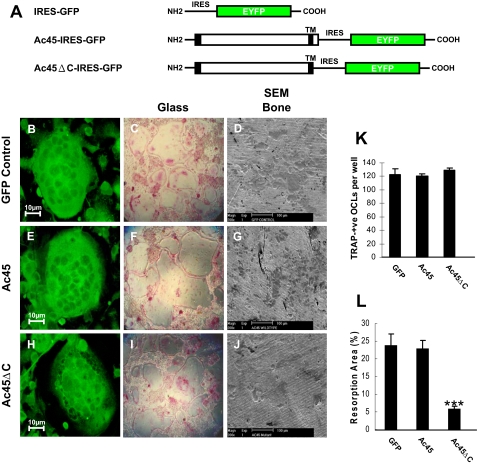
To explore this notion, retroviral constructs (Ac45-IRES-GFP and Ac45ΔC-IRES-GFP) were generated and retroviruses were produced using the plat E packaging cell line (Fig. 4A). Bone marrow cells were subsequently transduced with the virus, and up to 90% of cells were deemed to be GFP positive (+) for each group (data not shown). Following transduction, osteoclast formation was induced through the addition of M-CSF and RANKL. After 7 days, resulting multinucleated cells were fixed and stained for TRACP activity. Approximately 95% of all osteoclasts were GFP (+) in each group as judged by confocal microscopy (Fig. 4, B, E, and H). In addition, quantitative analysis of multinucleated (>3 nuclei/cell) TRACP positive cells yielded no significant difference among the three retrovirally transduced groups (Fig. 4, C, F, I, and K). Next, we examined whether overexpression of Ac45 or Ac45ΔC affected osteoclastic bone resorption. To this end, retrovirally transduced bone marrow cells were cultured on the dentine slices in the presence of M-CSF and RANKL. After 2 weeks, multinucleated cells were subsequently removed, and resorption lacunae were visualized under scanning electron microscopy. As shown in Fig. 4 (D, G, and L), no significant difference in bone resorption was observed in osteoclasts transduced with GFP or wild type Ac45. In contrast, osteoclasts expressing Ac45ΔC displayed a dramatic reduction in bone resorption capacity when compared with GFP and Ac45 groups (Fig. 4, J and L). Notably, resorption was not completely abolished, probably reflecting the presence of untransfected osteoclasts, given that the population is 95% GFP-positive. In addition, to explore the molecular mechanism of impaired osteoclastic bone resorption by Ac45ΔC, we tested whether overexpression of Ac45ΔC in osteoclasts could result in mislocalization of V0 a3 subunit. By using specific anti-a3 antibody, immunostaining analysis showed that there is no observable difference among the EYFP, Ac45-IRES-GFP, and Ac45ΔC-IRES-GFP osteoclasts (supplemental Fig. S2, A–I). Together, these data hint that the cytoplasmic tail of Ac45 may play an important role in osteoclastic bone resorption.
FIGURE 4.
The effect of overexpression of Ac45 or Ac45ΔC on osteoclast formation and osteoclastic bone resorption. A, schematic representation of GFP, Ac45-IRES-GFP, and Ac45ΔC-IRES-GFP constructs. Bone marrow cell derived osteoclasts transduced with GFP (B–D), Ac45-IRES-GFP (E–G), or Ac45ΔC-IRES-GFP (H–J) retroviruses. Transduced cells were seeded on glass coverslips or 96-well culture plates or bone slices in the presence of RANKL and M-CSF. After 7 days on the glass coverslips or the culture dishes, the cells were fixed and processed for confocal analysis (B, E, and H) or TRACP staining (C, F, and I). In 96-well culture plates, TRACP positive OCLs were counted, and no significant difference was observed between all groups (K). The cells on the bone slices were removed after 14 days of culture, and resorptive lacunae were examined by scanning electron microscopy (D, G, and J). Quantitative analysis shows the percentage of bone slice surface occupied by resorption lacunae (L). ***. p < 0.001.
