Abstract
Hominoid- and human-specific genes may have evolved to modulate signaling pathways of a higher order of complexity. TBC1D3 is a hominoid-specific oncogene encoded by a cluster of eight paralogs on chromosome 17. Initial work indicates that TBC1D3 is widely expressed in human tissues (Hodzic, D., Kong, C., Wainszelbaum, M. J., Charron, A. J., Su, X., and Stahl, P. D. (2006) Genomics 88,731 -736). In this study, we show that TBC1D3 expression has a powerful effect on cell proliferation that is further enhanced by epidermal growth factor (EGF) in both human and mouse cell lines. EGF activation of the Erk and protein kinase B/Akt pathways is enhanced, both in amplitude and duration, by TBC1D3 expression, whereas RNA interference silencing of TBC1D3 suppresses the activation. Light microscopy and Western blot experiments demonstrate that increased signaling in response to EGF is coupled with a significant delay in EGF receptor (EGFR) trafficking and degradation, which significantly extends the life span of EGFR. Moreover, TBC1D3 suppresses polyubiquitination of the EGFR and the recruitment of c-Cbl. Using the Ras binding domain of Raf1 to monitor GTP-Ras we show that TBC1D3 expression enhances Ras activation in quiescent cells, which is further increased by EGF treatment. We speculate that TBC1D3 may alter Ras GTP loading. We conclude that the expression of TBC1D3 generates a delay in EGFR degradation, a decrease in ubiquitination, and a failure to recruit adapter proteins that ultimately dysregulate EGFR signal transduction and enhance cell proliferation. Altered growth factor receptor trafficking and GTP-Ras turnover may be sites where recently evolved genes such as TBC1D3 selectively modulate signaling in hominoids and humans.
Hominoid- or human-specific genes are among the most important resources to have emerged from the completion of the human genome project. Still, we know little about this small group of genes, which presumably modulate or regulate signaling pathways that have evolved to a higher level of complexity and that distinguish hominoids and humans from less complex species. TBC1D3 belongs to a hominoid-specific gene family with no known orthologs outside of the primate lineage (1). TBC1D3 was originally identified by Pei et al. as a prostate and breast cancer oncogene (2). The TBC1D3 genes are arrayed along a region of human chromosome 17 that has undergone extensive intrachromosomal rearrangement and segmental duplication following its probable recent appearance within the hominoid lineage (3). Chromosome 17 has also been implicated in a wide variety of human genetic diseases and encodes genes involved in breast cancer (BRCA1), neurofibromatosis (NFI), and the DNA damage response (TP53). TBC1D3 (also known as PRC17), which encodes a protein containing a TBC (Tre-2, BUB2, cdc16) domain, was shown to induce tumors in nude mice and growth in low serum when exogenously expressed in mouse 3T3 fibroblasts (2). Previous work from our laboratory identified eight highly related TBC1D3 paralogs, organized in two clusters within the 17q12 genomic region. These genes potentially encode six individual TBC1D3 variants with differences in a handful of amino acids within the TBC domain. Interestingly, we reported a tissue-specific transcription pattern of TBC1D3 paralogs among normal human tissues and documented alterations in this pattern in several prostate tumors (1).
The EGF5 receptor (EGFR) is among the best studied receptor tyrosine kinases. Once EGFR is activated by its ligand, receptor dimerization and autophosphorylation occur (4), followed by the recruitment of multiple adaptor proteins. Activated EGFRs are ubiquitinated, internalized, and transported along the endocytic pathway while maintaining signaling. EGFR belongs to the ERBB receptor signal transduction network, extensively implicated in human cancer and a target for cancer therapeutics (5). Numerous aberrations have been shown in ERBB receptor genes (deletions, insertions, and point mutations) that may alter both signaling and membrane trafficking pathways (6, 7). Normal tissue morphogenesis, homeostasis, and growth depend on finely orchestrated cellular responses to growth factors and cytokines. Thus, it has been proposed that dysregulation of the endocytic machinery might lead to uncontrolled cell growth by disturbing the specificity and duration of one or more signaling pathways (8).
We speculated that TBC1D3 might modulate EGFR signaling and trafficking events triggering cell proliferation. In the present work, we examined the signaling, trafficking, and fate of the EGFR in murine and human cells expressing or depleted of TBC1D3. We show that the activation of Erk and protein kinase B/Akt pathways, the kinetics of receptor trafficking, and EGF-induced cell proliferation are modulated by TBC1D3 expression. Understanding the mode and mechanism of action of TBC1D3 may provide a unique insight in the “biological rationale” by which recently evolved genes modulate signaling pathways in a human-specific manner and thereby create models for exploring the role of human-specific genes in human metabolic control and physiology.
EXPERIMENTAL PROCEDURES
Reagents—125I-Labeled human EGF and [methyl-3H]thymidine were purchased from Amersham Biosciences. Antibodies against phospho-Akt, phospho-MAPK, total Akt, and total MAPK were from Cell Signaling Technology (Beverly, MA). Cbl antibody was from BD Transduction Laboratories (Rockville, MD). Monoclonal EGFR (Ab-5) antibody was purchased from Calbiochem (San Diego, CA). Monoclonal Ras and polyclonal EGFR antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal α-tubulin was purchased from Sigma-Aldrich. The ubiquitin antibody was from Zymed Laboratories Inc. (San Francisco, CA). The monoclonal antibody directed against the C-terminal 50 amino acids of TBC1D3 was generated by the Hybridoma Center at Washington University (St. Louis, MO). Alexa-Fluor conjugated antibodies were from Molecular Probes (Carlsbad, CA).
Construction of Recombinant Retroviruses and Stable Cell Lines Expressing TBC1D3—TBC1D3 paralog D cDNA (HGCN designation), kindly provided by Dr. Jing Li (Tularik Inc, San Francisco, CA), was subcloned into EcoRI/AccI restriction sites of the pBABE-puro vector using the forward primer 5′-CACCATGGACGTGGTAGAGGTCG-3′ and the reverse primer 5′-CTAGAAGCCTGGAGGGAACTG-3′. To make stable cell lines, 293T-packaging cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, penicillin, and streptomycin. When cells reached 80% confluence, they were transfected with either pBABE-puro:TBC1D3 or empty pBABE-puro along with pCLEco (replication-incompetent helper plasmid) for mouse cells or pUMVC3 plus pCVM-VSV-G for human cells. Retroviral supernatant was collected after 48 h and used with Polybrene (5 μg/ml) to infect either human DU145 prostate cancer cells or mouse NR6 fibroblasts already stably expressing the human EGFR (NR6:hEGFR) (9). After incubation for 48 h the infected cells were selected with medium containing puromycin (2 μg/ml). Single clones were grown to obtain clonal stable cell lines.
TBC1D3 Silencing—TBC1D3 was successfully knocked down using a commercial siRNA pool (SMARTpool, Dharmacon). Briefly, DU145 cells were transfected with TBC1D3 siRNA (50 nm final concentration) or irrelevant siRNA (scramble siRNA, Ambion) using Lipofectamine 2000 and were used 36 h later for the appropriate experiments. TBC1D3 depletion was evaluated by RT-PCR. Cells were subjected to RNA extraction in RNase-free conditions using TRIzol (Invitrogen). The RNA was then used to detect TBC1D3 transcript levels by RT-PCR.
