Abstract
Serotonin is involved in a variety of physiological processes in the central nervous system and the periphery. As the rate-limiting enzyme in serotonin synthesis, tryptophan hydroxylase plays an important role in modulating these processes. Of the two variants of tryptophan hydroxylase, tryptophan hydroxylase 2 (TPH2) is expressed predominantly in the central nervous system, whereas tryptophan hydroxylase 1 (TPH1) is expressed mostly in peripheral tissues. Although the two enzymes share considerable sequence homology, the regulatory domain of TPH2 contains an additional 41 amino acids at the N terminus that TPH1 lacks. Here we show that the extended TPH2 N-terminal domain contains a unique sequence involved in the regulation of enzyme expression. When expressed in cultured mammalian cells, TPH2 is synthesized less efficiently and is also less stable than TPH1. Removal of the unique portion of the N terminus of TPH2 results in expression of the enzyme at a level similar to that of TPH1, whereas protein chimeras containing this fragment are expressed at lower levels than their wild-type counterparts. We identify a region centered on amino acids 10–20 that mediates the bulk of this effect. We also demonstrate that phosphorylation of serine 19, a protein kinase A consensus site located in this N-terminal domain, results in increased TPH2 stability and consequent increases in enzyme output in cell culture systems. Because this domain is unique to TPH2, these data provide evidence for selective regulation of brain serotonin synthesis.
Serotonin (5-hydroxytryptamine (5HT)5) is involved in a wide range of functions throughout the body, including the regulation of vascular tone, appetite, wakefulness, and mood. Its role in mood regulation is supported by the successful treatment of a number of psychiatric disorders with drugs that regulate extracellular 5HT. Currently, this is accomplished by inhibiting either the serotonin transporter or monoamine oxidases, both of which increase the extracellular 5HT by blocking clearance of released transmitter (1–4). As the rate-limiting enzymes in 5HT synthesis, tryptophan hydroxylases (TPHs) provide another target for the regulation of 5HT levels. Two TPH isoforms, encoded by separate genes, have been identified in mammals (5). In adults, TPH1 is mostly expressed in non-neuronal cells and plays an essential role in peripheral 5HT synthesis (5–7). In contrast, TPH2 is expressed in neurons (6, 7) and controls brain 5HT synthesis (8). Since its discovery, genetic variants in the TPH2 gene have been identified in cohorts of patients with depression, suicidality, and bipolar disorder (9–18). Thus, mechanisms that specifically regulate TPH2 activity may be important for the etiology or management of psychiatric disorders.
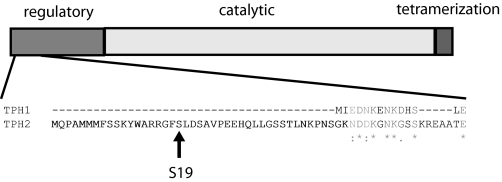
TPH2 is part of a family of amino acid hydroxylases that includes TPH1, tyrosine hydroxylase (TH), and phenylalanine hydroxylase. All members of this family share a similar structure composed of an N-terminal regulatory domain, a central catalytic domain, and a C-terminal tetramerization domain with considerable sequence homology across family members in all but the regulatory domains (19, 20). As closely related members of the hydroxylase family, however, TPH1 and TPH2 do share substantial homology in the regulatory domain with the exception of the first 41 amino acids at the N terminus of TPH2, which are unique to this enzyme (Fig. 1). The regulatory domains of other members of the family, such as TPH1 and TH, have been shown to modulate enzyme activity and thermostability through various mechanisms, including phosphorylation by protein kinases (21–31). In line with this, the extended N terminus of TPH2 contains a serine at position 19, which is a candidate for PKA phosphorylation (32) and has been recently shown to interact with 14-3-3 proteins in a phosphorylation-dependent manner to increase protein stability (33). This supports the hypothesis that the extended N-terminal domain of TPH2 may contain sites involved in regulating neuronal 5HT production.
FIGURE 1.
Schematic diagram of amino acid hydroxylases. Shown is a diagram of the major domains of amino acid hydroxylases and a sequence alignment (ClustalW) of the first 60 amino acids of human TPH2 with the corresponding residues of human TPH1. Identity is indicated with an asterisk, conserved substitutions are indicated with a colon, and semiconserved substitutions are shown with a period. The candidate PKA site, Ser19, is indicated.
In this study, we use a cell culture system to characterize the role of the extended N terminus of TPH2 in modulating the enzyme's properties. Our results show that this region contains sites that markedly reduce enzyme expression levels by altering both enzyme synthesis and stability. We also provide evidence that phosphorylation of TPH2 at Ser19 by PKA counteracts this reduced stability, resulting in an increase in enzyme expression with a concomitant increase in 5HT production.
MATERIALS AND METHODS
Cell Culture and Transfection—HEK 293 cells were grown in Eagle's minimal essential medium with Earle's salt supplemented with 10% (v/v) fetal bovine serum and a 50 μg/ml gentamicin solution (Invitrogen). PC12 cells were grown in Dulbecco's modified Eagle's medium supplemented with 5% (v/v) bovine calf serum and 5% (v/v) equine serum and 50 μg/ml gentamicin. Cells were transiently transfected by electroporation using 240 V and 1000 microfarads or 300 V and 1000 microfarads for HEK or PC12 cells, respectively, using a Gene Pulser II (Bio-Rad) and plated in 100- or 150-mm dishes (Molecular Technologies, St. Louis, MO). To facilitate detection of expressed proteins and eliminate potential confounds with variances in transcript initiation sequences, all constructs used in these studies were triple hemagglutinin (HA)-tagged at the N terminus and placed in a modified pcDNA3.1 vector (34) with the exception of the green fluorescent protein (GFP) fusion constructs, which also contained the triple HA tag but were generated in the pEGFP vector.
5HT Measurement—At 48–72 h post-transfection, PC12 cells were collected in a solution of phosphate-buffered saline with 1 mm EGTA and 1 mm EDTA and centrifuged at 1000 × g for 5 min at 4 °C. The resulting pellet was resuspended in 100 μl of 0.1 m perchloric acid for lysis using a tissue dismembranator (Fisher). The solution was then centrifuged at 15,000 × g for 10 min at 4 °C. The resulting supernatant was filtered using a 0.2-μm centrifuge filter tube (Millipore Corp., Billerica, MA) at 4 °C. 5 μl of the filtrate was analyzed by HPLC as previously described (35). Briefly, samples were separated on a microbore Unijet C18 reverse-phase column (5 μm, 1 × 150 mm; BAS, West Lafayette, IN) using a mobile phase of 50 mm monobasic sodium phosphate, 0.2 mm octyl sodium sulfate, 0.1 mm EDTA, 10 mm NaCl, and 10% methanol at pH 2.6. The applied electrode potential was +0.65 V. 5HT concentrations were normalized to endogenous dopamine levels to correct for variations in sample size (8).
