Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is a global scourge, and treatment options are becoming limited. The MRSA phenotype reverts to that of β-lactam-sensitive S. aureus when bacteria are grown at pH 5.0 in broth and, more importantly from a medical perspective (protracted, relapsing infections), after phagocytosis by macrophages, where the bacteria thrive in the acidic environment of phagolysosomes. The central factor for the MRSA phenotype is the function of the penicillin-binding protein (PBP) 2a, which maintains transpeptidase activity while being poorly inhibited by β-lactams because of a closed conformation of its active site. We document herein by binding, acylation/deacylation kinetics, and circular dichroism spectroscopy with purified PBP 2a that at acidic pH (i) β-lactams interact with PBP 2a more avidly; (ii) the non-covalent pre-acylation complex exhibits a lower dissociation constant and an increased rate of acyl-enzyme formation (first-order rate constant) without change in hydrolytic deacylation rate; and (iii) PBP 2a undergoes a conformational change in the presence of the antibiotic consistent with the opening of the active site from the closed conformation. These observations argue that PBP 2a most likely evolved for its physiological function at pH 7 or higher by adopting a closed conformation, which is not maintained at acidic pH. Although at the organism level the effect of acidic pH on other biological processes in MRSA could not be discounted, our report should provide the impetus for closer examination of the properties of PBP 2a at low pH and thereby identifying novel points of intervention in combating this problematic organism.
Methicillin-resistant Staphylococcus aureus (MRSA)3 is a difficult bacterial pathogen to treat and is presently a clinical scourge. This organism is resistant to all commercially available β-lactams antibiotics, which are currently mainstays of treatment of bacterial infections. MRSA is a global problem in hospitals, in nursing homes and in communities (1–5). It has now been found frequently in pets (6) or food animals (7–9), becoming a potential issue for humans in close contact with them (10, 11). MRSA represents also a therapeutic challenge because effective therapeutic options are becoming limited. MRSA, indeed, has a most remarkable ability in expanding its genome and in enhancing its ability to counter antibacterial agents (12). Thus, vancomycin, most commonly recommended for treating infections caused by the hospital-acquired strains of MRSA, becomes increasingly compromised by the spreading of variants with reduced susceptibility (vancomycin-intermediate S. aureus isolates) and, in some instances, the emergence of fully resistant organisms (13). Resistance to new antibiotics with approved indications for MRSA infections such as linezolid (an oxazolidinone), daptomycin (a lipopetide), or tigecycline (a modified tetracycline) has already been noted as well (14).
The major mechanism of resistance of MRSA to β-lactam antibiotics is due to the acquisition of the mecA gene encoding an additional penicillin-binding protein (PBP 2a; also referred to in the literature as PBP 2′) (15, 16) that can function as a transpeptidase. In the face of the challenge by β-lactam antibiotics, the other staphylococcal transpeptidases are inhibited, but PBP 2a retains its activity. In association with the transglycosylase domain of the native PBP 2, the activity of which is not inhibited by β-lactams (17), PBP 2a performs the critical cross-linking of the cell wall of the bacterium and as such serves a vital function for the organism.
Understanding of the properties of PBP 2a is at the forefront of restoring susceptibility to β-lactams, which is an important aim (12). Efforts toward this goal largely encompass work in the direction of discovery of novel β-lactams that inhibit both PBP 2a and other transpeptidases (18). It is of note, however, that susceptibility of MRSA to conventional β-lactams can be almost fully restored, if bacteria are grown at pH of 5.5 or lower. This phenomenon was originally described in the early 1970s for hospital-acquired MRSA growing in broth and exposed to β-lactamase-resistant penicillins or first generation cephalosporins (19). This observation has now been confirmed for other β-lactams, including carbapenems, and applies not only to hospital- but also community-acquired MRSA (20). It has also been extended to MRSA growing intracellularly in phagolysosomes of macrophages (where pH is in the acidic range) (21), giving it a broader significance than originally appreciated. This influence of acidic pH was first ascribed to a diminished copy numbers of PBP 2a (15). We showed, however, that growing bacteria at acidic pH (i) alters neither the expression of the PBP 2a-encoding gene (mecA) nor that of its regulatory genes and (ii) does not decrease the amount of the immunodetectable PBP 2a in whole bacteria (20). We also found that MRSA grown and exposed to [3H]penicillin G at acidic pH show a larger retention of radioactivity than if bacteria had been grown at neutral pH (this amount being actually brought to a level similar to what is observed with MSSA grown and exposed to [3H]penicillin G at neutral pH) (20).
An in-depth understanding of the molecular mechanism underlying restoration of susceptibility of MRSA to β-lactams by acidic pH is of profound significance. It would, indeed, not only explain the antibiotic susceptibility of the organism within phagolysosomes and other habitats where pH is low, but it might also reveal avenues of intervention in treatment of this problematic bacterium. In this report, we have examined the relationship between pH and the function of PBP 2a. Our data show that acidic pH causes a significant change in conformational behavior of PBP 2a, rendering it more susceptible to β-lactams. In terms of mechanistic consequence, we assert that these observations are quite similar to those recently reported for new cephalosporins that are capable of inhibiting PBP 2a effectively, arguing for an underlying principal that governs both cases (22–24).