Cell Proliferation Assay—Cell growth was measured by thymidine incorporation into DNA. The cells (1 × 105 cells/well) were serum-starved and incubated in the presence or absence of 100 ng/ml EGF for 36 h. 1 μCi/ml [methyl-3H]thymidine (2 Ci/mmol) was added in the last 6 h. The cells were then washed three times with phosphate-buffered saline (PBS). The incorporation of [3H]thymidine into DNA was determined after DNA precipitation with cold 10% (w/v) trichloroacetic acid and cell solubilization in 0.1 m NaOH. Tritium was measured by scintillation counting.
Ras Activation Assay—Raf-1 RBD (Ras binding domain) expressed as a GST fusion protein was purified from Escherichia coli by immobilization on glutathione-Sepharose beads and used to affinity precipitate the active form of Ras (Ras-GTP) from cell extracts (10). Active Ras precipitation was visualized by SDS-PAGE and Western blotting with a monoclonal pan-Ras antibody.
Lysate Preparation, SDS-PAGE, and Western Blotting—To prepare whole cell lysates, cell monolayers were washed with PBS and lysed in ice-cold lysis buffer (PBS, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 2 μg/ml pepstatin A, 2 μg/ml leupeptin, and 2 μg/ml aprotinin). The lysates were clarified by centrifugation, and protein concentrations were determined using the BCA Protein Assay Reagent Kit (Pierce). Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes, which were blocked and probed with the indicated antibodies. To determine relative protein amounts, three representative exposures for each sample were quantified using AlphaEaseFc software (Alpha Innotech Corp., San Leandro, CA).
Signaling and EGFR Degradation Assays—Akt and Erk1/2 activation and EGFR degradation were measured in cells serum-starved for 5 h. Cells were incubated in the presence of EGF (100 ng/ml) at 37 °C for different time points, washed with ice-cold PBS, and lysed as described above. Proteins were separated by SDS-PAGE and analyzed by Western blotting with phospho-Akt, phospho-Erk1/2, and EGFR antibodies.
Receptor Internalization—Cells expressing TBC1D3 or vector alone were serum-starved and then incubated at 4 °C for 1 h with 70 ng/ml 125I-EGF (750 Ci/mmol), washed with ice-cold PBS and warmed for different periods of time. At each time point, unbound ligand was removed by washing the monolayer five times with ice-cold PBS. Surface-bound and internalized ligands were measured as described previously (11). Briefly, surface-bound ligand was collected in ice-cold “acid strip” buffer (50 mm glycine-HCl, 100 mm NaCl, 1 mg/ml polyvinylpyrrolidone, pH 3.0), and internalized ligands were released in 0.1 n NaOH. 125I-EGF was quantified by scintillation counting. Nonspecific binding (∼3%) was tested in the presence of unlabeled human EGF (200 nm, Sigma) and subtracted from the total. EGF internalization rates were estimated as the ratio of internalized to surface-bound EGF.
EGFR Turnover—Cells were starved with media lacking cysteine and methionine plus 5% of dialyzed fetal bovine serum for 30 min at 37 °C and were pulse-labeled by incubation with 100 μCi/ml [35S]methionine (1175 Ci/mmol) in the same medium for 1 h. Cells were washed and incubated in cold medium supplemented with 2 mm each cysteine and methionine for the indicated times in the incubator. Thereafter cells were lysed in radioimmune precipitation assay buffer; EGFR was immunoprecipitated by incubation with monoclonal anti-EGFR antibody and resolved by SDS-PAGE. The gel was dried, and radioactivity was detected by autoradiography.
Immunoprecipitation—All immunoprecipitations were performed by lysing the cells in ice-cold immunoprecipitation buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100, 1 mm EGTA, 1 mm EDTA, 2.5 mm sodium pyrophosphate, 1 mm β-glycerolphosphate, 1 mm NaF, 1 mm sodium orthovanadate plus protease inhibitors). After clarification, protein concentrations were measured, and extracts were immunoprecipitated by incubation with the appropriate antibody followed by immobilization on Protein G-Sepharose beads (Sigma). Beads were resuspended in sample buffer and analyzed by SDS-PAGE and Western blot with the indicated antibodies.
Liquid Chromatography-Tandem Mass Spectrometry Analysis of EGFR Ubiquitination—To examine the ubiquitination pattern of the EGFR in cells that express TBC1D3, the ubiquitin-AQUA method was used (12). Briefly, NR6:hEGFR: TBC1D3 and NR6:hEGFR:vector cells were grown in 150-mm dishes and treated or untreated with 100 ng/ml EGF for 5 min at 37 °C. Lysates were generated by the addition of TGH solubilization buffer (Triton X-100/glycerol/HEPES) containing 1% sodium deoxycholate and 10 mm N-ethylmaleimide. EGFR was immunoprecipitated by a monoclonal EGFR antibody. After sequential washes with TGH/sodium deoxycholate buffer containing 500 mm, 100 mm, and no NaCl, samples were separated by SDS-PAGE and analyzed by mass spectrometry using isotope-labeled internal standard peptides as previously described.
Immunofluorescence—Cells were seeded onto glass coverslips (12 mm) at 0.5 × 105 per well, serum-starved, and cell-surface EGFR was saturated with 200 ng/ml EGF (Calbiochem) for 1 h at 4 °C. To follow internalization and trafficking of EGF, the cells were washed and placed in complete, pre-warmed (37 °C) media for different times. The coverslips were then washed with ice-cold PBS, and the cells were fixed for 20 min in PBS containing 3% (v/v) paraformaldehyde and autofluorescence quenched in 50 mm NH4Cl. The cells were permeabilized with PBS containing 0.05% (v/v) Triton X-100 for 10 min, and nonspecific binding sites were blocked with PBS containing 1% (w/v) bovine serum albumin and 2% goat serum. The coverslips were probed with primary antibodies followed by fluorophore-conjugated secondary antibodies and mounted onto slides with Fluorescent Mounting Medium (DakoCytomation, Carpinteria, CA). Images were captured using a microscope equipped with a MRC1024 confocal LSM scanhead (Bio-Rad Laboratories).
Statistical Analysis—All experiments presented were repeated a minimum of three times. The data represents the mean ± S.D. Student's t test was performed to calculate statistical significance.
RESULTS
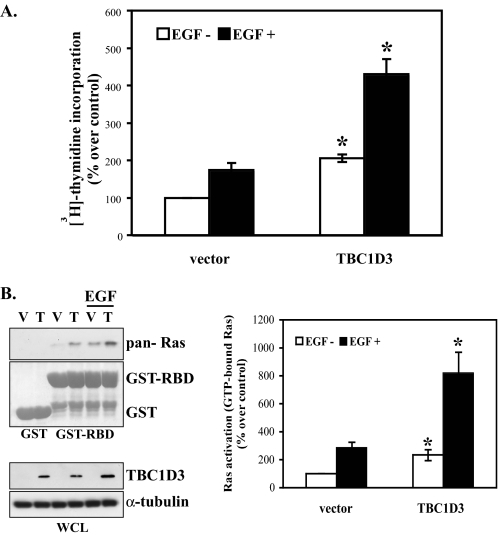
TBC1D3 Increases Cell Growth and Enhances the Proliferative Response to EGF—Initial reports identifying TBC1D3 as an oncogene (2) led us to investigate the effect of TBC1D3 expression and silencing on EGF-induced cell proliferation. The studies were carried out in DU145 cells, a well established human prostate carcinoma cell line. The cells were infected with retrovirus encoding TBC1D3D, and the resulting transductants were selected to obtain a clonal cell line expressing TBC1D3 (designated as DU145:TBC1D3). Similar results were obtained when different clonal cell lines were analyzed. DU145:vector (control) or DU145:TBC1D3 cells were incubated in the presence or absence of EGF for 36 h. The incorporation of [3H]thymidine into DNA was used as a measure of cell proliferation. The results (Fig. 1A) are expressed as the percentage of thymidine incorporation over control cells incubated without EGF (set at 100%). DU145:TBC1D3 cells grew more rapidly (∼2-fold), than control cells even in the absence of EGF. In the presence of EGF, both control and DU145:TBC1D3 cells displayed enhanced proliferation. However, EGF-treated DU145:TBC1D3 cells experienced a proliferative response that increased the amount of thymidine incorporation to ∼2.5-fold over that in vehicle-treated cells. Similar results were obtained using other human and mouse cell lines (data not shown). Experiments with murine lines correspond to a true null, because the gene is absent in the mouse genome. These results indicate that proliferation, the furthest downstream response to EGF, is augmented by the presence of TBC1D3, raising the question of how EGFR, TBC1D3, and proliferation are mechanistically linked.
