Abstract
The NADPH oxidases (Noxs) are a family of superoxide-generating enzymes implicated in a variety of biological processes. Full activity of Nox1, -2, and -3 requires the action of a Rac GTPase. A direct regulatory interaction of Rac with Nox2 has been proposed as part of a two-step mechanism for regulating electron transfer during superoxide formation. Using truncation analysis of Rac binding to the cytoplasmic tail of Nox2, along with peptides derived from this region in cell-free assays, we identify a Rac interaction site within amino acids 419–430 of Nox2. This region is required for binding Rac2 but not p47phox or p67phox cytosolic regulatory factors. A cell-permeant version of the peptide encompassing amino acids 419–430 specifically inhibits NADPH oxidase activation in intact human neutrophils. Mutational analysis of the putative Rac-binding site revealed specific residues, particularly Lys-421, Tyr-425, and Lys-426, individually required for Rac-dependent NADPH oxidase activity that are conserved in the Rac-regulated Nox1, Nox2, and Nox3 enzymes but not in Nox4 or Nox5. Mutation of the conserved residues in the Rac-binding site of Nox1 also result in the loss of Rac-dependent activity. Our data identify a functional Rac interaction site conserved in Rac-dependent Noxs and support a direct regulatory interaction of Rac GTPases to promote activation of these NADPH oxidases.
The phagocyte NADPH oxidase plays an important role in host innate immune responses to infection. Through its ability to catalyze the formation of superoxide anion via the one-electron reduction of molecular oxygen, the phagocyte NADPH oxidase initiates the formation of a variety of reactive oxygen species (ROS)2 that participate in the killing and destruction of ingested microorganisms (reviewed in Refs. 1, 2). Over the past several years, homologs of the leukocyte NADPH oxidase, termed Nox(s), have been identified in nonphagocytic cells. There are currently seven Nox family members, including Nox1–5 and Duoxs (dual oxidase) 1 and 2 (3–5). With the identification of these nonphagocyte Noxs has come the recognition that the generation of ROS by Nox has important roles in intracellular signaling in addition to host defense. Indeed, roles of Nox enzymes in the regulation of biological and pathological processes range from hearing and balance (Nox3), blood pressure regulation, inflammation, cell growth, and cancer (Nox1), angiogenesis and cellular differentiation (Nox4), and innate immunity in the respiratory and gut epithelium, as well as thyroid hormone synthesis (Duox1/2) (3).
The NADPH oxidase of phagocytic leukocytes, Nox2, is the best characterized oxidase system in terms of regulation, and serves as a prototype for understanding the control of the nonphagocyte Noxs. The redox center of the system is a heterodimeric flavocytochrome b558 composed of two integral membrane proteins, Nox2 (also known as gp91phox) and p22phox. The transfer of electrons from cytosolic NADPH to molecular oxygen to form superoxide anion is catalyzed by this Nox2/p22phox complex (1, 2). Assembly of the system and activation of electron transfer is controlled, however, by the action of cytosolic regulatory subunits, including p40phox, p47phox, p67phox, and Rac2 GTPase in human leukocytes (1, 2). Indeed, genetic deficiency in humans of Nox2 or any one of the regulatory oxidase components results in chronic granulomatous disease (CGD), a disorder of innate immunity characterized by severe, recurrent bacterial and fungal infections (6, 7).
The molecular events associated with NADPH oxidase assembly and activation have been partially established (reviewed in Refs. 8, 9); p40phox and p47phox are adapters containing protein-protein interaction PXXP and Src homology 3 motifs, as well as lipid/membrane-binding phox motifs. Although critical for the membrane assembly of the NADPH oxidase in response to cellular stimuli (10), these adapter proteins are not absolutely required for intrinsic activity of the in vitro system (11, 12). In contrast, p67phox and Rac GTPase are absolutely required for Nox2 activity. p67phox contains an “activation domain” that is necessary to stimulate electron transfer by Nox2 (13). In addition, p67phox contains a series of tetratricopeptide repeats that mediate its interaction with Rac GTPases (14, 15). Rac GTPase (Rac2 in human neutrophils (16, 17); Rac1 in human monocytes (18)) is required for activity of Nox2 in vivo, as evidenced by the lack of stimulated NADPH oxidase activity in the Rac2 knock-out mouse (19). In adherent leukocytes, Rac2 appears to act as the critical “molecular switch” for activating the NADPH oxidase (20). Although it has been proposed that Rac GTPases solely act to position the activation domain of p67phox on cytochrome b558 for regulation of electron transfer, there is also significant evidence that Rac itself interacts directly with the cytochrome b558 to catalyze the initial electron transfer step from NADPH to Nox2-bound FAD (reviewed in Ref. 21). In prior studies we provided evidence, using changes in the fluorescence of Rac2-N-methylanthraniloyl-GppNHp, for a direct interaction of Rac2 with partially purified cytochrome b558 (22), and we showed that GST-Rac1 or -Rac2 specifically bound cytochrome b558 from neutrophil membrane extracts (23). However, whether there is a direct regulatory interaction of Rac GTPase with Nox2 remains controversial (21, 24).
Similar to the phagocyte Nox2 system, recent evidence also indicates that the closely homologous Nox1 and Nox3 oxidases of nonphagocytic cells also require Rac GTPases for optimal activity (24–27). However, regulation by Rac GTPase does not appear to be required for the function of Nox4, Nox5, or the Duox family members (28–30). Studies to date have implicated the interaction of Rac1 with the NoxA1 regulatory component (the p67phox-homolog) as the means by which Nox function is regulated by Rac1 GTPase. However, the possible interaction of Rac1 directly with Nox1 or Nox3 has not been determined.
Here we use deletion analysis, site-directed mutagenesis, and in vitro/in vivo activity assays to identify a Rac-binding regulatory site on Nox2. This site is conserved on the Rac-dependent NADPH oxidases 1 and 3 but not on other Nox family members known to be insensitive to Rac GTPases. Peptides derived from the putative Rac interaction domain, as well as point mutations within the Rac binding region in the Nox proteins, disrupt Rac binding and Rac-dependent Nox2 and Nox1 activity in intact cell systems. Our results support a paradigm in which Rac GTPase directly interacts with Nox proteins to regulate ROS formation.
EXPERIMENTAL PROCEDURES
Materials—Phorbol 12-myristate 13-acetate (PMA), fMet-Leu-Phe (fMLP), horseradish peroxidase, luminol, cytochrome c (horse heart, type VI), p-iodonitrotetrazolium (INT), FAD, and NADPH were obtained from Sigma. ATP and GTPγS were from Roche Applied Science. Endofree plasmid maxi kit was purchased from Qiagen. Geneticin, isopropyl 1-thio-β-d-galactopyranoside, and molecular weight markers for SDS-PAGE were from Invitrogen. Nitrocellulose sheets for Western blotting were purchased from Amersham Biosciences. Green fluorescent protein antibody (A-11122) was purchased from Molecular Probes. Rabbit polyclonal anti-p67phox antibody (number 3958) was kindly provided by Dr. Sergio Catz (The Scripps Research Institute, La Jolla). Rac2 (R786), p47phox (R039), and Myc antibodies were generated in-house (31, 32). Rac1 monoclonal antibody (number 23A8) was from Upstate. A polyclonal antibody specific for Nox1 was a gift from Dr. David Lambeth (Emory University). Monoclonal antibodies 7D5, 44.1, and 54.1, specific for Nox2/gp91phox, were kindly provided by Dr. Mark Quinn (Montana State University). All Nox2 peptides and TAT epitope (YGRKKRRQRRR) peptides (33) were synthesized, purified, and sequence confirmed by the TSRI Peptide Core facility.
Expression and Purification of Recombinant Proteins—The recombinant cytosolic domain of Nox2/gp91phox (aa 290–570) and truncations thereof were expressed and purified as a GST fusion protein in Escherichia coli using standard isolation protocols. Rac2 proteins were also expressed and purified as GST fusion proteins in E. coli, but the GST tag was cleaved with thrombin prior to use in in vitro binding assays, per the manufacturer's directions. Purified Rac2 proteins were quantified by the binding of [35S]GTPγS and preloaded with guanine nucleotides as described previously (34, 35).
