Abstract
Small insertions and deletions of trinucleotide repeats (TNRs) can occur by polymerase slippage and hairpin formation on either template or newly synthesized strands during replication. Although not predicted by a slippage model, deletions occur preferentially when 5′-CTG is in the lagging strand template and are highly favored over insertion events in rapidly replicating cells. The mechanism for the deletion bias and the orientation dependence of TNR instability is poorly understood. We report here that there is an orientation-dependent impediment to polymerase progression on 5′-CAG and 5′-CTG repeats that can be relieved by the binding of single-stranded DNA-binding protein. The block depends on the primary sequence of the TNR but does not correlate with the thermodynamic stability of hairpins. The orientation-dependent block of polymerase passage is the strongest when 5′-CAG is the template. We propose a “template-push” model in which the slow speed of DNA polymerase across the 5′-CAG leading strand template creates a threat to helicase-polymerase coupling. To prevent uncoupling, the TNR template is pushed out and by-passed. Hairpins do not cause the block, but appear to occur as a consequence of polymerase pass-over.
Expansion of simple trinucleotide repeats (TNRs)4 is the underlying genetic defect of a number of human neurodegenerative diseases. TNR-associated pathophysiology in human diseases strictly depends on the number of trinucleotide repeats (reviewed in Refs. 1-3). Although expansion can occur in germ cells (4) and in somatic cells with age (5, 6), emerging evidence suggests that rapid cell division in the early embryo can reduce the size of the repeats and modulate disease potential (7). TNR deletion during rapid cell division may serve as a natural defense mechanism for keeping the length of the repeats in check at critical times in development. Reducing the length of the repeat tract in disease genes is being explored as a therapeutic strategy. However, the mechanisms by which deletions rather than expansions are favored during replication remain poorly understood.
DNA polymerase and strand slippage has been proposed as the primary mechanism for instability of trinucleotide repeats (1-3). During replication, the TNR units can misalign resulting in an extrahelical DNA loop that increases TNR length if it occurs on the daughter strand and decreases TNR length if it occurs on the template strand (1-3). In both prokaryotic and eukaryotic models, it is well established that TNR instability is sensitive to the length of the repeat (8-12), to its structure forming potential (13-17), and to its proximity to the origin of replication (18). The factors that regulate the probability and frequency of deletion events, however, remain poorly understood. By a polymerase slippage model, a deletion implies that hairpin formation has occurred on the template strand. Theoretically, for an identical sequence located either on the template or daughter strand, slippage and hairpin formation can occur with equivalent frequency on either strand. Yet, deletions are favored over insertions during replication by a factor of 102-105 in yeast and bacteria (14). A simple slippage model does not predict the deletion bias. Equally puzzling is the orientation dependence of deletion. 5′-CTG repeats, for example, have greater susceptibility to deletions when they serve as a template for lagging strand DNA synthesis. However, as the leading strand template, 5′-CTG repeats are relatively stable (10, 19). The same phenomenon has been observed for 5′-CGG/5′-CCG repeats with fewer contractions when the 5′-CCG sequence is the template for lagging strand DNA synthesis (20). Thus, the frequency of TNR instability and the extent of deletion vary with the direction of replication fork progression, despite the fact that the same template sequence is being replicated (1-3, 21).
Several models have been proposed to explain why 5′-CTG loops might preferentially form on the lagging strand template. First, biophysical (14, 17, 22-24) and structural (17, 23, 24-26) analyses have demonstrated that single-stranded 5′-CTG forms more stable hairpin structures than single-stranded 5′-CAG due to the presence of wobble pairing. Thus, the differential stability of 5′-CTG hairpins and their lifetimes may allow more frequent 5′-CTG loop entrapment and slippage during replication on the lagging strand template (23-27). If correct, however, it is not easy to explain why the thermodynamics of hairpin formation should apply only when 5′-CTG repeats are in the lagging strand template, and why hairpins should not be equally favored on the daughter strand. A second possibility is that the 5′-CTG template during lagging strand DNA synthesis inhibits the binding of single-stranded DNA binding (SSB) protein allowing 5′-CTG hairpins to exist transiently in a single-stranded state. The 5′-CTG hairpins might form more rapidly or unfold more slowly relative to 5′-CAG hairpins in the same orientation (8, 26). However, there are few biochemical data on the interaction of SSB with TNR tracts. Finally, TNR instability is influenced by transcription, which is inherently strand-specific. However, the effects of transcription are system-dependent. In cultured mammalian cells, long repeat tracts tend to delete (28). However, in animal models for 5′-CAG repeat diseases, TNR sequences tend to expand in somatic cells even though transcription is active (5, 6). In rapidly dividing bacterial and yeast cells, the rate of replication far exceeds that of transcription (7, 29-31), and the impact of transcription on deletions appears to be relevant only when cells pass through stationary phase and growth is slow (32). Despite extensive analysis of the length and structure dependence of TNR instability during replication (reviewed in Refs. 1, 3, 5, 33), none of these existing models adequately explains why hairpins form preferentially on the template rather than the daughter strand during replication, and why 5′-CTG undergoes more deletions as the template in the lagging strand.
In this report, we have addressed the underlying mechanism for orientation dependence of deletion using quantitative analyses of TNR instability. In these experiments, we have carefully controlled for effects of copying random repetitive elements. Quantitative analysis reveals previously unknown features relevant to orientation dependence of TRN deletion. TNRs present an impediment to polymerase passage. On the leading strand, 5′-CAG imposes a more severe impediment than 5′-CTG. However, SSB removes the impediment to polymerase passage on the lagging strand. Thus, the orientation dependence of deletion is driven by interaction of DNA polymerase with the 5′-CAG sequence during leading strand DNA synthesis rather than by 5′-CTG sequence during lagging strand DNA synthesis. We propose a “template-push” model in which the orientation dependence arises by a disparity in the polymerase speed and the helicase speed during leading strand synthesis. Deletion occurs in order to prevent uncoupling between the leading strand DNA polymerase and DNA helicase. These observations provide a plausible mechanism for the deletion bias associated with TNR instability and the orientation dependence of deletion during replication.
EXPERIMENTAL PROCEDURES
DNA Plasmids and Oligonucleotides—Plasmids used for the in vivo study are listed on Fig. 1A. Triplet repeats of different lengths and sequences were inserted into a plasmid vector pcDNA3 in both orientations to generate a series of increasing long repeat tracts. Constructions are named based on sequence of the leading template. The six plasmids derived from pcDNA3 and containing a trinucleotide repeat of various sequences and lengths used for the in vitro studies are listed and described on Table 1. The plasmid pEmpty is also derived from pcDNA3 but did not contain any repetitive sequence. Constructions are named based on sequence of the template strand. Oligonucleotide sequences are listed on Table 1. Their annealing position respective to the repetitive sequence is shown on Table 1. The p867/4ps oligonucleotide contained four phosphorothioate linkages at its 5′ extremity that made it resistant to the 5′→3′ T7 exonuclease.
FIGURE 1.
Orientation dependence of triplet deletion occurs between 30 and 120 repeats. A, triplet repeats of different lengths and sequences were inserted into a plasmid vector pcDNA3 in both orientations to generate a series of increasing long repeat tracts. The table shows the length and sequence of the tested plasmids. 5′-CAG and 5′-CTG are tracts that form secondary structures; 5′-CAA and 5′-GTT are tracts that do not form secondary structure. The starred sequences indicate plasmids that had additional interrupted repeats flanking the pure repeat tract, however, in all cases, the number in the repeat name indicates the longest pure repeat stretch. The sequence (5[prime]-CAA)87 is (5′-CAA)22A(5′-CAA)87; (5′-GTT)87 is (5′-GTT)87T(5′-GTT)22; (5′-CAG)120 is (5′-CAG)42CAC(5′-CAG)120; (5′-CAA)135 is (CAA)135A(CAA)50; and (5′-GTT)135 is (5′-GTT)50T(5′-GTT)135. B, gel representation of the deletion events that occur as a function of tract length, indicated by the bracketed numbers. Assays were reported for three to five different cultures after growth for 18 h. C, summary of the % deletion of structure-forming and non-structure-forming plasmids. The % deletions are taken as the intensity of bands with a smaller repeat tract divided by the total intensity of all the products multiplied by 100. Constructions are named based on sequence in the leading template. 5′-CAG (gray circles); 5′-CTG (open circles); 5′-GTT (black circles); 5′-GGT (striped circles). Solid arrows indicate repeat the threshold for deletion; the dotted arrow indicates the number at which orientation of dependence of deletion is lost.
TABLE 1.