FIGURE 1.
TBC1D3 increases EGF activation of cell growth through Ras activation. A, DU145 cells expressing vector alone or TBC1D3 were serum-starved and incubated in the presence or absence of 100 ng/ml EGF for 36 h. Incorporation of [methyl-3H]thymidine into DNA was determined as described under “Experimental Procedures.” Tritium was measured by scintillation counting. Results represent the mean ± S.D. of three independent experiments done by triplicate (*, p < 0.01). B, DU145 cells stably expressing TBC1D3 (T) or vector (V) were starved and stimulated or not for 10 min with EGF (100 ng/ml). Cells were then lysed and incubated with GST or GST Raf1-RBD glutathione-Sepharose beads for 1 h. The beads were washed and proteins were analyzed by Western blot using an anti-pan-Ras antibody. Data are expressed as percentage over non-stimulated vector (control) and represent the mean ± S.D. of three independent experiments (*, p < 0.01). WCL, whole cell lysate.
TBC1D3 Activates Ras and Enhances Ras Activation in Response to EGF—EGF receptor and other receptor tyrosine kinase are coupled to their downstream targets and to cell proliferation by Ras, which is constitutively activated in many human cancers (13). To determine whether Ras is activated in TBC1D3-expressing cells, GTP-Ras was monitored in cell lysates using the Raf-1 RBD pulldown assay (10). GTP-bound Ras was precipitated with GST-fused Raf-1 RBD from quiescent DU145 cells stably expressing TBC1D3 and control cells. GST alone was used as a control to estimate unspecific binding. The results showed that GTP-Ras was substantially increased (over 2-fold) in cells expressing TBC1D3 under steady-state conditions (Fig. 1B). To examine Ras following EGF stimulation, serum-starved TBC1D3 and control cells were stimulated for 10 min with 100 ng/ml EGF after which the lysates were incubated with GST or GST Raf1-RBD glutathione-Sepharose beads for 1 h. The Western blot in Fig. 1B demonstrates a robust increase in Ras activation in TBC1D3-expressing cells, which is enhanced almost 3-fold in response to EGF when compared with control cells. Similar activation profiles were obtained with other cell lines (data not shown).
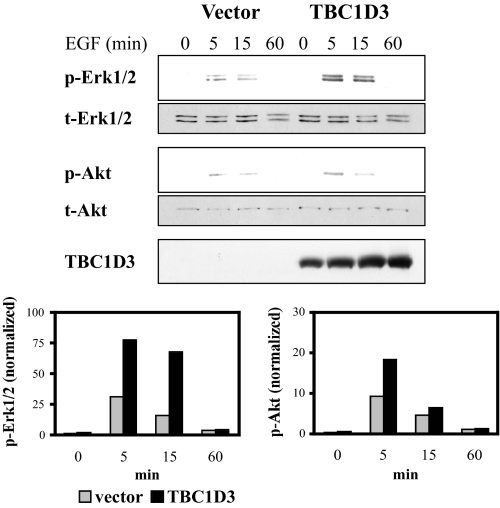
TBC1D3 and EGF Synergize in the Activation of the Erk1/2 and Akt Pathways—Signaling through Erk1/2 and protein kinase B/Akt are among the earliest post-EGF binding events associated with delayed apoptosis or cell proliferation. To investigate the effect of TBC1D3 on these two pathways, DU145 control or DU145:TBC1D3 cells were incubated with EGF at 37 °C, and levels of phospho-Erk1/2 and phospho-Akt were recorded by Western blotting. Total cellular contents of Erk1/2 or Akt were used to normalize the results in each sample. The data depicted in Fig. 2 show that following EGF stimulation both Erk1/2 and Akt activation were substantially elevated in cells expressing TBC1D3.
FIGURE 2.
TBC1D3 expression enhances activation of Akt and Erk1/2 following EGF stimulation. DU145 cells expressing vector alone or TBC1D3 were serum-starved and incubated at 37 °C in the presence of 100 ng/ml EGF. The cells were washed, lysed, and analyzed by Western blot. The bar graphs were derived from densitometric analysis and show p-Erk1/2 and p-Akt normalized to the actual protein loaded.
A parallel study, measuring dose response to EGF, was carried out with a well established mouse NR6 fibroblast line that stably expresses both human EGFR (9) and TBC1D3 (designated hereafter as NR6:hEGFR:TBC1D3). Murine cells do not express the hominoid-specific TBC1D3, and thus represent a null background, an optimal setting upon which to examine the effects of TBC1D3 expression. As shown in supplemental Fig. S1, control cells (vector alone) incubated with EGF, displayed activation kinetics that reached a maximum at 5 min for both Erk1/2 and Akt. NR6:hEGFR:TBC1D3 cells exhibited a heightened response to each concentration of EGF, also reaching a maximum at 5 min for both Erk1/2 and Akt. Additionally, Erk1/2 signaling in control cells subsided after 5 min, whereas TBC1D3-expressing cells sustained their Erk1/2 activation beyond 30 min. Thus, increases in both the magnitude and duration of Erk1/2 and Akt phosphorylation by TBC1D3 expression may account for the positive impact on cell proliferation observed following EGF stimulation.
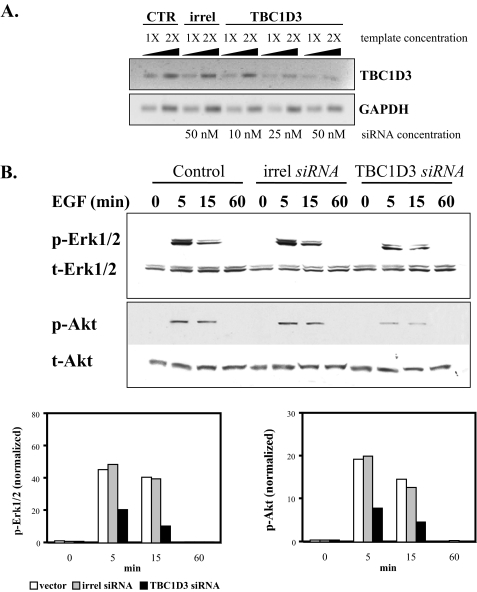
To rule out misleading phenotypes due to protein overexpression, we used RNA interference to suppress endogenous TBC1D3. DU145 cells were depleted of TBC1D3 using a specific commercial siRNA pool (Dharmacon). Transcript levels were evaluated by semi-quantitative RT-PCR 36 h later. A scrambled, irrelevant siRNA (Ambion) was used as a negative control. The extent of TBC1D3 silencing was ∼70% at the higher concentration assayed (Fig. 3A). Different concentrations of template demonstrated that the reactions fell within the linear range of template versus PCR product. In agreement with the results obtained when overexpressing TBC1D3, silencing TBC1D3 expression suppressed the activation of both Akt and Erk1/2 following EGF stimulation (Fig. 3B). Similar results were obtained with two additional in-house designed siRNAs (data not shown).