Circular Dichroism and Fluorescence Spectroscopy—For spectral analysis of the GST-gp91phox fusion constructs, the isolated proteins (in PBS) were centrifuged at 15,000 × g for 30 min at 4 °C, and the resulting protein concentrations were determined by absorption spectroscopy (λ280 based on the primary sequence using ProtParam). Far-UV CD measurements were collected at room temperature on a Jasco-J710 spectropolarimeter in 1-mm quartz cuvettes using protein concentrations from 50 to 120 μg/ml and the following instrument parameters: wavelength, 240–200 nm; bandwidth, 1 nm; scan rate, 50 nm/min; response time, 0.25 s; five acquisitions. The resulting CD spectra were smoothed and subtracted from buffer alone, and data were processed with software provided by the manufacturer. The secondary structure content of GST alone (in the absence of the C-terminal linker region that is encoded in the recombinant protein) was obtained on line using the Protein Data Bank file 1UA5. Fluorescence spectroscopy was conducted using a Quanta-Master QM-1 fluorometer from Photon Technologies at room temperature with the excitation monochromator set at 295 nm and intrinsic tryptophan fluorescence monitored from 305 to 400 nm. For fluorescence measurements, isolated proteins were diluted to 500 nm final in PBS or PBS, 3 m guanidine hydrochloride and incubated for at least 15 min at room temperature prior to analysis.
Pulldown Assays—2 μg of Rac2 protein (preloaded with either GTPγS or GDP) was incubated with 2 μg of GST-cytosolic domain of gp91phox (Nox2) or an equivalent amount of GST protein and 30 μl of glutathione-Sepharose beads in 0.5 ml of binding buffer (25 mm Tris, pH 7.4, 50 mm NaCl, 5 mm MgCl2, and protease inhibitor mixture) for 2 h with constant inversion at 4 °C. The complexes bound on the glutathione-Sepharose beads were washed with 1 ml of wash buffer (25 mm Tris, pH 7.4, 50 mm NaCl, 5 mm MgCl2, and 1% Nonidet P-40) three times and then eluted with 30 μl of Laemmli sample buffer prior to 12% SDS-PAGE and Western blot analysis using polyclonal Rac2 (R786) antibody at 1:1,000 dilution (23). A similar protocol was followed with p47phox or p67phox.
Cell Culture, Transient Transfection, and Western Blot–COS-p22 cells (a COS-7 cell line that stably expresses the phagocytic NADPH oxidase component p22phox (36)) were maintained in low glucose Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. For transfection, the cells were plated in 10-cm plates (Falcon) at 1.2 × 106 cells/ml on the day before transfection. Lipofectamine Plus (Invitrogen) was used to transfect 6 μg of the desired cDNA following the manufacturer's instructions. 21–24 h after transfection, the cells were harvested by trypsinization and lysed in RIPA buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1%, Nonidet P-40, 0.25% sodium deoxycholate, 1 mm EDTA) with protease inhibitor mixture (Sigma). Proteins (30 μg) were resolved by 12% SDS-PAGE and transferred to nitrocellulose membranes using the semi-dry electrophoretic transfer cell (Bio-Rad) at 8 V for 2 h. Following blocking with 3% bovine serum albumin, blots were probed using specific antibodies at the indicated dilutions (anti-p47phox antibody at 1:10,000; anti-p67phox antibody at 1:5,000; anti-Rac1 antibody at 1:1,000).
HEK293 cells were maintained in high glucose Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. For transfection, the 293 cells were plated in 6-well plates (Falcon) at appropriate density, grown overnight, and then transfected by using Lipofectamine 2000 following the manufacturer's instructions. 24 h after transfection, cells were washed twice in ice-cold PBS and then lysed in RIPA buffer with protease inhibitor mixture (Sigma). Proteins (20 μg) were resolved by 10% SDS-PAGE and transferred to polyvinylidene difluoride membrane using the semi-dry electrophoretic transfer cell (Bio-Rad) at 15 V for 1 h. Following blocking with nonfat dry milk (5%), blots were probed using specific antibodies at appropriate dilutions. Proteins were detected using standard Western blotting and visualized by ECL. The blots were stripped and re-probed as necessary.
Site-directed Mutagenesis of Nox2 and Nox1 Proteins—All mutations in Nox2 and Nox1 were created using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The sequences of all constructs were confirmed by DNA sequencing.
Detection of Nox Protein Expression—5 × 106 of COS-p22 cells were incubated with 1 μg of monoclonal antibody 7D5 (37, 38) or control monoclonal antibody 54.1 that recognizes the cytosolic domain of Nox2/gp91phox (39). After washing with HBSS three times, the cells were incubated with Alexa Fluor® 488 goat anti-mouse IgG (H+L) (Molecular Probes, Eugene, OR), and then flow cytometric analysis (FACSCalibur, BD Biosciences) was performed to detect cell surface protein expression. The expression of Nox2 and Nox1 proteins was also verified by Western blot using monoclonal antibody 54.1 for Nox2 detection (1:500 dilution) and Nox1-specific polyclonal antibody for Nox1 detection (1:2,000 dilution).
Cell-free Assay for Measurement of Electron Transfer—Human neutrophil membrane fractions (GSP) and neutrophil cytosolic fractions (GSS) were prepared as described (40). The cytochrome c reduction assay was as described previously (22) For determination of the rate of FAD reduction, 10 μg of purified neutrophil membrane and 100 μg of purified neutrophil cytosol were used for each reaction, as in Ref. 22. Iodonitrotetrazolium violet (INT) (100 μm, final), GTPγS (10 μm, final), FAD (10 μm, final), and SDS (90 μm, final) were then added to each reaction (22). Control wells also contained diphenyliodonium (2 μm, final). After the reaction mixture was incubated for 5 min at 25 °C, the reaction was initiated by the addition of nicotinamide adenine dinucleotide phosphate, the reduced form of NADPH (400 μm, final). The rate and extent of INT reduction was monitored for 8 min at 490 nm using a Versamax microplate reader (Molecular Devices). Initial rates were used to calculate Vmax values using Softmax Pro software (Molecular Devices).
Whole Cell Assay for Superoxide Production—Neutrophils were isolated from healthy human venous blood as in Ref. 41. 4 × 106 neutrophils were incubated with 100 μm cytochrome c in 200 μl of KRGH buffer (25 mm Hepes, pH 7.4, 1.2 mm KH2PO4, 118 mm NaCl, 4.7 mm KCl, 1 mm MgSO4, 1mm CaCl2, 5.5 mm dextrose) for 5 min at 37 °C. Superoxide production was induced with fMet-Leu-Phe (5 × 10–6 m) or PMA (2 μg/ml). For experiments using TAT peptides, the neutrophils were preincubated with the indicated concentration of each TAT peptide for 15 min at 37 °C prior to assay.
For COS-p22 cells, 2.5 × 105 cells were incubated with 50 μg/ml horseradish peroxidase and 72 μm luminol in 250 μl of HBSS buffer with calcium and magnesium (Invitrogen). The production of reactive oxygen species was monitored by chemiluminescence (42) upon the injection of PMA to the final concentration of 0.26 μg/ml (see below). For HEK293 cells, 2 × 105 cells in HBSS with calcium and magnesium were mixed with 250 μm luminol plus 1 unit of horseradish peroxidase in a 200-μl total volume in each well. Luminescence was quantified 30 min after the addition of the mixture by using a 96-well plate luminometer (Microplate LB 96V, Berthold), recording data every minute for 1 h.
RESULTS
Localization of Rac Binding to the Cytoplasmic Domain of Nox2 by Deletion Analysis—We have previously provided evidence for the direct interaction of Rac2 GTPase with Nox2 by showing changes in the fluorescence of N-methylanthraniloyl-GppNHp-labeled Rac2 GTPase upon interaction with partially purified cytochrome b (22), as well as by demonstrating the specific binding of GST-Rac1 or Rac2 to intact cytochrome b from human neutrophil lysates (23). In support of these results, we established that an expressed GST fusion protein containing aa 290–570 of the Nox2 C terminus, predicted to be the cytoplasmic portion of the Nox2 (or gp91phox) subunit, is capable of specifically binding Rac2 (Fig. 1B). Binding was detected with both Rac1 and Rac2 and was largely independent of the nucleotide state of the GTPase (not shown). These results confirm what we have previously observed with the full-length proteins (22, 23).