Description of plasmids and oligonucleotides used in in vitro studies
E. coli Strains—SURE and BL21(DE3pLysE) bacteria were from Stratagene and Novagen, respectively.
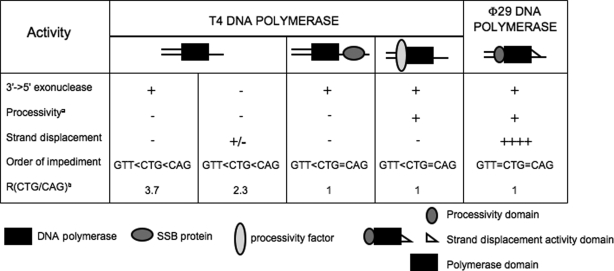
Proteins—Escherichia coli single-stranded DNA-binding protein was from GE Healthcare. Sequenase Version 2.0 DNA polymerase was from USB. Herculase was from Stratagene. T7 exonuclease; N. AlwI nicking enzyme; EcoRI, NdeI, and XbaI restriction endonucleases; T7 DNA polymerase; and T4 polynucleotide kinase were from New England Biolabs. ϕ29 DNA polymerase was either a gift from Dr. M. Salas or purchased from New England Biolabs. The T4 DNA polymerases, gp43, 3′→5′ exonuclease deficient or proficient, the T4 single-stranded DNA-binding protein, gp32, the sliding clamp, gp45, were prepared as previously described (35). To remove the DNA polymerase activity that contaminated previous protein stocks, the T4 clamp loader, gp44/62 complex, was purified as described previously (35) with the following modifications. The ssDNA cellulose column was replaced by a 40-ml hydroxyapatite (HA, Hydroxyapatite Bio-Gel HTP from Bio-Rad) and a 3-ml macro-prep HighS (support from Bio-Rad) columns. Both columns were performed subsequently to the DE52 (Whatman) and P11 (Whatman) columns. The low salt HA buffer contained 20 mm potassium phosphate, pH 7, 2.5 mm Mg(SO4)2, 10% glycerol, 5 mm β-mercaptoethanol. The high salt HA buffer contained 200 mm potassium phosphate, pH 7, 2.5 mm Mg(SO4)2, 10% glycerol, 5 mm β-mercaptoethanol. The low salt macroprep HighS buffer contained 30 mm MOPS, pH 7, 1 mm Mg(SO4)2, 10% glycerol, 5 mm β-mercaptoethanol. The high salt macroprep HighS buffer contained 30 mm MOPS, pH 7, 750 mm NaCl, 2.5 mm Mg(SO4)2, 10% glycerol, 5 mm β-mercaptoethanol. gp44/62 containing fractions eluted from the P11 column were dialyzed overnight against 2 liters of low salt HA buffer and loaded onto the HA column at a flow rate of 2 ml/min. Column was next washed with 40 ml of low salt HA buffer, and the gp44/62 protein complex was eluted by a 400-ml salt (20-200 mm potassium phosphate) gradient. The column was finally washed with 40 ml of high salt HA buffer. gp44/62-containing fractions were dialyzed overnight against 2 liters of low salt macroprep HighS buffer and loaded onto the macroprep HighS column at a flow rate of 0.5 ml/min. Column was next washed with 5 ml of low salt macroprep HighS buffer, and the gp44/62 protein complex was eluted by a 25-ml salt (0-750 mm NaCl) gradient. The column was finally washed with 5 ml of high salt macroprep HighS buffer. gp44/62-containing fractions were dialyzed overnight against 1 liter of prestorage buffer (50 mm potassium phosphate, pH 7, 5 mm Mg(SO4)2, 25% glycerol, 0.5 mm DTT) and for the next 24 h against 1 liter of storage buffer (50 mm potassium phosphate, pH 7, 5 mm Mg(SO4)2, 50% glycerol, 0.5 mm DTT). Protein complex was at least 98% pure as judged by a gel Coomassie Brilliant Blue R staining. Protein stocks were kept at -20 °C. All protein concentrations were determined by UV absorbance at 280 nm, using molar extinction coefficients of 1.3 × 105 m-1cm-1, 1.91 × 104 m-1cm-1, 1.23 × 105 m-1cm-1, and 3.7 × 105 m-1cm-1 for monomeric gp43 (3′→5′ exonuclease deficient or proficient), monomeric gp45, gp44/62 complex, and monomeric gp32, respectively. DNA polymerase and ssDNA-binding protein concentrations are given in monomeric units, gp44/62 complex concentration is given in protein complex units (one protein complex contains four subunits of gp44 and one subunit of gp62), and gp45 concentration is given in trimeric units.
Other Materials—dNTPs, ATP, and [γ-32P]ATP were from GE Healthcare.
TNR Plasmids and Stability Assay in E. coli—Plasmids containing triplet repeats of different sizes were generated by overlap PCR. Oligonucleotides containing 25 triplet repeat units were denatured, reannealed, and extended using Taq polymerase. Each round of denaturation generated asymmetric pairing of the oligonucleotides, and, after extension with Taq polymerase, repeats of multiple lengths were generated. Repeats were subcloned into pcDNA3 vectors, and the resulting plasmids were used to create bacterial stocks in SURE cells (Stratagene). Only plasmids containing uninterrupted repeats were used in the analysis (Fig. 1A). The stability of repeat sequences in the plasmids was assayed after growth in culture (36). Plasmids were isolated by cesium chloride gradients, restricted using EcoRI, and the lengths of the inserts were determined after separation in a DNA sequencing gel relative to DNA molecular weight markers. For transcription studies, plasmids of known repeat size and sequence were transformed into BL21(DE3LysE) cells. Isolated colonies were grown 18 h in 200 ml of LB in the presence or absence of 0.6 mm IPTG, and plasmids were then extracted. Plasmid yield was quantified by UV spectroscopy, and DNA was stored at -20 °C until further use. Repeat size after growth was evaluated by sizing 5′-end-labeled restriction fragments (NdeI and XbaI) containing the triplet repeat. Labeling was performed using [γ-32P]ATP. Fragments were resolved by gel electrophoresis as previously described (37). Fragments were detected using a PhosphorImager 425 (Amersham Biosciences), and deletion was quantified from digital images using IP Lab Gel software (Signal Analytics). Percent deletion was calculated by dividing the signal from the deletion products by the total signal (original insert size plus deletion products).
Plasmid Sequencing—Plasmid sequencing using the Sequenase version 2.0 DNA polymerase was performed as recommended by the manufacturer (USB).
Primer Extension Assay—Plasmid denaturation was performed by incubating 2.2 mg of plasmid in a denaturing solution (0.1 mg/ml plasmid, 2.3 mm EDTA, 200 mm NaOH) for 30 min at 37 °C and heating the sample for 2.5 min at 85 °C. The sample was next put on ice, and denatured plasmid was precipitated by adding EtOH and NaOAc and leaving the sample overnight at -20 °C. Precipitated plasmid was recovered by centrifugation (30 min, 4 °C, 14,000 rpm), washed with EtOH 100% and dried. Radiolabeled primer was annealed to the denatured plasmid by resuspending the denatured plasmid in 13.75 ml of primer mix (100 nm radiolabeled p821, 80 mm Tris-HCl, pH 7.5, 100 mm NaCl, 4 mm MgCl2) and incubating the sample at 85 °C for 1 min followed by 30 min at 37 °C. Sample was diluted two times with H2O, and each of the four dNTPs and DTT were added to a final concentration of 0.240 mm and 2.5 mm, respectively. Primer extension was initiated by adding the DNA polymerase. T4 DNA polymerases, 3′→5′-proficient and -deficient, were used at a final concentration of 35 nm and 50 nm, respectively. F29 DNA polymerase and T7 DNA polymerase were used at a final concentration of 0.5 and 8 milliunits/ml, respectively. When used, SSB proteins were allowed to coat the ssDNA by a 10-min incubation period at 37 °C before adding the DNA polymerase. E. coli SSB and T4 SSB proteins were used at a final concentration of 6 mm and 14 mm, respectively. When processive T4 DNA polymerase was used, NaCl concentration was raised to 250 mm to prevent non-processive DNA polymerase to work. ATP, gp44/62, and gp45 were added to a final concentration of 4 mm, 70 nm, and 600 nm, respectively. Samples containing DNA, gp44/62, and gp45 were incubated 5 min at 37 °C to allow clamp loading before adding the T4 wildtype DNA polymerase. In all cases, primer extension reactions proceeded at 37 °C and were quenched at various time points by adding a mixture of SDS and EDTA to a final concentration of 0.25% and 50 mm, respectively. Sample proteins were degraded by a proteinase K treatment (proteinase K at 0.67 mg/ml, 10 min at 37 °C, 10 min at 65 °C). Denaturing loading blue solution (97.5% deionized formamide, 10 mm EDTA, 0.3% bromphenol blue, 0.3% xylene cyanol-FF) was added, and samples were heated (3 min at 85 °C) before being loaded onto a 6% sequencing gel (19:1 = acrylamide:bisacrylamide mass ratio; TBE (Tris borate/EDTA) 1×;7 m urea) containing 30% formamide. The gel was run at 40 watts for 4 h, dried, and exposed on a PhosphorImager screen. The screen was scanned by using an Amersham Biosciences Storm 820 PhosphorImager, and Image-QuaNT software was used to quantify the results.