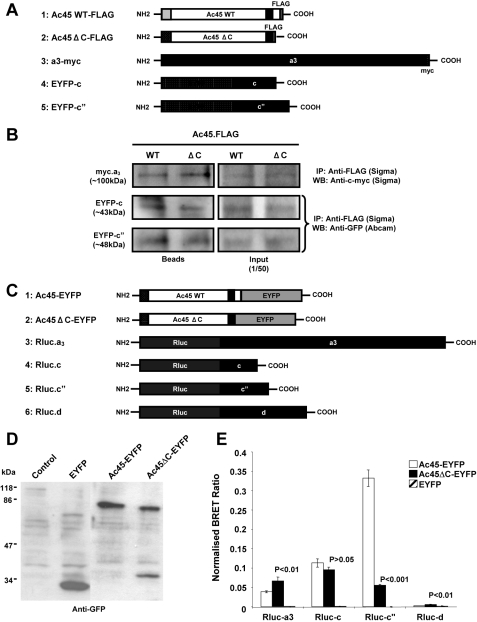
Ac45 Specifically Associates with V0 Subunits of the V-ATPase Complex—Ac45 has been previously reported to co-purify with the V0 sector of the V-ATPase complex (28). However, its precise association with specific subunits of the V0 domain of the V-ATPase complex has not yet been established. Because of Ac45 apparent function in osteoclast bone resorption, we sought to clarify its association with the V-ATPase complex in an effort to shed further insight into its potential role. To this end, we examined the association of Ac45 with other V0 subunits. Having established that Ac45 co-localizes with a3 in osteoclasts, we initially confirmed this physical association by co-immunoprecipitation. For this purpose COS-7 cells were co-transfected with Ac45-FLAG or Ac45ΔC-FLAG and a3-Myc, EYFP-c, or EYFP-c″, and immunoprecipitation was carried out 48 h post-transfection. Following immunoprecipitation (IP), bound a3, c, or c″, proteins was detected using an anti-Myc or anti-GFP antibody, respectively. As shown in Fig. 5B, both Ac45 and Ac45ΔC interacted with a3, suggesting that the C terminus is not required for its association with the a3 subunit. Similarly, the c and c″ subunits were also immunoprecipitated with Ac45 as well as with the C-terminal deletion mutant. These data indicate that both Ac45 wild type and the C-terminal truncation mutant Ac45ΔC is incorporated into the V0 complex. Considering that all these V0 subunits are membrane-bound proteins, we also co-transfected Ac45 or Ac45ΔC with Rab3D gene encoding a membrane bound vesicle protein (3) into COS-7 cells to exclude the possibility of nonspecific interactions shown in these IP results. No interaction was detected between either Ac45 or Ac45ΔC and Rab3D proteins (data not shown), indicating that the IP observed for a3, c, and c″ with Ac45 and Ac45ΔC was a specific interaction.
FIGURE 5.
A, schematic representation of Ac45-FLAG, Ac45ΔC-FLAG, a3-cMyc, EYFP-c, and EYFP-c″ constructs used for immunoprecipitation assays. B, immunoprecipitation analysis showing that Ac45 and Ac45ΔC interact with a3, c, and c″, respectively. C, schematic representation of Ac45-EYFP, Ac45ΔC-EYFP, Rluc-a3, Rluc-c, Rluc-c″, and Rluc-d1 constructs used for BRET assays. D, Western blot analysis of COS-7 cells transfected with EYFP, Ac45-EYFP, or Ac45ΔC. Transfected COS-7 cells were lysed in standard sample buffer, and cell lysates (30 μl) were subjected to analysis by SDS-PAGE and immunoblotting with anti-GFP rabbit polyclonal antibody. E, BRET assays showing the levels of interaction of Ac45 with V0 subunits a, c, c″, and d1. COS-7 cells co-expressing Ac45-EYFP or Ac45ΔC-EYFP and either Rluc tagged a3, c, c″, or d1 subunits were assayed following the addition of coelenterazine and the BRET ratio relative to the Rluc alone expressing cells determined (normalized BRET ratio). Similarly, the normalized BRET ratio was determined for cells co-expressing EYFP with each of the Rluc-tagged subunits. The data represent the means from six independent experiments ± S.E. p values < 0.05 indicate significant differences between Ac45-EYFP and EYFP alone control. p values < 0.05 indicate the significant differences between Ac45 mutant and wild type (WT) Ac45.