5-Hydroxytryptophan (5HTP) Measurement—At 48 h posttransfection, cells were treated with 100 μm NSD-1015 (Sigma) to block amino acid decarboxylase (AADC) activity and either 2.5 μm forskolin (Sigma) or an equal volume of DMSO (control) for 16 h. Cells were then prepared as described above for 5HT measurement, and 5HTP concentrations were measured using the same HPLC conditions. Because we experimentally determined that in the presence of NSD-1015, forskolin did not measurably alter accumulated DA levels (data not shown), 5HTP concentrations were normalized to endogenous dopamine levels to correct for variations in sample size.
Western Blotting—Cells were collected as noted above for HPLC assays and lysed in a solution of 20 mm Tris (pH 7.5), 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, and 1% Triton X-100 plus 1× Complete, EDTA-free protease inhibitor mixture (Roche Applied Science) for 15 min. The lysate was centrifuged at 20,000 × g for 10 min at 4 °C, and the resulting supernatant was collected, and its protein content was assessed using a Bio-Rad DC protein assay kit. Samples were diluted to 4 μg of protein/μl in lysis buffer and further diluted to 2 μg/μl in Laemmli sample buffer supplemented with 5% β-mercaptoethanol. Samples were incubated for 20 min at 55 °C before they were loaded onto a 10% SDS-PAGE gel (Invitrogen) and run at 110 V. Transfers were performed using the X-Cell blot module (Invitrogen) at 25 V overnight at 4 °C. HA-tagged proteins were analyzed using the anti-HA antibody (Covance, Inc., Princeton, NJ) diluted at 1:3000. Anti-actin (Chemicon International, Inc., Temecula, CA), diluted 1:1000, anti-neomycin phosphotransferase (Upstate Cell Signaling Solutions, Lake Placid, NY), diluted 1:500, and anti-vimentin (Chemicon), diluted 1:1000, antibodies were used as controls. All primary antibody incubations took place overnight. Blots were then incubated for 1 h with sheep anti-mouse or goat anti-rabbit secondary antibodies conjugated to horseradish peroxidase (Amersham Biosciences) and revealed using the Pico Lum ECL kit (Pierce) and Eastman Kodak Co. BioMax film.
Messenger RNA Levels—Total RNA was prepared using the TRIZOL Reagent (Invitrogen). First strand cDNA was then generated using the iScript cDNA synthesis kit (Bio-Rad). Quantitative reverse transcriptase PCR was run for 40 cycles using SYBR green dye on a Lightcycler 2.0 system (Roche Applied Science). The following primers were used at a concentration of 1 μm: GFP forward, 5′-TGACCCTGAAGTTCATCTGCACCA-3′; GFP reverse, 5′-AAGAAGTCGTGCTGCTTCATGTGG-3′; TPH2 forward, 5′-TGGAGCAGGGTTACTTTCGTCCAT-3′; TPH2 reverse, 5′-AGTAAGCGTCCTGAAAGGTGGTGA-3′; TPH1 forward, 5′-TCCTCTCTTGGCTGAACCCAGTTT-3′; and TPH1 reverse, 5′-TCTTGTTTGCACAGCCCAAACTCC-3′. The second derivative maximum was used to derive the cycle number for each sample. Sample cycle numbers were then normalized to actin to account for differences in cell number.
Solubility—Cells were prepared as described above for Western blotting except that after removal of the postlysis supernatant (soluble fraction), the resulting pellet (insoluble fraction) was then denatured using a mixture of 8 m urea and 0.5% SDS for 15 min on ice. This new sample was then centrifuged again at 20,000 × g for 10 min, and both supernatant fractions were processed as described above for Western blotting.
Pulse Chase—Cells were placed in Met- and Cys-free Dulbecco's modified Eagle's medium for 1 h prior to treatment with 300 μCi of [35S]cysteine and methionine EXPRE35S35S Easy Tag (PerkinElmer Life Sciences) for 45 min. Medium was then removed and replaced with complete Dulbecco's modified Eagle's medium. Cells were collected at the indicated times and lysed as described above for Western blotting. Samples were then immunoprecipitated using anti-HA-conjugated agarose beads (Sigma) and run on a 10% SDS-polyacrylamide gel at 110 V. The gel was then dried overnight on a Savant slab gel dryer (ThermoFisher Scientific, Waltham, MA). Following drying, gels were exposed to Kodak BioMax MR film for 3–6 days at -70 °C. Band intensities were quantified and fit to a first order decay function to estimate the half-life of each enzyme for each experiment. These values were then averaged to obtain the mean half-life for each enzyme.
Cellular Protein Stability—Cells were initially serum-starved for 4 h and then treated with 10 μg/ml cycloheximide to arrest protein translation. Cells were then harvested at the stated time points and lysed, and the TPH contents remaining were analyzed by Western blotting as described above. Similar to the pulse-chase experiments, half-lives were estimated by curve fitting for each individual experiment and averaged to obtain the mean half-life for each enzyme.
In Vitro Translation— 0.5 μg of the cDNA used to transfect cells mentioned above was used with the TNT®T7 coupled reticulocyte lysate in vitro translation system (Promega, Madison, WI). Samples were then diluted 1:5 in Laemmli sample buffer and run on a 10% SDS-PAGE, and protein levels were analyzed by Western blot as described above.
Phosphorylation Assays—A peptide with the sequence corresponding to amino acids 11–24 of TPH2 with an N-terminal HA tag (YPYDVPDYAKYWARRGFSLDSAV) and a control peptide with an alanine substitution at position 19 (YPYDVPDYAKYWARRGFALDSAV) were obtained from Sigma and used as substrates for a PKA assay kit (Upstate Cell Signaling Solutions). Briefly, 5 μg of peptide was incubated with recombinant PKA, cAMP, and both cold and [32P]ATP in the presence of inhibitors of PKC and calcium- and calmodulin-dependent protein kinase for 10 min at 30 °C, and the resulting mixture was aliquoted on P81 paper. The paper was then washed several times, and the levels of incorporated 32P were determined using a Packard 1900 TR liquid scintillation counter.
PKA Activation—Cells were treated with 2.5 μm forskolin for 4 h at 37 °C, and changes in protein expression were assessed by Western blot as described above.
Statistical Analysis—A one-tailed t test was used to compare the mean half-lives of TPH1 and TPH2 and differences in translation efficiency between TPH1 and TPH2. All other data were compared by analysis of variance with Tukey's post hoc analysis. p < 0.05 was considered statistically significant. GraphPad Prism version 3.02 was used to generate all graphs and to perform all statistical analyses (GraphPad Software, San Diego, CA).
