Abstract
Cells are the building blocks of biological complexity. They are complex systems sustained by the coordinated cooperative dynamics of several biochemical networks. Their replication, adaptation and computational features emerge as a consequence of appropriate molecular feedbacks that somehow define what life is. As the last decades have brought the transition from the description-driven biology to the synthesis-driven biology, one great challenge shared by both the fields of bioengineering and the origin of life is to find the appropriate conditions under which living cellular structures can effectively emerge and persist. Here, we review current knowledge (both theoretical and experimental) on possible scenarios of artificial cell design and their future challenges.
Keywords: cells, cell cycle, cell membrane, metabolism, information, synthetic biology
1. Introduction
Cellular life cannot be described in terms of only DNA (or any other information-carrying molecule) nor as metabolism or as compartment (cell membrane) alone. Cellular life emerges from the coupling among these three components. A container is a prerequisite of biological organization in order to at least confine reactions in a limited space, where interactions are more likely to occur (Deamer et al. 2002). It also provides a spatial location able to effectively facilitate division of labour among reactions. Moreover, in modern cells, the membrane is an active cell component, channelling nutrients in and waste products out of the cell by means of specialized transport catalysts (Pohorille et al. 2005). A metabolism (Smith & Morowitz 2004) provides the source of non-equilibrium and a means of energy storage required in order to build and maintain cellular components. It is also required to allow cell growth to occur and eventually force splitting into two different (but similar) copies.
If cells must adapt to a changing environment, information carriers will be also needed, as well as their coupling with metabolic reactions. The fundamental problem, of course, is how to obtain the coupling in such a way that self-reproduction of identical structures is achieved and self-maintained. Even though the scientific community has reached a consensus on the requirements and properties of a minimal living system (Pohorille & New 2001; Deamer 2005), the materialization of this vision into a concrete laboratory prototype is still incomplete.
In this review, we explore some of the methods, theories and perspectives related to the goal of constructing simple artificial cellular systems. The research programme to which this special issue is dedicated and which we name synthetic protocell biology (SPB) aims at the construction of a chemical life-like ensemble in the form of an artificial cell system able to self-maintain, self-reproduce and potentially evolve. The article is organized as follows: as SPB can be considered as a field intersecting synthetic biology, which is a general framework encompassing systems biology and bioengineering, we shall place the former in the context of the latest developments and major objectives of the latter. Subsequently, we shall move from general to particular, from this overall synthetic perspective to the specific requirements of a minimal protocell. We shall review advances in the modelling approaches and also comment briefly on the experimental techniques that have drawn us closer than ever to achieving the goal of SPB.
2. Synthetic biology
As a direct consequence of recent advances in molecular biology techniques and following the direction of the increasing simulation–experiment feedback, the discovery-driven biology of the last century is being transformed into a hypothesis-driven biology. This approach has made possible the control and design of new cellular functions and genetic circuits (Sprinzak & Elowitz 2005) and has marked the beginning of a new well-defined research field: synthetic biology (Benner & Sismour 2005). It reflects the idea that the best way to test the accuracy of current biological knowledge is to modify or construct a different version of a biological system and compare its behaviour with theoretical expectations. As an inheritance from systems biology (Ideker et al. 2001) and computational cell biology (Slepchenko et al. 2002), synthetic biology uses a wide variety of mathematical descriptions including graph theory, Boolean logic, ordinary, partial and stochastic differential equations. It is the solid feedback between an accurate and realistic modelling of the subject under investigation and the precise reproduction of the genetic or chemical design in the laboratory that has recently granted synthetic biology a high-profile attention in the scientific community (McDaniel & Weiss 2005; Andrianantoandro et al. 2006).
There is still no clear distinction in the scientific community between synthetic biology and somewhat older research fields, such as systems biology, biological or biomedical engineering and recombinant DNA techniques (Brent 2004). However, one might argue that the main distinctive feature of synthetic biology seems to regard the emphasis on design and testing via simulation of new living biochemical systems endowed with complex behaviour, followed by their experimental implementation. From this point of view, both synthetic biology and SPB follow the same course of action.
Two main research branches can be delimited within the field of synthetic biology. The first one concerns the SPB: the assembly of synthetic units into chemical systems endowed with biological properties, that is reproduction, inheritance and evolution (Pohorille & Deamer 2002). The second branch aims at assembling biological units extracted from living systems and obtaining a modified or even an improved version of a (an existing) biological system. This second branch confers a clear engineering feature to synthetic biology by dealing with the creation and rewriting of genetic circuits using building blocks (Endy 2005).