EXPERIMENTAL PROCEDURES
Strains and Susceptibilities—A methicillin-sensitive β-lactamase-negative S. aureus (ATCC 25923; American Type Culture Collection, Manassas, VA), a methicillin-resistant β-lactamase-negative S. aureus (COL, referenced as NRS100; Network on Antimicrobial Resistance in S. aureus; Focus Technologies, Inc., Herndon, VA), and a methicillin-resistant β-lactamase-positive S. aureus (ATCC 33591) were used thorough these studies. MICs were measured by microdilution (log2 progression) in Muller Hinton broth supplemented by 2% NaCl for MRSA (United States Clinical Laboratory Standards Institute, Wayne, PA) and adjusted to appropriate pH by the careful addition of 1 n HCl (we checked in previous work (20) that the final pH levels of the broths at the end of the 24-h incubation period were close to the original one at all antibiotic concentrations equal to the MIC or above).
Binding of Bocillin FL to Whole Cells—Bocillin FL is a fluorescent derivative of penicillin V (Ref. 25; see also supplemental material) that has been validated for PBP binding studies (26). We used it here to examine the penicillin binding properties of bacteria growing at acidic versus neutral pH, following a procedure used previously to study the binding of radiolabeled penicillin G (20, 26). A 10-ml portion of growing bacteria (5-h culture; OD660 nm =∼0.5) were exposed to Bocillin FL (25 μg) for 30 min at pH 7.4 or 5.5. Bacteria were then collected by centrifugation, washed four times with phosphate-buffered saline, and lysed by three successive freeze-thawing cycles (5 min at –80 °C followed by 5 min at 37 °C). The amount of fluorescent material was measured in a Packard Fluorocount Microplate Reader instrument (PerkinElmer Life Sciences and Analytical Science, Inc., Waltham, MA) using excitation and emission wavelengths set at 485 and 530 nm, respectively, with linearity obtained between 0.002 and 2.5 mg/liter; R2 > 0.998) and expressed by reference to the sample protein content.
Purification and Determination of the Solubility of PBP 2a—The mecA gene was cloned previously, and a general procedure for protein purification was used as described earlier (27). The solubility of the protein was assessed as described (22), except that the protein was incubated for up to 72 h in 25 mm HEPES (pH 7.0) or in 25 mm phosphate buffer (pH 5.5; the pH was carefully checked prior to all experiments), both supplemented with 1 m NaCl. The results for each condition of pH indicated that the protein remained fully soluble for up to 3 days.
Bocillin FL-PBP 2a Binding Assay—We adapted a previously described method (28) but using Bocillin FL as a reporter antibiotic. The purified protein was first diluted for 10 min in pH-adjusted buffer containing 1 m NaCl (pH 7.0, 25 mm HEPES buffer; pH 5.0–7.0, 25 mm phosphate buffer; we checked that the results obtained at pH 7.0 were not influenced by the buffer used) and then incubated with Bocillin FL for various time intervals. Aliquots were collected, proteins were separated by SDS-PAGE, and the acyl-enzyme complex was detected by fluorescence (Storm 840® Fluorimager; Amersham Biosciences). The results are expressed as relative emission intensities. Equiloading of the gels was checked in all experiments by Coomassie Blue staining. For control purposes, (i) the binding of Bocillin FL was also examined by immunodetection using an antibody raised against the fluorescent moiety of the molecule (anti-BODIPY® FL rabbit IgG) and goat horseradish peroxidase-labeled anti-rabbit IgG (both from Invitrogen); (ii) we also looked at the binding of 3H-labeled penicillin G, following the same approach as for Bocillin FL with assay of radioactivity in the PBP 2a band observed after SDS-PAGE.
Determination of the Kinetic Parameters for Interactions of Nitrocefin with PBP 2a—Acylation and deacylation studies (using nitrocefin for acylation and Bocillin FL for deacylation as reporter molecules, respectively) were performed as described earlier (27).
Circular Dichroism—CD spectra of PBP 2a were recorded on a stopped flow circular dichroism spectrometer (Aviv model 202 SF; Aviv Biomedical Inc., Lakewood, NJ) with a 2-mm path length at 25 °C in the absence or presence of 30 μm oxacillin (22).
Materials—Penicillin G, penicillin V, oxacillin, and nitrocefin were purchased from Sigma-Aldrich, and Bocillin FL was from Invitrogen (the structures of these compounds are shown in the supplemental material). Unless stated otherwise, all other reagents were obtained from Merck AG or Sigma-Aldrich.
Statistical Analyses—Curve fitting analyses were made using GraphPad Prism® version 4.03 for Windows (GraphPad Prism Software, San Diego, CA). The significance of the differences between paired values was tested by Student's t test (Excel 2000; Microsoft, Inc., Bellevue, WA).
RESULTS
MRSA is known to produce two main resistance factors for β-lactam antibiotics: PBP 2a and a class A β-lactamase. The β-lactamase was a first response to the early penicillins, which was successfully coped with by the use of β-lactamase-resistant penicillins and cephalosporins, and β-lactamase inhibitors. However, the consequence of the expression of PBP 2a in MRSA was more profound, because it conferred resistance to all commercially available β-lactam antibiotics (12).