Although IP provides important confirmation of protein-protein interaction, it does not quantitatively reflect the protein association in living cells. Therefore we examined this interaction in live COS-7 cells by BRET analysis. EYFP-tagged Ac45 or Ac45ΔC and Rluc-tagged V0 a3, c, c″, or d fusion proteins (Fig. 5C) were co-expressed in COS-7 cells, and the BRET ratios were determined by measuring the Rluc activity and EYFP activity. As shown in Fig. 5E, the ratios of Ac45-EYFP and Rluc-a3 were significant higher compared with EYFP control, again confirming Ac45 association with the a3 subunit. In addition, Ac45-EYFP wild type was found to associate with Rluc-c and Rluc-c″, but not Rluc-d. The BRET signals were highest between Ac45 and the c″ subunit followed by subunits c and a3, suggesting that Ac45 is most tightly associated with the c″ subunit of the V0 domain. Like Ac45-EYFP, Ac45ΔC-EYFP also interacted with V0 subunit c; however, this association is markedly lower with subunit c″ as compared with wild type Ac45, suggesting that the cytoplasmic tail of Ac45 maybe part of a specific binding element that is required for efficient interaction with subunit c″, hence a reduction but not total attenuation of interaction between the two subunits. Furthermore this reduced association of Ac45ΔC with the c″ subunit may also account for the impaired resorptive activity of osteoclasts overexpressing Ac45ΔC. Interestingly, however, Ac45ΔC showed a slight increase in BRET ratio with a3 and d1 as compared with wild type. This enhancement may reflect conformational changes in Ac45 because of a deletion at its C terminus. Again to test for specificity of the BRET assay, EYFP-Rab3D was used as a control. EYFP-Rab3D did not interact with any of the Rluc-tagged V0 subunits (data not shown).
DISCUSSION
Generation of a continuous proton gradient across the osteoclastic ruffled border domain is critical to its resorptive function (2, 5). Previous studies have shown that various V-ATPase subunits play important roles in osteoclast differentiation and function (9, 24, 39). Targeted disruption of the ATP6I gene in mouse that encodes the osteoclast specific a3 subunit of the vacuolar proton pump, termed OC116, confers a severe osteopetrotic phenotype because of the inability of the osteoclast-like cells to carry out resorption lacunae acidification (9). Furthermore, mutations in the TCIRG1 gene, which encodes for the human a3 subunit, have been reported in patients affected by infantile malignant osteopetrosis, a heterogeneous autosomal recessive disorder of bone metabolism (12–15). In addition, recent study has shown that V0 subunit d2 is important for osteoclast fusion (37). Although these studies highlight the importance of V-ATPase activity in osteoclast function, the precise role(s) of the individual subunits that comprises the V-ATPase complex in osteoclast differentiation and function remains to be elucidated.
In this study, using a subtractive hybridization approach, we identified accessory V-ATPase subunit Ac45 in osteoclasts. Furthermore, Ac45 was found to be differentially expressed during osteoclastogenesis, albeit at a lesser degree than that of V-ATPase subunits a3 and d2. By immunofluorescence, Ac45 was found to co-localize with pH-dependent transferrin, consistent with previous reports indicating that V-ATPases are enriched on endocytic compartments (18, 40). This finding is also in line with previous localization studies of Ac45 in the Xenopus (38). In addition, Ac45 was found associate with the ruffled border and sealing zone of resorbing osteoclasts as evidenced by its significant co-localization with the a3 V-ATPase subunit and F-actin. These findings are consistent with the notion that V-ATPases are transported to the ruffled border via actin filaments (2, 41–43). Studies by Nakamura et al. (42) demonstrated that V-ATPases associate with the detergent-insoluble actin cytoskeleton of osteoclast and that this interaction is crucial for osteoclast function (42). In addition, V-ATPases have been shown to bind to actin filaments and to be transported to the polarized ruffled border membrane by actin-myosin II contraction (41). More recently, V1 subunits B and C have also been shown to directly associate with filamentous actin in osteoclasts (44, 45). Whether Ac45 targeting is also actin-dependent will be the focus of future investigations.