FIGURE 3.
TBC1D3 depletion decreases Akt and Erk1/2 activation. A, semi-quantitative RT-PCR. TBC1D3 was suppressed in DU145 cells using a commercial siRNA pool (Dharmacon) as described under “Experimental Procedures.” Transcript levels of TBC1D3 (top panel) were determined by semi-quantitative RT-PCR from control cells (lanes 1 and 2), cells transfected with irrelevant siRNA (lanes 3 and 4) and cells transfected with TBC1D3 siRNA (lanes 5-10). B, DU145 cells were transfected with TBC1D3 siRNA (50 nm), irrelevant siRNA (irrel) or untransfected (Ctr). 36 h later the cells were serum-starved, incubated with EGF (100 ng/ml) at 37 °C for different time points, and analyzed by Western blot. The bar graphs were derived from densitometric analysis and show p-Erk1/2 and p-Akt normalized to the actual protein loaded.
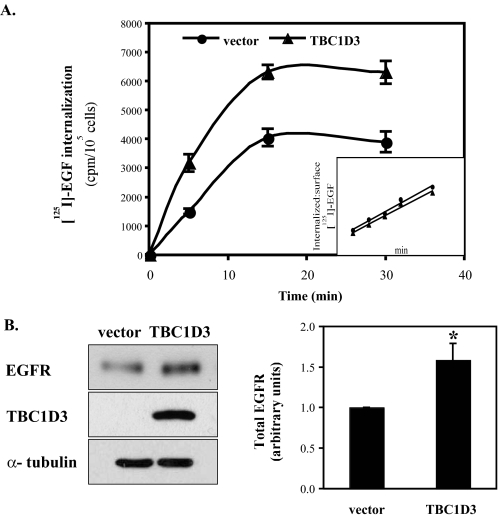
EGF Binding and Internalization Are Enhanced by TBC1D3—EGF-EGFR internalization and degradation represent a well studied paradigm for receptor tyrosine kinase trafficking (14, 15). To examine whether TBC1D3 regulates EGFR-mediated endocytosis, 125I-EGF uptake was measured in NR6:hEGFR: vector and NR6:hEGFR:TBC1D3 cells. Cells were serum-starved and then incubated with 125I-EGF (70 ng/ml) at 4 °C to allow ligand binding. After the cells were washed with ice-cold PBS, EGF uptake was evaluated by shifting the cells to 37 °C for different times. As shown in Fig. 4A, EGF accumulation, which plateaued at 15 min, was significantly greater in cells expressing TBC1D3. However, the rate of EGF internalization, expressed as the ratio of internalized to surface-bound EGF, was unaffected by the presence of TBC1D3 (inset in Fig. 4A). When cells were incubated with 125I-EGF for 3 h at 4 °C and surface-bound ligand was released, a 2-fold increase in EGF binding was recorded in cells expressing TBC1D3 (supplemental Fig. S2). Increased cell surface EGF binding and enhanced EGF internalization may be explained by higher levels of EGFR in cells that express TBC1D3. Indeed, TBC1D3-expressing cells contain ∼50% more EGFR at steady state than control cells (Fig. 4B). The data in Fig. 4B (right panel) show the densitometric analysis of multiple Western blots where EGFR signals were normalized to the total protein loaded. Similar results were obtained using human cell lines transiently transfected with TBC1D3 (data not shown).
FIGURE 4.
TBC1D3 increases EGF internalization. A, NR6:hEGFR cells expressing TBC1D3 were serum-starved and incubated at 4 °C for 1 h with 125I-EGF (70 ng/ml). Internalization at 37 °C was quantified as described under “Experimental Procedures.” Inset: EGF internalization rate was estimated by the ratios of internalized to surface-bound radioactivity. B, EGFR levels are enhanced in cells expressing TBC1D3. NR6:hEGFR cells expressing TBC1D3 or vector alone were lysed and analyzed by Western blot. The bar graph (right panel) was derived from densitometric analysis and shows EGFR signals normalized to the actual protein loaded, using the α-tubulin signal (*, p < 0.01). The data are presented as means ± S.D. of three independent experiments.
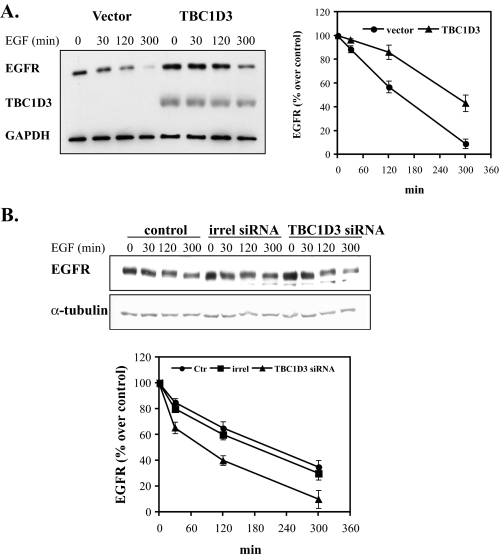
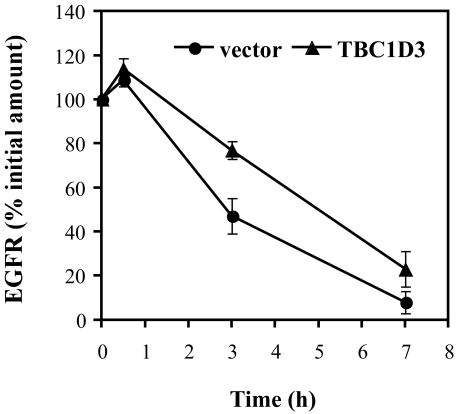
EGFR Degradation Is Delayed by TBC1D3—Enhanced levels of EGFR in cells expressing TBC1D3 could be due to increased receptor synthesis, delayed receptor degradation, or both. Normally, EGF-EGFR binding results in the internalization and targeting of the receptor-ligand complex to multivesicular endosomal compartments followed by lysosomal degradation of the complex. One explanation for enhanced EGFR levels in cells expressing TBC1D3 could be a change in the kinetics of EGFR degradation. To test whether TBC1D3 slows this process, DU145 cells were transiently transfected with pCMV-Myc-TBC1D3 or vector alone. After 48 h the cells were serum-starved and incubated with EGF (100 ng/ml) at 37 °C for the indicated times. Total cellular levels of EGFR were quantified by Western blot (Fig. 5A). Cycloheximide (25 μg/ml) was added to suppress de novo EGFR synthesis. Control cells rapidly degraded EGFR following internalization; after 120 min of incubation with EGF, over 40% of the EGFR signal was lost (see Fig. 5A, right panel, for quantitation). In contrast, in cells expressing TBC1D3, EGFR degradation was significantly delayed: merely 15% of the receptor population was degraded after 120 min, and only after >4 h was there a substantial reduction in EGFR (∼40%). On the contrary, when TBC1D3 was suppressed with siRNA, EGFR degradation was significantly increased (Fig. 5B). Whereas control cells expressing endogenous levels of TBC1D3 displayed ∼40% receptor degradation after 120 min, TBC1D3-depleted cells showed a faster degradation rate (∼60%) at the same time point. Comparable data were obtained with mouse NR6:hEGFR cells expressing TBC1D3 (supplemental Fig. S3). Moreover, we obtained similar results using a fluorescent ligand for the EGFR and observing cells by light microscopy; after 2 h of internalization of Alexa488-EGF (200 ng/ml), EGF was barely detectable in control cells, whereas the signal was readily apparent in cells expressing TBC1D3 (data not shown). Finally, the delayed degradation of EGFR was confirmed by a pulse-chase experiment following metabolic labeling of cells with [35S]methionine (Fig. 6). EGFR was immunoprecipitated following various chase periods with non-radioactive medium and was quantified by densitometry of the autoradiograms. These experiments indicated that, while NR6:hEGFR:vector cells retained ∼50% of the radiolabeled receptor after a 3-h chase, 80% of the radiolabeled EGFR was still detectable in cells expressing TBC1D3. In addition to slowed receptor degradation, it is formally possible that the heightened EGFR expression in TBC1D3-expressing cells could be in part due to enhanced receptor synthesis. However, this is very unlikely, because the amount of receptor biosynthesis measured after a 30-min chase period appeared similar in cells expressing and lacking TBC1D3 (Fig. 6).