FIGURE 1.
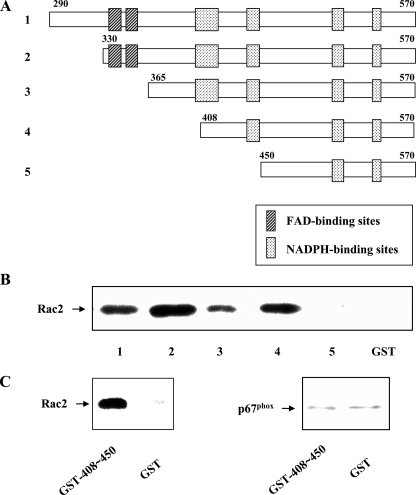
Deletional analysis identifies a Rac2 GTPase binding region on the Nox2 cytoplasmic tail. A, schematic diagram showing the series of truncated Nox2 cytoplasmic tail (aa 290–570) constructs used for the Rac2 binding studies. Predicted FAD and NADPH binding regions are indicated. B, binding of Rac2 to the indicated Nox2 cytoplasmic tail construct was determined as described under “Experimental Procedures.” Binding activity was lost upon deletion of aa 408–450 (construct 5). Binding to GST alone was minimal. Results shown are representative of more than three experiments. C, GST fusion protein containing aa 408–450 of Nox2 is sufficient to bind Rac GTPase (left panel) but is unable to bind p67phox (right panel) nor p47phox (not shown). Representative of n = 2 experiments.
To narrow down the region involved in the binding of Rac GTPase to the cytoplasmic domain of Nox2, we generated a series of deletion constructs in which increasingly larger N-terminal portions of the cytoplasmic domain were removed (see Fig. 1A). Using these purified fusion protein constructs in pulldown assays, we observed that Rac2 binding was maintained until the loss of the segment spanning aa 408–450, whereupon Rac binding was lost (Fig. 1B). To provide a more detailed characterization of the GST-gp91phox fusion constructs used in this study, the isolated proteins were examined by both CD and fluorescence spectroscopy. Characterization of the serially truncated GST-gp91phox constructs used in Fig. 1B demonstrated far-UV CD spectra (see Fig. 4D) that were qualitatively similar, and intrinsic tryptophan fluorescence spectra that demonstrated a similar red-shift from ∼337 nm in phosphate-buffered saline to ∼348 nm in the presence of 3 m guanidine hydrochloride (data not shown). These spectral data support the hypothesis that loss of Rac binding was thus not simply because of structural changes in the cytoplasmic domain constructs, as there was no evidence for extensive protein unfolding as the C-terminal tail was sequentially deleted (43, 44). These data thus suggest that the aa 408–450 region is critical for the interaction of Nox2 with Rac2.
FIGURE 4.
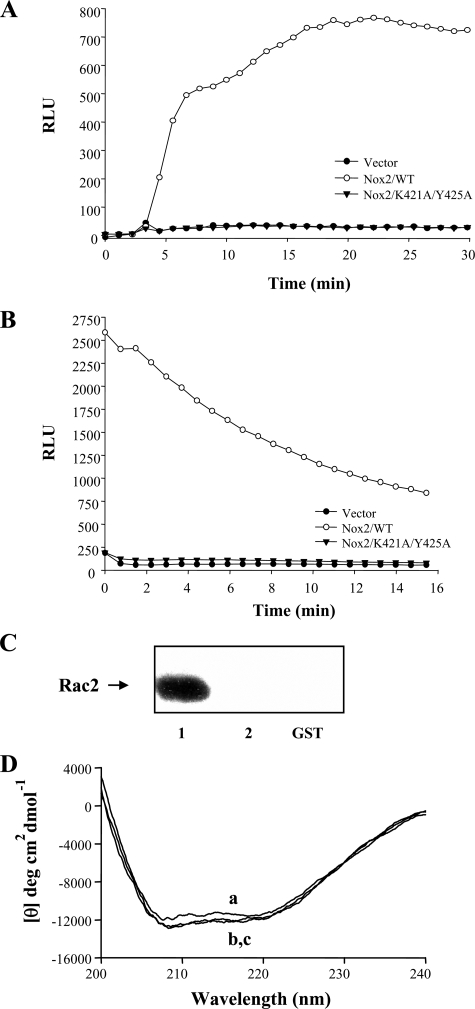
Analysis of the activity of a double point mutant in the Nox2 Rac2 binding domain is associated with inhibition of Rac2 binding in vitro. A, Nox2 K421A/Y425A double mutant protein was expressed in COS-p22 cells, along with p47phox and p67phox, and PMA-stimulated superoxide formation was determined, as described under “Experimental Procedures.” Controls for protein expression are shown in supplemental Fig. S2. B, Nox2 K421A/Y425A double mutant protein was expressed in COS-p22 cells and transfected Rac1Q61L-stimulated superoxide formation determined as described under “Experimental Procedures.” Controls for protein expression are shown in supplemental Fig. S2. RLU, relative light units. C, K421A/Y425A double mutation was made in the GST-Nox2 cytoplasmic tail fusion protein (aa 330–570), and Rac2 binding was determined by pulldown, as described under “Experimental Procedures.” Results shown in A–C are representative of n = 2 or more experiments. D, point mutations that eliminate Rac binding do not significantly alter the secondary structure content of gp91phox(330–570). To investigate the structural consequences of the K421A/Y425A mutations in GST-gp91phox (330–570), the isolated proteins were analyzed by far-UV circular dichroism spectroscopy as follows: curve a, GST alone; curve b, GST-gp91phox(330–570); and curve c, GST-gp91phox(330–570 K421A/Y425A). Although a precise secondary structure estimate was beyond the scope of the present study, it of interest to note that spectra obtained for the above gp91phox constructs were similar to that obtained for GST alone (which contains 50% α-helix and 9% β-sheet excluding the linker region encoded in our recombinant protein), and demonstrated no major changes in overall secondary structure content as a result of the K421A/Y425A point mutations. Similar results were obtained with the deletion constructs shown in Fig. 1A.
We verified that the isolated fragment encompassing aa 408–450 was indeed able to specifically bind Rac2 on its own by direct pulldown assay (Fig. 1C). In contrast, this fragment did not bind to p47phox or p67phox (Fig. 1C), even though these regulatory proteins specifically bind to the intact Nox2 cytoplasmic domain (see Fig. 2C). The region within Nox2 encompassing aa 408–450 is bordered by two (aa 403– 417 and 441– 450) of four total domains that have been hypothesized to make up the NADPH-binding site of Nox2, based upon homology to consensus NADPH-binding sites of the ferredoxin-NADP+ reductase family (45– 47). However, a substantial portion of aa 408–450 is not predicted to be involved in NADPH binding. When tested in a cell-free NADPH oxidase (Nox2) assay system, the aa 408–450 peptide was able to cause a partial inhibition of SDS-induced Nox2 activity at concentrations as low as 50 nm (data not shown). These results encouraged us to further localize the putative Rac2 interaction site within aa 408–450.
FIGURE 2.