Preparation of ssDNA Fragments Carrying a TNR Sequence—A PCR reaction using the adequate TNR-containing plasmid or pEmpty plasmid, the p867/4ps, and phosphorylated p1033 oligonucleotides was performed using the following program: cycle 1 (2 min at 95 °C); cycle 2 repeated 25 times (30 s at 95 °C, 30 s at 60 °C, and 25 s at 72 °C); cycle 3 (7 min at 70 °C); and cycle 4 (4 °C throughout reaction). Plasmid and oligonucleotides were at a final concentration of 0.2 ng/ml and 0.5 mm, respectively. Each dNTP was at a final concentration of 250 mm. Herculase DNA polymerase was used at a final concentration of 50 milliunits/ml and was added last. The reaction was done in the Herculase buffer provided by the manufacturer. The PCR product was next radiolabeled with T4 DNA polynucleotide kinase, treated with T7 exonuclease (5 min at 30 °C with 0.35 milliunit/ml enzyme for 5′-GTT53, 5′-CAG60, and random substrates and 40 min at 30 °C with 0.35 milliunit/ml enzyme for 5′-CTG60 substrate). The resulting ssDNA fragment was purified on a 5% acrylamide (29:1 = acrylamide:bisacrylamide mass ratio; TBE 1×) native gel. The band corresponding to the ssDNA was excised, eluted from the gel in TBE 1× buffer (overnight, 37 °C), precipitated and resuspended in TE buffer (Tris/EDTA) supplemented with 100 mm NaCl.
Bandshift Assay—Interaction between ssDNA fragments carrying a TNR sequence or a random sequence and SSB proteins were performed in a buffer containing 40 mm Tris-HCl, 50 mm NaCl, 2 mm MgCl2, 4 mm DTT. Protein concentration was as indicated in the appropriate figure legends. After a 10-min incubation period at 25 °C native loading blue (5% glycerol, 0.04% bromphenol blue, 0.04% xylene cyanol-FF, 8 mm EDTA) was added to the samples. Protein-DNA complexes and DNA were separated by electrophoresis (100 V, 24 °C, 2 h) on a 3% or 10% native acrylamide (29:1 = acrylamide:bisacrylamide mass ratio; TBE 1×) gel. After electrophoresis the gel was dried and exposed on a PhosphorImager screen. The screen was scanned using a Storm 820 PhosphorImager.
Preparation of a Radiolabeled Nicked 373-bp Long Double-stranded DNA Fragment—A PCR reaction on the pEmpty plasmid (p660 and p1033 oligonucleotides) was performed using the following program: cycle 1 (2 min at 95 °C); cycle 2 repeated 25 times (30 s at 95 °C, 1 min at 60 °C, 1 min at 74 °C); cycle 3 (7 min at 70 °C); cycle 4 (4 °C, throughout reaction). Plasmid and oligonucleotides were at a final concentration of 0.06 ng/ml and 0.5 mm. Each of the four dNTPs was at a final concentration of 250 mm. Herculase DNA polymerase was used at a final concentration of 50 milliunits/ml and was added last. The reaction was carried out in the Herculase buffer provided by the manufacturer. After the PCR reaction, sample was precipitated. The PCR product was purified by electrophoresis on a 2% agarose gel made in TBE 1×. After electrophoresis, gel was stained by ethidium bromide, DNA band corresponding to the PCR product (373 bp) was excised, and DNA material was electro-eluted. After electro-elution, DNA was precipitated and resuspended in TE buffer. To introduce the nick, the 373-bp long PCR product was treated with the nicking enzyme N. AlwI (0.4 milliunit/ml, 1 h, 37 °C). The nicked DNA was radiolabeled with T4 polynucleotide kinase.
Strand Displacement DNA Synthesis Assay—Strand displacement DNA synthesis activity of T4 DNA polymerase 3′→5′ exonuclease proficient or deficient was tested by incubating the radiolabeled nicked 373-bp long PCR fragment with the DNA polymerase in a buffer containing 40 mm Tris-HCl, 50 mm NaCl, 2 mm MgCl2, 2.5 mm DTT, and the four dNTPs. Each of the four dNTPs was present at 250 mm concentration. DNA polymerase concentration ranged from 70 nm to 2.5 mm. Reaction proceeded at 37 °C and was quenched at various time points by adding a mixture of SDS and EDTA to a final concentration of 0.25% and 50 mm, respectively. Sample proteins were degraded by a proteinase K treatment (proteinase K at 0.67 mg/ml, 10 min at 37 °C, 10 min 65 °C). Denaturing loading blue was added, and samples were heated (5 min at 85 °C) before being loaded on a 6% sequencing gel (19:1 = acrylamide:bisac-rylamide mass ratio; TBE 1×, 7 m urea) containing 30% formamide. Gel was run at 40 watts, dried, and exposed on a PhosphorImager screen. The screen was scanned by using a Storm 820 PhosphorImager and ImageQuaNT software was used to quantify the results.
RESULTS
Orientation Dependence of TNR Instability Cannot Be Distinguished from Random Deletions above 110 Repeats—We undertook in vivo experiments to define parameters with which to construct an in vitro model for the orientation dependence of TNR instability. The length dependence of TNR instability has been extensively investigated (between 35 and 250 repeats). However, at long TNRs, structure-independent deletions will occur spontaneously, particularly at longer tracts. Thus, quantifying deletions relevant to 5′-CTG/5′-CAG tracts in vivo requires separating structure-specific effects from spontaneous deletions at all lengths of the repeat. Toward this end, we created a series of plasmids with tracts of pure 5′-CTG/5′-CAG and a closely matched series of repetitive, but structure-incapable 5′-GTT/5′-AAC tracts (Fig. 1A). To define the relevant lengths where structure-dependent deletions were resolved from random ones, we normalized deletion in 5′-CTG/5′-CAG tracts with that of 5′-GTT/5′-AAC tracts of equivalent length. Plasmids containing defined TNR lengths were grown in E. coli SURE cells, and, after growth, were restricted at sites flanking the repeat insert to estimate their size (Fig. 1B). Consistent with other reports, deletions were far more prevalent than expansions, the 5′-CTG sequence was more unstable as a lagging strand template relative to the leading strand template, and deletions were greater for structure-forming than for non-structure-forming repeats (Fig. 1C). However, when deletion in 5′-CTG/5′-CAG tracts was normalized for deletion in non structure-forming repetitive 5′-GTT/5′-AAC of matching length, we found that the orientation dependence was lost above 110 TNRs. Thus, only TNR tract below 110 TNRs provided sufficient resolution for quantitative analysis relevant to the orientation dependence of 5′-CAG/5′-CTG deletion.
Within the relevant range, we determined that the transcriptional status of TNR had little effect on 5′-CAG and 5′-CTG deletions during exponential growth (Fig. 2). We constructed two sets of plasmids using a pcDNA3 vector containing 25-120 repeats. The plasmids were oriented within the T7 transcriptional unit of pcDNA3, containing a unidirectional origin, such that we could monitor RNA polymerase traveling toward or away from the DNA polymerase (Fig. 2, A and B). Because plasmids harboring TNR sequences of intermediate lengths have no selective growth advantage in bacteria (38), the effects of transcription could be cleanly resolved from those induced by replication. Each plasmid was transformed into BL21(DE3pLysE) bacteria that contain an IPTG-inducible T7 RNA polymerase under the control of the lac repressor (Fig. 2A). We found that the orientation dependence was maintained at TNR lengths between 30 and 120 with or without IPTG induction (Fig. 2B). Deletion was also independent of whether RNA and DNA polymerase traveled in the same or in opposite directions (Fig. 2B). As a positive control for T7-driven gene expression, we monitored, in parallel experiments, the IPTG induction of an unrelated gene, the transcription factor cAMP-response element-binding protein (CREB). CREB expression levels, monitored by Western blot before and after induction with IPTG, demonstrated that transcriptional activity was robust at the concentration of IPTG (0.6 mm) used in the experiment (Fig. 2C). These data suggested that the orientation dependence of TNR instability and the deletion bias primarily arose in dividing cells from some properties of TNR sequence and its interaction with the replication machinery. To further dissect the mechanism of orientation-dependent deletion, we constructed an in vitro replication system designed to model both the leading and lagging strand DNA synthesis.