FIGURE 5.
EGFR degradation is delayed in cells expressing TBC1D3. A, DU145 cells transfected with pCMV-Myc-TBC1D3 or vector alone were serum-starved and incubated with 100 ng/ml EGF at 37 °C for the indicated times. The cells were washed, and lysates (10 μg of protein each) were analyzed by Western blotting. B, TBC1D3 depletion accelerates EGFR degradation. DU145 cells were transfected with a commercial siRNA pool (Dharmacon) (50 nm), irrelevant siRNA (irrel) or untransfected (Ctr). 36 h later the cells were serum-starved and incubated with 100 ng/ml EGF at 37 °C for the indicated times. The cells were lysed and analyzed by Western blotting. Data are presented as a ratio of the amount of EGFR present at each time point over the amount in each sample at 0 min.
FIGURE 6.
TBC1D3 alters EGFR turnover. EGFR turnover was assayed by metabolically labeling the cells with [35S]methionine for 1 h as described under “Experimental Procedures.” Cells were chased in complete medium supplemented with cysteine and methionine. At different times cells were lysed, and EGFR was immunoprecipitated, resolved by SDS-PAGE, and detected by autoradiography.
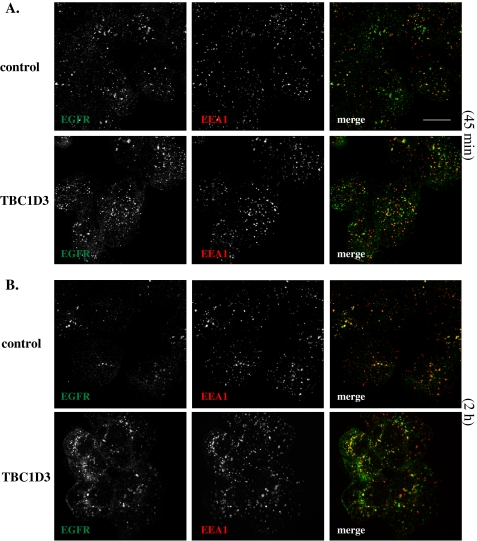
To better understand how TBC1D3 may be altering the intracellular fate of EGFR, we evaluated the effects of TBC1D3 expression on EGFR trafficking by confocal microscopy. DU145 cells stably expressing TBC1D3 or control cells were serum-starved, incubated with 100 ng/ml EGF at 4 °C for 1 h, and then shifted to 37 °C, after which they were fixed and processed for immunofluorescence microscopy. No apparent differences were detected in the cell surface labeling of EGFR or in the co-localization with the early endosomal marker EEA1, between TBC1D3-expressing and control cells at 0 and 10 min, respectively (data not shown). Following 45 min of internalization, there exists little overlap between EGFR (green) and EEA1 (red) in control cells (Fig. 7A, upper panel), suggesting that the ligand had transited through early endosomes and progressed to a later endocytic compartment (presumably late endosomes or lysosomes). However, in TBC1D3-expressing cells at the same time point, a significant portion of EGFR still appears to be co-localized with EEA1 (Fig. 7A, lower panel). Two hours later, the majority of the EGFR appeared to be degraded in control cells (Fig. 7B, upper panel). Meanwhile, as shown before, a significant portion of internalized EGFR is still traveling through the endocytic compartment and a subset of EGFR still co-localizes with EEA1 (Fig. 7B, lower panel). These results were corroborated by fractionation experiments on sucrose density gradients (data not shown). This study indicates that TBC1D3 delays the exit of EGFR from early endosomes, possibly extending the lifetime of the receptor. Moreover, these findings suggest that the post-translational modifications that normally accompany EGFR trafficking and mediate its degradation may be modulated by TBC1D3.
FIGURE 7.
EGFR trafficking is altered by TBC1D3 expression. DU145 cells stably expressing TBC1D3 or vector alone were starved and incubated with EGF (100 ng/ml) for 1 h at 4°C. To follow EGF internalization and trafficking, cells were washed and incubated at 37 °C for 45 min (A) or 2 h (B). The coverslips were then fixed, probed for EEA1 with a monoclonal antibody (red) and EGFR with polyclonal antibody (green), and processed for confocal microscopy as described under “Experimental Procedures.” Merged images demonstrate that after 2 h some EGFR molecules still co-localize with EEA1-positive organelles in cells expressing TBC1D3. Confocal data are representative of at least three independent experiments. Scale bar, 10 μm.
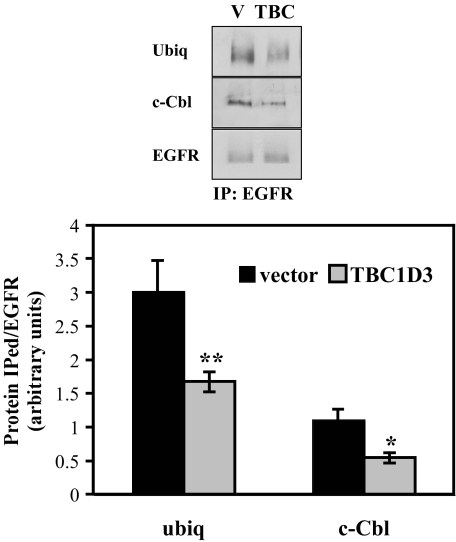
TBC1D3 Delays the Recruitment of Cbl and Suppresses EGFR Ubiquitination—Signal transducing receptors such as the EGFR are commonly ubiquitinated following activation as part of the mechanism for targeting to the lysosomal or proteasomal-degradative pathways (16). Delayed degradation of EGFR in cells expressing TBC1D3 may result from failure of activated EGFR to be properly ubiquitinated. To examine this possibility, we analyzed the ubiquitination state of the EGFR in TBC1D3-expressing (or control) cells after ligand binding. Cells were stimulated with EGF for 5 min and lysed in buffer containing 10 mm N-ethylmaleimide (to prevent loss of the ubiquitination signal). EGFR was immunoprecipitated, resolved by SDS-PAGE, and transferred to membranes that were probed with ubiquitin and EGFR antibodies. The latter was used to normalize the relative intensity of the EGFR signal so that direct comparisons could be made. The results shown in Fig. 8 confirm that EGFR ubiquitination is reduced by >30% when TBC1D3 is expressed. Similar results were obtained using cells transiently transfected with TBC1D3 (data not shown). Ubiquitination of EGFR is mediated by Cbl (17), an E3 ubiquitin ligase that is recruited to activated EGFR, both directly via EGFR-SH2 domain interactions, and indirectly via a second adaptor protein, Grb2 (18). To determine whether the interaction of EGFR with Cbl was disrupted by TBC1D3, co-immunoprecipitation experiments were performed. As shown in Fig. 8, the recruitment of Cbl to EGFR was substantially reduced in cells expressing TBC1D3. Fig. 8 (lower panel) summarizes the quantification of three independent Western blots where the signals were normalized to the amount of EGFR precipitated in each case. Equal amounts of c-Cbl were recovered in cells expressing TBC1D3 or vector alone (data not shown).