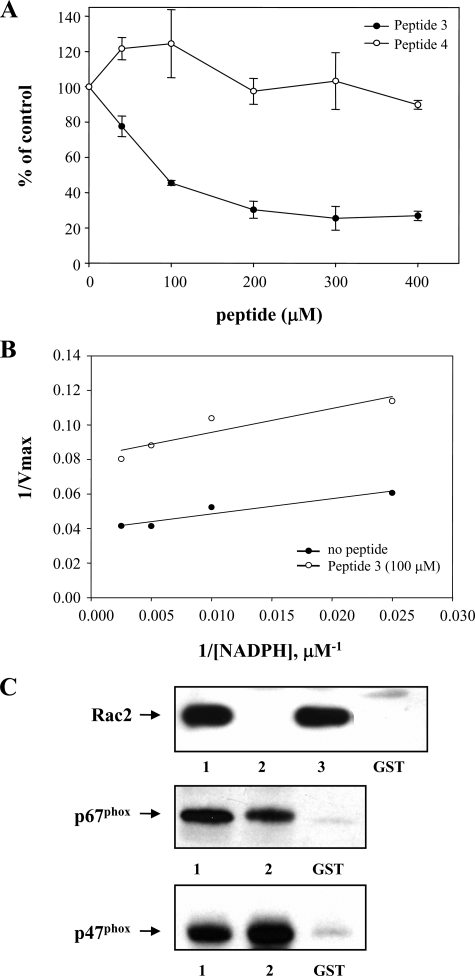
Inhibition of cell-free NADPH oxidase (Nox2) activity by a peptide encompassing the Rac2-binding site correlates with inhibition of Rac2 binding. A, Peptide 3 (aa 419–430 of Nox2), but not Peptide 4 (aa 424–435 of Nox2), exhibits concentration-dependent inhibition of cell-free, Rac2-dependent INT reduction by Nox2. Results shown represent the mean ± S.E. of n = 2 experiments. B, Km value for NADPH in a cell-free NADPH oxidase INT assay (22) in the absence (closed circles) or presence (open circles) of 100 μm Peptide 3 was derived from the slope of the double-reciprocal plot shown. Representative of n = 2 similar experiments. C, specific binding of Rac2, p67phox, or p47phox to the GST-Nox2 cytoplasmic tail (aa 290–570) was determined (as described under “Experimental Procedures”) in the presence or absence as follows: lane 1, no peptide; lane 2, 200 μm Peptide 3; lane 3, 200 μm Peptide 4; or GST alone, as indicated. The binding of p47phox to the Nox2 cytoplasmic tail fusion protein was specific and did not require either phosphorylation or the presence of ionic detergents. Representative of n = 2 similar experiments.
To do so, we generated a series of 12-amino acid peptides that sequentially overlapped throughout aa 408–450, and we tested these for their ability to antagonize Rac2-dependent NADPH oxidase activity. As shown in Table 1, Peptide 3 (aa 419–430) was the only peptide that significantly inhibited cell-free NADPH oxidase activity when tested at a concentration of 400 μm. Over a concentration range from 0 to 400 μm, Peptide 3 produced a concentration-dependent inhibition of cell-free NADPH oxidase activity measured by cytochrome c reduction, up to a maximum inhibition of ∼75%, with an approximate Ki of 80 μm (data not shown). In contrast, neither Peptide 1, Peptide 4, nor a reverse-sequence Peptide 3 had a comparable inhibitory effect. Similar results are shown in Fig. 2A using the INT assay to measure the initial electron transfer step from NADPH to Nox2-bound FAD in the cell-free assay, which we have previously verified to be a Rac-dependent reaction (22). Thus, Peptide 3, encompassing the putative Rac-interaction domain on the Nox2 cytoplasmic tail, inhibits the initial Rac-dependent electron transfer step in NADPH oxidase activation, the transfer of electrons from NADPH to Nox2-bound FAD. In contrast, a peptide derived from the adjacent region on the Nox2 cytoplasmic tail, Peptide 4 (see Table 1), had no inhibitory effect at the concentrations examined. We had difficulty maintaining the solubility of a reversed sequence (or scrambled sequence) Peptide 3 in this assay, thus precluding a precise determination of soluble peptide concentration, but the level of inhibition observed appeared to be substantially less than with wild type Peptide 3 (not shown).
TABLE 1.
Effects of peptides spanning the putative Rac interaction domain on step 1 of NADPH oxidase activation measured by INT reduction in a cell-free human neutrophil membrane/cytosol system
| Control activity | |
|---|---|
| % | |
| Control | 100.00 |
| Peptide 1 (406LVGAGIGVTPFA417) | 105.4 ± 4.4 |
| Peptide 2 (412GVTPFASILKSV423) | 98.6 ± 3.1 |
| Peptide 3 (419ILKSVWYKYCNN430) | 26.9 ± 2.6 |
| Peptide 4 (424WYKYCNNATNLK435) | 95.8 ± 4.7 |
| Peptide 5 (429NNATNLKLKKIY440) | 114.96 ± 8.2 |
| Peptide 6 (435KLKKIYFYWLCR446) | Insoluble |
All peptides (400 mm) were added into the cell-free system before addition of SDS. Control activity (no peptide) was set to 100%, and data are presented as mean +/- S.E. of n = 3 experiments.
Because the regions close to Peptide 3 contain portions of the predicted NADPH binding domains on Nox2, we considered the possibility that the observed effects on oxidase activity could be a result of interference with NADPH binding. To assess this possibility, we determined the Km value for NADPH in the cell-free assay in the presence or absence of 100 μm Peptide 3. As shown in Fig. 2B, there was no significant difference in the Km value for NADPH in the presence of this ∼50% inhibitory concentration of Peptide 3 (Km = 17 μm with peptide versus 22 μm without peptide, n = 2). Increasing the Peptide 3 concentration had no additional effect. These data indicate that Peptide 3 does not interfere with the binding of NADPH to Nox2.
Finally, to verify that Peptide 3 selectively antagonized Rac binding to Nox2, we examined the effect of various peptides on Rac2 binding to the Nox2 cytoplasmic tail fusion protein. As shown in Fig. 2C, Peptide 3 was effective at inhibiting the binding of Rac2 to the cytoplasmic tail of Nox2. In contrast, other peptides, including the adjacent Peptide 4 (see Fig. 2C), did not inhibit Rac2 binding. Additionally, the effect of Peptide 3 was selective, as it had no effect on the binding of p67phox or p47phox to the Nox2 cytoplasmic tail in vitro (Fig. 2C). Taken together, these results demonstrate the selective antagonism of Nox2 Rac-dependent activity by Peptide 3, encompassing aa 419–430 of the cytoplasmic tail of Nox2, through its ability to antagonize Rac2 binding.
Point Mutations in the Rac Binding Domain of Nox2 Disrupt NADPH Oxidase Activity—In addition to Nox2, a number of other Nox proteins have been shown to require Rac GTPase for full activity, including Nox1 and Nox3 (24). At least a portion of the effect of Rac GTPases on Nox 1–3 function appears to be due to interactions with p67/NoxA1 regulatory components via their tetratricopeptide domains (26). In contrast, the activity of Nox4, Nox5, and the Duox proteins has been reported to be independent of Rac GTPase.
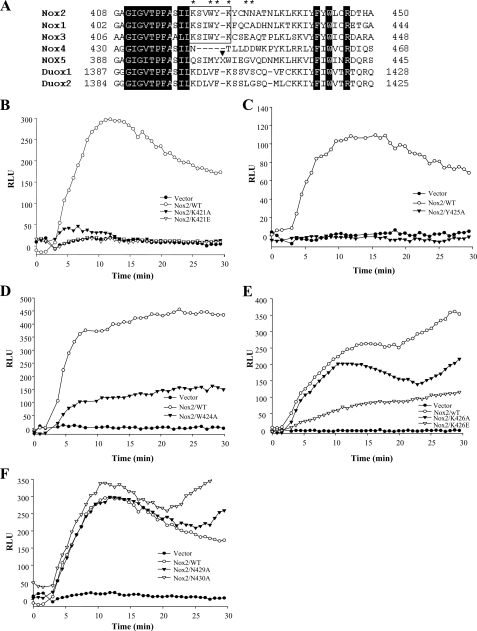
We aligned the sequences of Nox family oxidases surrounding the putative Rac binding region (aa 419–430 of Nox2; Nox2 numbering used throughout) that we had identified. The surrounding (predicted) NADPH-binding motifs (aa 403– 417 and 441– 450) were highly conserved between the Nox1–5 proteins, as well as in Duox1/2 (Fig. 3A). In addition, we noted that there was significant conservation of homology within the aa 419–430 region of Nox1–3 and Duox1/2 but that there were substantial differences in Nox4 and Nox5 in this region. Indeed, Nox5 contained a large peptide insertion just after the amino acid residue equivalent to Nox2 Tyr-425 (Fig. 3A, indicated by arrowhead). To examine the contribution to Rac-dependent NADPH oxidase activity of individual amino acid residues in this domain that were conserved in the Rac-dependent Nox1–3 oxidases, but that differed in Nox4 and 5, we generated individual point mutations in the putative Rac binding region of full-length Nox2, as indicated by the stars in Fig. 3A.