One of the main goals pursued by both research branches consists in constructing, modifying and using biological mechanisms into performing desired functions, in other words, in obtaining a programmable plug-in genetic device or biological entity (Kobayashi et al. 2004). Either by a rational complete design (Weiss et al. 2003) or by directed evolution techniques (using combinatorial synthesis: Yokobayashi et al. 2002), programming artificially designed living protocells appears as the worldwide synthetic biology objective. In addition to programmable features, SPB also aims at integrating a property unique to living matter: evolvability. Even though there still is a long way to go before achieving this objective, there is active ongoing research on the evolutionary potential of protocellular and prebiotic structures (Yokobayashi et al. 2003; Száthmary et al. 2005). Moreover, the concept of computation is intimately connected to both evolvability and programmability features and latest studies point to it as a fundamental characteristic of biological systems (Fernández & Solé 2003). We shall discuss momentarily these concepts and the relationship between them in the framework of SPB.
3. Building protocells
The major works dedicated to SPB are organized in table 1. Even though SPB is a relatively young research field, it is related to as well as a continuation of several research fields and thus its frontiers are not clearly established. For this reason, the task of choosing among the most significant and relevant results obtained so far is extremely difficult. As a general rule, we chose to cite mainly review papers that incorporate direct citations of particular and detailed works.
Table 1.
Protocell studies in the literature. (Review works appear in italics.)
| approach | container | |
|---|---|---|
| bottom-up | (1) lipid world | Segré et al. (2001) |
| (2) self-catalytic lipid aggregate formation. Theory and simulations | Mavelli & Luisi (1996) | |
| (3) self-catalytic lipid aggregate formation. Experiments | Bachman et al. (1992); Berclaz et al. (2001a,b); Takakura et al. (2003); Chen & Szostak (2004); Hanczyc & Szostak (2004); Takakura & Sugawara (2004) | |
| (4) self-catalytic lipid aggregate formation. Simulations | Noguchi & Takasu (2002); Nilsson et al. (2003); Chen & Kim (2004); Lyubartsev (2005) | |
| (5) increasing bilayer permeability | Monnard & Deamer (2001) | |
| channels: Noireaux & Libchaber (2004); Pohorille et al. (2005) | ||
| microtubulation: Roux et al. (2002) | ||
| alternating electric field: Fischer et al. (2002) | ||
| (6) thermodynamics of self-assembling of lipid aggregates | Mayer et al. (1997) | |
| (7) fusion of liposomes/vesicles | Pantazatos & MacDonald (1999) | |
| (8) photoinduced vesicle formation | Veronese et al. (1998) | |
| biochemical reactions in container | ||
| reconstruction | (9) conceptual framework of macromolecules encapsulation | Szostak et al. (2001) |
| (10) protein expression in liposomes | Oberholzer et al. (1999) | |
| (11) increased efficiency of gene expression (transcription of T7 DNA) in liposomes | Tsumoto et al. (2001); Nomura et al. (2003) | |
| (12) two-stage genetic network encapsulated in liposomes | Ishikawa et al. (2004) | |
| (13) enzyme-driven vesicle replication | Walde et al. (1994b); Szostak et al. (2001); Hanczyc et al. (2003) | |
| (14) coupled RNA-template and vesicle replication. Experiments | Oberholzer et al. (1995); Chen et al. (2004) | |
| (15) homeostatic behaviour | Zepik et al. (2001) | |
| (16) encapsulation of E. coli cell-free expression system in liposomes | Noireaux & Libchaber (2004) | |
| (17) coordinated shell-core growth. Simulations | Munteanu et al. (2007); Serra et al. (in press) | |
| (18) photochemically driven shell-core growth & replication | Rasmussen et al. (2003) | |
| information (I)/metablosm (M)/container (C) | ||
| (19) MC system | Cronhjort & Bloomberg (1997); Gánti (2003) | |
| (20) CI system | Szostak et al. (2001); Fontanari et al. (2006) | |
| (21) MCI system. Simulations | SCM, Fernando & Di Paolo (2004); Munteanu et al. (2007) | |
| evolutionary potential | ||
| (22) general implications of autocatalytic subsystems | Kaneko (2005) | |
| (23) minimal protocell is as unit of evolution | Szathmáry & Maynard-Smith (1997) | |
| (24) evolving replicators in limited dispersion | Szabó et al. (2002); Zintzaras et al. (2002) | |
| top-down | minimal cell | |
| (25) generalized kinetic model of E. coli metabolic mapping | Browning & Shuler (2001) | |
| (26) conceptual framework of minimal gene set | Koonin (2000); Luisi et al. (2002, 2006); Gil et al. (2004) | |
| (27) experiments on minimal gene set | Mushegian & Koonin (1996); Hutchinson et al. (1999) | |
| (28) comparative genomics | Koonin (2003) | |
| computational potential | ||
| (29) protein molecules as computational elements | Bray (1995) | |
| (30) design of genetic circuits | Savageau (2001); Françis & Hakim (2004) | |
| (31) programmable functions | You et al. (2004) | |