FIGURE 2.
Deletion does not depend on transcription. A, schematic of IPTG-dependent transcriptional regulation of pcDNA3 plasmids. We constructed plasmids using a pcDNA3 vector in which the TNRs are placed within a T7 transcriptional unit. BL21(DE3pLysE) cells contain a T7 polymerase, whose expression is inhibited by the lac repressor. In the presence of IPTG, repression is relieved, the lac repressor dissociates and T7 polymerase is transcribed, which in turn, initiates transcription of the TNR sequences. B, the % deletion of 5′-CAG and 5′-CTG as leading strand templates in replicating cells in the absence (-IPTG; filled symbols) or presence (+IPTG; open symbols) of transcription. Symbols indicate 5′-CAG or 5′-CTG templates in which replication (R) and transcription (T) with respect to the repeats proceeded in opposite (opposing arrows) or the same (unidirectional arrows) directions. C, expression in BL21(DE3pLysE) cells from the pET vector plasmid harboring a transcription factor CREB. The CREB cDNA is driven by the T7 promoter and regulated by IPTG. The concentration of IPTG used in the growth media is indicated. IPTG optimally activates expression between 0.6 and 1.0 mm. -, uninduced cultures.
5′-CAG and 5′-CTG Primary Sequences Pose a Differential Impediment to DNA Polymerase—To model leading strand DNA synthesis, we took advantage of a primer extension assay. TNR plasmids harboring short or long complementary 5′-CAG or 5′-CTG sequences were denatured and annealed to radiolabeled p821 primers, which recognized the sequence 45 bp upstream of the TNR (Fig. 3A). We tested whether the deletion bias and its orientation dependence might arise from the differential stability of the 5′-CTG and 5′-CAG hairpins and their effects on polymerase passage. If hairpins inhibited polymerase progression, then we predicted that the speed of polymerase passage on 5′-CTG templates would be slower than on 5′-CAG templates. Our goal was to systematically and quantitatively measure DNA polymerase passage across a specific TNR sequence. Toward that end, all data for short or long 5′-CTG and 5′-CAG sequences were 1) normalized for the effects of polymerase passage on non-structured 5′-GTT repeats of equivalent lengths and 2) normalized for polymerase speeds on the random plasmid DNA before synthesis of the TNR region.
FIGURE 3.
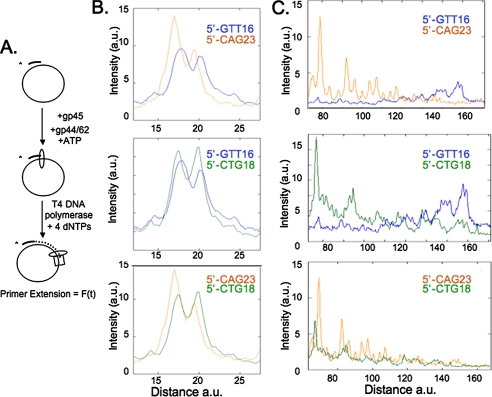
Short tract of structure-forming 5′-CAG repeats imposes a stronger impediment to polymerase passage than does 5′-CTG or 5′-GTT of equivalent lengths. A, experimental scheme of primer extension kinetics. The star indicates 32P-labeled primer. B, primer extension kinetics of T4 3′→5′ exonuclease-proficient DNA polymerase across single-stranded 5′-GTT16, 5′-CTG18, and 5′-CAG23 templates. Sequencing gels showing primer extension kinetics with single-stranded 5′-GTT16, 5′-CTG18, and 5′-CAG23 templates and T4 3′→5′ exonuclease-proficient DNA polymerase. Kinetic time points indicated by black triangles are 0.5, 1, 2.5, 5, and 7.5 min. Double arrows indicate the TNR units. C, quantification method. Black rectangles indicate bands used for quantification of polymerase speed. D and E, quantification results from B. Plots indicate the DNA synthesized before (D) and after (E) the TNRs. Leading strand templates: gray circles, 5′-CAG23; white circles, 5′-CTG18, and black circles, 5′-GTT16. Sequences of plasmids are indicated as ATCG.
DNA substrates harboring short repeats, p-5′-GTT16, p-5′-CTG18, or p-5′-CAG23, were incubated with a non-processive T4 wild-type DNA polymerase, which is proficient in its 3′→5′ exonuclease activity (T4 (+exo)) but lacks strand displacement activity (Fig. 3B). The lack of strand displacement activity of T4 (+exo) ensured that elongation is initiated only from a primer annealed to a single-stranded template (and not from a primer embedded into a potential D-loop). To specifically evaluate the effect of the TNR sequence, we estimated the speed of replication before and after passage through the TNR unit. The speed before the TNR was determined by loss of signal intensity of primers elongated by 18-33 nucleotides (black rectangle “Before TNR,” Fig. 3C). Because all TNR sequences shared the same flanking sequence, the choice to monitor extension products elongated by 18-33 nucleotides was arbitrary. The speed of the polymerase through the TNR was quantified and was normalized to the speed before encountering the TNRs. This process corrected for any effects due to the plasmid sequence and its interface with different TNRs. Extended primers of this length decreased with time, because they were intermediate synthesis products of larger extension products (Fig. 3D). To assess the effects of passage through the TNR region, we estimated the amount of elongated primers after DNA polymerase passage (black rectangle “Past TNR,” Fig. 3C). Elongation products that contained the full-length TNR unit increased over time, because the products accumulated as they were synthesized (Fig. 3E).
DNA polymerase was indeed sensitive to the TNR sequence even for short repeats (Fig. 3, D and E). Polymerase passage through the plasmid DNA prior to the TNR was influenced by the harbored TNR (suggesting conformational effects of the TNR), but the speed did not depend on its potential to form DNA secondary structure (Fig. 3D). The T4 wild-type DNA polymerase traveled three times faster on the template containing 23 5′-CAG repeats relative to the non-structure forming template containing 16 5′-GTT repeats (Fig. 3D). Speed on template containing 5′-CTG18 repeats was intermediate between the two (Fig. 3D). The situation was reversed after passage through the TNRs (Fig. 3E). Little accumulation of extension product was observed on the 5′-CAG23 template, whereas the 5′-GTT16 template accumulated the highest amount of extension product (Fig. 3E). After normalization for the speed before encountering the TNR, it was clear that the 5′-GTT repeat template was the easiest for DNA polymerase passage, the 5′-CAG repeat template posed the most severe impediment, and the 5′-CTG template was intermediate between the two (Fig. 3E). The 5′-CAG23-containing template inhibited the polymerase passage by 10-fold relative to the 5′-GTT16 sequence, and the rate of polymerase passage through the 5′-CAG23 sequence was 4-fold slower relative to the 5′-CTG18 sequence (Table 2). The same pattern has been found with the Klenow fragment.5 Thus, TNRs with the capability to form secondary structures were more difficult to replicate. However, inhibition of replication in the leading strand model did not correlate with the thermodynamic stability of the hairpins.
TABLE 2.
Polymerase activities and their effects on orientation dependence of TNR deletion
a Number of base pairs synthesized before a dissociation event.
b R(CTG/CAG) = DNA synthesized past 5′-CTG18 repeat unit/DNA synthesized past 5′-CAG23 repeat unit. This reflects the differential impediment to DNA synthesis across 5′-CTG18 and 5′-CAG23 repeats.
The impediment to polymerase passage imposed by the TNR sequences was amplified at longer tracts, but the trends were the same (Fig. 4). We performed primer extension kinetics with radiolabeled p821 primer annealed to denatured p-5′-GTT53, p-5′-CTG60, or p-5′-CAG60 plasmids (Fig. 4A). The progression of T4 wild-type DNA polymerase along the three TNR containing templates was measured during 7.5 min of DNA synthesis (Fig. 4). The scans clearly indicated that T4 wild-type DNA polymerase was able to synthesize through most of the repeat unit of the 5′-GTT53 containing template in the 7.5-min time frame (Fig. 4B, top panel). As with the shorter repeats, 5′-CAG60 posed the most severe impediment (Fig. 4B, bottom panel). The T4 wild-type DNA polymerase synthesized little product within the reaction time and was unable to pass beyond half of the template length (extensions on the average were around 12 repeats) (Fig. 4B, bottom panel). Long 5′-CTG tracts also imposed an impediment to the polymerase. However, the speed of the DNA polymerase through the 5′-CTG60 containing template was significantly faster than on 5′-CAG60, (Fig. 4B, middle panel), and more similar to the 5′-GTT53 template. Thus, the 5′-CAG and 5′-CTG were not equivalent templates: 5′-CAG was a poorer template than 5′-CTG in vitro under all conditions. Thermodynamically, a 5′-CTG hairpin is more stable than a 5′-CAG hairpin, yet the 5′-CAG sequence imposed a more serious impediment to the DNA polymerase than a 5′-CTG. Taken together, these data suggested that the polymerase interaction with the primary TNR sequence, not hairpin formation, caused the impediment in the leading strand model.