FIGURE 8.
TBC1D3 inhibits EGFR ubiquitination and c-Cbl recruitment. HeLa cells stably expressing TBC1D3 or vector alone (V) were incubated with 100 ng/ml EGF for 5 min at 37 °C. Cells were washed and lysed, and EGFR was immunoprecipitated as described under “Experimental Procedures.” Proteins were separated by SDS-PAGE and probed using antibodies specific for ubiquitin, EGFR, and c-Cbl. The bar graph (lower panel) was derived from densitometric analysis of Western blots normalized to EGFR total amounts. The data are presented as means ± S.D. of three independent experiments (*, p < 0.01; **, p < 0.001).
EGFR Polyubiquitination Is Not Affected by TBC1D3—Different types of ubiquitin conjugates are involved in the regulation of distinctive cellular processes (19). Whereas mono-ubiquitination has been implicated in endocytosis and membrane trafficking (20), polyubiquitin chains formed via ubiquitin Lys-48 act as targeting signals for proteasome degradation, and chains linked via Lys-63 are proposed to be involved in DNA repair and endocytosis (21, 22). EGFR is modified by both mono- and polyubiquitination (19, 23, 24). Our finding, that ubiquitination of activated EGFR is substantially reduced in cells expressing TBC1D, led us to investigate whether the nature of the ubiquitin linkage was modified as well. To quantify the forms of polyubiquitin bound to the EGFR in NR6: hEGFR:TBC1D3 cells, we used the ubiquitin-AQUA method recently developed by Kirkpatrick et al. (12). This strategy relies on the use of a synthetic internal standard peptide that is introduced at a known concentration to the cell lysates during digestion. Control and TBC1D3-expressing cells were stimulated with EGF for 5 min, and EGFR was immunoprecipitated from cell lysates and resolved by SDS-PAGE. The gel was stained with Coomassie Blue, and the region immediately above the EGFR was excised and analyzed. The results are presented in Table 1. Polyubiquitin chains linked through Lys-63 were more abundant than all the other linkages, comprising almost 35% of the total EGFR ubiquitination. In addition, Lys-48 linkage was also identified, representing ∼10% of the ubiquitinated EGFR. The other ubiquitin forms (i.e. Lys-29, Lys-11, Lys-6, Lys-27, and Lys-33) were below the limit of quantitation. The data clearly indicate that, despite suppressed ubiquitination, the linkage pattern for EGFR in TBC1D3-expressing cells was nearly identical to that observed in control cells. We conclude that the impact of TBC1D3 expression on the fate of internalized EGFR is not due to alterations in the type of receptor ubiquitination.
TABLE 1.
TBC1D3 does not affect EGFR polyubiquitination
NR6:hEGFR cells expressing TBC1D3 or vector alone were stimulated with EGF for 5 min, EGFR was immunoprecipitated, and ubiquitination was explored by Ub-AQUA analysis. Values represent % total EGFR-associated ubiquitin ± S.D. (n = 3).
|
EGFR |
||
|---|---|---|
| Control | TBC1D3 | |
| % | ||
| Lys-63 | 32.67 ± 1.46 | 33.23 ± 1.49 |
| Lys-48 | 10.20 ± 0.64 | 9.83 ± 1.56 |
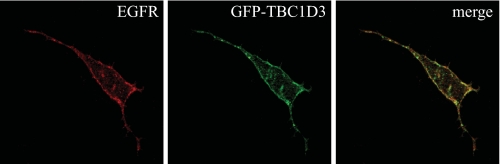
TBC1D3 Is Localized to the Plasma Membrane—To localize TBC1D3 by confocal microscopy, green fluorescent protein-TBC1D3 was transiently transfected into HeLa cells. After 36 h post-transfection, the cells were fixed and processed for immunofluorescence. A monoclonal anti-EGFR antibody was used to visualize EGFR localization. The results indicate that TBC1D3 mainly localized to structures at the plasma membrane (Fig. 9, middle panel). These findings are consistent with previous work on TRE17 (also known as Tre-2 or USP6), a novel Arf6 regulator that contains a similar TBC domain, which also showed a membrane localization (25). The merged panel (Fig. 9, right panel) clearly shows that TBC1D3 significantly co-localized with EGFR at the plasma membrane, supporting the notion that it may interfere with the ability of c-Cbl to be recruited to the receptor.
FIGURE 9.
TBC1D3 localizes at the plasma membrane. HeLa cells were transfected with green fluorescent protein-TBC1D3. After 36 h post-transfection, cells were fixed and processed for microscopy as described. EGFR was detected using a mouse monoclonal antibody followed by an anti-mouse Alexa594 conjugated antibody (red). Images are representative of three independent experiments.
DISCUSSION
Understanding the cell and molecular biology of hominoid- and human-specific genes and proteins may provide a unique opportunity to better understand physiologic regulatory mechanisms in humans. Our studies here suggest a potential mechanism by which TBC1D3 may act as a regulator of signal transduction and as an oncoprotein. We have focused our efforts on TBC1D3D, although experiments on a second paralog (TBC1D3) have produced results indistinguishable to those reported here.
Growth factor receptor signaling is a major determinant of cell proliferation in normal cell and tissue development and in cancer. Recent work has shown that growth factor receptor activation and endocytic trafficking are linked (26, 27). We report that expression of TBC1D3, initially identified as a prostate and breast cancer oncogene, increased the basal level of cellular growth and enhanced the proliferative response to EGF. Enhanced cell proliferation was correlated with a substantial increase in EGF binding and a decrease in EGFR degradation. EGFR degradation studies demonstrated that the lifetime of internalized EGFR is extended in cells expressing TBC1D3, by causing a delay in the exit of the receptor from an early endosome compartment en route to the degradative compartment. TBC1D3 is active in both human and mouse cells, and siRNA studies in human cells indicated that silencing TBC1D3 depresses EGFR signaling and enhances EGFR degradation. TBC1D3 appears to be expressed in human cells at very low levels, and although available antibodies readily detect exogenously expressed TBC1D3 in mouse or human cells, routine detection and quantification of endogenous by Western blotting were not reliable. We used semi-quantitative RT-PCR to evaluate the effects of siRNA silencing of TBC1D3. To evaluate EGFR degradation in TBC1D3-expressing cells, we used three different approaches: quantification of the levels of receptor remaining after EGF stimulation by Western blotting, measurement of EGFR synthesis and turnover using radiolabeled methionine, and light microscopy to follow receptor localization. Our findings reveal delayed EGFR degradation and altered trafficking rather than enhanced biosynthesis. The endosomal compartment where internalized EGFR accumulates in the presence of TBC1D3 is reminiscent of that reported by Zerial and colleagues, who found that a fraction of internalized EGFR was diverted to a compartment that recruited APPL1, a multidomain-containing protein that physically carries the signal from the EGFR signaling sites into the nucleus (28). Similarly, both APPL1 and TBC1D3 interact with the small GTPase Rab5, a well known regulator protein in several growth factor signaling and trafficking cascades. Further investigation is required to elucidate the nature and origin of this endosomal compartment.