FIGURE 3.
Analysis of activity of individual point mutations in the putative Rac binding domain of full-length Nox2. A, amino acid sequence alignment of the regions in Nox1 (GenBank™ accession number AF127763), Nox3 (accession number AF190122), Nox4 (accession number NM_016931), Nox5 (accession number AF317889), Duox1 (accession number AF213465), and Duox2 (accession number AF267981), equivalent to aa 408–450 of Nox2 (accession number NM000397), was carried out by ClustalW (54). The black blocks indicate residues conserved among the Nox family members, the white boxes indicate the residues conserved between Noxs1 and -3 in the Rac binding region, and the residues in this region mutated for activity studies are indicated by the stars. The position of a 15-aa insertion in Nox5 (X = RHQKRKHTCPSCQHS) is indicated by the black arrowhead. B–F, indicated Nox2 mutant proteins were expressed in COS-p22 cells and PMA-stimulated superoxide formation determined as described under “Experimental Procedures.” Controls for protein expression are shown in supplemental Fig. S1. Mutations of K421A/K421E or Y425A resulted in the almost total loss of NADPH oxidase activity; mutations of W424A or K426E resulted in 60–75% reductions in NADPH oxidase activity compared with wild type (WT) controls; and mutations of the nonconserved N429A or N430A residues produced no significant loss of NADPH oxidase activity. Results shown are representative of two or more experiments.
These Nox2 mutants were transiently expressed, along with p47phox and p67phox, in COS-7 cells stably expressing the p22phox Nox2-associated membrane subunit (COS-p22 cells (36)). As shown in supplemental Fig. S1, these Nox2 mutant constructs were expressed as full-length post-translationally modified proteins and were present at the cell surface in substantial and essentially equal amounts. Additionally, the expression of the Nox2 mutants had no effect on the expression levels of p47phox, p67phox, p22phox, or Rac1 in these cells (supplement Fig. S1B). We therefore examined NADPH oxidase activity stimulated by PMA in cells expressing wild type versus mutant Nox2 proteins at similar levels. As shown in Fig. 3, B–F, the mutations exhibited various levels of activity, ranging from about 100% of wild type Nox2 (N429A and N430A), to partial loss of activity (W424A and K426E), to almost total loss of activity (K421A/K421E, Y425A). Mutation to either an Ala or a Glu residue caused similar losses of activity, except for aa Lys-426, where mutation to Glu caused substantial loss of activity but mutation to Ala did not.
We noted that the mutations causing substantial loss of Nox2 activity were grouped within aa 421–426 (aa Lys-421, Trp-424, Tyr-425, and Lys-426). Significantly, these amino acids were not conserved in Nox4, and Nox5 had a 15-amino acid insertion just after Tyr-425 (Fig. 3A). A full-length Nox2 protein with the double K421A/Y425A mutation exhibited total loss of NADPH oxidase activity in response to stimulation with either PMA (Fig. 4A) or co-transfection with active RacQ61L (Fig. 4B). This was not because of differences in expression of the Nox2 mutant protein at the cell surface nor to changes in other NADPH oxidase regulatory components (supplemental Fig. S2). We attempted to evaluate the translocation of endogenous Rac1 versus p47phox and p67phox to the plasma membrane in PMA-stimulated COS-p22 cells expressing the Nox2 Rac-binding site mutations, but we were technically unable to observe quantitatively consistent translocation.
Correlating with the loss of Nox2 activity, however, mutation of the K421A/Y425A sites in the Nox2 cytoplasmic tail fusion protein abrogated Rac2 binding in vitro (Fig. 4C). Fig. 4D shows representative far-UV CD spectra collected for both GST-gp91phox(330–570) (curve b) and GST-gp91phox(330–570, K421A/Y425A) (curve c). Importantly, spectra for these constructs were nearly superimposable, which suggests that the loss of Rac binding by the GST-gp91phox(330–570, K421A/Y425A) construct did not result from major structural perturbations induced by the point mutations (43). Additional support for this hypothesis was obtained by measuring intrinsic tryptophan fluorescence (which is dominated by the gp91phox(330–570) component), where the fluorescence emission maximum of both GST-gp91phox(330–570) and GST-gp91phox(330–570 K421A/Y425A) was red-shifted from ∼337 nm in phosphate-buffered saline to ∼348 nm in the presence of 3 m guanidine hydrochloride (data not shown). These data indicate that both constructs contain a similar population of buried Trp residues that become solvent-exposed upon protein denaturation (44).
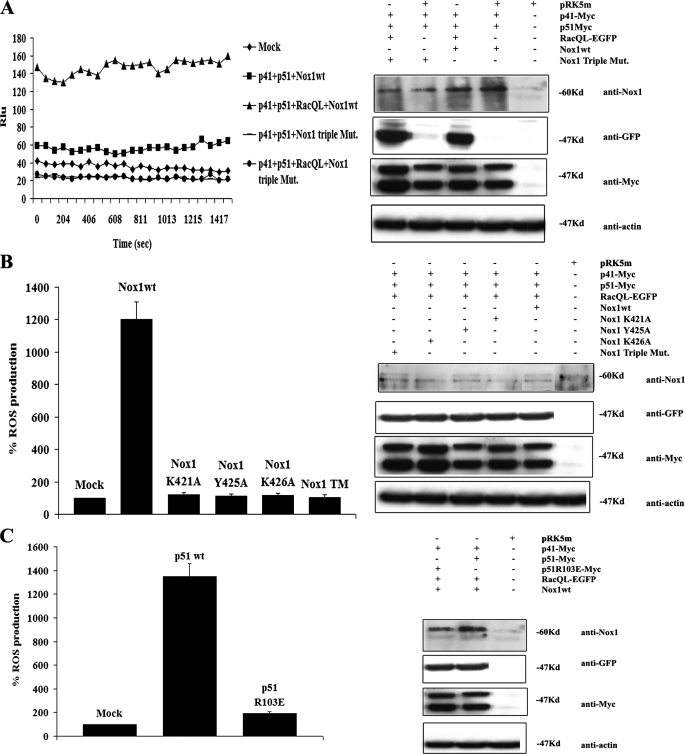
Mutations in the Conserved Rac Binding Domain of Nox1 Cause Loss of Rac1-dependent Nox1 Activity–As noted above, the activity of Nox1–3 have all been shown to be regulated by Rac GTPase, whereas Nox4 and Nox5 are not. The conservation of the Rac binding domain we identified on Nox2 in the Nox1 and Nox3 family members suggests that Rac may also directly regulate the activity of these Nox enzymes. To test this, we mutated the conserved residues K421A/Y425A/K426A in Nox1, and we expressed the triple mutant protein in HEK293 cells to assess the ability of active Rac1 to stimulate Nox1-dependent ROS formation. As shown in Fig. 5A, in the presence of NoxO1 (or p41) and NoxA1 (or p51) regulatory components, active Rac1Q61L stimulated formation of superoxide anion. Nox1 activity was completely inhibited in the presence of 2–10 μm diphenyliodonium (not shown). In contrast, when the Nox1 triple mutant was substituted for the wild type protein and was expressed at similar levels, it did not support the generation of ROS. This was not because of effects on the expression levels of any of the regulatory components or Rac1Q61L itself (Fig. 5), nor was activity restored by increasing the expression of NoxO1, NoxA1, or Rac1Q61L (not shown).
FIGURE 5.