RESULTS
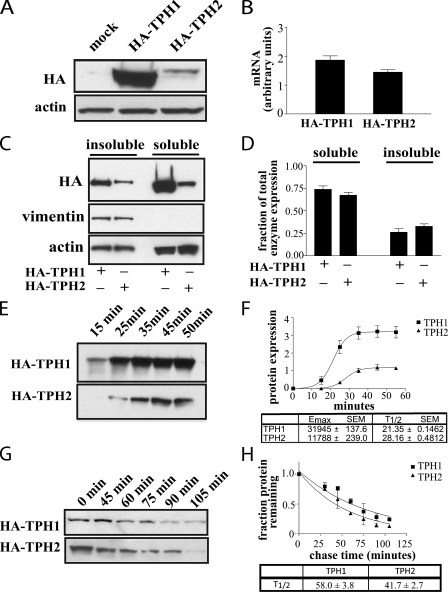
Differences in Expression of TPH1 and TPH2—TPH1 and TPH2 share considerable homology across the majority of their sequences with the exception of the additional 41 amino acids at the N terminus of TPH2 that are lacking in TPH1 (Fig. 1). Because these amino acids lie in the regulatory domain of the enzyme, our first step in characterizing the role of these residues was to identify differences in the regulation of TPH2 versus TPH1. To this end, we generated stable cell lines expressing each of the two hydroxylases in PC12 cells. When protein expression levels were examined through Western blotting of whole cell lysates, we found that TPH1 was expressed at a much higher level than TPH2 (Fig. 2A) despite similar mRNA levels as measured using quantitative reverse transcription-PCR (Fig. 2B) (p = 0.07). In addition, analysis of the insoluble/noncytosolic fraction of the cell lysate revealed that TPH1 and TPH2 exhibit similar levels of solubility, with ∼70% of both enzymes appearing in the soluble/cytosolic fraction of the lysate (Fig. 2, C and D). Thus, the difference in expression levels between TPH1 and TPH2 results from differences in either their translational efficiency or stability. To address the first possibility, plasmids containing the cDNA for both HA-TPH1 and HA-TPH2 were used for in vitro translation, which revealed markedly less synthesis of TPH2 compared with TPH1 (Fig. 2, E and F) (p < 0.01). The two enzymes differed in both maximum enzyme production and rate of synthesis with a ∼2.5-fold greater maximum expression of TPH1 over TPH2 and an ∼25% faster rate of synthesis. To address the second possibility, that differences in stability underlie the differences in enzyme expression, we examined degradation of both enzymes in cells following inhibition of new protein synthesis with cyclohexamide as well as pulse-chase experiments. TPH1 exhibited a half-life of 59.0 ± 3.9 min, and the half-life of TPH2 was 43.5 ± 3.7 min (Fig. 2G) (p < 0.05). Similarly, in the pulse-chase experiments, TPH1 exhibited a half-life of 58.0 ± 3.8 min, whereas that of TPH2 was 41.7 ± 2.7 min (Fig. 2H) (p < 0.05). These results suggest that TPH2 is degraded more rapidly than TPH1. Thus, it appears that differences in the rate of enzyme synthesis coupled with differences in enzyme stability both contribute to the observed differences in expression levels.
FIGURE 2.
The extended N terminus of TPH2 contains a regulatory determinant. A, PC12 cells stably expressing HA-TPH1 exhibited higher levels of hydroxylase expression than those stably expressing HA-TPH2 despite similar levels of mRNA (B) as assessed by quantitative reverse transcription-PCR (p = 0.07). C and D, analysis of the soluble/cytosolic and insoluble/noncytosolic fractions of HA-TPH1 and HA-TPH2 from PC12 cell lysates shows similar patterns of soluble versus insoluble expression, with both enzymes roughly 70% soluble and 30% insoluble (i.e. associated with the particulate fraction). The intermediate filament protein vimentin was used as a positive control for the insoluble/noncytosolic fraction. E and F, results of an in vitro translation performed on cDNA in a modified pcDNA vector show that nearly 3-fold more HA-TPH1 is synthesized than HA-TPH2 and that HA-TPH1 is synthesized ∼25% faster than HA-TPH2. This difference is significant (p < 0.01). G, a representative blot of n = 3 assays using 10 μg/ml cycloheximide to block new protein synthesis in PC12 cells stably transfected with HA-TPH1 or HA-TPH2, which show that TPH2 is degraded more rapidly than TPH1 (t½ is 59.0 ± 3.9 min for TPH1 and 43.5 ± 3.7 min for TPH2) (p < 0.05). H, a graph displaying the average results obtained from n = 6 pulse-chase assays reveals that TPH2 is degraded more rapidly than HA-TPH1 in stably transfected PC12 cells (t½ is 58.0 ± 3.8 min for TPH1 and 41.7 ± 2.7 min for TPH2) (p < 0.05). Cells were labeled with [35S]methionine and [35S]cysteine and then returned to regular medium for the indicated times. Labeled HA-TPH was immunoprecipitated, and the amount of labeled protein was assessed by SDS-PAGE and autoradiography. The half-life was calculated for each individual experiment and is presented as means ± S.E. for each enzyme. Graphical data presented represent means ± S.E.
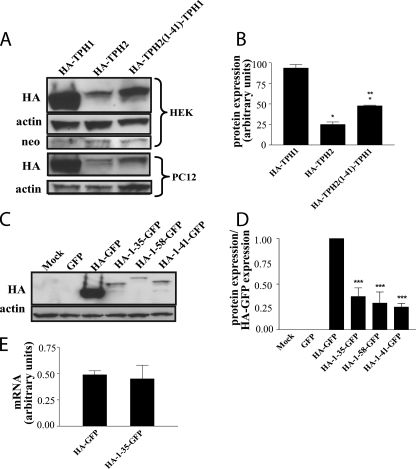
Role of TPH2 Extended N-terminal Domain in Modulating Enzyme Expression—After examining both the rates of synthesis and degradation between TPH1 and TPH2, we sought to assess whether the extended N-terminal domain of TPH2 may contribute to the apparent differences between the two enzymes. To accomplish this, we generated a chimeric protein composed of the first 41 amino acids of TPH2 appended to the N terminus of TPH1 (HA-TPH2-(1–41)-TPH1) and expressed this protein in two different cell lines (HEK293 and PC12). As seen in Fig. 3, A and B, the expression level of the chimeric protein was significantly decreased compared with TPH1 in both cell lines (p < 0.01 in both cases) and was more similar to that of TPH2. These results suggest that the extended N terminus of TPH2 is responsible for the bulk of the difference in expression levels observed between TPH1 and TPH2. To assess the specific effect of this segment, we appended HA-tagged TPH2 N-terminal fragments (amino acids 1–35, 1–41, and 1–58) to the N terminus of GFP. These three regions represent the sequence encoded by the first exon of the gene, the number of residues in TPH2 not present in TPH1, and the entire region of the TPH2 N-terminal that contains no homology to TPH1, respectively. As illustrated in Fig. 3, C and D, the addition of each of these three fragments to GFP significantly reduced the expression levels of the chimeric proteins in cells compared with the HA-GFP construct without any of the fragments (p < 0.001 in all cases). The fact that all three fragments reduced HA-GFP expression to a similar extent indicates that the sequence contained in the first exon of the N terminus of TPH2 alone is sufficient to reduce protein expression without altering mRNA levels (Fig. 3E) (p = 0.35).