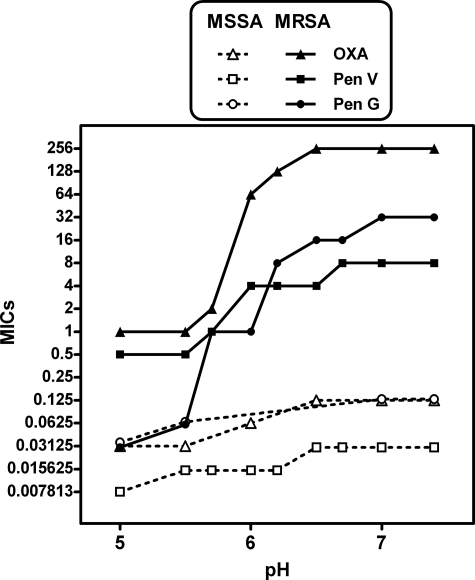
In studying the mechanism of β-lactam resistance by MRSA, we already have reported that acidic pH markedly decreases the MIC of cloxacillin (a β-lactamase-resistant penicillin) and meropenem (a carbapenem) for the β-lactamase-positive MRSA strain ATCC 33591 (20). In the context of the present study, we repeated these experiments with the β-lactamase-negative MRSA COL strain in comparison with the β-lactamase-negative MSSA strain ATCC 25923, using penicillin G (also known as benzylpenicillin, selected as reference β-lactam), penicillin V (Bocillin FL used for our binding and kinetic studies is a fluorogenic derivative of penicillin V (25, 29)), and oxacillin (a β-lactamase-resistant penicillin recommended for the treatment of infections caused by MSSA and also used by us in subsequent studies). The results presented in Fig. 1 reveal that, as a general trend, decreasing the pH from 7.4 to 5.0 lowered the MICs for all three antibiotics. The results were quite dramatic for penicillin G, with MICs for the MRSA COL strain experiencing attenuation of 11 log2 dilutions, reaching values identical to those observed for this antibiotic when tested against the MSSA ATCC 25923 strain (for which acidic pH caused only a modest decrease (1 log2 dilution) compared with neutral pH). For oxacillin, the decrease in MICs for the MRSA COL strain reached 9 log2 dilutions for the same change of pH. The decrease in MICs for penicillin V for the MRSA COL strain was less marked but still quite significant (4 log2 dilutions). The MICs of oxacillin and penicillin V at pH 5 were similar (0.25–1 mg/liter) but ∼4–5 log2 dilutions higher than those of penicillin G. Thus, the restoration of absolute susceptibility of the MRSA COL strain to oxacillin and penicillin V was less marked than for penicillin G, a point that will need to be taken into account for correct interpretation of the results of our subsequent studies.
FIGURE 1.
Influence of pH on the MIC of penicillin G (Pen G, circles), penicillin V (Pen V, squares), and oxacillin (OXA, triangles) on MSSA ATCC 25923 (open symbols) and MRSA COL (closed symbols) strains.
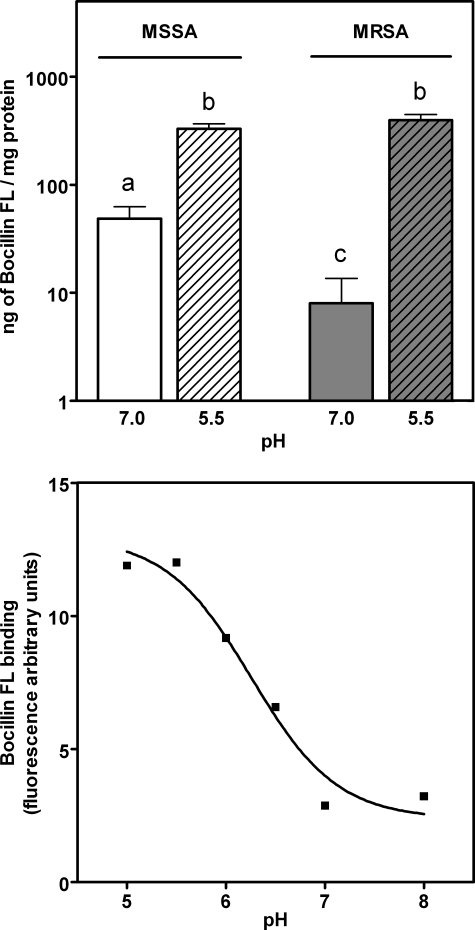
We then compared the influence of pH on total penicillin binding properties of growing MRSA (COL strain) and MSSA (ATCC 25923 strain) by exposing them to Bocillin FL in broth and measuring thereafter the amount of Bocillin FL associated with cell extracts. Fig. 2 (upper panel) shows that the binding of Bocillin FL was considerably less for MRSA in comparison with MSSA when bacteria had been grown and binding had been performed at pH 7. Binding to MRSA was increased ∼50-fold when the experiment was performed at pH 5.5. Most interestingly, no significant difference was noted between MSSA and MRSA for bacteria grown and tested at that pH.
FIGURE 2.
Influence of pH on the binding of Bocillin FL to whole cells and to purified PBP 2a. Upper panel, growing bacteria were incubated in broth at 37 °C with Bocillin FL for 30 min at the pH indicated in the abscissa, and the samples were prepared for fluorescence measurement. White bars, MSSA ATCC 25923; gray bars, MRSA COL. The values are the means ± S.D. (n = 3). Bars with different letters are significantly different from all others (p < 0.01). Lower panel, Bocillin FL (0.2 μg) was mixed for 20 min with 3 μm purified PBP 2a in 50 mm phosphate buffers adjusted to different pH values before being applied to gel for electrophoretic separation (the value recorded in HEPES buffer at pH 7.0 was not significantly different from that shown for the corresponding phosphate buffer here).