Parallel to the effects on endocytic trafficking, TBC1D3 substantially suppressed the ubiquitination of activated EGFR. Reduced EGFR ubiquitination, in turn, appears to correlate with suppressed recruitment of c-Cbl, the E3 ligase that catalyzes EGFR ubiquitination. Moreover, in pulldown experiments, TBC1D3 was found to interact either directly or indirectly with c-Cbl (data not shown), and this interaction may hamper the efficient recruitment and binding of the E3 ligase to EGFR. This notion is supported, in part, by the localization of TBC1D3 in structures located at the plasma membrane. We cannot exclude the possibility that TBC1D3 stimulates the action of deubiquitinating enzymes that remove ubiquitin from proteins such as EGFR and prevent ubiquitin-mediated degradation of proteins. There is a large collection of deubiquitinating enzymes encoded in the human genome (29). UBPY (also called USP8) is known to prevent EGFR degradation, to activate Erk signaling when overexpressed, and to block progression of the cell cycle when deleted (30, 31). Finally, TBC1D3 may also affect ubiquitin-like proteins, such as NEDD8, that covalently couple EGFR after ligand stimulation in a Cbl-dependent fashion (32). NEDD8 residues targeted to the receptor may serve a redundant function to ensure EGFR degradation. Inhibition of EGFR neddylation by TBC1D3 might explain the reduced ubiquitination of the receptor. We cannot rule out the possibility of EGFR recycling to the cell surface in the absence of ubiquitination in TBC1D3-expressing cells as another cause for extended EGFR signaling. However, more extensive studies should be made to address this point.
To establish whether TBC1D3 alters the quality of EGFR polyubiquitination as an explanation for delayed degradation, we analyzed EGFR ubiquitin linkages using the recently developed ubiquitin-AQUA methodology. The results, however, did not reveal significant qualitative differences between cells expressing TBC1D3 and the controls. These observations are consistent with previous reports using the EGFR mutant Y1045F. This point mutation abrogates phosphorylation of residue 1045; however, while the ubiquitination of the receptor was impaired and c-Cbl binding diminished, the forms of ubiquitin conjugated to EGFR were of identical composition to those seen on wild-type EGFR (24). A plausible explanation for these findings is that in both cases (i.e. by expressing the Y1045F mutant or aberrantly expressing TBC1D3) when EGFR gets modified, it gets fully modified. The main physiological effect (increased/prolonged EGFR activity) results not from a difference in the composition of the ubiquitin signal on any given ubiquitinated EGFR, but rather from a decrease in the proportion of total modified receptor. However, we cannot rule out the co-existence of various EGFR pools with different ubiquitination levels as the outcome of expressing TBC1D3.
In addition to the extended lifetime of the internalized EGFR, elevated signaling by the EGFR in cells expressing TBC1D3 may be due to enhanced priming of the Ras activation pathway. We noted elevated levels of activated Ras in cells expressing TBC1D3 without EGF stimulation. GTP-Ras levels in cells expressing TBC1D3 are further enhanced by EGF stimulation. It is possible that TBC1D3 acts directly or indirectly to alter the mechanism of GTP-Ras production, either by elevating guanine nucleotide exchange on Ras or by inhibiting GTP-Ras hydrolysis. Experiments to resolve this issue are underway.
Lastly, TBC1D3 contains a TBC domain that was initially reported to be a Rab5 GAP (2). Using a GAP assay, including RN-Tre, a well known Rab5 GAP as a positive control, we found that TBC1D3 has no GAP activity (data not shown). Moreover, Tre-2, a hominoid-specific chimeric protein, linking an ancient ubiquitin ligase domain with a domain possessing 89% identity with the TBC domain of TBC1D3, has also been found to have no GAP activity (33, 34). GAP domains have a motif characterized by an “arginine finger” required for their catalytic activity, as contained in RN-Tre and RabGap5 (35), two proven Rab5 GAPs. TBC1D3 lacks the arginine that is critical for GTP hydrolysis. Inspection of the TBC domain of each of the TBC1D3 paralogs revealed that this key arginine residue has been replaced by a glycine (Gly-151). Although TBC1D3 binds to Rab5 in pulldown experiments and interacts with Rab5 in translation experiments in vitro (data not shown), we believe that TBC1D3 operates as a Rab5 effector, not a Rab5 GAP. This function may be similar to a recent finding by De Camilli and colleagues (36), who found that OCRL, a phosphoinositide phosphatase regulating endocytosis, binds Rab5 and APPL1, the Rab5 effector.
Hominoid- and human-specific genes are more likely to behave as regulators of complex signaling pathways as opposed to defining new pathways de novo. TBC1D3 and its paralogs may play diverse roles in receptor signaling during development and in regulation of cellular events during normal hominoid or human physiology, as well as being a participant in a raft of proliferative diseases. Future work will focus on the function of these paralogs as well as the regulation of their transcription and expression. Understanding the mode and mechanism of action of TBC1D3 may provide a unique insight into recently evolved genes that modulate signaling pathways in a human-specific manner and provide a platform for exploring the role of human-specific genes in human physiology and pathogenesis.
Addendum—While the manuscript was under review another report on TBC1D3 appeared “ahead of print” (Frittoli, et al. (37)).
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Jing Li for providing TBC1D3 cDNA; Dr. Ignacio Rubio for providing GST-Raf-1 RBD; E. Peters for excellent technical assistance; D. Owyoung for editing assistance; and Didier Hodzic and Xiong Su for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM42259 (to P. D. S.) and R01 GM67945 (to S. P. G.). This work was also supported by the Jose Carreras International Leukemia Foundation (to M. A. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S3 (Data 1-3).
Footnotes
The abbreviations used are: EGF, epidermal growth factor; EGFR, EGF receptor; Erk, extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase; siRNA, small interference RNA; RT, reverse transcription; PBS, phosphate-buffered saline; RBD, Ras binding domain; GST, glutathione S-transferase; CMV, cytomegalovirus; E3, ubiquitin-protein isopeptide ligase; EEA1, early endosomal antigen 1.