FIGURE 3.
The N-terminal regulatory determinant is transferable. A, attaching the first 41 amino acids of TPH2 to the N terminus of HA-TPH1 results in a chimeric protein with expression levels most similar to those of wild-type HA-TPH2 in both HEK293 and PC12 cells. Because the vector used to make the constructs contains a neomycin resistance gene (neomycin phosphotransferase), a blot for this protein was used as a transfection control in HEK293 cells. The graph shown in B is the result of a densitometric analysis of blots from HEK293 cells (n = 3) with results presented in arbitrary units normalized to neomycin phosphotransferase levels. *, p < 0.01 compared with HA-TPH1; **, p < 0.05 compared with HA-TPH2. C and D, insertion of the first 35, 41, or 58 amino acids of TPH2 after the HA epitope of an HA-GFP construct results in decreased HA-GFP expression levels in PC12 cells. Unlabeled GFP was included to assess antibody specificity. Results presented are normalized to HA-GFP levels (n = 4). ***, p < 0.001 compared with HA-GFP. E, there is no difference in mRNA levels between the HA-GFP construct and the representative chimera, HA-(1–35)-GFP, as measured by quantitative reverse transcription-PCR (n = 4). All graphical data represent means ± S.E.
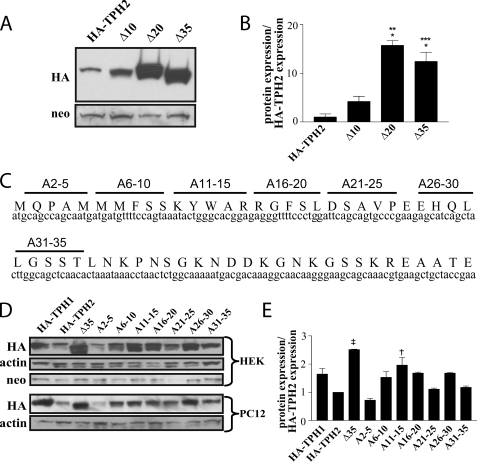
Functional Mapping of the TPH2 Extended N-terminal Domain—Because the addition of the first 35 amino acids of TPH2 proved sufficient to reduce expression of GFP, we generated a series of N-terminal deletion mutants, progressively removing the nucleotides corresponding to 10 or 15 amino acids at a time, with the goal of identifying specific stretches of sequence responsible for the difference in protein expression. Analysis of protein levels in cells transfected with constructs containing deletions of nucleotides corresponding to the first 10, 20, and 35 amino acids revealed that although removal of the first 10 amino acids had only a small effect on protein expression, deletion of the first 20 and the first 35 amino acids resulted in 15- and 10-fold increases, respectively, in protein expression compared with full-length TPH2 (p < 0.001 in both cases) (Fig. 4, A and B). This indicates that, although there is some effect on protein expression of removing the first 10 amino acids, residues 11–20 comprise the bulk of the sequence responsible for the lower expression of TPH2. To confirm this finding and ensure that the observed difference in expression was due to a loss of specific nucleotides or amino acids and not a general loss of residues, we performed an alanine-scanning mutagenesis of the first 35 amino acids of TPH2, altering the cDNA such that the sequence corresponding to five amino acids at a time would instead code for alanine (Fig. 4C). The results presented in Fig. 4, D and E, show that mutation of amino acid residues 6–10 (nucleotides 16–30), 11–15 (nucleotides 31–45), or 16–20 (nucleotides 46–60) to those coding for alanine all resulted in increased protein expression, although the mutation corresponding to residues 6–10 produced a smaller effect than the other two, and the mutation corresponding to residues 11–15 was the only one that reached statistical significance (p < 0.01). This mirrors the results seen with the deletion mutants, supporting the hypothesis that the mRNA and/or amino acids corresponding to the region of residues 10–20 are responsible for the bulk of the reduction in expression of proteins containing the extended N-terminal domain of TPH2.
FIGURE 4.
Isolation of the residues containing the regulatory determinant. A and B, Western blots from HEK293 cells expressing HA-TPH2 constructs containing truncations of 10, 20, and 35 residues reveal the bulk of the sequence required for the decreased protein expression lies within the sequence coding for amino acids 10–20. B, results presented in the graph were normalized to neomycin phosphotransferase expression and presented as a ratio to full-length HA-TPH2 expression (n = 3). *, p < 0.001 compared with HA-TPH2; **, p < 0.001 compared with Δ10; ***, p < 0.01 compared with Δ10. C, a schematic illustrating the residues mutated in an alanine-scanning mutagenesis. D, Western blots from both HEK293 and PC12 cells showing expression levels of HA-TPH2 proteins with the indicated residues mutated to alanine support the importance of the sequence coding for residues 10–20 in mediating the decreased protein expression. E, the accompanying graph represents average protein levels in HEK293 cells normalized to neomycin expression and compared with wild type HA-TPH2 expression (n = 3). †, p < 0.001 compared with HA-TPH2; ‡, p < 0.01 compared with HA-TPH2. Data are means ± S.E.
There was also an additional region between residues 26 and 30 (nucleotides 76–90), which, when mutated, also resulted in an increase in protein stability, although this effect did not reach statistical significance.