Knock-out of specific V-ATPase subunits commonly result in embryonic lethality (46–49), pointing to an essential role for the V-ATPases in early embryonic development. Targeted disruption of the Ac45 gene in mice has been reported to inhibit blastocyst development (50), indicating that Ac45 is indispensable for cell functioning and survival. It has previously been shown that the cytoplasmic tail of Ac45 contains autonomous targeting information that is capable of mediating endocytosis of Ac45 from the cell surface to vacuolar structures (38). Therefore in an effort to investigate the potential function of Ac45 in osteoclasts, we overexpressed a deletion mutant of Ac45 lacking the C-terminal domain in osteoclasts and examined its effect on bone resorption. Strikingly, overexpression of Ac45ΔC was found to dramatically reduce osteoclastic bone resorption, indicating that the C-terminal domain is an important requirement for osteoclastic function. Unfortunately because of the lack of appropriate reagents, at this stage we are unable to determine whether the deletion of the C-terminal domain impairs the targeting of Ac45 to the ruffled border or the proton translocation of the V-ATPase itself. Immunostaining analysis using specific antibody to V0 subunit a3 suggests that this C-terminal deletion of Ac45 seemed not affect the intracellular localization of a3. This may indicate a more direct role of the C-terminal truncation mutant of Ac45 itself in the V-ATPase function in osteoclasts. Although further investigation comparing the V-ATPase activity of wild type and C-terminal truncation mutant Ac45 will need to be carried out to draw any conclusion, our data support a role for Ac45 in the resorptive process of the osteoclasts.
Previous studies indicate that Ac45 co-purifies with the V0 domain of the V-ATPase complex in bovine chromaffin granules (28). However, the precise localization and subunit(s) association of Ac45 within the V-ATPase complex remains unclear. Ultrastructural analysis of the V-ATPase complex has previously suggested that Ac45 may represent a globular density orientated toward the luminal side V0 domain subcomplex (51). This proposed structure was found to be associated and centered in the asymmetric ring composed of six c subunits and one c″ subunit, with the a subunit seen as peripheral density next to ring of c/c″ subunits (51). In this study, our immunoprecipitation data showed that both Ac45 and Ac45ΔC co-precipitated with these V0 subunits in vitro. The IP data indicate that both the wild type Ac45 and the C-terminal truncation mutant Ac45ΔC can interact with the V0 subunits and are incorporated into the V0 subcomplex. A recent study suggested that tagged a3 subunit did not exhibit the corresponding localization as the authentic protein (24), raising the concern that tagged V-ATPase subunits may cause misfolding or mislocalization. However, other groups have demonstrated that GFP-tagged a3 and other a isoforms localize to the proper intracellular compartments (52, 53). Moreover, studies have used epitope-tagged V-ATPase subunits to rescue the various defective phenotypes caused by knock out of the corresponding V-ATPase subunits in yeast and fly (54) and osteoclasts (37) as well as in immunoprecipitation assays for the identification of interacting V-ATPase subunits (55–59).
V-ATPases are large multisubunit complexes where their correct assembly into functional units requires dedicated assembly factors for orchestrating the assembly events within the endoplasmic reticulum (60). In yeast, several endoplasmic reticulum assembly factors, including Vma12, Vma21, and Vma22, are responsible for V0 domain assembly, an event that occurs independently from V1 assembly event (60, 61). However, at present, the mammalian homologues of these chaperones remain to be identified. In addition, how different V0 subunits associate under physiological conditions remains unclear. Although our data suggest that Ac45 is incorporated into the V0 complex through its association with subunits a3, c, and c″, the fact of an incomplete co-localization between Ac45 and a3 could not conclude that Ac45 is a functional essential subunit of the osteoclastic V-ATPases.