Two fundamental approaches have been considered towards the synthesis of a protocell (figure 1). The first class, so-called top-down approach (Luisi et al. 2002) involves the creation of a minimal cell by means of reducing the genome of modern cells. Since numerous genes are involved in cell–cell communication while others have been shown to be non-essential to cell functioning, it was earlier suggested that it would be possible to reduce genome complexity to a minimal set of genes able to sustain (under given external conditions) cell life and reproduction. In this context, using the parasitic bacterium Mycoplasma genitalium as a case study, Mushegian & Koonin (1996) showed that approximately 256 genes seem to be needed to build a minimal gene set that is necessary and sufficient to sustain the existence of a modern-type cell. No matter how small, cell genomes must contain all the information necessary for the cells to perform essential (housekeeping) functions allowing them to maintain metabolic homeostasis, reproduce and evolve, the three main properties of living cells. Moya and co-workers (Gil et al. 2004) have also studied this problem using both genome comparison and computational modelling approaches, further reducing the list of essential genes to only 206 (see also Gabaldo´n et al. 2007).
Figure 1.
The two main approaches to protocell building: top-down and bottom-up. Three potential classes of synthetic protocells can result: self-maintained (non-replicating), replicating but not evolving and fully evolvable protocells (see text).
In this paper, we restrict our attention to the bottom-up approach. Contrary to the top-down approach, it starts from scratch: a life-like entity is built by self-assembly molecular components (Rasmussen et al. 2003). These can be of biological nature or instead completely ad hoc chemical components. In both cases, a compartment is required, formed of some type of amphiphilic molecules characterized by a natural tendency to make aggregates to which further cell components can join and couple. Polymers that mimic lipid amphiphilicity can also assemble into vesicles, offering a much wider spectrum of properties (Discher & Eisenberg 2002).
To detail even further, Luisi et al. (2006) also suggest the term reconstruction as the intermediate between the bottom-up and the top-down approaches. This term refers to the encapsulation of nucleic acids and enzymes in liposomes defining thus a ‘semi-artificial cell’ (Pohorille & Deamer 2002) whose self-replication is again sought in experimental (Oberholzer et al. 1995) and theoretical works (Szostak et al. 2001).
As indicated in figure 1, the final outcome of both the approaches does not need to be a self-replicating, evolving cell. The artificial cell (Acell) can replicate but not evolve, or it might even be unable to replicate itself: such a simple self-maintained Acell would be able to persist under given conditions, exchanging energy and matter from its surroundings without growing. Although this seems a fairly limited situation, it is actually an interesting one: one could design or lead into self-assembling a protocell able to perform some given functions or computations in predefined ways (Pohorille & Deamer 2002). These two major concepts, self-assembly and self-replication, pervade the field of SPB and origin of life and we shall discuss them below in more detail.
(a) A self-replicating machine
Since building a protocell using a bottom-up approach is not limited to biological, evolved ingredients, we should ask ourselves if very different combinations of the previous three ingredients (or none of them) might lead to life-like entities. To answer this question requires formulating the problem from a theoretical perspective. Such view was taken by the Hungarian mathematician John von Neumann in the 1940s (von Neumann 1966). In his seminal work, von Neumann considered the logical conditions under which an abstract—but embodied—automaton would be able to reproduce itself. The approach was fully computational: the reproducing system was viewed as a machine equipped with building blocks and instructions. The solution found was innovative and visionary. A basic scheme is shown in figure 2. Here, the basic building blocks are indicated, including the following:
the constructor (A), able to build a physical new system by using the available raw material in the surroundings,
the blueprint or instructions containing information on what has to be performed by the constructor,
a duplicator (B) which takes the instructions and duplicates them accurately, and
the controller (C) required to guarantee that the whole process takes place in some well-defined sequence.
Figure 2.
The logic of the self-reproducing machine: von Neumann's self-replicating automaton components closely related to those found in living cells. See text for more details.
Although not explicitly said in the previous description, the automaton envisioned by von Neumann had a physical embodiment: he specifically thought about how to build a physical system able to perform the whole replication cycle. In other words, von Neumann's system was a machine. He designed a system which should have 29 states and estimated that on the order of 105 elements would be required. However, smaller versions of the system have been obtained even at the hardware level (Restrepo et al. 2000).