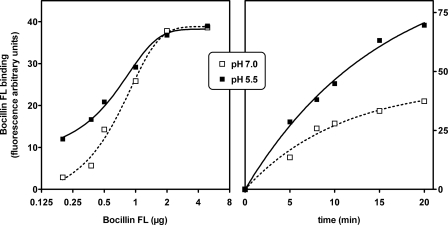
These experiments prompted us to directly measure the binding of Bocillin FL to purified PBP 2a. Fig. 2 (lower panel) shows that binding was pH-dependent with values ∼5-fold larger at pH 5 compared with pH 7, under the experimental condition used. This indicates that the covalent chemistry that PBP 2a experiences and, by necessity, the access to the active site by the antibiotic are enhanced at lower pH. In subsequent experiments, the influence of pH on binding of Bocillin FL to PBP 2a was further characterized with respect to concentration and time. The results presented in Fig. 3 (left panel) show that more Bocillin FL could bind to PBP 2a at low concentrations at pH 5.5 compared with pH 7.0, denoting potentially a higher affinity. The experiment in the right panel of Fig. 3 reveals that acylation of the enzyme is achieved more readily at lower pH within the 20 min of the experimental time frame, which is consistent with and relevant to the doubling time of most staphylococci. This indicates, as hypothesized in this study, that PBP 2a is more susceptible to inhibition by a β-lactam antibiotic within the growth time frame of the organisms. Given unlimited time for the requisite conformational change of PBP 2a to take place, the fluorescence that results from covalent chemistry caused by the ultimate 100% protein modification at the two experimental pH points should converge.
FIGURE 3.
Influence of pH on the binding of Bocillin FL to purified PBP 2a. Incubations were performed at room temperature in 1 m NaCl buffered at 5.5 (25 mm phosphate) or at 7.0 (25 mm HEPES) with PBP 2a (3 μm). Left panel, binding measured after 25 min in the presence of increasing amounts of Bocillin FL (logarithmic scale) Right panel, binding measured over a 20 min time frame in the presence of 0.5 μg of Bocillin FL.
Two types of controls were made to check against possible artifacts because of the use of the fluorescent tracer. First, we examined whether acidic pH would modify the fluorescence of Bocillin FL (tested at two dilutions) and found no effect. Second, we looked for Bocillin FL binding by immunochemical detection rather than by fluorescence and for 3H-labeled-penicillin G (using another sample of soluble PBP 2a (kindly provided by J. Coyette)) and found also an increased binding.
The kinetics of PBP 2a acylation and deacylation by β-lactams at neutral and acidic pH were then undertaken using nitrocefin and Bocillin FL as reporter molecules for acylation and deacylation, respectively. We took advantage of the distinctive change in the absorption spectrum of nitrocefin upon cleavage of the amide bond in the β-lactam ring by enzyme acylation and the capacity of Bocillin FL to bind PBP 2a as the acyl-enzyme species experienced deacylation (see Ref. 27 for experimental design). We first investigated nitrocefin for the purpose of this work by measuring its MIC toward the MRSA COL strain at pH 5.5 versus 7.4 and observed values of 0.25 and 2 mg/liter, respectively. Subsequently, we determined certain kinetic parameters for interactions of the antibiotic with PBP 2a (Table 1). The minimal interaction scheme assumes binding of nitrocefin (N) to the enzyme in an initial non-covalent complex (EN), which proceeds to acylate the active site serine of PBP 2a, to give rise to the covalent complex E-N (Table 1). This covalent complex enjoys substantial longevity, but nonetheless ultimately experiences hydrolytic deacylation to liberate PBP 2a from inhibition. Table 1 shows the pertinent dissociation constant (Kd) of the nitrocefin-PBP 2a non-covalent complex (EN), the PBP 2a acylation rate constant (k2) to yield the acyl-enzyme species (E-N), and the deacylation rate constant from E-N (k3). It appears that at the acidic pH the dissociation constant for the pre-acylation complex was lowered by 2-fold, and the rate constant for enzyme acylation was enhanced by somewhat more than the same magnitude. We hasten to add that PBP 2a does not have the catalytic machinery for the deacylation of the acyl-enzyme species, and this is the reason that once the acyl-enzyme species has formed, it enjoys substantial longevity. As such, we note that the values for the deacylation rate constants at the two pH values remain unchanged within the error limits recorded for the determinations. This is exactly as would have been expected. As a result, the apparent second-order rate constant for the process between PBP 2a and nitrocefin (k2/Kd), a measure of the overall efficiency of PBP 2a inhibition by the β-lactam, experienced an increased of 5-fold in acidic pH, in agreement with the change in MIC. We also checked for the maximal amplitude of the reaction at the pH 5.5 and 7.0 and found no significant difference, indicating that PBP 2a was not intrinsically resistant to acylation at pH 7 versus pH 5.5 but rather required larger drug concentration to achieve saturation of the enzyme. This is also indicated by the elevated MIC values at higher pH.
TABLE 1.
Kinetic parameters for interactions of nitrocefin and PBP 2a at pH 5.0 and 7.0
The parameters apply to the minimal kinetic scheme E + N ⇄ EN → E-N → E + P, where E is the PBP 2a, N is nitrocefin, EN is the noncovalent complex between PBP 2a and nitrocefin, E-N is the covalent complex between PBP 2a and nitrocefin (acyl-enzyme), and P is the product of hydrolysis of nitrocefin. The ⇄ arrows refer to the reversible formation of the non-covalent complex EN (with dissociation constant Kd), the first → arrow refers to the formation of the acylated enzyme (with rate constant k2), and the second → arrow refers to the deacylation reaction (with rate constant k3). k2/Kd is the second order rate constant for the bimolecular encounter between nitrocefin and PBP 2a.