FIGURE 4.
Long tract of structure-forming 5′-CAG repeats imposes a stronger impediment to polymerase passage than does 5′-CTG or 5′-GTT of equivalent lengths. A, sequencing gels showing the primer extension kinetics of T4 3′→5′ exonuclease-proficient DNA polymerase across denatured p-5′-GTT53, p-5′-CTG60, and p-5′-CAG60 plasmids. Conditions are as described in Fig. 3. Kinetic time points are 0.5, 1, 2.5, 5, and 7.5 min. Sequences of plasmids are indicated as ATCG. Double arrows indicate the TNR units. B, scan of extension products across the TNR unit at the 7.5-min time point for the indicated templates.
SSB Protein Stimulates DNA Polymerase and Alleviates the Impediment to Polymerase Passage across TNR Sequences—We next tested whether hairpin stability had effects on the lagging strand. To model lagging strand DNA synthesis, we prepared ssDNA from structure-capable (5′-CTG60 or 5′-CAG60) or non structure-forming (5′-GTT53) TNRs. TNR sequences were incubated with purified SSB proteins from either T4 bacteriophage or E. coli (Fig. 5, A and B), and each species (DNA and protein-DNA complexes) was resolved on a native gel. In vivo, SSB prevents secondary structure formation and nuclease attack on the lagging strand template. SSB only protects the lagging strand, because the binding polarity of SSB on the fork DNA prohibits its recruitment to the leading template (39). Thus, the orientation dependence of deletion might arise if the 5′-CTG repeats inhibited binding of SSB protein on the lagging strand template to a greater extent than does 5′-CAG. Under these conditions, the more stable 5′-CTG loops would be preferentially trapped in the lagging strand template leading to a higher deletion rate. In contrast to the latter prediction, however, the binding of neither T4 nor E. coli SSB protein was affected by the TNR sequence (Fig. 5, A-C). T4 SSB protein is active as a monomer and has a binding site of seven nucleotides. We found that the T4 SSB protein was able to bind to single-stranded 5′-CAG and 5′-CTG with roughly the same affinity (250 nm) (Fig. 5, A and B), and as efficiently as single-stranded 5′-GTT or random sequence (Fig. 5C). Similar results were observed with E. coli SSB protein, which is active as a tetramer and has a binding site of 35 nucleotides, and binds either 5′-CAG or 5′-CTG with an approximate affinity of 50 nm (Fig. 5, A and B).
FIGURE 5.
T4 and E. coli SSB proteins bind single-stranded 5′-CAG60, 5′-CTG60, and 5′-GTT53 or random DNA with comparable affinity. Gel mobility shift assay of 5′-CTG60 (A) or 5′-CAG60 (B) ssDNA templates bound to either T4 SSB proteins (left panels) or E. coli SSB proteins (right panels). The protein concentration in nanomolar is indicated and given in monomeric units; species are separated on a 10% acrylamide gel. C, gel mobility shift assay for T4 SSB protein binding to 5′-CTG60, 5′-CAG60, 5′-GTT53, and random sequence DNA. Species are resolved on a 3% acrylamide gel. T4 SSB protein concentrations indicated by black triangles are 0, 55 nm, 550 nm, 5.5 mm, and 55 mm.
Because all of the TNR templates bound SSB equally well, we tested whether binding of SSB protein had an impact on the rate of polymerase passage on 5′-CAG and 5′-CTG repeats relative to 5′-GTT repeats of equivalent lengths (Figs. 6 and 7). Primer extension kinetics were performed with radiolabeled p821 primer annealed to denatured p-5′-GTT53, p-5′-CTG60, or p-5′-CAG60 plasmids with or without SSB proteins (Fig. 6A). We found that inclusion of either E. coli (Fig. 6, B and C, shown only for E. coli SSB proteins and 5′-CAG template) or T4 SSB proteins (Fig. 7) at a 1:1 binding site molar ratio greatly stimulated DNA synthesis at all three TNR sequences. To give a quantitative view of the T7 DNA polymerase stimulation by E. coli SSB protein, we scanned the 7.5-min time point lane from the beginning of the 5′-CAG unit to the top of the gel (Fig. 6C). Elongated primers containing the full-length TNR unit represented only 2.7% of the elongated primers in the absence of E. coli SSB proteins, and this reached 50% in the presence of E. coli SSB proteins. Thus, binding of E. coli SSB proteins reduced the impediment to polymerase passage and facilitated synthesis of longer DNA products. The same results were obtained using T4 SSB proteins and T4 wildtype DNA polymerase (Fig. 7). SSB protein stimulated polymerase progression through 5′-CTG60 and 5′-CAG60 (Fig. 7A).
FIGURE 6.
E. coli SSB proteins stimulate T7 DNA polymerase progression across 5′-CAG60 repeats. A, schematic diagram of assay. The star indicates 32P-labeled primer. B, sequencing gel showing the primer extension kinetics of T7 DNA polymerase across 5′-CAG60 template without (-) or with (+) E. coli SSB proteins. The kinetic time points indicated by black triangles are 0.5, 1, 2.5, 5, and 7.5 min. Sequences (A, T, C, and G) of the plasmid are indicated. Double arrow indicates the TNR unit. Stoichiometric amounts of SSB protein relative to plasmid were added assuming a binding site size of tetrameric E. coli SSB of 35 nucleotides. C, scans of gels from B at 7.5 min in the absence (black line) or presence (gray line)of E. coli SSB proteins. The heavy arrow on the x-axis of the plot indicates the end of the TNR tract. The light arrow on the x-axis of the plot indicates the beginning of the repeat tract.
FIGURE 7.
T4 SSB proteins stimulate T4 DNA polymerase progression across long TNR sequences and removes the differential impediment imposed by 5′-CAG and 5′-CTG templates. A, sequencing gels showing the primer extension kinetics of T4 DNA polymerase across 5′-CAG60, 5′-CTG60, and 5′-GTT53 templates without (-) or with (+) T4 SSB proteins. The black triangles indicate four time points, 0.5, 1, 2.5, and 5 min. Stoichiometric amounts of SSB proteins relative to plasmid were added assuming a binding site size of monomeric T4 SSB protein of 7 nucleotides. Sequences (A, T, C, and G) of the plasmid are also shown. Double arrows indicate the TNR unit. B-D, scans of primer extension products on 5′-GTT53 (B), 5′-CTG60 (C), and 5′-CAG60 (D) templates at 5 min as indicated. Heavy arrows on the x-axis of the plots indicate the end of the TNR tracts. Light arrows on the x-axis of the plots indicate the beginning of the repeat tracts. E, quantification of the scanned lanes from B-D. Plotted are the % extension product containing a partial (white squares) or full-length (dotted squares) extension products at 5 min for the indicated templates.
Surprisingly, however, upon binding SSB protein, the orientation dependence of polymerase passage between 5′-CAG and 5′-CTG was lost. For example, in the presence of T4 SSB proteins, T4 wild-type DNA polymerase passage was almost complete after 5 min along a non structure-forming 5′-GTT53 sequence and 94.5% of the elongated primers contained the full-length TNR unit (Fig. 7B). However, binding of SSB protein rendered polymerase passage on a 5′-CTG60 template equivalent to that of the 5′-CAG template (Fig. 7, C and D). Without T4 SSB protein, the T4 wild-type DNA polymerase did not reach the end of the 5′-CTG60 and 5′-CAG60 containing templates during the time frame monitored (7.5 min, see Fig. 4B). However, for both 5′-CAG and 5′-CTG sequences, the entire TNR sequence template was synthesized in the presence of T4 SSB protein during the same period (Fig. 7, C and D). For either 5′-CTG60 and 5′-CAG60 templates, ∼67% of the elongated primers contained the full-length TNR unit (63.5% and 71.5% for 5′-CTG60 and 5′-CAG60, respectively, Fig. 7E). Thus, binding of SSB proteins eliminated the differential impediment of 5′-CAG and 5′-CTG sequences observed in the leading strand DNA synthesis model.