One great implication of von Neumann's solution was the need for a copy mechanism for the instruction set. If no such mechanism were present, an infinite recurrence problem would arise: the instructions would have to contain a replica of themselves which would be passed to the new machine. Since he investigated the required logic of replication, he was neither interested in nor had the necessary tools to build a working machine at the biochemical or genetic level. Remember that in his time DNA had not yet been discovered as nature's genetic material. The fact that von Neumann's approach was so close to current cell organization is revealing. It suggests that universal principles might pervade the ways information, metabolism and container need to be coupled. We can identify the components of the automaton with those of living cells as follows: (i) the instruction set is the DNA molecule, (ii) the duplicator is provided by the DNA polymerase and other components of the cell's replication machinery, (iii) the constructor corresponds to the RNA polymerase and the translation machinery (allowing proteins to be formed), and (iv) the controller is nothing but the regulation of transcription and translation.
The road initiated by von Neumann has been followed by many other researchers over the last few decades of the twentieth century. As molecular biology advanced, new tools and methods allowed the consideration of a different approach to the problem of self-replication: replacing machines by biological components. In this context, several important advances were obtained within the context of self-replicating molecules, both conceptually (von Kiedrowski 1993; Szathmáry & Maynard-Smith 1997) and experimentally (see Paul & Joyce 2004, and references therein). These include different small-sized molecules able to display a closed replication cycle.
The machine's self-reproduction envisioned by von Neumann has an equivalent picture within the context of protocell replication. The question here is: what are the conditions allowing a simple artificial protocell to reach reliable reproduction? von Neumann's picture includes two key components of a complex adaptive system able to process information: hardware and software. In modern cells, software is carried by DNA, whereas proteins play the role of cellular hardware. But in order to achieve reproduction, no software is required (Dyson 1999): if the appropriate mechanisms of molecular assembly are in place and the system is able to spontaneously grow out from equilibrium, replication can occur without the designed concurrence of all the von Neumann's components. In this context, the physical and chemical properties of molecules are able to define a replicating entity without using the machine-like picture of sequential operations. In this self-organized picture, the basic cell cycle includes two steps: growth and division, summarized in figure 3. Both steps need the presence of non-equilibrium conditions unless some externally driven mechanism (such as shear forces) is present. They are also affected by both internal and external fluctuations, here indicated as noise sources (ξ). Basic physical and chemical constraints are the main players in this process, connected with the stability of membrane shape. To a large extent, it is the active breaking of such stability towards non-spherical vesicle shapes which predates the problem of protocell reproduction. This has been explored by a number of authors (Svetina & Žekš 1989; Seifert et al. 1991; Jung et al. 2001; Božič & Svetina 2004; Du et al. 2006) and is formulated in terms of the bending energy function b associated to the closed vesicle. If we indicate b[S] as the free energy density at a given point S on the surface S of the lipid bilayer, the total bending energy will be the integral over the cell's surface S,
| (3.1) |
Figure 3.
A schematic self-reproduction cycle of a protocell, requiring growth (G), deformation (D) and replication (R) phases in order to be completed. When dealing with a nanoscale scenario, both internal and external noisy fluctuations (ξ) are expected to affect the reliability of the whole process.
In its simplest form and considering low temperatures (i.e. thermal fluctuations are ignored), we have
| (3.2) |
where κ[S] is the bending modulus and C(S)−C0(S) is the mean curvature of the vesicle surface at S. The result of the minimization of such energy function, i.e. the solutions of
| (3.3) |
allow us to explain a number of basic vesicle shapes, from red blood cells (Zhong-can & Helfrich 1987; Seifert et al. 1991; Jülicher 1996; Waugh 1996; Piotto & Mavelli 2004; Du et al. 2006) to potential conditions for self-reproduction (Svetina & Žekš 1989; Božič & Svetina 2004). In a more general context, it can be easily shown that energy configurations forbid spontaneous splitting of a spherical vesicle to occur under the absence of other energy sources (Solé et al. 2006). Below we will explore some of the potential scenarios able to allow cell division to take place. Evolution of cell division and other membrane remodelling mechanisms has led to sophisticated mechanisms of organization and self-assembly that make them more and more independent of external fluctuations (Shapiro & Benenson 2005). However, much simpler protocells face a rather different situation, where the presence of noise can be an inevitable component to be taken into account, particularly when dealing with nanoscale systems (Fellermann & Solé 2007).