References
- 1.Hodzic, D., Kong, C., Wainszelbaum, M. J., Charron, A. J., Su, X., and Stahl, P. D. (2006) Genomics 88 731-736 [DOI] [PubMed] [Google Scholar]
- 2.Pei, L., Peng, Y., Yang, Y., Ling, X. B., Van Eyndhoven, W. G., Nguyen, K. C., Rubin, M., Hoey, T., Powers, S., and Li, J. (2002) Cancer Res. 625420 -5424 [PubMed] [Google Scholar]
- 3.Zody, M. C., Garber, M., Adams, D. J., Sharpe, T., Harrow, J., Lupski, J. R., Nicholson, C., Searle, S. M., Wilming, L., Young, S. K., Abouelleil, A., Allen, N. R., Bi, W., Bloom, T., Borowsky, M. L., Bugalter, B. E., Butler, J., Chang, J. L., Chen, C. K., Cook, A., Corum, B., Cuomo, C. A., de Jong, P. J., DeCaprio, D., Dewar, K., FitzGerald, M., Gilbert, J., Gibson, R., Gnerre, S., Goldstein, S., Grafham, D. V., Grocock, R., Hafez, N., Hagopian, D. S., Hart, E., Norman, C. H., Humphray, S., Jaffe, D. B., Jones, M., Kamal, M., Khodiyar, V. K., LaButti, K., Laird, G., Lehoczky, J., Liu, X., Lokyitsang, T., Loveland, J., Lui, A., Macdonald, P., Major, J. E., Matthews, L., Mauceli, E., McCarroll, S. A., Mihalev, A. H., Mudge, J., Nguyen, C., Nicol, R., O'Leary, S. B., Osoegawa, K., Schwartz, D. C., Shaw-Smith, C., Stankiewicz, P., Steward, C., Swarbreck, D., Venkataraman, V., Whittaker, C. A., Yang, X., Zimmer, A. R., Bradley, A., Hubbard, T., Birren, B. W., Rogers, J., Lander, E. S., and Nusbaum, C. (2006) Nature 4401045 -104916625196 [Google Scholar]
- 4.Schlessinger, J. (2000) Cell 103211 -225 [DOI] [PubMed] [Google Scholar]
- 5.Citri, A., and Yarden, Y. (2006) Nat. Rev. Mol. Cell Biol. 7505 -516 [DOI] [PubMed] [Google Scholar]
- 6.Lynch, T. J., Bell, D. W., Sordella, R., Gurubhagavatula, S., Okimoto, R. A., Brannigan, B. W., Harris, P. L., Haserlat, S. M., Supko, J. G., Haluska, F. G., Louis, D. N., Christiani, D. C., Settleman, J., and Haber, D. A. (2004) N. Engl. J. Med. 3502129 -2139 [DOI] [PubMed] [Google Scholar]
- 7.Paez, J. G., Janne, P. A., Lee, J. C., Tracy, S., Greulich, H., Gabriel, S., Herman, P., Kaye, F. J., Lindeman, N., Boggon, T. J., Naoki, K., Sasaki, H., Fujii, Y., Eck, M. J., Sellers, W. R., Johnson, B. E., and Meyerson, M. (2004) Science 3041497 -1500 [DOI] [PubMed] [Google Scholar]
- 8.Crosetto, N., Tikkanen, R., and Dikic, I. (2005) FEBS Lett. 5793231 -3238 [DOI] [PubMed] [Google Scholar]
- 9.Chen, P., Gupta, K., and Wells, A. (1994) J. Cell Biol. 124547 -555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor, S. J., Resnick, R. J., and Shalloway, D. (2001) Methods Enzymol. 333333 -342 [DOI] [PubMed] [Google Scholar]
- 11.Sorkin, A., Waters, C., Overholser, K. A., and Carpenter, G. (1991) J. Biol. Chem. 2668355 -8362 [PubMed] [Google Scholar]
- 12.Kirkpatrick, D. S., Hathaway, N. A., Hanna, J., Elsasser, S., Rush, J., Finley, D., King, R. W., and Gygi, S. P. (2006) Nat. Cell Biol. 8700 -710 [DOI] [PubMed] [Google Scholar]
- 13.Bos, J. L. (1989) Cancer Res. 494682 -4689 [PubMed] [Google Scholar]
- 14.Chen, W. S., Lazar, C. S., Lund, K. A., Welsh, J. B., Chang, C. P., Walton, G. M., Der, C. J., Wiley, H. S., Gill, G. N., and Rosenfeld, M. G. (1989) Cell 59 33-43 [DOI] [PubMed] [Google Scholar]
- 15.Wells, A., Welsh, J. B., Lazar, C. S., Wiley, H. S., Gill, G. N., and Rosenfeld, M. G. (1990) Science 247962 -964 [DOI] [PubMed] [Google Scholar]
- 16.Resat, H., Ewald, J. A., Dixon, D. A., and Wiley, H. S. (2003) Biophys. J. 85 730-743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levkowitz, G., Waterman, H., Zamir, E., Kam, Z., Oved, S., Langdon, W. Y., Beguinot, L., Geiger, B., and Yarden, Y. (1998) Genes Dev. 123663 -3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waterman, H., Katz, M., Rubin, C., Shtiegman, K., Lavi, S., Elson, A., Jovin, T., and Yarden, Y. (2002) EMBO J. 21303 -313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haglund, K., Di Fiore, P. P., and Dikic, I. (2003) Trends Biochem. Sci. 28598 -603 [DOI] [PubMed] [Google Scholar]
- 20.Hicke, L. (2001) Nat. Rev. Mol. Cell Biol. 2195 -201 [DOI] [PubMed] [Google Scholar]
- 21.Pickart, C. M. (2001) Annu. Rev. Biochem. 70503 -533 [DOI] [PubMed] [Google Scholar]
- 22.Weissman, A. M. (2001) Nat. Rev. Mol. Cell Biol. 2169 -178 [DOI] [PubMed] [Google Scholar]
- 23.Mosesson, Y., Shtiegman, K., Katz, M., Zwang, Y., Vereb, G., Szollosi, J., and Yarden, Y. (2003) J. Biol. Chem. 27821323 -21326 [DOI] [PubMed] [Google Scholar]
- 24.Huang, F., Kirkpatrick, D., Jiang, X., Gygi, S., and Sorkin, A. (2006) Mol. Cell 21 737-748 [DOI] [PubMed] [Google Scholar]
- 25.Martinu, L., Masuda-Robens, J. M., Robertson, S. E., Santy, L. C., Casanova, J. E., and Chou, M. M. (2004) Mol. Cell Biol. 249752 -9762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polo, S., and Di Fiore, P. P. (2006) Cell 124897 -900 [DOI] [PubMed] [Google Scholar]
- 27.Sorkin, A., and Von Zastrow, M. (2002) Nat. Rev. Mol. Cell Biol. 3600 -614 [DOI] [PubMed] [Google Scholar]
- 28.Miaczynska, M., Christoforidis, S., Giner, A., Shevchenko, A., Uttenweiler-Joseph, S., Habermann, B., Wilm, M., Parton, R. G., and Zerial, M. (2004) Cell 116445 -456 [DOI] [PubMed] [Google Scholar]
- 29.Millard, S. M., and Wood, S. A. (2006) J. Cell Biol. 173463 -468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naviglio, S., Mattecucci, C., Matoskova, B., Nagase, T., Nomura, N., Di Fiore, P. P., and Draetta, G. F. (1998) EMBO J. 173241 -3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alwan, H. A., and van Leeuwen, J. E. (2007) J. Biol. Chem. 2821658 -1669 [DOI] [PubMed] [Google Scholar]
- 32.Oved, S., Mosesson, Y., Zwang, Y., Santonico, E., Shtiegman, K., Marmor, M. D., Kochupurakkal, B. S., Katz, M., Lavi, S., Cesareni, G., and Yarden, Y. (2006) J. Biol. Chem. 28121640 -21651 [DOI] [PubMed] [Google Scholar]
- 33.Martinu, L., Santiago-Walker, A., Qi, H., and Chou, M. M. (2002) J. Biol. Chem. 27750996 -51002 [DOI] [PubMed] [Google Scholar]
- 34.Bizimungu, C., Thomas, A., Brasseur, R., and Vandenbol, M. (2007) Biotechnol. Lett. 291927 -1937 [DOI] [PubMed] [Google Scholar]
- 35.Haas, A. K., Fuchs, E., Kopajtich, R., and Barr, F. A. (2005) Nat. Cell Biol. 7 887-893 [DOI] [PubMed] [Google Scholar]
- 36.Erdmann, K. S., Mao, Y., McCrea, H. J., Zoncu, R., Lee, S., Paradise, S., Modregger, J., Biemesderfer, D., Toomre, D., and De Camilli, P. (2007) Dev. Cell 13 377-390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frittoli, E., Palamidessi, A., Pizzigoni, A., Lanzetti, L., Garrè, M., Troglio, F., Troilo, A., Fukuda, M., Di Fiore, P. P., Scita, G., and Confalonieri, S. (January 16,2008. ) Mol. Biol. Cell 10.1091/mbc.E07-06-0594 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.