Nevertheless, using BRET analysis, we demonstrated that Ac45 is in close proximity with V0 subunits c″> c > a3 > d. These findings are in line with the notion that Ac45 is tightly affiliated with the proteolipid c/c″ ring proposed by Wilkens and Forgac (62) based on their three-dimensional structure analysis of the V0 domain. The high BRET signal for subunit c″ may reflect the close association of Ac45 within this ring. Interestingly deletion of the c-terminal of Ac45 (Ac45ΔC) dramatically decreased its association with c″. Significant reduction, albeit not complete inhibition, in the interaction between Ac45ΔC and c″ suggests that the C-terminal domain may only account for part of the Ac45-c″ interacting domain. Additional interacting elements within Ac45ΔC may enable c″ to remain in close proximity, although not as efficient as the wild type Ac45. In addition, the deletion of the C-terminal of Ac45 also altered the affinity for subunits a3 and d, suggesting that the loss of the C-terminal may also alter the conformation of Ac45, thus increasing its proximity and association with subunits a3 and d. Future investigations will focus on elucidating the precise Ac45 structural domains required for V-ATPase subunit interactions.
Previous modeling studies have demonstrated that the asymmetric protein ring (c, c′, c″, and the C terminus of a) of the V0 domain exists with two small openings on the luminal side and one large opening on the cytoplasmic side (62). A globular protein and two cross-linked and elongated proteins have been predicted to cover the central pore of luminal side and cytoplasmic opening, respectively. In the same study, Ac45 has been suggested to be a mass located and covering the luminal side, whereas the N terminus of a3 and d is localized in a way that covers the gap on the cytoplasmic side of asymmetric proteolipid ring (62). In addition, the rotation of the proteolipid ring relative to the a subunit drives the active transport of protons across the limiting membrane, an event of which is precisely manipulated by conformational rearrangements of the subunits (a and d) and the c/c″ proteolipid rotary ring (60, 62). The three-dimensional resolution of this model of the structure of V0 domain, together with our BRET results, suggests that the C terminus of Ac45 may play an important role in covering the luminal opening of the proteolipid ring and is required for the correct conformational rearrangement of V0 domain during proton translocation. Taken as such, the C-terminal deletion of Ac45, although it does not affect the assembly of the V0 domain, may actually impair the V0 rotary mechanism, thus perturbing the stability and efficiency of the V-ATPase complex. This may account for the reduction, albeit not complete inhibition, of osteoclastic bone resorption observed in vitro.
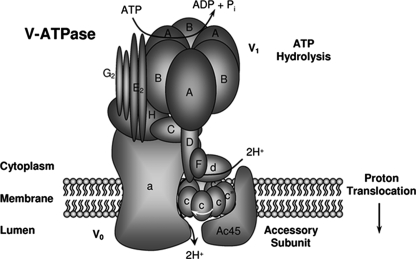
Overall, based on these findings together with the previously predicted model of the V0 domain (18), we proposed a revised structural model of Ac45 with other subunits of the V0 domain (Fig. 6). Ac45 appears to closely associate with the c″ subunit through its C-terminal tail, whereas the Ac45 N-terminal region, which has been predicted to be located within the lumen (51), may serve to anchor Ac45 with a3. According to current models, rotational catalysis requires relative rotation of the c/c″ proteolipid ring relative to the a subunit. The interaction between Ac45 and c″, as modeled in Fig. 6 may modulate rotation of the c/c″ ring and rotational catalysis. Truncation of the C-terminal tail of Ac45 results in an altered association configuration with the V0 complex that might account for malfunctions of V-ATPase during osteoclastic bone resorption.
FIGURE 6.
Schematic diagram of the predicted localization and close proximity of the Ac45 with other V0 subunits in the V-ATPase complex.
Supplementary Material
Acknowledgments
We thank Drs. Lin Huang and Karin Kroeger for expert technical assistance.
This work was supported by the National Health and Medical Research Council. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: V-ATPase, vacuolar H+-adenosine triphosphatase; BRET, bioluminescence resonance energy transfer; EYFP, enhanced yellow green fluorescent protein; Rluc, Renilla luciferase; TRACP, tartrate-resistant acid phosphatase; PBS, phosphate-buffered saline; GST, glutathione S-transferase; RT, reverse transcriptase; GFP, green fluorescent protein; PBS, phosphate-buffered saline; TBS, Tris-buffered saline; M-CSF, macrophage colony-stimulating factor; OCL, osteoclast-like cell; IP, immunoprecipitation.