In von Neumann's picture, self-reproduction was based on a deterministic sequence of events where the software and hardware of the machine interacted in a predictable way. When dealing with wet machines, we must take into account the spontaneous contribution of self-assembly processes and the constraints derived from them. We can also move away from the machine-carrying instructions to be copied. Here, such information is embedded in the container and its interactions with metabolism and information, but the latter can also be removed from the system. Nevertheless, ongoing experimental efforts have explicitly considered von Neumann's approach by using a small set of genes to be enclosed within a vesicle and truly acting as a software system able to sequentially control a chain of events eventually leading to cell replication (Noireaux et al. 2005).
(b) Self-assembly versus design
Most of these potentially feasible levels of protocell complexity are linked to vesicle containers. Thus, special attention has been dedicated to exploring the behaviour of these self-organized structures. Three decades ago, a first crucial step towards the bottom-up synthesis of life consisted of the experiments on self-assembly phenomena leading to microscopic gel structures: Oparin's coacervates (Oparin & Gladilin 1980) and Fox's protenoid microspheres. Early continuations of these scenarios of self-assembling prebiotic containers have focused on surfactant assemblies into micelles and vesicles. By now, it is clear that the lipid vesicles are the meeting point of the top-down and bottom-up approaches, as the idea of cellular life evolving from within a compartment is universally accepted (Luisi et al. 1999). More precisely, experimental work (Monnard & Deamer 2002) complemented by computer simulations of self-assembly of amphiphiles (Fellermann & Solé 2007) made the object of study in the bottom-up approach, while the top-down and reconstruction studies focused on the more biological-like membrane structure called liposomes (Oberholzer & Luisi 2002).
Liposomes are lipid vesicles (bilayers) typically prepared from phospholipids, the components of today's cell membrane. Besides being improbable if not impossible that such molecules existed in the prebiotic environment, they confer a low permeability to the constituting membrane and thus a high resistance to the uptake of nutrients (Deamer et al. 2002). Even in the absence of the evolved transport mechanisms of today's living cells, there are solutions to improve liposomes' permeability (Monnard & Deamer 2001). From the point of view of both origin of life and life synthesis, the use of fatty acids (surfactants) in membrane structures instead of phospholipids allows a higher permeability to ionic solutes and thus seams more plausible and efficient.
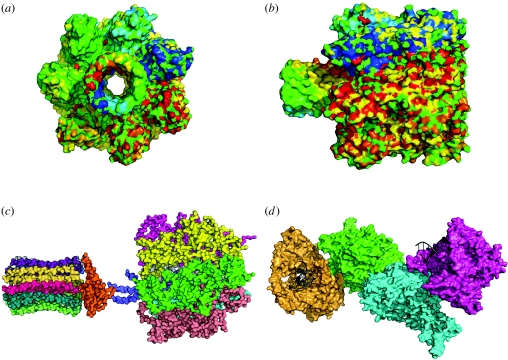
In order to achieve the desired behaviour, researchers have also used a number of basic building blocks in most experimental designs. Some of them are shown in figure 4. These include transmembrane proteins able to facilitate the active flow of precursors, special enzymes and polymerases taken from viruses and bacteria and other well-characterized molecules. These are of course only a few items from a potentially huge universe of possible molecular structures, and the ongoing progress of synthetic biology, with interacting sets of molecules defining oscillators or switches, will help designing protocells able to perform complex functional tasks.
Figure 4.
Some macromolecules used in experimental approaches to the synthesis of protocells based on vesicles as containers. (a,b) Two views of the α-hemolysin pore protein are shown. It has been used as a membrane channel to create selective permeability conditions in phospholipid vesicles. (c) The ATP synthase enzyme, spontaneously incorporated to liposome membranes and able to pump protons inside them, thus allowing a simple metabolism to be maintained. (d) The T7 RNA polymerase structure is shown, with a small piece of the DNA chain also indicated.
(c) Building scenarios
As previously mentioned, the endpoint of SPB is the building of artificial cells able to behave in a life-like manner. This includes self-reproduction, self-maintenance and evolvability. But different intermediate stages can be also relevant, even if they do not incorporate all the previous ingredients. Before we present the different approaches taken, a tentative list of artificial protocell types can be defined as follows.
Self-maintaining protocells. In this type of synthetic protocell, neither growth nor replication would take place (e.g. Zepik et al. 2001). However, active transfer of energy and matter can occur by means of appropriate energy-transducing mechanisms (such as transmembrane proteins pumping protons)—table 1(5). Although this cell is unable to replicate itself, it might have desirable properties such as acting as a nanomachine able to process matter or information under given external conditions. A biosensor or a drug delivery system can be potentially implemented at this level.