|
Parameter
|
pH
|
|
|---|---|---|
| 7.0 | 5.5 | |
| Kd (μm) | 195 ± 28 | 100 ± 30 |
| k2 (s-1 × 103) | 6.0 ± 0.6 | 15.2 ± 1.5 |
| k3 (s-1 × 106) | 2.5 ± 0.2 | 2.8 ± 0.2 |
| k2/Kd (s-1m-1) | 31 ± 3 | 150 ± 14 |
The results reported above argue for a general improved access by the antibiotic to the active site. This is manifested in the lowering of the dissociation constant for the pre-acylation complex, which indicates a more favorable formation of the complex at the lower pH. In addition, we note a more favorable process for the acylation of the enzyme. These observations are telling in view of the available x-ray structure for PBP 2a in the native state and in complex with two antibiotics, including nitrocefin (30). The existing structural information reveals PBP 2a to exist in a state that is best described as a closed conformation for the active site for both the native and the acyl-enzyme species (27). This closed active site is the reason that the existing β-lactam antibiotic cannot inhibit the enzyme. Yet, we know that the enzyme is catalytically competent in cross-linking of the cell wall. Previous work disclosed that PBP 2a has an allosteric site for its substrate, the cell wall (31). Binding of the cell wall at this allosteric site triggers a conformational change in the protein that leads to the opening of the active site and access for the substrate to it. Fuda et al. (27) have reported that for β-lactam antibiotics to gain access to the active site, given ample time and favorable establishment of equilibria, the protein has to undergo a conformational change to open the active site before the antibiotic has the ability to acylate the serine. Hence, conformational changes are necessary both for the physiological function of the enzyme in cross-linking of the cell wall and for its inhibition by β-lactams.
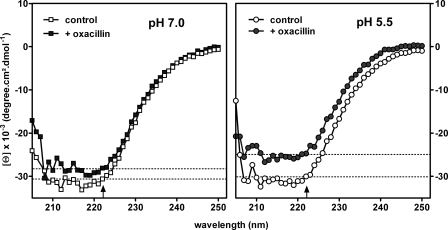
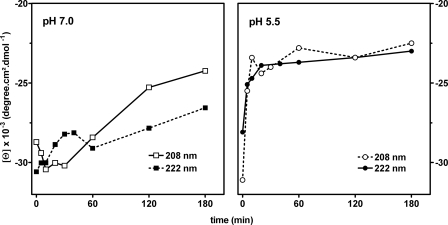
Edified by this information, we wondered whether the pH of the medium played a role in the conformational changes of PBP 2a in the presence of the antibiotic. This possibility was explored using CD spectroscopy of the protein in the presence and in the absence of oxacillin. As revealed in Fig. 4, the spectra of PBP 2a at pH 7.0 and 5.5 were virtually the same in the absence of antibiotic. On exposure of PBP 2a to oxacillin, however, we noted a change in the conformation of the protein, as was reported in an earlier study (27). Whereas this conformational flexibility was seen at both pH values, the magnitude of the effect was larger at pH 5.5. Moreover, Fig. 5 shows that the change (as detected by the increase in the ellipticity of the minima at both 208 and 222 nm) occurred within minutes of exposure to oxacillin at pH 5.5, with the new value remaining quite stable for at least 3 h. In contrast, this change developed only slowly at pH 7.0.
FIGURE 4.
Circular dichroic spectra of PBP 2a at pH 7. 0 (left panel) and pH 5.5 (right panel) in the absence (open symbols) and in the presence (closed symbols) of oxacillin (30 μm) for 30 min at 25 °C. The thin dotted lines in each graph represent minima of PBP 2a molar ellipticity at 222 nm (vertical arrow on the abscissa) for each condition. The spectrum of oxacillin has been subtracted from all data points.
FIGURE 5.
Change in PBP 2a molar ellipticity observed over time at pH 7. 0 (left panel) or pH 5.5 (right panel) upon exposure to oxacillin and recorded at 208 (open symbols) or 222 nm (closed symbols). The reactions were carried out with 30 μm oxacillin at 25 °C. The spectrum of oxacillin has been subtracted from all data points.
DISCUSSION
The importance of MRSA as a human pathogen is increasingly significant and costly (5, 32). Hence, it bodes poorly for humanity to lose β-lactams, the most important and generally accepted class of antibiotics (33), for this type of infection.
Restoration of activity of β-lactams toward MRSA at acidic pH was successively ascribed to (i) a reduced or even outright absence of the expression of PBP 2a (15), which is poorly inhibited by β-lactams (34), and (ii) a lack of transpeptidase activity of PBP 2a at low pH (20). The first possibility can be dismissed, because expression of mecA and the immunodetection of the encoded protein are both unaffected by acidic pH (20). Although the second hypothesis cannot be ruled out, the data presented in this report offer an alternative explanation. We propose that at acidic pH the active site of PBP 2a becomes more accessible to β-lactams when exposed to these antibiotics. The active site of this PBP is, indeed, sheltered under normal conditions, which is the reason for the lack of effective inhibition of the enzyme by conventional β-lactams. Improved accessibility at lower pH within the time frame relevant for the growth of MRSA was documented in this study by (i) an increase in the apparent rate and extent of binding of Bocillin FL, (ii) a change in protein conformation, (iii) an attenuation of the dissociation constant for the antibiotic, and (iv) an enhancement of the rate of enzyme acylation in the presence of the antibiotic. Thus, we suggest that conformational transition allowing acylation upon exposure to β-lactams becomes more facile at lower pH. Another explanation could simply be that the reaction of the β-lactam within the active site is stimulated by an acidic functional group on the protein (or antibiotic) that begins to become protonated at pH 5. This, however, is unlikely because the approach of the serine hydroxyl to the β-lactam carbonyl in the active site results in a nucleophilic addition reaction (to form the tetrahedral intermediate), which will be enhanced only at higher pH values (opposite to what is observed) (35). Nevertheless, titration of amino acid residues on the protein could enhance the conformational flexibility of PBP 2a at lower pH, resulting not only in accelerated rate of reaction but also in an overall increased exposition of the active site. This would also explain why more Bocillin FL could be bound to PBP 2a at acidic versus neutral pH, in which case the data of Fig. 3 (right panel) should be interpreted as reflecting differences in accessibility rather than in true equilibrium.