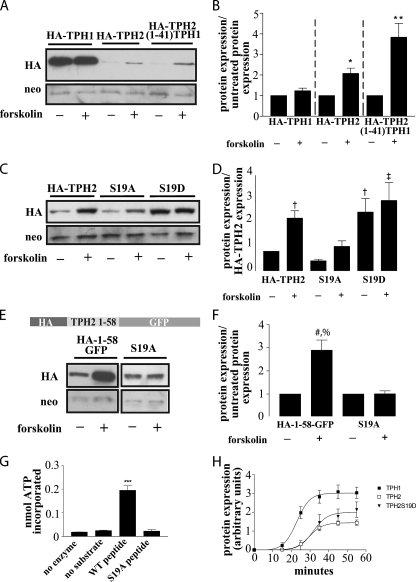
Phosphorylation of Serine 19 Regulates TPH2 Protein Expression—Another feature within the extended N terminus of TPH2 is a putative PKA consensus phosphorylation site at Ser19. To confirm the ability of PKA to phosphorylate Ser19, we assessed the phosphorylation of the residue corresponding to Ser19 in a peptide consisting of an HA tag followed by residues 11–24 of TPH2, whereas a similar peptide with an S19A mutation was used as a negative control. As Fig. 5G indicates, PKA phosphorylated the peptide containing Ser19, whereas it failed to phosphorylate the control peptide with an alanine substitution at Ser19 (S19A) (p < 0.01). Upon confirming the ability of PKA to phosphorylate Ser19, we sought to assess the effect of this phosphorylation on protein expression. To do so, we treated cells expressing either TPH1, TPH2, or the HA-TPH2-(1–41)-TPH1 chimera with forskolin to increase PKA activity through increased cAMP production. As shown in Fig. 5, A and B, this forskolin-mediated activation of PKA resulted in a significant increase in expression of both TPH2 (p < 0.05) and the chimera (p < 0.001) with little or no increase in TPH1 expression. To validate the role of Ser19 phosphorylation in modulating TPH2 expression levels, we generated a S19A mutant to prevent phosphorylation of this site and an S19D mutant to mimic its phosphorylation. Comparison of the S19A mutant with wild type TPH2 showed no appreciable difference in baseline expression, but the forskolin-induced increase in expression was reduced by roughly 50%, and the significance of the forskolin effect was lost in cells transfected with the mutant construct (Fig. 5, C and D). Conversely, cells expressing the S19D mutant exhibited base-line protein levels 2–3 times higher than the cells expressing wild-type TPH2, and there was essentially no further effect of forskolin (p < 0.05 and 0.01, respectively) (Fig. 5, C and D). Additionally, the pseudophosphorylated TPH2 S19D protein was expressed at levels similar to the wild-type TPH2 in cells treated with forskolin (Fig. 5, C and D), suggesting that phosphorylation at Ser19 is sufficient to produce the increased expression seen with forskolin-mediated PKA activation. The residual response to forskolin treatment seen with the S19A mutant may be attributable to phosphorylation of Ser104, a site homologous to Ser58 in TPH1, which has been previously identified as a PKA target (24).
FIGURE 5.
Phosphorylation of Ser19 modulates TPH2 expression. A, a Western blot from HEK293 cells expressing HA-TPH1, HA-TPH2, or the HA-TPH2-(1–41)-TPH1 chimera with and without treatment with 2.5 μm forskolin for 4 h demonstrates a PKA-mediated increase in TPH2 and TPH2-(1–41)-TPH1 expression with no appreciable effect on TPH1 expression. B, bar graph showing the result of n = 5 experiments. *, p < 0.05 compared with untreated control; **, p < 0.001 compared with untreated control. The data in graph B are normalized to neomycin expression and compared with their respective untreated controls. C, a Western blot from HEK293 cells expressing HA-TPH2(S19A) to remove the putative PKA phosphorylation site or S19D to create a pseudophosphorylated protein supports the hypothesis that phosphorylation of Ser19 leads to an increase in protein expression. The responses to forskolin treatment (described above) support this, since the pseudophosphorylated S19D construct is not appreciably affected by forskolin-mediated PKA activation, and the S19A response to forskolin is blunted compared with wild type TPH2. The graph shown in D represents n = 3 experiments. †, p < 0.05 compared with HA-TPH2; ‡, p < 0.01 compared with HA-TPH2. The data in graph D are normalized to neomycin expression and compared with untreated HA-TPH2 expression. E and F, to remove the possible confound of additional PKA phosphorylation sites, the S19A mutation was generated in the HA-(1–58)-GFP construct. Although the expression of the HA-(1–58)-GFP construct increased in response to the previously outlined forskolin treatment, expression of the S19A mutant remained unchanged. The results shown in F represent n = 3 experiments with treated samples normalized to neomycin and compared with the untreated partner sample. #, p < 0.01 compared with HA-(1–58)-GFP; %, p < 0.01 compared with S19A + forskolin. G, phosphorylation of a peptide corresponding to amino acids 10–20 and that same peptide with the S19A mutation confirm the status of Ser19 as a PKA phosphorylation site (n = 3). ***, p < 0.01 compared with “no enzyme,” “no substrate,” and “S19A peptide” controls. H, in vitro translation performed on cDNA in a modified pcDNA vector shows that the S19D mutation does not dramatically alter TPH2 translational efficiency (n = 4). All results are means ± S.E.
To remove the confounding effects of this secondary site from our characterization and to, once again, assess the transferability of the effect, we generated an S19A mutant in the HA-(1–58)-GFP hybrid construct. In this case, mutation of Ser19 to alanine resulted in a complete loss of response to forskolin, whereas the nonmutated hybrid exhibited increased expression in response to forskolin treatment similar in magnitude to that seen when wild-type TPH2 was treated with forskolin (Fig. 5, E and F) (p < 0.01). Finally, in vitro translation results indicate that the higher expression of the S19D mutant does not result from changes in its translational efficiency (Fig. 5H) (p = 0.126 compared with TPH2).
Phosphorylation of TPH2 Ser19 Increases 5HT Synthesis—To assess the functional outcome of the observed changes in TPH2 stability, we measured 5HT production in PC12 cells expressing wild type TPH2 and the TPH2 S19A mutant with and without forskolin treatment as well as the pseudophosphorylated TPH2 S19D construct. Because PC12 cells do not contain any endogenous TPH, they do not produce any endogenous 5HT or 5HTP. They do, however, contain TH and, thus, contain all the cellular machinery necessary to support amino acid hydroxylase function. To differentiate between the products of TPH2 activity before and after forskolin treatment, both control and forskolin-treated cells were treated with NSD-1015 to block AADC function at the time of forskolin treatment. Because endogenous AADC is highly efficient, pre-NSD-1015 treatment levels of 5HTP are undetectable. This means that all 5HTP measured here was produced during the treatment period.
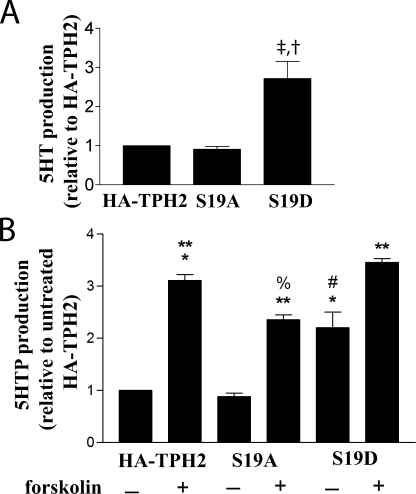
The data in Fig. 6 indicate that differences in 5HT levels and 5HTP accumulation as measured by HPLC are consistent with the observed changes in protein levels associated with these mutants. Specifically, there were no appreciable differences in either the protein expression or 5HT concentrations between untreated wild type and S19A samples. In contrast, the S19D samples exhibited a roughly 2.5-fold higher level of 5HT (Fig. 6A), mirroring the roughly 2.5-fold increase in protein expression observed with this construct (Fig. 5, C and D). Furthermore, treatment of cells transfected with HA-TPH2 with forskolin produced a greater than 3-fold increase in 5HTP production (p < 0.001), which was reduced (p < 0.05) but not abolished by the mutation of S19A. This is not entirely unexpected, since the S19A mutation also did not completely block the effects of forskolin on TPH2 expression (Fig. 5, C and D). It is notable that neither forskolin treatment of the S19A mutant nor the pseudophosphorylation in the S19D mutant was able to completely mimic the effect of forskolin treatment on 5HTP production of wild-type HA-TPH2. However, forskolin treatment of the S19D mutant brought 5HTP to levels that were nearly identical to those obtained with the forskolin-stimulated wild-type construct. Thus, although phosphorylation of Ser19 does appear to increase 5HTP production by increasing enzyme expression, additional targets also probably contribute to the overall increase in 5HTP production in response to PKA activation.