Growing vesicles. Owing to active transformations of external precursors, growth can take place over some transient time. These systems eventually stop growing (once an equilibrium between internal metabolism and matter exchange is reached), but can be helpful in exploring through both experiments and simulations the mechanisms that allow self-aggregates and artificially designed vesicles to grow—(Chen & Szostak 2004) table 1(1–8). This system can also be a first step towards cell division by using extrusion mechanisms through membrane pores—table 1(13).
Replicating vesicles. If vesicles or micelles are driven out from equilibrium and keep growing, they can eventually reach unstable configurations favouring membrane asymmetries, deformation and eventually division (Hanczyc & Szostak 2004). The simplest scenario does not include information-carrying molecules and thus does not consider evolution. However, in spite of the limitations imposed by evolution-free systems, it might actually be important in a number of applications (such as biomedicine) where evolvability is actually something to be avoided.
Replicating protocells. The ‘reconstruction’ approach is probably one of the most active directions in SPB, where macromolecules are encapsulated in vesicles or liposomes and catalyse metabolic functions necessary in the life cycle of the protocell. One such example is Szostak's theoretical minimal RNA cell, not yet implemented in the laboratory in the exact composition as prescribed by Szostak et al. (2001). However, different versions of the RNA cell have been created in the laboratory (Bartel & Unrau 1999; Chen et al. 2004). In the long list of experimental work, an important place is held by the work of Walde et al. (1994a) and Oberholzer et al. (1995) showing ‘core-shell’ replication in designed protocells. This coordinated replication of both container and metabolites (and/or genetic material) is necessary in order to avoid death by dilution after several generations of protocells. Different from the reconstruction approach, the Los Alamos Bug model (Rasmussen et al. 2003) and versions of it, based on the self-assembling approach, have been proved to fulfil this condition, an emergent property of the catalytic coupling of protocell's subsystems—table 1(17). Opposed to these catalysis-based protocell models, a stoichiometric model of a protocell, the chemoton model (Gánti 2003), accomplishes the coordinated growth of all its components by means of a precise imposed stoichiometry in the transformation nutrients–metabolites–waste.
Evolving protocells. If the macromolecular organization of the SPB includes an information carrier coupled to metabolic and container dynamics, the whole assembly can experience evolutionary changes and Darwinian selection (Száthmary et al. 2005). For example, in the stoichiometrically coupled chemoton model (Gánti 2003), the information is carried by polymers of a given length, and thus changes in the number of constituent monomers can induce changes in the efficiency of protocell dynamics and thus a genotype–phenotype mapping.
4. Protocell models
In the context of the scenarios mentioned previously, several models of synthetic protocell can be envisioned (Mavelli & Ruiz Mirazo 2007). In this section, we enumerate some possible examples of systems that allow cell self-replication by using different potential mechanisms. Some of these systems have been explored from the mathematical point of view.
Enzyme-driven, information-free protocells. Based on an early suggestion by the Russian biomathematician Nicolas Rashevsky, it has been shown that the stability of a spherical closed membrane can be lost by providing an appropriate set of metabolic reactions (Rashevsky 1960). Specifically, Rashevsky conjectured that a vesicle having enzymes allowing metabolic reactions to occur inside the compartment could experience a destabilization eventually leading to cell division. It has been recently shown that this is the case for a very simple model involving, for simplicity, two enzyme molecules, placed at the two opposite poles of the cell (figure 5a). If these enzymes catalyse a reaction transforming a given precursor (R in figure 5a) into a new molecule G, then close to the location of each enzyme, the concentration of G would increase and eventually trigger a heterogeneous pressure distribution along the membrane (Macía & Solé 2007a). Additionally, surrounding lipid molecules L become incorporated into the vesicle as membrane bounded molecules (here indicated as Lμ). Since the process is necessarily linked to enzymatic activity, the splitting is a single event, not able to be repeated.
- Turing-like protocells. These model protocells introduce a very simple coupling between a reaction–diffusion (RD) system defining a metabolism and membrane dynamics (figure 5b). In this scenario, there is no need for spatially localized enzymes. Instead, externally provided precursors R1, R2 are supplied, entering the membrane and being transformed into new molecules through a set of simple reactions. The reactions inside and outside the cell are represented by a set of n RD equations (Murray 1989), namely
with i=1, …, n, the index associated to the ith morphogen having a local concentration Ci. Each term Φi describes how the ith molecular species reacts to the other molecules. The last term on the right hand side is the diffusion term accounting for the spontaneous, random movement of molecules through space. However, the formalism needs to be extended by incorporating a changing boundary which now acts as a permeable membrane, also coupled to the reactions described by Φi. These reactions will define the protocell metabolism. Since osmotic pressures are associated with differences in molecular concentrations, active mechanisms generating spatial heterogeneity are expected to create changing pressure fields. These instabilities can break the osmotic pressure symmetry along the membrane, and after division the reactions defining the metabolism must be able to trigger a new growth–division cycle (Macía & Solé 2007b).(4.1)
Figure 5.