Strong Strand Displacement Activity Associated with DNA Synthesis Relieves the Impediment to Polymerase Passage at Structure-forming TNRs—Together, these data raised the possibility that the orientation dependence of TNR deletion arose on the leading strand driven by a differential impediment to polymerase passage on the 5′-CAG and 5′-CTG primary sequences. Indeed, instability has been observed in vivo during rolling circle replication from a phage f1 origin (40). To better define a mechanism by which the TNRs might inhibit polymerase passage, we evaluated which activities associated with the DNA polymerases might remove the orientation-dependent impediment to passage at TNRs in a leading strand model.
In the first experiment, we tested whether strand displacement activity alone could remove the differential impediment to passage at TNRs. To test this hypothesis, we utilized T4 (-exo) polymerase. T4 (+exo) is a non-processive enzyme. However, loss of its exonuclease activity (T4 (-exo)) confers a weak strand displacement activity without affecting processivity. We established several key controls to quantitatively assess the effects of strand displacement on polymerase progression through the TNRs. In the T4 (-exo) mutant, the 3′→5′ exonuclease domain is present but the aspartic acid residue at position 219 has been changed to alanine (Asp-219 provides one of the four essential carboxylate residues for the two-metal ion exonuclease reaction). To control for structural and functional effects due to mutation and loss of the exonuclease activity, we determined the enzyme concentration at which the primer extension kinetics on a control template lacking any TNR sequence (pEmpty) was equivalent to that of the T4 (+exo) polymerase. These experiments controlled for the effects of removing the 3′→5′ exonuclease activity on copying random sequences. We determined that, at a concentration of 50 nm, T4(-exo) gave the same kinetic profile on pEmpty-denatured plasmid as 35 nm concentration of T4 (+exo) (data not shown). Using these conditions, we were able to quantitatively measure only the effects of a weak strand displacement activity on polymerase progression through the TNRs. As a second control, we checked for differences in strand displacement activity of the two polymerases as a function of polymerase concentration during strand displacement DNA synthesis (schematically shown, Fig. 8A). We were unable to observe any strand displacement DNA synthesis of T4 (-exo) on a nicked 200-bp fragment at a concentration of <100 nm. Because extension of a primer embedded into a D-loop does not take place under the reaction conditions used in the primer extension assays (50 nm DNA polymerase), this control ensured that the observed products arose from the primer annealed to a template in a single-stranded state.
FIGURE 8.
Non structure-forming and structure-forming TNR sequences are equivalent templates in the presence of a strong displacement activity associated with the DNA synthesis activity. A, schematic representation of the strand displacement DNA synthesis assay. Stars indicate 32P-labeled DNA ends. B, amount of DNA synthesized past the TNR unit as a function of time. The kinetics of primer extension by T4 (-exo) through 5′-GTT16 (black circles), 5′-CTG18 (white circles), and 5′-CAG (gray circles) are shown. Increasing the strand displacement activity of the non-processive T4 DNA polymerase is not sufficient to overcome the impediment to DNA polymerase passage across the TNR. C and D, effect of strand displacement activity on the impediment to polymerase passage across 5′-CAG23 template. Non-processive T4 (+exo) DNA polymerase with no strand displacement activity (black circles); T4 (-exo) DNA polymerase with weak strand displacement activity (gray circles). The strand displacement activity in T4 (-exo) is not needed to unwind DNA before the TNR tract (C), but stimulates the passage through the TNR region (D). E, schematic diagram of primer extension assay with F29 DNA polymerase, a processive DNA polymerase with strong strand displacement activity. F, sequencing gels showing the progression of the F29 DNA polymerase across 5′-GTT16, 5′-CTG18, and 5′-CAG23 templates over time. The kinetic time points are 0.5, 1, 2.5, 5, 7.5 min. Bracket indicates the TNR units. Sequences (A, T, C, and G) of all three plasmids are also shown. G, sequencing gels showing the progression of the F29 DNA polymerase across 5′-GTT53, 5′-CTG60, and 5′-CAG60 templates over time. The kinetic time points are 0.5, 1, 2.5, 5, and 7.5 min. Brackets indicate the TNR units. Sequences (A, T, C, and G) of all three plasmids are also shown.
Having established these quantitative conditions, we tested the importance of weak strand displacement activity on polymerase progression through short TNRs (Fig. 8, B-D, and Table 2). We measured the primer extension rate of T4 (-exo) on the p-5′-GTT16, p-5′-CTG18, or p-5′-CAG23 templates. As with T4 (+exo) (Fig. 3), we found that strand displacement activity of T4 (-exo) did not remove the impediment to polymerase passage, nor did it alter its sequence context (Fig. 8B). Stimulation of the T4 (-exo) synthesis activity on the 5′-GTT template was independent of the TNR (Table 3). For the 5′-CAG template the 3′→5′ proofreading and weak strand displacement activities were not needed before reaching the TNR tract (Fig. 8C). However, after the 5′-CAG repeat tract, the synthesis rate of T4 (-exo) was enhanced by 2-fold relative to that of the T4 (+exo) enzyme (Fig. 8D and Table 3). The stimulation of synthesis activity on the 5′-CTG template was intermediate, with only a 1.5-fold increase relative to that of the T4 (+exo) enzyme past the 5′-CTG unit (Table 3). Thus, removal of the 3′→5′ exonuclease activity and the accompanying increase in strand displacement activity did not change the relative order of DNA polymerase speeds. The 5′-GTT was the best template, 5′-CAG was the worst template, and 5′-CTG was intermediate between the two (Fig. 8B and Table 2). Overall, we found that weak strand displacement activity, even in the presence of strong DNA synthesis activity, was not sufficient to overcome the differential polymerase impediment on 5′-CAG or 5′-CTG sequences.
TABLE 3.
DNA synthesis of T4 (-exo) and T4 (+exo) before and after the TNR unit
Ext. (T4 −exo)/Ext. (T4 +exo) = extent of DNA synthesis of T4 (−exo) divided by extent of DNA synthesis of T4 (+exo). The ratio of DNA synthesis from T4 polymerase with (+) or without (+) exonuclease activity for three TNR units (5′-GTT, 5′-CTG and 5′-CAG) before or after the repetitive sequence. They have been calculated by averaging the extent of DNA synthesis at 1, 2.5, 5, and 7.5 min as illustrated in Figure 3E (T4 +exo) and BB (T4 −exo).
We next tested whether a strong strand displacement activity coupled to DNA synthesis was needed for unimpeded passage through the TNR sequences. To test this hypothesis, we repeated the primer extension reaction on the TNR plasmids using ϕ29 DNA polymerase (Fig. 8E). The strong strand displacement activity associated with the DNA synthesis activity of ϕ29 DNA polymerase abrogates the need of a replicative helicase, and such an enzyme mimics a “perfect” and “undisruptable” polymerase-helicase couple. For all six TNR templates (p-5′-GTT16, p-5′-CTG18, p-5′-CAG23, p-5′-GTT53, p-5′-CTG60, or p-5′-CAG60) harboring short (Fig. 8F) or long (Fig. 8G) repeats, the major DNA synthesis products that accumulated over time were high molecular weight products. The polymerase synthesis rates among the TNR sequences were indistinguishable. Thus, strong strand displacement activity coupled to processive DNA synthesis could overcome the orientation-dependent impediment presented by TNR sequences during replication (Table 2). These data suggested that the TNRs might cause deletion by disrupting the coupling of the polymerase with the helicase at the fork.
Strand displacement activity alone was not sufficient to overcome the differential block unless it was coupled to strong processivity. Thus, in a last set of experiments, we tested whether the processivity alone contributed significantly to the orientation dependence. In these experiments, we assembled the T4 wild-type DNA polymerase at the primer/template junction in the presence of gp44/62, the T4 clamp loader, gp45, the T4 sliding clamp, and ATP. Thus, the effect of increasing processivity could be monitored in the absence of strand displacement activity (Fig. 9A). Experimental conditions had to be adjusted in terms of salt concentrations and incubation times to make sure that non-processive DNA polymerase was inactive, and only the effects of the processivity were measured. We tested the effects of T4 accessory proteins on the DNA synthesis rate of T4 DNA polymerase on p-5′-GTT16, p-5′-CTG18, and p-5′-CAG23 plasmids up to 20 min. The reaction products were separated on polyacrylamide gels, and the resulting synthesis products were quantified (scans in Fig. 9). To correct for loading differences among lanes, we normalized the scans using the doublet bands present on the vector sequence before the TNR unit (Fig. 9B). As before, we found that the non-structure-forming repeat (5′-GTT16) was a better template for synthesis relative to 5′-CTG18 or 5′-CAG23 repeat templates (Fig. 9C, top and middle panels, and Table 2). However, in the presence of the T4 processivity factor, the differential impediment to DNA synthesis by T4 wild-type DNA polymerase on 5′-CTG18 and 5′-CAG23 templates was no longer present. Although minor differences in peak intensity existed, overall, DNA polymerase progressions on 5′-CAG and 5′-CTG were equivalent (Fig. 9, bottom panel, and Table 2). Thus, the differential impediment on 5′-CTG18 and 5′-CAG23 was overcome if the polymerase processivity was strong (Fig. 9, bottom panel, and Table 2).