We observed a 50-fold enhancement of Bocillin FL binding with whole bacteria. It is difficult to single out a single binding event at the entire organismal level as the key event in any study. This issue is further complicated with the example in hand, because Bocillin FL, a β-lactam molecule, binds to all PBPs. It is known that inhibition of a mere fraction of PBP of any one kind is sufficient to kill the organism (36). Hence, at pH 7.0 a smaller fraction of PBPs are likely inhibited, but the function of PBP 2a is the reason that the organism survives in the face of the antibiotic challenge. We speculate that at pH 5.5 a much larger fraction of PBPs is modified by Bocillin FL; hence the effect might not be limited to only inhibition of PBP 2a. It is now shown that at least two PBPs have closed and open conformations (31, 37). One wonders whether additional examples might be found among PBPs of S. aureus to explain the rather large fluorescence reading at the whole cell level.
If PBP 2a shows facilitated acylation by β-lactams at acidic pH, we need, however, to explain why the protein could not be detected by autoradiographic analysis of PBPs prepared from bacteria grown at acidic pH in the presence of [3H]benzylpenicillin (15). An ongoing work suggests that PBP 2a could be unstable under the conditions used for sample preparation, and it therefore escapes detection. This is why we examined the binding of Bocillin FL not only in growing bacteria (where it could bind to all PBPs) but also in purified PBP 2a. The increase in Bocillin FL binding, when moving from neutral to acidic pH in the latter condition, parallels nicely with the decrease in MICs of penicillin V under a similar pH shift.
The magnitude of the lowering of the dissociation constant and the enhancement of the rate of acylation upon exposure of PBP 2a to nitrocefin at acidic versus neutral pH might seem modest at first glance. However, analysis of the β-lactam-resistant Streptococcal PBP 2x in comparison with the natural, sensitive PBP 2 and of a reverse, sensitive mutant (Sp328 PBP 2x) has shown that switching from resistance to susceptibility and vice versa can be achieved by variations in the acylation rate constants of merely 2–6-fold (38). The magnitude of the changes in the kinetic parameters measured here need also be examined in the context of the diminution of MIC values for nitrocefin, when switching pH from 7 to 5 (3 log2 decrease). Moreover, nitrocefin itself proved already fairly active against MRSA at neutral pH (MIC of 2 mg/liter). Other molecules such as penicillin G (this study) or meropenem or cloxacillin (32), which show a large decrease of MIC when tested at pH 5.5 versus pH 7.4, are nonchromogenic and therefore more difficult to use in this type of experiments. In contrast to Bocillin FL, also, these molecules are somewhat labile at acidic pH, which makes the experiments much more difficult to interpret. We also need to compare the values of the second-order rate constant (k2/Kd) at acidic pH (150 ± 14 s–1 m–1) with those recorded by Fuda et al. (22) (145 and 205 s–1 m–1) for three experimental anti-MRSA cephalosporins with MICs of 1–8 mg/liter.
The comparison between the influence of acidic pH on the binding of β-lactams to PBP 2a and the mode of action of the anti-MRSA cephalosporins also finds parallels in that they both manifest conformational changes in the protein. Our data show that incubation of PBP 2a with oxacillin at acidic pH induces conformational changes consistent with a loss of α-helical content, as was observed with three experimental anti-MRSA cephalosporins (22) and with ceftobiprole (24), the first clinically developed anti-MRSA cephalosporin (39). Of note is that (i) the changes seen here occur almost immediately upon the addition of oxacillin and remained stable thereafter subsequent to protein acylation and (ii) experiments were performed under conditions that were shown earlier to exclude artifacts that could have affected the circular dichroism spectra such as precipitation of the acyl-enzyme species or aggregation or oligomerization of the protein. Although the occurrence of conformational change during exposure of PBP 2a to β-lactam is now well documented, we hesitate to quantify this effect for reasons explained by Fuda et al. (31). Nevertheless, our data indicate a commonality of effects of a global environmental variable such as pH and the binding of a specific ligand on PBP 2a conformation for facilitating access to its active site. It would be interesting to examine whether (i) similar conformational changes could be initiated by other environmental variables or other ligands (which might be the case for the peptidoglycan itself (27)) and (ii) the effect of acidic pH and an anti-MRSA cephalosporin could be additive or perhaps even synergic on the killing of MRSA.
The influence of acidic pH on MRSA susceptibility probably has a broad significance, because (i) it is seen for several β-lactams (20)4; (ii) survival of S. aureus in phagocytic cells, where pH is acidic, is probably a significant cause of relapses and recurrences of many staphylococcal infections (40, 41); and (iii) pH may reach sufficiently low values in extracellular habitats such as skin surfaces (pH 4.2–5.9), mouth (pH 5–7), vagina (pH 4.2–6.6), and urinary tract (pH 4.6–7). Interestingly, the transglycosylase activity of PBP 2, needed for biosynthesis of peptidoglycan in MRSA prior to the action of PBP 2a in cross-linking two existing strands of it, exhibits maximal catalysis at pH 4.5–5.5 (42). Hence, a full knowledge of chemistry of both catalysis and inhibition under acidic pH is quite relevant to elucidation of the properties of this important bacterial pathogen.