Examples of protocell models involving a well-defined membrane separating the external environment from the inner volume, where reactions take place. These include: (a) simple systems with a single replication cycle, (b) Turing protocells, exploiting spatial instabilities as a source of membrane change, (c) the chemoton, (d) synthetic cells involving ribozymes, (e) minimal protocells including an internal compartment performing energy transduction and (f) minimal cells with all the components found in complex cells.
Chemoton-like protocells. The chemoton introduced by Gánti (1975) consists of three functionally dependent autocatalytic subsystems: the metabolic chemical network; the template polymerization; and the membrane subsystem enclosing them all (figure 5c). The self-reproducing metabolic network transforms the external nutrients into the chemoton's internal material necessary for template replication and membrane growth. The correct functioning of the chemoton lies in the precise stoichiometric coupling of the three subunits, more precisely the coordination between the accumulation of molecules and the surface increase in order to achieve an equilibrium of the osmotic pressure relative to the environment. The model imposes a closed stoichiometric coupling of the autocatalytic cycles such that the number of membrane molecules necessary for surface doubling is equal to the number of polymerization iterations needed for a complete replication of all double-stranded template molecules. As shown by recent work (table 1—17), stoichiometric coupling is not necessary for fulfilling coordinated growth as the catalytic coupling of the subsystems can ensure their coordinated growth, and thus the doubling of their components prior to division, which is the condition for a viable replication cycle.
Ribocells. Bartel & Unrau (1999) and Szostak et al. (2001) have suggested possible RNA protocells under the form of minimal ribo-organism: one encapsulated ribozyme would synthesize the vesicle membrane component, and a second ribozyme would replicate itself and the first one (figure 5d). The components necessary for the RNA implementation as seen in these works are not yet available, and there are clues suggesting that a DNA cell might appear easier to implement experimentally than ‘simpler’ RNA cell (Luisi et al. 2002). However, a different RNA cell has been proposed and implemented in the laboratory by Chen et al. (2004) showing that RNA replicating within vesicles could increase membrane growth rate by creating internal osmotic pressure. An even bigger step towards the prototype of the minimal protocell was accomplished by Ishikawa et al. (2004) through the laboratory implementation of a two-level cascading protein expression in liposomes. The simple structure of ribocells allows them to be fully described by means of whole-protocell simulation models (e.g. Flamm et al. 2007).
Transduction-driven protocells. Efficient protocellular systems incorporating both DNA and RNA (figure 5e) could be obtained by incorporating appropriate energy transduction systems (Pohorille et al. 1996). One such subsystem appears indicated within the protocell in figure 5e. It consists of two proteins, bacteriorhodopsin (BR) and the F0F1 ATPase from the thermophile Bacillus PS3, embedded in liposomes (Richard et al. 1995). BR is a light-driven proton pump. It generates the transmembrane proton gradient required for the ATP-making activity of the ATPase. It has been shown to have high turnover and stability. This would be a minimal protocell incorporating compartments inside its structure, thus allowing metabolism to be effectively associated with a special protocell substructure.
Minimal cells. The concept of the minimal cell (figure 5f) is the target of the work employing the top-down approach, as mentioned at the beginning of the previous section. Even though it appears as a more promising and straight-forward strategy in the search of a minimal protocell, when compared with bottom-up approaches, there is still a lot of experimental work needed in order to establish tenable and incontestable rules of thumb for the behaviour of genetic modules within the genetic regulatory networks. The latest advances in synthetic biology have revealed opposing facts: that there is an enormous potential for designing and building programmable genetic devices, on one hand, and that surprises are to be expected when passing from simple circuits to devising multimodular or hierarchical genetic networks as a consequence of emergent new behaviours of numerous interacting agents, on the other. Thus, the step from simple genetic circuits to a new artificial cellular entity has to deal with both the unlimited behavioural diversity and the precision in reaching the programmed target intended for the artificial cell.