The integrity of the Lys-421, Tyr-425, and Lys-426 residues of Nox1 are necessary for Nox1-dependent ROS generation stimulated by Rac1. A, HEK293 cells were co-transfected with empty vector or with the expression vectors for NoxO1(p41), NoxA1(p51), and (where indicated) with the constitutive active form of Rac1 (RacQL) and with Nox1 wild type or triple mutant (bearing the mutations K421A, Y425A, and K426A). 24 h after transfection, ROS formation was measured (left panel) by chemiluminescence (see “Experimental Procedures”). The expression of each transfected protein was determined by Western blot (right panel). B, HEK293 cells were co-transfected with empty vector or with the expression vectors for NoxO1(p41), NoxA1(p51), RacQL, and (where indicated) with wild type or individual single Nox1 mutants. 24 h after transfection, ROS formation was measured (left panel) by chemiluminescence. The expression of each transfected protein was determined by Western blot (right panel). C, HEK293 cells were co-transfected with empty vector or with the expression vectors for NoxO1(p41), RacQL, Nox1 wild type (wt), and (where indicated) with wild type NoxA1 or with NoxA1 R103E, the NoxA1 mutant unable to bind Rac1. 24 h after transfection, ROS generation was measured (left panel), and the expression of each transfected protein was determined by Western blot (right panel).
Fig. 5B shows the results of analysis of individual mutations in/near the Rac binding domain of Nox1. The data show that mutations that resulted in the loss of Rac2-dependent Nox2 activity also caused a similar loss of Rac1-dependent Nox1 activity. As with Nox2, mutations at nearby nonconserved residues did not affect ROS formation (not shown). To further test our hypothesis that the Nox1 residues Lys-421, Tyr-425, and Lys-426 are responsible for the Rac1 binding, we generated a Nox1 chimera in which the Nox1 region between Lys-421 and Lys-426 was substituted with the corresponding region of Nox4, a member of the Nox family whose activity has been clearly shown to be Rac GTPase-independent. As shown in supplemental Fig. S3, such a Nox1 chimera dramatically loses responsiveness to Rac1QL, even though it is expressed at levels equivalent to the wild type Nox2 at the cell surface. Although the mechanism of the loss of responsiveness was not further characterized here, these results support the idea that the region, including Lys-421, Tyr-425, and Lys-426 residues of Nox1, may be fundamental for Rac1-mediated activity.
Because prior studies have established that at least part of the ability of Rac GTPase to regulate Nox1 activity is because of its interaction with the conserved tetratricopeptide domain of NoxA1, we examined the effect of a non-Rac1-binding NoxA1 on Nox1 activity stimulated by Rac1Q61L (Fig. 5C). The presence of a NoxA1 R103E mutation that prevents the interaction of NoxA1 with Rac GTPase (15) also dramatically decreased Nox1 activity. These results indicate that the ability of Rac GTPase to interact with Nox1 directly, as well as with the regulatory cofactor NoxA1 is required for full NADPH oxidase activity. Overall, our data are consistent with an NADPH oxidase regulatory mechanism involving the direct interaction of Rac GTPase with Nox subunits, as proposed previously (21, 22).
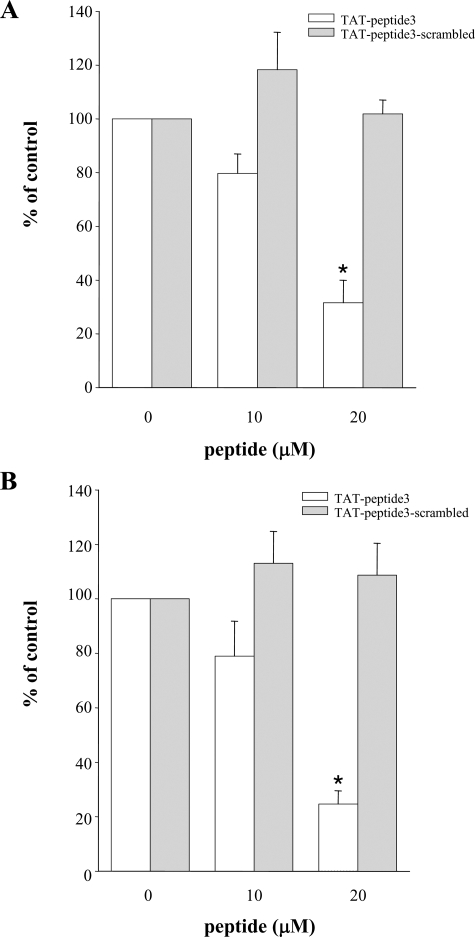
Peptides Derived from the Rac Binding Region Inhibit Intact Neutrophil NADPH Oxidase Activity—To test the requirement for the putative Nox2 Rac interaction site for ROS production in intact neutrophils, we generated a cell-permeant TAT peptide that encompassed the Rac binding domain, aa 419–430 (equivalent to Peptide 3 of Table 1). This peptide exhibited a concentration-dependent inhibition of ROS formation stimulated by the chemoattractant fMLP (Fig. 6A) or by the phorbol ester PMA (Fig. 6B). In contrast, a scrambled sequence Peptide 3 was inactive. Both fMLP and PMA are known to activate Nox2 through Rac2-dependent mechanisms in human neutrophils (34). These data demonstrate that the Rac interaction site we have identified on Nox2 is required for stimulated NADPH oxidase activity in intact leukocytes.
FIGURE 6.
A peptide encompassing the Rac GTPase-binding domain of Nox2 inhibits stimulus-dependent cellular NADPH oxidase activity. A, human neutrophils were incubated with the indicated concentration of TAT-Peptide 3, as described under “Experimental Procedures,” and then stimulated with 5 μm fMLP. B, human neutrophils were incubated with the indicated concentration of TAT-Peptide 3 as described under “Experimental Procedures,” and then stimulated with 2 μg/ml PMA. The initial rates of superoxide production for control incubations with no peptide were set to 100%. White bars = TAT-Peptide 3 (ILKSVWYKYCNN); gray bars = TAT-scrambled sequence Peptide 3 s(NKNWCVYISKLY). Results shown represent the mean ± S.E. of n = 3 experiments. * indicates significant difference at p < 0.05.
DISCUSSION
Nox2 Contains a Rac GTPase-binding Regulatory Site—We establish here the existence of a Rac GTPase-binding regulatory site directly on the Nox2 protein, and we provide supporting evidence that this site is also conserved and functional in Nox1 and perhaps in Nox3 and Duox proteins as well. Through deletion analysis and the use of inhibitory peptides overlapping the putative Rac-binding site, we show that this region participates in the binding and functional interaction of Nox2 with Rac GTPases (Figs. 1, 2, 3; Table 1). We have shown previously using a fluorescence-based approach that the interaction of Rac with Nox2 requires the insert domain of the Rac GTPase (22). Consistent with this, binding to the cytoplasmic domain fusion protein is largely independent of whether Rac2 is in a GDP or a GTP form (not shown). However, activation of the NADPH oxidase activity of Nox2 does require Rac2 to be in the GTP-bound active state (21, 48).
The Rac binding domain on Nox2 is flanked by two regions that have been hypothesized to participate in the binding of NADPH to the Nox proteins. Significantly, a peptide overlapping the Rac binding domain of Nox2 (aa 419–430), which inhibits both Rac2 binding and cell-free NADPH oxidase activity, does not affect the Km value for NADPH in the cell-free assay (Fig. 2B). It is unlikely that the Rac binding domain affects the binding of FAD either, as it is not near the putative FAD interaction site (45–47). Finally, the aa 419–430 peptide does not affect the specific interaction of the Nox2 cytoplasmic tail with either p47phox or p67phox in vitro (Fig. 2C).
Further supporting the relevance of the Rac-interaction domain for Nox2 activity, we showed that the peptide encompassing this region, aa 419–430, selectively inhibited cell-free NADPH oxidase activity, both determined with INT reduction (electron transfer from NADPH to FAD) and by cytochrome c reduction (Table 1 and Fig. 2A). The INT reduction assay has been shown to be dependent on the separate activities of both Rac2 and 67phox (22). Additionally, when point mutations were generated in the full-length Nox2 within conserved residues of the Rac binding domain, these mutations (K421A/K421E, Y425A, and K426E) dramatically reduced Rac GTPase-dependent NADPH oxidase activity in a COS-p22 cell system stimulated either with PMA or by expression of RacQ61L itself (Figs. 3 and 4). In contrast, mutation of nonconserved residues within this region did not affect NADPH oxidase activity (e.g. Fig. 3), suggesting that the structural integrity of this region was not particularly sensitive to mutations in this portion of the Nox2 protein. This is also supported by the lack of substantial secondary structure disruptions as the Nox2 cytoplasmic tail was sequentially truncated, as determined by CD and fluorescence analysis.