FIGURE 6.
5HT and 5HTP production in PC12 cells. PC12 cells were transfected with wild-type HA-TPH2, HA-TPH2 S19A, or HA-TPH2 S19D, and 48–72 h post-transfection, 5HT was measured, or cells were treated with 100 μm NSD-1015 to block the activity of AADC and either 2.5 μm forskolin or an equal volume of DMSO for 16 h to assess accumulation of 5HTP. A, 5HT content in untreated cells was measured by HPLC and normalized to the endogenously produced dopamine content produced by the cells to control for differences in the number of cells used to produce the lysates. ‡, p < 0.05 compared with HA-TPH2; †, p < 0.05 compared with S19A. B, similar to the results in A, 5HTP content was measured in cells transfected with each of the three constructs and treated with 100 μm NSD-1015 for 16 h to block AADC as described above (*, p < 0.001 compared with HA-TPH2). 5HTP accumulation was also assessed in cells treated with forskolin and NSD-1015 for 16 h (**, p < 0.001 compared with untreated controls; %, p < 0.05 compared with HA-TPH2 + forskolin; #, p < 0.001 compared with HA-TPH2 + forskolin). 5HTP content in the cell lysates was also normalized to the endogenously produced dopamine content produced by the cells before the initiation of treatment to control for differences in cell number used to produce the lysates. Data are means ± S.E. (n = 4).
DISCUSSION
The results presented here demonstrate that TPH1 and TPH2 differ markedly in their expression levels in cultured mammalian cells. This appears to be primarily due to differences in the efficiency of expression of the proteins, although a difference in the post-translational stability of the enzymes may also play a role. The ability of the N terminus of TPH2 to reduce the expression of both TPH1 and GFP in protein chimeras suggests that the unique extended N terminus of TPH2 alone is sufficient to alter protein expression. Furthermore, the region surrounding amino acids 11–20 appears essential for producing this effect, but other factors certainly contribute as well.
Examination of the extended N terminus of TPH2 also revealed single amino acid residues with the potential to play a role in the regulation of TPH2 expression. The first of these residues, Ser19, is a PKA consensus phosphorylation site (32). Mutation of this residue to aspartate to mimic phosphorylation results in increased expression of the enzyme similar to that seen when cells containing wild type TPH2 are treated with forskolin to activate PKA. This increase in enzyme expression correlates with an increase in 5HT levels in cells transfected with the pseudophosphorylated S19D mutant. Our results also demonstrate that PKA phosphorylation of Ser19 appears to explain at least part of the observed increase in 5HTP production in forskolin-treated cells expressing wild-type TPH2.
Phosphorylation-mediated changes in function are not unprecedented among hydroxylases. In the case of TH, catecholamine binding to Gi-coupled autoreceptors results in a reduction in monoamine production through PKA-mediated changes in TH function (21, 25). It is thus possible that a similar principle may underlie the changes in 5HT synthesis in response to extracellular 5HT levels. The enzymatic mechanism behind the changes in synthesis, however, would differ. Although PKA-mediated phosphorylation of TH alters catecholamine production through changes in TH activity, phosphorylation of TPH2 S19 appears to alter 5HT production by altering the level of TPH2 expression, an effect that was recently attributed to an interaction with 14-3-3 proteins (33). Given the lack of homology between the N-terminal domains of these two enzymes, it is not surprising that despite the fact that PKA phosphorylates the regulatory domains of both enzymes, the results of these phosphorylation events differ. The physiological demands on these enzymes may also contribute to the different effects of phosphorylation, since TH operates at full capacity in the brain, whereas TPH2 does not (reviewed in Ref. 36). In either case, the presence of the hydroxylase at the synaptic terminal (37) means that regulatory mechanisms affecting either enzyme function or stability can rapidly affect the production of neurotransmitters for the releasable pool.
Regardless of the mechanism behind this effect, the finding that PKA-mediated phosphorylation of Ser19 modulates TPH2 stability raises the possibility that multiple PKA-mediated signaling events may have the potential to alter brain 5HT production, in addition to alterations in PKA activity in response to autoreceptor activity. Studies of TH have revealed a similar pattern whereby several non-catecholaminergic stimuli, including angiotensin (38), estrogen (39), and intermittent hypoxia (40), have been shown to alter TH activity through PKA-mediated pathways, resulting in changes in catecholamine synthesis and release. Because PKA plays a prominent role in so many signaling pathways, it is therefore possible that other inputs may also modulate brain 5HT levels by altering the phosphorylation state of Ser19 and, therefore, the expression of TPH2. Moreover, there is also the possibility that other kinases, including CamKII, may phosphorylate this site (41), adding yet another dimension to the possible signaling pathways that may alter the phosphorylation state of Ser19 to modulate 5HT production by altering TPH2 expression.