References
- 1.Teitelbaum, S. L. (2000) Science 289 1504-1508 [DOI] [PubMed] [Google Scholar]
- 2.Blair, H. C., Teitelbaum, S. L., Ghiselli, R., and Gluck, S. (1989) Science 245 855-857 [DOI] [PubMed] [Google Scholar]
- 3.Pavlos, N. J., Xu, J., Riedel, D., Yeoh, J. S. G., Teitelbaum, S. L., Papadimitriou, J. M., Jahn, R., Ross, F. P., and Zheng, M. H. (2005) Mol. Cell. Biol. 25 5253-5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao, H., Laitala-Leinonen, T., Parikka, V., and Vaananen, H. K. (2001) J. Biol. Chem. 276 39295-39302 [DOI] [PubMed] [Google Scholar]
- 5.Baron, R., Neff, L., Louvard, D., and Courtoy, P. J. (1985) J. Cell Biol. 101 2210-2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baron, R. (1989) Anat. Rec. 224 317-324 [DOI] [PubMed] [Google Scholar]
- 7.Vaananen, H. K., Zhao, H., Mulari, M., and Halleen, J. M. (2000) J. Cell Sci. 113 377-381 [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee, D., Chakraborty, M., Leit, M., Neff, L., Jamsa-Kellokumpu, S., Fuchs, R., and Baron, R. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 6257-6261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, Y. P., Chen, W., Liang, Y., Li, E., and Stashenko, P. (1999) Nat. Genet. 23 447-451 [DOI] [PubMed] [Google Scholar]
- 10.Sundquist, K. T., and Marks, S. C., Jr. (1994) J. Bone Miner. Res. 9 1575-1582 [DOI] [PubMed] [Google Scholar]
- 11.Xu, J., Feng, H. T., Wang, C., Yip, K. H., Pavlos, N., Papadimitriou, J. M., Wood, D., and Zheng, M. H. (2003) J. Cell. Biochem. 88 1256-1264 [DOI] [PubMed] [Google Scholar]
- 12.Frattini, A., Orchard, P. J., Sobacchi, C., Giliani, S., Abinun, M., Mattsson, J. P., Keeling, D. J., Andersson, A.-K., Wallbrandt, P., Zecca, L., Notarangelo, L. D., Vezzoni, P., and Villa, A. (2000) Nat. Genet. 25 343-346 [DOI] [PubMed] [Google Scholar]
- 13.Kornak, U., Schulz, A., Friedrich, W., Uhlhaas, S., Kremens, B., Voit, T., Hasan, C., Bode, U., Jentsch, T. J., and Kubisch, C. (2000) Hum. Mol. Genet. 9 2059-2063 [DOI] [PubMed] [Google Scholar]
- 14.Michigami, T., Kageyama, T., Satomura, K., Shima, M., Yamaoka, K., Nakayama, M., and Ozono, K. (2002) Bone 30 436-439 [DOI] [PubMed] [Google Scholar]
- 15.Scimeca, J.-C., Quincey, D., Parrinello, H., Romatet, D., Grosgeorge, J., Gaudray, P., Philip, N. A., and Georges F. Carle, F. (2003) Hum. Mutat. 21 151-157 [DOI] [PubMed] [Google Scholar]
- 16.Xu, J., Cheng, T., Feng, H. T., Pavlos, N. J., and Zheng, M. H. (2007) Histol. Histopathol. 22 443-454 [DOI] [PubMed] [Google Scholar]
- 17.Stevens, T. H., and Forgac, M. (1997) Annu. Rev. Cell Dev. Biol. 13 779-808 [DOI] [PubMed] [Google Scholar]
- 18.Forgac, M. (1999) J. Biol. Chem. 274 12951-12954 [DOI] [PubMed] [Google Scholar]
- 19.Grinstein, S., Nanda, A., Lukacs, G., and Rotstein, O. (1992) J. Exp. Biol. 172 179-192 [DOI] [PubMed] [Google Scholar]
- 20.Swallow, C. J., Grinstein, S., Sudsbury, R. A., and Rotstein, O. D. (1993) J. Cell. Physiol. 157 453-460 [DOI] [PubMed] [Google Scholar]