The Rac Regulatory Site Is Conserved on Nox1 and Nox3–The region involved in Rac GTPase binding to Nox2 is encompassed by at least aa residues 419–430. It is of interest that this region appears to be more highly conserved in Nox1, Nox2, and Nox3 than in other members of the Nox family (Fig. 3A) (4, 49). Nox1–3 contain identical or highly conserved amino acid residues from 419 to 427 (using the Nox2 numbering). In contrast, Nox4 contains only two conserved residues within this same 9-amino acid region. We note that Nox4 actually has a gap in the alignment from aa 422 to 425, whereas Nox5 contains a large 15-amino acid insertion in this region that is likely to disrupt Rac binding (50). Mutation of several of the highly conserved residues (Lys-421, Trp-424, Tyr-425, and Lys-426) found in Nox1–3 to either alanine or glutamic acid demonstrated that these residues were critical for Rac binding and Rac-dependent NADPH oxidase activity by both Nox2 (Figs. 3 and 4) and Nox1 (Fig. 5). We are aware that these residues, except for Trp-424, which is a Val in Duox1/2, are also conserved in both Duox1 and -2, suggesting the possibility that Duox1/2 might interact with Rac GTPases. However, there are other differences in the 419–427-aa region (three of nine residues are nonconserved in Duox1/2) that may preclude Duox regulation by Rac, as has been reported (29). This remains to be examined in greater detail. Overall, our data support the idea that a direct Rac GTPase regulatory site exists on certain members of the Nox family, and that the presence of this site contributes to their susceptibility to regulation by Rac GTPases. It is also intriguing that only Nox1–3 are known to be positively regulated by a NoxA1/p67phox-like regulatory protein, which may additionally contribute to the Rac-dependent regulatory mechanism (see below).
Regulation of Nox Function by Rac GTPase, Implications for the Two-step Mechanism—The basis for Nox regulation by Rac GTPase has been the subject of much debate. In the phagocyte NADPH oxidase, several groups have suggested that Rac GTPase does not interact directly with cytochrome b558, but rather it inserts into the plasma membrane via its prenylated and polybasic C terminus, where it aids in the positioning of p67phox on the cytochrome b558 to promote electron transfer (reviewed in Ref. 21). In contrast, we have previously provided evidence that Rac GTPase must interact functionally with cytochrome b558 to promote electron transfer and that this involves a direct binding of Rac to Nox2 (22). Furthermore, we have proposed that these data indicate a two-step mechanism through which electron transfer is regulated by the concerted actions of Rac GTPase and p67phox. The current identification of a Rac GTPase-binding site on Nox2 provides strong support for this model of NADPH oxidase regulation. The high degree of conservation of the Rac-binding site on Nox1 and Nox3, both known to be regulated by Rac GTPases, supports the relevance of this interaction domain to Nox function. Furthermore, the demonstration that both the Rac binding domain on Nox1 and the Rac-binding site on NoxA1 are necessary to support superoxide formation (Figs. 3, 4, 5) is consistent with the two-step regulatory mechanism for electron transfer that we originally proposed for Nox2 in human neutrophils (22). Our current results thus also indicate that this mechanism is likely to be operative with Nox1, Nox2, and Nox3. Indeed, the lack of this Rac regulatory site on the Nox4 and Nox5 members of the Nox family strongly suggests that the basic mechanism(s) underlying the regulation of their electron transfer activities may be dramatically different from the Rac-regulated NADPH oxidases.
The Nox2 Rac Binding Domain, a Role in CGD?—Intriguingly, several patients with CGD have been identified with mutations that lie within (or near) the Rac binding domain of Nox2 that we have identified (6). A patient identified with the X-linked form of the disease had a mutation at L420P and was unable to generate superoxide (51). Neutrophils from this patient, however, were shown to be lacking significant levels of Nox2 protein. A second patient was identified with a form of X-linked CGD associated with an S422P mutation (52). Nox2 protein levels were not assessed in this patient. Finally, nearby Pro-415 mutations have been identified in several variant X-linked CGD patients in which Nox2 heme spectrum was similar to controls, suggesting that expression of Nox2 protein was normal (6). It has been proposed that these patients may be impaired in their ability to interact with a cytosolic regulatory component. Based upon the results presented here, we suggest the possibility that this deficient regulatory interaction might involve Rac GTPase.
Conclusion—Overall, our data provide evidence for a direct Rac GTPase regulatory site on the Rac GTPase-regulated NADPH oxidases, Nox1, Nox2, and Nox3. We show that mutation of conserved residues in this site are sufficient to disrupt Nox1 and Nox2 activity, correlating with the loss of Rac GTPase binding, but not binding of p47phox or p67phox, to the Nox2 cytoplasmic tail. It is noteworthy that the Rac interaction domain lies immediately adjacent to two portions of the predicted NADPH-binding site of Nox2. In particular, it has been suggested that a region encompassing the Rac GTPase binding domain we have identified might act as a molecular “cap” to interfere with the binding of NADPH to Nox2 protein (53). It will be of interest in future studies to examine the mechanism by which Rac GTPase binding at this site modulates NADPH oxidase activity.
Supplementary Material
Acknowledgments
We thank Bruce Fowler for expert technical assistance. We appreciate the advice of Dr. Becky Diebold (Emory University) for initiating some of these studies.
This work was supported, in whole or in part, by National Institutes of Health Grant HL48008 (to G. M. B.). This work was also supported by a grant from the Chronic Granulomatous Disorder Research Trust (to Y.-Y. K. and G. M. B.). This is Manuscript 19141 from the Department of Immunology, The Scripps Research Institute. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: ROS, reactive oxygen species; aa, amino acid; GST, glutathione S-transferase; PBS, phosphate-buffered saline; PMA, phorbol 12-myristate 13-acetate; HBSS, Hanks' balanced salt solution; INT, p-iodonitrotetrazolium; fMLP, fMet-Leu-Phe; GTPγS, guanosine 5′-3-O-(thio)triphosphate; CGD, chronic granulomatous disease; GppNHp, guanosine-5-(β-imino)triphosphate.