Because the global effect of the change of pH at the organism level might manifest itself at other sites than merely PBP 2a, other possibilities to explain our observations on the susceptibility of MRSA to β-lactams cannot be ruled out at this time. However, our detailed analyses from a number of distinct but convergent experiments lead us to conclude that the effects on PBP 2a play a major effort in the phenotypic outcome. We have shown here for Bocillin FL and in a previous report for [3H]radiolabeled penicillin G (20) that acidic pH increases the binding of the antibiotic to the bacterium to a much larger extent than what is seen with purified PBP 2a. Acidic pH might therefore also favor binding of β-lactams to other PBPs. Yet, based on the copy numbers for PBPs in S. aureus (43) and the observations made in this report, the influence of pH should primarily involve PBP 2a because it is the most abundant. Moreover, PBP 2a function is pivotal for MRSA survival.
Although the quest for other antibiotic targets is important, the observations with PBP 2a reported herein should provide the impetus for additional study in effective control of MRSA. Ironically, indeed, establishing that MRSA is the offending organism leads to discontinuation ofβ-lactams by clinicians in favor of other antibiotics such as vancomycin or linezolid, which we know, today, to be poorly effective against intracellular forms of S. aureus (44, 45).4 Future studies will need to critically examine whether maintaining aβ-lactam in combination with these drugs could allow for faster and more extensive eradication of MRSA.
Supplementary Material
Acknowledgments
We thank J. Coyette for the kind gift of a sample of PBP 2a used for performing control experiments incorporated in response to a referee comment. We are grateful to Drs. S. Carryn, Y. Jossin, M. Suvorov, and S. Vakulenko for scientific discussions and to Dr. M. Page for critical reading of our manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants AI33170 and GM 61629 in the United States. This work was also supported in Belgium by Fonds de la Recherche Scientifique Médicale Grant 3.4.597.06, Fonds de la Recherche Scientifique travel fellowships (to S. L.), and Belgian Federal Science Policy Office Research Project P5/33 (research action P5). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. SP1.
Footnotes
The abbreviations used are: MRSA, methicillin-resistant S. aureus; MSSA, methicillin-susceptible S. aureus; PBP, penicillin-binding protein; MIC, minimum inhibitory concentration; Bocillin FL, Boron-difluoro-[2-[N-[3-[2-[(3,5-dimethyl-1H-pyrrol-2-yl-κN)methylene]-2H-pyrrol-5-yl-κN]-1-oxopropylamino]-penicillin V.
S. Lemaire, A. Olivier, F. Van Bambeke, P. M. Tulkens, P. C. Appelbaum, and Y. Glupczynski, submitted for publication.
References
- 1.Appelbaum, P. C. (2006) Clin. Microbiol. Infect. 12 (Suppl. 2) 3–10 [DOI] [PubMed] [Google Scholar]
- 2.Wijaya, L., Hsu, L. Y., and Kurup, A. (2006) Ann. Acad. Med. Singapore 35 479–486 [PubMed] [Google Scholar]
- 3.File, T. M., Jr. (2007) Cleve. Clin. J. Med. 74 (Suppl. 4) S6–S11 [DOI] [PubMed] [Google Scholar]
- 4.Anderson, D. J., Sexton, D. J., Kanafani, Z. A., Auten, G., and Kaye, K. S. (2007) Infect. Control Hosp. Epidemiol. 28 1047–1053 [DOI] [PubMed] [Google Scholar]
- 5.Klevens, R. M., Morrison, M. A., Nadle, J., Petit, S., Gershman, K., Ray, S., Harrison, L. H., Lynfield, R., Dumyati, G., Townes, J. M., Craig, A. S., Zell, E. R., Fosheim, G. E., McDougal, L. K., Carey, R. B., and Fridkin, S. K. (2007) J. Am. Med. Assoc. 298 1763–1771 [DOI] [PubMed] [Google Scholar]
- 6.Weese, J. S. (2005) J. Am. Anim. Hosp. Assoc. 41 150–157 [DOI] [PubMed] [Google Scholar]
- 7.Lee, J. H. (2003) Appl. Environ. Microbiol. 69 6489–6494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Normanno, G., Corrente, M., La Salandra, G., Dambrosio, A., Quaglia, N. C., Parisi, A., Greco, G., Bellacicco, A. L., Virgilio, S., and Celano, G. V. (2007) Int. J. Food Microbiol. 117 219–222 [DOI] [PubMed] [Google Scholar]
- 9.de Neeling, A. J., van den Broek, M. J., Spalburg, E. C., Santen-Verheuvel, M. G., Dam-Deisz, W. D., Boshuizen, H. C., van de Giessen, A. W., van Duijkeren, E., and Huijsdens, X. W. (2007) Vet. Microbiol. 122 366–372 [DOI] [PubMed] [Google Scholar]
- 10.Huijsdens, X. W., van Dijke, B. J., Spalburg, E., Santen-Verheuvel, M. G., Heck, M. E., Pluister, G. N., Voss, A., Wannet, W. J., and de Neeling, A. J. (2006) Ann. Clin. Microbiol. Antimicrob. 5 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wulf, M. W., Sorum, M., van Nes, A., Skov, R., Melchers, W. J., Klaassen, C. H., and Voss, A. (2008) Clin. Microbiol. Infect. 14 29–34 [DOI] [PubMed] [Google Scholar]