FIGURE 9.
5′-CAG and 5′-CTG are equivalent templates in the presence of T4 gp44/62 clamp loader and gp45 processivity factor. A, schematic diagram of assay. The star indicates 32P-labeled primer. The clamp loading phase followed by the DNA synthesis phase are shown. B, scans of the doublet bands present before the TNR unit and used to correct for loading differences among lanes. Top, middle, and bottom scans are for 5′-GTT16 and 5′-CAG23, 5′-GTT16 and 5′-CTG18, and 5′-CAG23 and 5′-CTG18 couples, respectively. Kinetic time point for the scan is 7.5 min. C, scans at 7.5 min of primer extension from the beginning of the TNR unit to the top of the gel. Top, middle, and bottom scans are for 5′-GTT16 and 5′-CAG23, 5′-GTT16 and 5′-CTG18, and 5′-CAG23 and 5′-CTG18 couples, respectively.
DISCUSSION
It has been widely observed, and our data confirm (Fig. 1), that TNR units, in vivo, tend to delete rather than to expand in rapidly dividing cells. Furthermore, the 5′-CTG sequences undergo frequent deletion events as the lagging strand template, but it is relatively stable as the leading strand template. To explain these observations, it has been suggested that instability arises when stable 5′-CTG hairpins are formed during lagging strand synthesis and are subsequently deleted. Our data reported here suggest an alternative mechanism. We found that the primary 5′-CAG sequence inhibits polymerase progression on naked DNA and that SSB protein eliminates the impediment. 5′-CAG hairpins are promoted under these conditions. These in vitro data are consistent with the in vivo observation that deletion is observed when 5′-CTG is in the lagging strand template. However, deletion would arise from removal of 5′-CAG hairpins formed on the leading strand template rather than 5′-CTG hairpins formed on the lagging strand template. In support of that hypothesis, we found that the 5′-CTG template only modestly inhibits polymerase progression on naked DNA and imposes no impediment upon binding SSB protein. Consequently, 5′-CTG hairpins are infrequent, consistent with the in vivo observation that 5′-CTG template is relatively stable in the leading strand orientation. Thus, SSB creates an asymmetry that drives orientation-dependent instability by relieving replication stress on the lagging strand.
Our quantitative in vitro data can be used to distinguish among proposed models for TNR deletion. It has been postulated that protein-protein complexes or transient generation of single strands on the lagging strand template preferentially promote the formation of the 5′-CTG hairpins relative to 5′-CAG hairpins. If correct, then this model makes the prediction that 5′-CTG hairpins form before DNA polymerase passage and more rapidly than 5′-CAG hairpins and/or differentially inhibit SSB protein binding. As a consequence, 5′-CTG but not 5′-CAG hairpins would be preferentially trapped during lagging strand DNA synthesis. However, three findings argue against such a model.
First, we found that the binding of SSB protein to the 5′-CAG and 5′-CTG templates is equally robust and indistinguishable from binding to non-structure-forming TNR or random sequences (Fig. 5). At least in vitro, 5′-CAG and 5′-CTG strands do not appear to differentially exist in a single-stranded state or inhibit binding of SSB protein. Furthermore, although 5′-CTG hairpins are more stable than 5′-CAG hairpins, kinetic studies indicate that 5′-CAG and 5′-CTG hairpins will reform duplex molecules at equal rates under pseudo first order conditions suggesting that the kinetic lifetime of both hairpins in vitro are similar (16).
Second, we found that the binding of SSB protein removes the differential impediment to polymerase progression across 5′-CAG and 5′-CTG templates (Figs. 6 and 7). Thus, SSB proteins appear to unfold 5′-CAG and 5′-CTG strands to the same extent.
Third, we found that the orientation-dependent impediment to polymerase passage does not depend on hairpin thermodynamic stability. Rather, we found that the differential impediment imparted by the 5′-CAG and 5′-CTG sequences arises from an interaction of the DNA polymerase with the DNA primary sequences. Why 5′-CAG preferentially inhibits polymerase progression is not known. However, it is widely documented that DNA primary sequence can influence and have measurable effects on any of the steps involved in the dNTP incorporation (41). This includes polymerase binding to DNA, dNTP binding to the binary complex, transition from open to closed conformation of the DNA polymerase, and catalysis (42, 43). Regardless of the step involved, the differential impediment to polymerase progression promotes strand-biased slippage when 5′-CAG is the leading strand template.
We found that the orientation dependence of deletion is strongly influenced by polymerase processivity. The differential impediment is partially overcome in vitro if strand displacement is weak and totally overcome if the processivity or the strand displacement activity is strong (Tables 2 and 3). Thus, the impediment imposed by the primary 5′-CAG sequence tends to uncouple unwinding and leading strand DNA synthesis activities. Such a model is consistent with recent findings that DNA polymerase proofreading activity (44) and weak strand displacement activity (45) contribute to orientation dependence of 5′-CAG/5′-CTG TNR instability in E. coli. Overall, these data argue against a model in which the hairpins form ahead of the polymerase and initiate the polymerase impediment. Rather, hairpins are more likely to form during or after polymerase passage across the TNR sequence. Intrastrand hydrogen bonding within the 5′-CAG hairpin can recover the energetic costs of “unpairing” the duplex and provides a kinetic stability to the looped state.
We propose a template-push model for the deletion bias of TNRs and its orientation dependence (Fig. 10). In vitro experiments have shown that if the DNA synthesis rate becomes sufficiently slower than the helicase unwinding rate, then helicase and DNA polymerase become functionally uncoupled (34). This circumstance can occur when polymerase encounters a difficult sequence, such as the TNRs. The difference in the progression rate of the polymerase and that of the helicase has the potential to create a gap of unreplicated TNR template and a functional uncoupling of helicase and leading DNA polymerase at the fork. At 5′-CAG/5′-CTG repeats, the 5′-CAG sequence poses a greater impediment on the leading strand template. To avoid uncoupling, the DNA polymerase pushes away and passes over a small tract of 5′-CAG DNA in the leading strand template, saving synthesis time, and the unreplicated 5′-CAG loop is trapped in the process. Uncoupling stress is relieved when the fork has moved through the entire repeat region and resumes copying random sequence DNA. By this mechanism, polymerase-helicase coupling is maintained but at the expense of hairpin formation in the template strand. In such a view, deletion occurs to avoid polymerase-helicase uncoupling. Deletion would not depend on the hairpin thermodynamic stability, as we observe, because 5′-CAG and 5′-CTG hairpins form during or after loop entrapment when polymerase passes over the TNR. However, it would depend on the interaction between DNA polymerase and template primary sequence.
FIGURE 10.
Template-push model for deletion of repetitive sequences as visualized on an E. coli replication fork. CAG poses a larger impediment to polymerase progression. a, on the leading strand, before reaching the CAG sequence, the replicative polymerase and helicase are tightly coupled (symbolized by the closely spaced green and orange balls; green is the polymerase, orange is the helicase, pink is primase, colored bars are the TNR sequence, gray bars are random DNA base pairs, and gray balls are single strand binding proteins). b, when the polymerase encounters the CAG tract on the leading strand, polymerase progression is slowed while the helicase speed is unaffected. The differential speeds threaten their coupling (symbolized by space between green and orange balls). The presence of SSB proteins prevents the differential speed of polymerase and helicase on the lagging strand. c, leading and lagging strand synthesis can remain coupled if the leading strand polymerase pushes away and by-passes a small segment of unreplicated CAG template on the leading strand. d, coupling stress is relieved when the fork has moved through the entire CAG repeat region and resumes copying random DNA sequence. Processing of the loop results in deletion of the CAG tract.
The template-push model provides a mechanism to explain both the orientation dependence of deletion and its bias during replication. The orientation dependence of TNR instability arises because the 5′-CAG sequence, as a leading template, creates a greater impediment to DNA polymerase progression than a 5′-CTG sequence, whereas in the lagging strand template, SSB protein removes the differential impediment independently of the sequence context. Deletion bias occurs because the tight coupling between the leading DNA polymerase and helicase prevents release of the daughter strand. Thus, there is little opportunity to generate the ssDNA precursor needed for hairpin formation that would be required for an expansion event. Although a simple slippage model predicts equal opportunity for slippage in the template and daughter strand, hairpins would preferentially form in the template. The template-push model will be further tested using a system in which the polymerase and helicase activities are coupled to create a functional replication fork.