As noted previously, although phosphorylation of Ser19 in the extended N terminus of TPH2 may play a role in regulating its function, other mechanisms may also contribute to the overall regulation of its physiological function. In addition to the region surrounding amino acids 11–20, there is a region corresponding to residues 26–30, which, when mutated, also resulted in a slight increase in TPH2 expression. The mechanism behind this effect and whether these two regions functionally interact to regulate TPH2 stability or synthesis, however, remain unclear. Another residue in this region, Lys11, is a candidate for ubiqitination. However, mutation of this residue to either alanine or arginine had no effect on protein stability (data not shown). In addition, analysis of the mRNA sequence corresponding to the first 40 amino acids of TPH2 using two mRNA secondary structure prediction programs (MFOLD (42, 43) and CARNAC (44, 45)) suggests that it may be part of an mRNA stem-loop structure and contains sequences that may also interact with mRNA-binding proteins. This provides a possible explanation, at least in part, for the observed differences in translational efficiency and also suggests a potential site for post-transcriptional regulation of TPH2 expression. TPH2-specific regulation is particularly interesting in the context of a number of psychiatric disorders. There is evidence for TPH2 involvement in mood disorders; for instance, a number of TPH2 polymorphisms have been associated with various illnesses, such as depression, bipolar disorder, and anxiety disorders (9, 10, 12–18). One such polymorphism identified in humans has recently been engineered in the mouse Tph2 gene to create a model of marked serotonin deficiency (46). Interestingly, although no polymorphisms have been found to date in the residues highlighted in this study, there have been a few coding mutations identified in the TPH2 N terminus, including a deletion of Met7 and two substitutions of Leu36 (L36V and L36P) (47, 48). There was also a functional polymorphism (S41Y) identified in a Han Chinese population in conjunction with an increased risk for bipolar disorder (49), suggesting that the N terminus of TPH2 represents a candidate site for functional disease-related mutations. Current treatments for 5HT-related psychiatric disorders increase 5HT levels by blocking 5HT degradation with monoamine oxidase inhibitors or blocking 5HT reuptake through the serotonin transporters. Both treatments appear to increase extracellular serotonin levels 2–3-fold (50, 51). Although these drugs are effective in treating a sizeable number of patients, both of these approaches have their drawbacks. Among these, selective serotonin reuptake inhibitors can induce a host of peripheral side effects, including issues with changes in blood pressure, sleep disturbance, gastrointestinal disturbance, and weight gain. Thus, the presence of a unique regulatory domain in the brain enzyme, TPH2, may present a target for a novel type of pharmacological treatment for mood disorders with specificity for the central 5HT system. Successful manipulation of the mechanisms underlying the regulatory function of the N-terminal domain, which affect a 2–3-fold increase in TPH2, could potentially increase brain 5HT to levels similar to those seen with reuptake blockers and monoamine oxidase inhibitors. TPH2-based therapy, however, might avoid many of the specificity-related side effects seen with the currently available 5HT-based therapies. Before this is a real possibility, however, a thorough understanding of the mechanisms behind these effects must be acquired such that we can specifically manipulate these properties to produce the desired effects.
Acknowledgments
We thank Ava Sweeney for help in generating DNA constructs.
This work was supported, in whole or in part, by National Institutes of Health Grants MH-40159 and MH-79201 (to M. G. C.). This work was also supported by a grant from the Lennon Family Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: 5HT, 5-hydroxytryptamine; TPH, tryptophan hydroxylase; TPH1 and TPH2, tryptophan hydroxylases 1 and 2; TH, tyrosine hydroxylase; HA, hemagglutinin; 5HTP, 5-hydroxytryptophan; AADC, amino acid decarboxylase; HPLC, high pressure liquid chromatography; GFP, green fluorescent protein.
References
- 1.Blier, P., and Abbott, F. V. (2001) J. Psychiatry Neurosci. 26 37-43 [PMC free article] [PubMed] [Google Scholar]
- 2.Courtet, P., Jollant, F., Castelnau, D., Buresi, C., and Malafosse, A. (2005) Am. J. Med. Genet. C Semin. Med. Genet. 133 25-33 [DOI] [PubMed] [Google Scholar]
- 3.Hasler, G., Drevets, W. C., Manji, H. K., and Charney, D. S. (2004) Neuropsychopharmacology 29 1765-1781 [DOI] [PubMed] [Google Scholar]
- 4.Lesch, K. P. (2004) J. Psychiatry Neurosci. 29 174-184 [PMC free article] [PubMed] [Google Scholar]
- 5.Walther, D. J., and Bader, M. (2003) Biochem. Pharmacol. 66 1673-1680 [DOI] [PubMed] [Google Scholar]
- 6.Patel, P. D., Pontrello, C., and Burke, S. (2004) Biol. Psychiatry 55 428-433 [DOI] [PubMed] [Google Scholar]
- 7.Walther, D. J., Peter, J. U., Bashammakh, S., Hortnagl, H., Voits, M., Fink, H., and Bader, M. (2003) Science 299 76. [DOI] [PubMed] [Google Scholar]
- 8.Zhang, X., Beaulieu, J. M., Sotnikova, T. D., Gainetdinov, R. R., and Caron, M. G. (2004) Science 305 217. [DOI] [PubMed] [Google Scholar]
- 9.De Luca, V., Voineskos, D., Wong, G. W., Shinkai, T., Rothe, C., Strauss, J., and Kennedy, J. L. (2005) Psychiatry Res. 134 195-198 [DOI] [PubMed] [Google Scholar]
- 10.Harvey, M., Shink, E., Tremblay, M., Gagne, B., Raymond, C., Labbe, M., Walther, D. J., Bader, M., and Barden, N. (2004) Mol. Psychiatry 9 980-981 [DOI] [PubMed] [Google Scholar]
- 11.Mossner, R., Walitza, S., Geller, F., Scherag, A., Gutknecht, L., Jacob, C., Bogusch, L., Remschmidt, H., Simons, M., Herpertz-Dahlmann, B., Fleischhaker, C., Schulz, E., Warnke, A., Hinney, A., Wewetzer, C., and Lesch, K. P. (2006) Int. J. Neuropsychopharmacol. 9 437-442 [DOI] [PubMed] [Google Scholar]