- 21.Arai, H., Terres, G., Pink, S., and Forgac, M. (1988) J. Biol. Chem. 263 8796-8802 [PubMed] [Google Scholar]
- 22.Perin, M. S., Fried, V. A., Stone, D. K., Xie, X. S., and Sudhof, T. C. (1991) J. Biol. Chem. 266 3877-3881 [PubMed] [Google Scholar]
- 23.Nishi, T., and Forgac, M. (2000) J. Biol. Chem. 275 6824-6830 [DOI] [PubMed] [Google Scholar]
- 24.Toyomura, T., Oka, T., Yamaguchi, C., Wada, Y., and Futai, M. (2000) J. Biol. Chem. 275 8760-8765 [DOI] [PubMed] [Google Scholar]
- 25.Oka, T., Murata, Y., Namba, M., Yoshimizu, T., Toyomura, T., Yamamoto, A., Sun-Wada, G. H., Hamasaki, N., Wada, Y., and Futai, M. (2001) J. Biol. Chem. 276 40050-40054 [DOI] [PubMed] [Google Scholar]
- 26.Nelson, N. (1992) J. Exp. Biol. 172 149-153 [DOI] [PubMed] [Google Scholar]
- 27.Adachi, I., Puopolo, K., Marquez-Sterling, N., Arai, H., and Forgac, M. (1990) J. Biol. Chem. 265 967-973 [PubMed] [Google Scholar]
- 28.Supek, F., Supekova, L., Mandiyan, S., Pan, Y. C., Nelson, H., and Nelson, N. (1994) J. Biol. Chem. 269 24102-24106 [PubMed] [Google Scholar]
- 29.Xu, J., Tan, J. W., Huang, L., Gao, X. H., Laird, R., Liu, D., Wysocki, S., and Zheng, M. H. (2000) J. Bone Miner. Res. 15 2178-2186 [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., 18.12-18.15, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 31.Lange, P. F., Wartosch, L., Jentsch, T. J., and Fuhrmann, J. C. (2006) Nature 440 220-223 [DOI] [PubMed] [Google Scholar]
- 32.Morita, S., Kojima, T., and Kitamura, T. (2000) Gene Ther. 7 1063-1066 [DOI] [PubMed] [Google Scholar]
- 33.Yip, K. H., Feng, H., Pavlos, N. J., Zheng, M. H., and Xu, J. (2006) Am. J. Pathol. 169 503-514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kroeger, K. M., Hanyaloglu, A. C., Seeber, R. M., Miles, L. E., and Eidne, K. A. (2001) J. Biol. Chem. 276 12736-12743 [DOI] [PubMed] [Google Scholar]
- 35.Holthuis, J. C., Jansen, E. J., Schoonderwoert, V. T., Burbach, J. P., and Martens, G. J. (1999) Eur. J. Biochem. 262 484-491 [DOI] [PubMed] [Google Scholar]
- 36.Hung, H., Tsai, M. J., Wu, H. C., and Lee, E. H. (2000) Brain Res. Mol. Brain Res. 75 330-336 [DOI] [PubMed] [Google Scholar]
- 37.Lee, S. H., Rho, J., Jeong, D., Sul, J. Y., Kim, T., Kim, N., Kang, J. S., Miyamoto, T., Suda, T., Lee, S. K., Pignolo, R. J., Koczon-Jaremko, B., Lorenzo, J., and Choi, Y. (2006) Nat. Med. 12 1403-1409 [DOI] [PubMed] [Google Scholar]
- 38.Jansen, E. J., Holthuis, J. C., McGrouther, C., Burbach, J. P., and Martens, G. J. (1998) J. Cell Sci. 111 2999-3006 [DOI] [PubMed] [Google Scholar]
- 39.Laitala-Leinonen, T., and Vaananen, H. K. (1999) Antisense Nucleic Acid Drug Dev. 9 155-169 [DOI] [PubMed] [Google Scholar]
- 40.Nelson, N. (1992) J. Bioenerg. Biomembr. 24 407-414 [DOI] [PubMed] [Google Scholar]
- 41.Lee, B. S., Gluck, S. L., and Holliday, L. S. (1999) J. Biol. Chem. 274 29164-29171 [DOI] [PubMed] [Google Scholar]