References
- 1.Cross, A. R., and Segal, A. W. (2004) Biochim. Biophys. Acta 1657 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morel, F., Doussiere, J., and Vignais, P. V. (1991) Eur. J. Biochem. 201 523–546 [DOI] [PubMed] [Google Scholar]
- 3.Bedard, K., and Krause, K. H. (2007) Physiol. Rev. 87 245–313 [DOI] [PubMed] [Google Scholar]
- 4.Cheng, G., Cao, Z., Xu, X., van Meir, E. G., and Lambeth, J. D. (2001) Gene (Amst.) 269 131–140 [DOI] [PubMed] [Google Scholar]
- 5.Bokoch, G. M., and Knaus, U. G. (2003) Trends Biochem. Sci. 28 502–508 [DOI] [PubMed] [Google Scholar]
- 6.Heyworth, P. G., Curnutte, J. T., Rae, J., Noack, D., Roos, D., van Koppen, E., and Cross, A. R. (2001) Blood Cells Mol. Dis. 27 16–26 [DOI] [PubMed] [Google Scholar]
- 7.Smith, R. M., and Curnutte, J. T. (1991) Blood 77 673– 686 [PubMed] [Google Scholar]
- 8.DeLeo, F. R., and Quinn, M. T. (1996) J. Leukocyte Biol. 60 677– 691 [DOI] [PubMed] [Google Scholar]
- 9.Groemping, Y., and Rittinger, K. (2005) Biochem. J. 386 401– 416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinn, M. T., and Gauss, K. A. (2004) J. Leukocyte Biol. 76 760–781 [DOI] [PubMed] [Google Scholar]
- 11.Freeman, J. L., and Lambeth, J. D. (1996) J. Biol. Chem. 271 22578–22582 [DOI] [PubMed] [Google Scholar]
- 12.Koshkin, V., Lotan, O., and Pick, E. (1996) J. Biol. Chem. 271 30326–30329 [DOI] [PubMed] [Google Scholar]
- 13.Han, C. H., Freeman, J. L., Lee, T., Motalebi, S. A., and Lambeth, J. D. (1998) J. Biol. Chem. 273 16663–16668 [DOI] [PubMed] [Google Scholar]
- 14.Koga, H., Terasawa, H., Nunoi, H., Takeshige, K., Inagaki, F., and Sumimoto, H. (1999) J. Biol. Chem. 274 25051–25060 [DOI] [PubMed] [Google Scholar]
- 15.Lapouge, K., Smith, S. J., Walker, P. A., Gamblin, S. J., Smerdon, S. J., and Rittinger, K. (2000) Mol. Cell 6 899–907 [DOI] [PubMed] [Google Scholar]
- 16.Kim, C., and Dinauer, M. C. (2001) J. Immunol. 166 1223–1232 [DOI] [PubMed] [Google Scholar]
- 17.Knaus, U. G., Heyworth, P. G., Evans, T., Curnutte, J. T., and Bokoch, G. M. (1991) Science 254 1512–1515 [DOI] [PubMed] [Google Scholar]
- 18.Zhao, X., Carnevale, K. A., and Cathcart, M. K. (2003) J. Biol. Chem. 278 40788–40792 [DOI] [PubMed] [Google Scholar]
- 19.Roberts, A. W., Kim, C., Zhen, L., Lowe, J. B., Kapur, R., Petrynlak, B., Spaetti, A., Pollock, J., Borneo, J. B., Bradford, G. B., Atkinson, S. J., Dinauer, M. C., and Williams, D. A. (1999) Immunity 10 1–20 [DOI] [PubMed] [Google Scholar]
- 20.Zhao, T., Benard, V., Bohl, B. P., and Bokoch, G. M. (2003) J. Clin. Investig. 112 1732–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bokoch, G. M., and Diebold, B. A. (2002) Blood 100 2692–2696 [DOI] [PubMed] [Google Scholar]
- 22.Diebold, B. A., and Bokoch, G. M. (2001) Nat. Immunol. 2 211–215 [DOI] [PubMed] [Google Scholar]
- 23.Diebold, B. A., Fowler, B., Lu, J., Dinauer, M. C., and Bokoch, G. M. (2004) J. Biol. Chem. 279 28136–28142 [DOI] [PubMed] [Google Scholar]
- 24.Miyano, K., and Sumimoto, H. (2007) Biochimie (Paris) 89 1133–1144 [DOI] [PubMed] [Google Scholar]
- 25.Cheng, G., Diebold, B. A., Hughes, Y., and Lambeth, J. D. (2006) J. Biol. Chem. 281 17718–17726 [DOI] [PubMed] [Google Scholar]
- 26.Miyano, K., Ueno, N., Takeya, R., and Sumimoto, H. (2006) J. Biol. Chem. 281 21857–21868 [DOI] [PubMed] [Google Scholar]
- 27.Ueyama, T., Geiszt, M., and Leto, T. L. (2006) Mol. Cell. Biol. 26 2160–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamiguti, A. S., Serrander, L., Lin, K., Harris, R. J., Cawley, J. C., Allsup, D. J., Slupsky, J. R., Krause, K. H., and Zuzel, M. (2005) J. Immunol. 175 8424–8430 [DOI] [PubMed] [Google Scholar]
- 29.Fortemaison, N., Miot, F., Dumont, J. E., and Dremier, S. (2005) Eur. J. Endocrinol. 152 127–133 [DOI] [PubMed] [Google Scholar]
- 30.Martyn, K. D., Frederick, L. M., von Loehneysen, K., Dinauer, M. C., and Knaus, U. G. (2006) Cell. Signal. 18 69–82 [DOI] [PubMed] [Google Scholar]
- 31.Martyn, K. D., Kim, M. J., Quinn, M. T., Dinauer, M. C., and Knaus, U. G. (2005) Blood 106 3962–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quinn, M. T., Evans, T., Loetterle, L. R., Jesaitis, A. J., and Bokoch, G. M. (1993) J. Biol. Chem. 268 20983–20987 [PubMed] [Google Scholar]
- 33.Vocero-Akbani, A., Chellaiah, M. A., Hruska, K. A., and Dowdy, S. F. (2001) Methods Enzymol. 332 36–49 [DOI] [PubMed] [Google Scholar]
- 34.Benard, V., Bohl, B. P., and Bokoch, G. M. (1999) J. Biol. Chem. 274 13198–13204 [DOI] [PubMed] [Google Scholar]
- 35.Knaus, U. G., Heyworth, P. G., Kinsella, B. T., Curnutte, J. T., and Bokoch, G. M. (1992) J. Biol. Chem. 267 23575–23582 [PubMed] [Google Scholar]
- 36.Price, M. O., McPhail, L. C., Lambeth, J. D., Han, C. H., Knaus, U. G., and Dinauer, M. C. (2002) Blood 99 2653–2661 [DOI] [PubMed] [Google Scholar]
- 37.Nakamura, M., Kobayashi, S., Sendo, S., Koga, T., and Kanegasaki, S. (1988) Acta Paediatr. Hung. 29 179–183 [PubMed] [Google Scholar]
- 38.Nakamura, M., Murakami, M., Koga, T., Tanaka, Y., and Minakami, S. (1987) Blood 69 1404–1408 [PubMed] [Google Scholar]
- 39.Burritt, J. B., Foubert, T. R., Baniulis, D., Lord, C. I., Taylor, R. M., Mills, J. S., Baughan, T. D., Roos, D., Parkos, C. A., and Jesaitis, A. J. (2003) J. Immunol. 170 6082–6089 [DOI] [PubMed] [Google Scholar]
- 40.Curnutte, J. T., Kuver, R., and Scott, P. J. (1987) J. Biol. Chem. 262 5563–5569 [PubMed] [Google Scholar]
- 41.Heyworth, P. G., Bohl, B. P., Bokoch, G. M., and Curnutte, J. T. (1994) J. Biol. Chem. 269 30749–30752 [PubMed] [Google Scholar]
- 42.Cheng, G., and Lambeth, J. D. (2004) J. Biol. Chem. 279 4737–4742 [DOI] [PubMed] [Google Scholar]
- 43.Kelly, S. M., Jess, T. J., and Price, N. C. (2005) Biochim. Biophys. Acta 1751 119–139 [DOI] [PubMed] [Google Scholar]
- 44.Lakowicz, J. R. (1999) Principles of Fluorescence Spectroscopy, Kluwer Academic/Plenum, New York
- 45.Rotrosen, D., Yeung, C. L., Leto, T. L., Malech, H. L., and Kwong, C. H. (1992) Science 256 1459–1462 [DOI] [PubMed] [Google Scholar]
- 46.Segal, A. W., West, I., Wientjes, F., Nugent, J. H., Chavan, A. J., Haley, B., Garcia, R. C., Rosen, H., and Scrace, G. (1992) Biochem. J. 284 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sumimoto, H., Sakamoto, N., Nozaki, M., Sakaki, Y., Takeshige, K., and Minakami, S. (1992) Biochem. Biophys. Res. Commun. 186 1368–1375 [DOI] [PubMed] [Google Scholar]
- 48.Heyworth, P. G., Knaus, U. G., Xu, X., Uhlinger, D. J., Conroy, L., Bokoch, G. M., and Curnutte, J. T. (1993) Mol. Biol. Cell 4 261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lambeth, J. D. (2004) Nat. Rev. Immunol. 4 181–189 [DOI] [PubMed] [Google Scholar]
- 50.Banfi, B., Molnar, G., Maturana, A., Steger, K., Hegedus, B., Demaurex, N., and Krause, K. H. (2001) J. Biol. Chem. 276 37594–37601 [DOI] [PubMed] [Google Scholar]
- 51.Kaneda, M., Sakuraba, H., Ohtake, A., Nishida, A., Kiryu, C., and Kakinuma, K. (1999) Blood 93 2098–2104 [PubMed] [Google Scholar]
- 52.Roos, D. (1996) Immunol. Today 17 517–521 [DOI] [PubMed] [Google Scholar]
- 53.Taylor, W. R., Jones, D. T., and Segal, A. W. (1993) Protein Sci. 2 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994) Nucleic Acids Res. 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.