This work was supported, in whole or in part, by National Institutes of Health Grants GM 066359 and NS40738 (to C. T. M.). This work was also supported by the Mayo Foundation, CNRS, and Institut Curie (Réplication, Instabilité Chromosomique et Cancer; to G. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TNR, trinucleotide repeat; HA, hydroxyapatite; ssDNA, single-stranded DNA; SSB, ssDNA-binding protein; MOPS, 4-morpholinepropanesulfonic acid; DTT, dithiothreitol; IPTG, isopropyl 1-thio-β-d-galactopyranoside; CREB, cAMP-response element-binding protein.
E. Delagoutte, personal communication.
References
- 1.Mirkin, S. M. (2007) Nature 447 932-940 [DOI] [PubMed] [Google Scholar]
- 2.Bichara, M., Wagner, J., and Lambert, I. B. (2006) Mutat. Res. 598 144-163 [DOI] [PubMed] [Google Scholar]
- 3.Pearson, C. E., Nichol Edamura, K., and Cleary, J. D. (2005) Nat. Rev. Genet. 6 729-742 [DOI] [PubMed] [Google Scholar]
- 4.Kovtun, I. V., and McMurray, C. T. (2001) Nat. Genet. 27 407-411 [DOI] [PubMed] [Google Scholar]
- 5.Kovtun, I. V., Liu, Y., Bjoras, M., Klungland, A., Wilson, S. H., and McMurray, C. T. (2007) Nature 447 447-452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mangiarini, L., Sathasivam, K., Mahal, A., Mott, R., Seller, M., and Bates, G. P. (1997) Nat. Genet. 15 197-200 [DOI] [PubMed] [Google Scholar]
- 7.Kovtun, I. V., Thornhill, A. R., and McMurray, C. T. (2004) Hum. Mol. Genet. 13 3057-3068 [DOI] [PubMed] [Google Scholar]
- 8.Malter, H. E., Iber, J. C., Willemsen, R., de Graaff, E., Tarleton, J. C., Leisti, J., Warren, S. T., and Oostra, B. A. (1997) Nat. Genet. 15 165-169 [DOI] [PubMed] [Google Scholar]
- 9.Kang, S., Jaworski, A., Ohshima, K., and Wells, R. D. (1995) Nat. Genet. 10 213-218 [DOI] [PubMed] [Google Scholar]
- 10.Freudenreich, C. H., Kantrow, S. M., and Zakian, V. A. (1998) Science 279 853-856 [DOI] [PubMed] [Google Scholar]
- 11.Freudenreich, C. H., Stavenhagen, J. B., and Zakian, V. A. (1997) Mol. Cell. Biol. 17 2090-2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rolfsmeier, M. L., Dixon, M. J., Pessoa-Brandao, L., Pelletier, R., Miret, J. J., and Lahue, R. S. (2001) Genetics 157 1569-1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon, M. J., and Lahue, R. S. (2004) Nucleic Acids Res. 32 1289-1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spiro, C., Pelletier, R., Rolfsmeier, M. L., Dixon, M. J., Lahue, R. S., Gupta, G., Park, M. S., Chen, X., Mariappan, S. V., and McMurray, C. T. (1999) Mol. Cell 4 1079-1085 [DOI] [PubMed] [Google Scholar]
- 15.Moore, H., Greenwell, P. W., Liu, C. P., Arnheim, N., and Petes, T. D. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 1504-1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miret, J. J., Pessoa-Brandao, L., and Lahue, R. S. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 12438-12443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gacy, A. M., Goellner, G., Juranic, N., Macura, S., and McMurray, C. T. (1995) Cell 81 533-540 [DOI] [PubMed] [Google Scholar]
- 18.Cleary, J. D., Nichol, K., Wang, Y. H., and Pearson, C. E. (2002) Nat. Genet. 31 37-46 [DOI] [PubMed] [Google Scholar]
- 19.Kang, S., Ohshima, K., Shimizu, M., Amirhaeri, S., and Wells, R. D. (1995) J. Biol. Chem. 270 27014-27021 [DOI] [PubMed] [Google Scholar]
- 20.Balakumaran, B. S., Freudenreich, C. H., and Zakian, V. A. (2000) Hum. Mol. Genet. 9 93-100 [DOI] [PubMed] [Google Scholar]
- 21.Rossi, M. L., Purohit, V., Brandt, P. D., and Bambara, R. A. (2006) Chem. Rev. 106 453-473 [DOI] [PubMed] [Google Scholar]
- 22.Paiva, A. M., and Sheardy, R. D. (2004) Biochemistry 43 14218-14227 [DOI] [PubMed] [Google Scholar]
- 23.Zheng, M., Huang, X., Smith, G. K., Yang, X., and Gao, X. (1996) J. Mol. Biol. 264 323-336 [DOI] [PubMed] [Google Scholar]
- 24.Petruska, J., Arnheim, N., and Goodman, M. F. (1996) Nucleic Acids Res. 24 1992-1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, G. K., Jie, J., Fox, G. E., and Gao, X. (1995) Nucleic Acids Res. 23 4303-4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Usdin, K., and Woodford, K. J. (1995) Nucleic Acids Res. 23 4202-4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohshima, K., and Wells, R. D. (1997) J. Biol. Chem. 272 16798-16806 [DOI] [PubMed] [Google Scholar]
- 28.Lin, Y., Dion, V., and Wilson, J. H. (2006) Nat. Struct. Mol. Biol. 13 179-180 [DOI] [PubMed] [Google Scholar]
- 29.Kovtun, I. V., Spiro, C., and McMurray, C. T. (2004) Methods Mol. Biol. 277 309-320 [DOI] [PubMed] [Google Scholar]
- 30.Liu, B., and Alberts, B. M. (1995) Science 267 1131-1137 [DOI] [PubMed] [Google Scholar]
- 31.Elias-Arnanz, M., and Salas, M. (1997) EMBO J. 16 5775-5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowater, R. P., Jaworski, A., Larson, J. E., Parniewski, P., and Wells, R. D. (1997) Nucleic Acids Res. 25 2861-2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gatchel, J. R., and Zoghbi, H. Y. (2005) Nat. Rev. Genet. 6 743-755 [DOI] [PubMed] [Google Scholar]
- 34.Delagoutte, E., and von Hippel, P. H. (2001) Biochemistry 40 4459-4477 [DOI] [PubMed] [Google Scholar]
- 35.Nossal, N. G., Hinton, D. M., Hobbs, L. J., and Spacciapoli, P. (1995) Methods Enzymol. 262 560-584 [DOI] [PubMed] [Google Scholar]
- 36.Maniatis, T., Fritsch, E. F., and Sambrook, J. (1989) Molecular Cloning: A Laboratory Manual, 5.68-65.71, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 37.Goellner, G. M., Tester, D., Thibodeau, S., Almqvist, E., Goldberg, Y. P., Hayden, M. R., and McMurray, C. T. (1997) Am. J. Hum. Genet. 60 879-890 [PMC free article] [PubMed] [Google Scholar]
- 38.Schumacher, S., Fuchs, R. P., and Bichara, M. (1998) J. Mol. Biol. 279 1101-1110 [DOI] [PubMed] [Google Scholar]
- 39.Jones, C. E., Mueser, T. C., and Nossal, N. G. (2004) J. Biol. Chem. 279 12067-12075 [DOI] [PubMed] [Google Scholar]
- 40.Iyer, R. R., and Wells, R. D. (1999) J. Biol. Chem. 274 3865-3877 [DOI] [PubMed] [Google Scholar]
- 41.Richetti, M., and Buc, H. (1990) EMBO J. 9 1583-1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minetti, C. A., Remeta, D. P., Miller, H., Gelfand, C. A., Plum, E., Grollman, A. P., and Breslauer, K. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 14719-14724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhuang, P., Kolbanovskiy, A., Amin, S., and Geacintov, N. E. (2001) Biochemistry 40 6660-6669 [DOI] [PubMed] [Google Scholar]
- 44.Zahra, R., Blackwood, J. K., Sales, J., and Leach, D. R. (2007) Genetics 176 27-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hartenstine, M. J., Goodman, M. F., and Petruska, J. (2002) J. Biol. Chem. 277 41379-41389 [DOI] [PubMed] [Google Scholar]