- 12.Fuda, C. C., Fisher, J. F., and Mobashery, S. (2005) Cell Mol. Life Sci. 62 2617–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Appelbaum, P. C. (2006) Clin. Microbiol. Infect. 12 (Suppl. 1) 16–23 [DOI] [PubMed] [Google Scholar]
- 14.Pantosti, A., Sanchini, A., and Monaco, M. (2007) Fut. Microbiol. 2 323–334 [DOI] [PubMed] [Google Scholar]
- 15.Hartman, B. J., and Tomasz, A. (1984) J. Bacteriol. 158 513–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgopapadakou, N. H., Smith, S. A., and Bonner, D. P. (1982) Antimicrob. Agents Chemother. 22 172–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinho, M. G., Filipe, S. R., de Lencastre, H., and Tomasz, A. (2001) J. Bacteriol. 183 6525–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guignard, B., Entenza, J. M., and Moreillon, P. (2005) Curr. Opin. Pharmacol. 5 479–489 [DOI] [PubMed] [Google Scholar]
- 19.Sabath, L. D., Wallace, S. J., and Gerstein, D. A. (1972) Antimicrob. Agents Chemother. 2 350–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemaire, S., Van Bambeke, F., Mingeot-Leclercq, M. P., Glupczynski, Y., and Tulkens, P. M. (2007) Antimicrob. Agents Chemother. 51 1627–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohkuma, S., and Poole, B. (1978) Proc. Natl. Acad. Sci. U. S. A. 75 3327–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuda, C., Hesek, D., Lee, M., Heilmayer, W., Novak, R., Vakulenko, S. B., and Mobashery, S. (2006) J. Biol. Chem. 281 10035–10041 [DOI] [PubMed] [Google Scholar]
- 23.Davies, T. A., Page, M. G., Shang, W., Andrew, T., Kania, M., and Bush, K. (2007) Antimicrob. Agents Chemother. 51 2621–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovering, A., Danel, F., Page, M. G. P., and Strynadka, N. J. (2006) Clin. Microbiol. Infect. 12 Suppl. 4, 1586 (abst.) [Google Scholar]
- 25.Gee, K. R., Kang, H. C., Meier, T. I., Zhao, G., and Blaszcak, L. C. (2001) Electrophoresis 22 960–965 [DOI] [PubMed] [Google Scholar]
- 26.Chambers, H. F., and Miick, C. (1992) Antimicrob. Agents Chemother. 36 656–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuda, C., Suvorov, M., Vakulenko, S. B., and Mobashery, S. (2004) J. Biol. Chem. 279 40802–40806 [DOI] [PubMed] [Google Scholar]
- 28.Preston, D. A., Wu, C. Y., Blaszczak, L. C., Seitz, D. E., and Halligan, N. G. (1990) Antimicrob. Agents Chemother. 34 718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao, G., Meier, T. I., Kahl, S. D., Gee, K. R., and Blaszczak, L. C. (1999) Antimicrob. Agents Chemother. 43 1124–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim, D., and Strynadka, N. C. (2002) Nat. Struct. Biol. 9 870–876 [DOI] [PubMed] [Google Scholar]
- 31.Fuda, C., Hesek, D., Lee, M., Morio, K., Nowak, T., and Mobashery, S. (2005) J. Am. Chem. Soc. 127 2056–2057 [DOI] [PubMed] [Google Scholar]
- 32.Gould, I. M. (2006) Int. J. Antimicrob. Agents 28 379–384 [DOI] [PubMed] [Google Scholar]
- 33.Moellering, R. C., and Eliopoulos, G. M. (2007) in Principles and Practice of Infectious Diseases (Mandell, G. L., Bennett, J. E., and Dolin, R., eds) Elsevier Inc.
- 34.Berger-Bachi, B., Strassle, A., and Kayser, F. H. (1986) Eur. J. Clin. Microbiol. 5 697–701 [DOI] [PubMed] [Google Scholar]
- 35.Meroueh, S. O., Fisher, J. F., Schlegel, H. B., and Mobashery, S. (2005) J. Am. Chem. Soc. 127 15397–15407 [DOI] [PubMed] [Google Scholar]
- 36.Macheboeuf, P., Fischer, D. S., Brown, T., Jr., Zervosen, A., Luxen, A., Joris, B., Dessen, A., and Schofield, C. J. (2007) Nat. Chem. Biol. 3 565–569 [DOI] [PubMed] [Google Scholar]
- 37.Lovering, A. L., De Castro, L., Lim, D., and Strynadka, N. C. (2006) Protein Sci. 15 1701–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mouz, N., Gordon, E., Di Guilmi, A. M., Petit, I., Petillot, Y., Dupont, Y., Hakenbeck, R., Vernet, T., and Dideberg, O. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 13403–13406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Page, M. G. (2007) Expert. Opin. Emerg. Drugs 12 511–524 [DOI] [PubMed] [Google Scholar]
- 40.Foster, T. J. (2005) Nat. Rev. Microbiol. 3 948–958 [DOI] [PubMed] [Google Scholar]
- 41.Lowy, F. D. (2000) Trends Microbiol. 8 341–343 [DOI] [PubMed] [Google Scholar]
- 42.Barrett, D., Leimkuhler, C., Chen, L., Walker, D., Kahne, D., and Walker, S. (2005) J. Bacteriol. 187 2215–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fisher, J. F., Meroueh, S. O., and Mobashery, S. (2005) Chem. Rev. 105 395–424 [DOI] [PubMed] [Google Scholar]
- 44.Barcia-Macay, M., Seral, C., Mingeot-Leclercq, M. P., Tulkens, P. M., and Van Bambeke, F. (2006) Antimicrob. Agents Chemother. 50 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barcia-Macay, M., Lemaire, S., Mingeot-Leclercq, M. P., Tulkens, P. M., and Van Bambeke, F. (2006) J. Antimicrob. Chemother. 58 1177–1184 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.