- 42.Nakamura, I., Takahashi, N., Udagawa, N., Moriyama, Y., Kurokawa, T., Jimi, E., Sasaki, T., and Suda, T. (1997) FEBS Lett. 401 207-212 [DOI] [PubMed] [Google Scholar]
- 43.Toyomura, T., Murata, Y., Yamamoto, A., Oka, T., Sun-Wada, G. H., Wada, Y., and Futai, M. (2003) J. Biol. Chem. 278 22023-22030 [DOI] [PubMed] [Google Scholar]
- 44.Vitavska, O., Wieczorek, H., and Merzendorfer, H. (2003) J. Biol. Chem. 278 18499-18505 [DOI] [PubMed] [Google Scholar]
- 45.Holliday, L. S., Lu, M., Lee, B. S., Nelson, R. D., Solivan, S., Zhang, L., and Gluck, S. L. (2000) J. Biol. Chem. 275 32331-32337 [DOI] [PubMed] [Google Scholar]
- 46.Dow, J., Davies, S., Guo, Y., Graham, S., Finbow, M., and Kaiser, K. (1997) J. Exp. Biol. 200 237-245 [DOI] [PubMed] [Google Scholar]
- 47.Inoue, H., Noumi, T., Nagata, M., Murakami, H., and Kanazawa, H. (1999) Biochim. Biophys. Acta 1413 130-138 [DOI] [PubMed] [Google Scholar]
- 48.Oka, T., and Futai, M. (2000) J. Biol. Chem. 275 29556-29561 [DOI] [PubMed] [Google Scholar]
- 49.Sun-Wada, G.-H., Murata, Y., Yamamoto, A., Kanazawa, H., Wada, Y., and Futai, M. (2000) Dev. Biol. 228 315-325 [DOI] [PubMed] [Google Scholar]
- 50.Schoonderwoert, V. T., and Martens, G. J. (2002) Mol. Membr. Biol. 19 67-71 [DOI] [PubMed] [Google Scholar]
- 51.Wilkens, S., Vasilyeva, E., and Forgac, M. (1999) J. Biol. Chem. 274 31804-31810 [DOI] [PubMed] [Google Scholar]
- 52.Dettmer, J., Hong-Hermesdorf, A., Stierhof, Y. D., and Schumacher, K. (2006) Plant Cell 18 715-730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wassmer, T., Kissmehl, R., Cohen, J., and Plattner, H. (2006) Mol. Biol. Cell 17 917-930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du, J., Kean, L., Allan, A. K., Southall, T. D., Davies, S. A., McInerny, C. J., and Dow, J. A. (2006) J. Cell Sci. 119 2542-2551 [DOI] [PubMed] [Google Scholar]
- 55.Hurtado-Lorenzo, A., Skinner, M., El Annan, J., Futai, M., Sun-Wada, G. H., Bourgoin, S., Casanova, J., Wildeman, A., Bechoua, S., Ausiello, D. A., Brown, D., and Marshansky, V. (2006) Nat. Cell Biol. 8 124-136 [DOI] [PubMed] [Google Scholar]
- 56.Ohira, M., Smardon, A. M., Charsky, C. M. H., Liu, J., Tarsio, M., and Kane, P. M. (2006) J. Biol. Chem. 281 22752-22760 [DOI] [PubMed] [Google Scholar]
- 57.Sambade, M., and Kane, P. M. (2004) J. Biol. Chem. 279 17361-17365 [DOI] [PubMed] [Google Scholar]
- 58.Norgett, E. E., Borthwick, K. J., Al-Lamki, R. S., Su, Y., Smith, A. N., and Karet, F. E. (2007) J. Biol. Chem. 282 14421-14427 [DOI] [PubMed] [Google Scholar]
- 59.Sun-Wada, G.-H., Yoshimizu, T., Imai-Senga, Y., Wada, Y., and Futai, M. (2003) Gene (Amst.) 302 147-153 [DOI] [PubMed] [Google Scholar]
- 60.Forgac, M. (2007) Nat. Rev. Mol. Cell. Biol. 8 917-929 [DOI] [PubMed] [Google Scholar]
- 61.Nishi, T., and Forgac, M. (2002) Nat. Rev. Mol. Cell. Biol. 3 94-103 [DOI] [PubMed] [Google Scholar]
- 62.Wilkens, S., and Forgac, M. (2001) J. Biol. Chem. 276 44064-44068 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.