- 12.Sheehan, K., Lowe, N., Kirley, A., Mullins, C., Fitzgerald, M., Gill, M., and Hawi, Z. (2005) Mol. Psychiatry 10 944-949 [DOI] [PubMed] [Google Scholar]
- 13.Zhang, X., Beaulieu, J. M., Gainetdinov, R. R., and Caron, M. G. (2006) Cell Mol. Life Sci. 63 6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang, X., Gainetdinov, R. R., Beaulieu, J. M., Sotnikova, T. D., Burch, L. H., Williams, R. B., Schwartz, D. A., Krishnan, K. R., and Caron, M. G. (2005) Neuron 45 11-16 [DOI] [PubMed] [Google Scholar]
- 15.Zhou, Z., Roy, A., Lipsky, R., Kuchipudi, K., Zhu, G., Taubman, J., Enoch, M. A., Virkkunen, M., and Goldman, D. (2005) Arch. Gen. Psychiatry 62 1109-1118 [DOI] [PubMed] [Google Scholar]
- 16.Zill, P., Baghai, T. C., Zwanzger, P., Schule, C., Eser, D., Rupprecht, R., Moller, H. J., Bondy, B., and Ackenheil, M. (2004) Mol. Psychiatry 9 1030-1036 [DOI] [PubMed] [Google Scholar]
- 17.Zill, P., Buttner, A., Eisenmenger, W., Moller, H. J., Bondy, B., and Ackenheil, M. (2004) Biol. Psychiatry 56 581-586 [DOI] [PubMed] [Google Scholar]
- 18.Walitza, S., Renner, T. J., Dempfle, A., Konrad, K., Wewetzer, C., Halbach, A., Herpertz-Dahlmann, B., Remschmidt, H., Smidt, J., Linder, M., Flierl, L., Knolker, U., Friedel, S., Schafer, H., Gross, C., Hebebrand, J., Warnke, A., and Lesch, K. P. (2005) Mol. Psychiatry 10 1126-1132 [DOI] [PubMed] [Google Scholar]
- 19.Fitzpatrick, P. F. (1999) Annu. Rev. Biochem. 68 355-381 [DOI] [PubMed] [Google Scholar]
- 20.Martinez, A., Knappskog, P. M., and Haavik, J. (2001) Curr. Med. Chem. 8 1077-1091 [DOI] [PubMed] [Google Scholar]
- 21.Dunkley, P. R., Bobrovskaya, L., Graham, M. E., von Nagy-Felsobuki, E. I., and Dickson, P. W. (2004) J. Neurochem. 91 1025-1043 [DOI] [PubMed] [Google Scholar]
- 22.Jiang, G. C., Yohrling, G. J. t., Schmitt, J. D., and Vrana, K. E. (2000) J. Mol. Biol. 302 1005-1017 [DOI] [PubMed] [Google Scholar]
- 23.Johansen, P. A., Jennings, I., Cotton, R. G., and Kuhn, D. M. (1995) J. Neurochem. 65 882-888 [DOI] [PubMed] [Google Scholar]
- 24.Kumer, S. C., Mockus, S. M., Rucker, P. J., and Vrana, K. E. (1997) J. Neurochem. 69 1738-1745 [DOI] [PubMed] [Google Scholar]
- 25.Lehmann, I. T., Bobrovskaya, L., Gordon, S. L., Dunkley, P. R., and Dickson, P. W. (2006) J. Biol. Chem. 281 17644-17651 [DOI] [PubMed] [Google Scholar]
- 26.Stenfors, C., and Ross, S. B. (2002) J. Neural Transm. 109 1353-1363 [DOI] [PubMed] [Google Scholar]
- 27.Carkaci-Salli, N., Flanagan, J. M., Martz, M. K., Salli, U., Walther, D. J., Bader, M., and Vrana, K. E. (2006) J. Biol. Chem. 281 28105-28112 [DOI] [PubMed] [Google Scholar]
- 28.Kleppe, R., and Haavik, J. (2004) FEBS Lett. 565 155-159 [DOI] [PubMed] [Google Scholar]
- 29.Sakowski, S. A., Geddes, T. J., and Kuhn, D. M. (2006) J. Neurochem. 96 758-765 [DOI] [PubMed] [Google Scholar]
- 30.Tenner, K., Walther, D., and Bader, M. (2007) J. Neurochem. 102 1887-1894 [DOI] [PubMed] [Google Scholar]
- 31.Winge, I., McKinney, J. A., Knappskog, P. M., and Haavik, J. (2007) J. Neurochem. 100 1648-1657 [DOI] [PubMed] [Google Scholar]
- 32.McKinney, J., Knappskog, P. M., and Haavik, J. (2005) J. Neurochem. 92 311-320 [DOI] [PubMed] [Google Scholar]
- 33.Winge, I., McKinney, J. A., Ying, M., D'Santos, C. S., Kleppe, R., Knappskog, P. M., and Haavik, J. (2007) Biochem. J. 410 195-204 [DOI] [PubMed] [Google Scholar]
- 34.Zhang, X., Kim-Miller, M. J., Fukuda, M., Kowalchyk, J. A., and Martin, T. F. (2002) Neuron 34 599-611 [DOI] [PubMed] [Google Scholar]
- 35.Xu, F., Gainetdinov, R. R., Wetsel, W. C., Jones, S. R., Bohn, L. M., Miller, G. W., Wang, Y. M., and Caron, M. G. (2000) Nat. Neurosci. 3 465-471 [DOI] [PubMed] [Google Scholar]
- 36.Fernstrom, J. D. (1983) Physiol. Rev. 63 484-546 [DOI] [PubMed] [Google Scholar]
- 37.Pickel, V. M., Joh, T. H., and Reis, D. J. (1977) Brain Res. 131 197-214 [DOI] [PubMed] [Google Scholar]
- 38.Ma, F. Y., Grattan, D. R., Bobrovskaya, L., Dunkley, P. R., and Bunn, S. J. (2004) J. Neurochem. 90 431-441 [DOI] [PubMed] [Google Scholar]
- 39.Chaube, R., and Joy, K. P. (2005) Gen. Comp. Endocrinol. 141 116-125 [DOI] [PubMed] [Google Scholar]
- 40.Kumar, G. K., Kim, D. K., Lee, M. S., Ramachandran, R., and Prabhakar, N. R. (2003) J. Appl. Physiol. 95 536-544 [DOI] [PubMed] [Google Scholar]
- 41.Kuhn, D. M., Sakowski, S. A., Geddes, T. J., Wilkerson, C., and Haycock, J. W. (2007) J. Neurochem. 103 1567-1573 [DOI] [PubMed] [Google Scholar]
- 42.Mathews, D. H., Sabina, J., Zuker, M., and Turner, D. H. (1999) J. Mol. Biol. 288 911-940 [DOI] [PubMed] [Google Scholar]
- 43.Zuker, M. (2003) Nucleic Acids Res. 31 3406-3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perriquet, O., Touzet, H., and Dauchet, M. (2003) Bioinformatics 19 108-116 [DOI] [PubMed] [Google Scholar]
- 45.Touzet, H., and Perriquet, O. (2004) Nucleic Acids Res. 32 W142-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beaulieu, J. M., Zhang, X., Rodriguiz, R. M., Sotnikova, T. D., Cools, M. J., Wetsel, W. C., Gainetdinov, R. R., and Caron, M. G. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 1333-1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nielsen, D. A., Barral, S., Proudnikov, D., Kellogg, S., Ho, A., Ott, J., and Kreek, M. J. (2008) Behav. Genet. 38 133-150 [DOI] [PubMed] [Google Scholar]
- 48.Breidenthal, S. E., White, D. J., and Glatt, C. E. (2004) Psychiatr. Genet. 14 69-72 [DOI] [PubMed] [Google Scholar]
- 49.Lin, Y. M., Chao, S. C., Chen, T. M., Lai, T. J., Chen, J. S., and Sun, H. S. (2007) Arch. Gen. Psychiatry 64 1015-1024 [DOI] [PubMed] [Google Scholar]
- 50.Anderson, G. M. (2004) Int. J. Dev. Neurosci. 22 397-404 [DOI] [PubMed] [Google Scholar]
- 51.Kinney, G. G., Taber, M. T., and Gribkoff, V. K. (2000) Mol. Neurobiol. 21 137-152 [DOI] [PubMed] [Google Scholar]