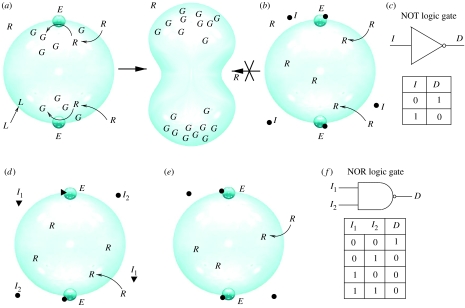
Most of these basic cell models can be used to implement computational tasks. Here, computation means some sort of predictable response to external signals. In figure 6, we show two simple examples of explicit designs of two basic logic gates (the NOT and NOR gates, respectively) from the first model described in this section. Here, the external signals correspond to some type of inhibitor of enzyme activity. As a measure of the output, we use cell division: the output D will be one if division takes place and zero otherwise. In other possible scenarios, the output can correspond to some type of produced molecule resulting from cell reactions.
Figure 6.
Protocells could be used to perform simple types of computations. Here, two very simple examples are given, for illustrative purposes only. Computation is linked to cell division (D): as the output to some given inputs, the cell can (D=1) or cannot (D=0) replicate itself. Here, we use a protocell model (Macía & Solé 2007a) where two enzyme molecules (E) are used as catalytic systems, transforming an external precursor R into a new molecule G. If the enzymes can be repressed by using given inhibitor molecules (I), then simple switches can be made. In (a, b), we show how to implement a NOT gate (c), whereas the second example (d,e) illustrates a more complex, two-input gate (f) (the NOR gate).
The NOT gate can be obtained by using an enzyme having a single inhibitor I: under the presence of I no division occurs, whereas if absent the vesicle experiences reproduction. A simple generalization of this using two enzyme inhibitors (here indicated as I1 and I2) leads to a NOR gate: unless both are absent, no reproduction can occur. Although these are rather trivial examples, they illustrate possible ways of designing simple types of computational cell structures. Once we incorporate as inputs the produced molecules or alternatively introduce different types of cells responding to different signals, it is not difficult to generate (at least at this theoretical level) more complex systems able to describe switches or even memory structures. It is worth mentioning that very complex computational devices (including Turing machines) are currently being developed experimentally at the molecular level (Shapiro & Benenson 2006). These molecular automata might benefit in the future from the current advances in SPB.
5. Discussion
The previous models and theoretical approaches to protocellular systems define a range of complexity levels from non-evolving, non-replicating structures to fully reproducing and evolving entities. So far, none of these systems has been shown to exhibit autonomous reproduction. Achieving such a goal would represent a great leap in biology: it would provide the logical basis for understanding the nature and requirements of life at its simplest level. All the evolutionary advantages of cellular life (from compartmentalization to division of labour) would enhance the potential advantages of synthetic molecular systems.
It is important to mention that possible scenarios of vesicle change in nature are not restricted to whole cell changes associated to reproduction. A wide diversity of mechanisms of membrane formation and processing exist inside complex cells, representing a variety of possible forms of building vesicles. These include transport dynamics using Golgi vesicles, endosomes, lysosomes and clathrin-coated vesicles (Alberts et al. 2002). Inspiration from such processes might help in the design of new types of protocellular system. Some of these processes include the participation of the ribosome, a complex nanomachine involved in reading RNA molecules and translating them into proteins. The computational power of ribosomes, combined with designed protocellular systems exploiting spontaneous pathways of vesicle dynamics might help to expand the current state of the art in this area. Additionally, vesicles obtained from non-biological polymers have been shown to allow interfacing with biological structures (Discher & Eisenberg 2002). These polymersomes can mimic many biological processes, including encapsulation of relevant molecules and transfer-loading through membrane proteins. Their enormous versatility allows the potential exploration of a highly diverse universe of hybrid biomembrane designs.
The success of any of the previous model approaches (and perhaps others not considered here) will provide the basis for a new field at the crossroads between biology and computation. Travelling from non-living to living matter means crossing a twilight zone: some transition domain where the preconditions for reliable cell replication (and thus life) exist. Although some steps need to be completed and some key processes are not yet understood, we are likely to see the success of synthetic cellular life soon at work over the next decade.
Acknowledgments
The authors would like to thank the members of the Complex Systems Lab for useful discussions. Special thanks to Jurgen F. Sebastian for useful discussions. This work has been supported by grants FIS2004-0542, IST-FET PACE project of the European Community founded under EU contract FP6-0022035 (Programmable Artificial Cell Evolution) and by the Santa Fe Institute.
Footnotes
One contribution of 13 to a Theme Issue ‘Towards the artificial cell’.
References
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Garland; New York, NY: 2002. Molecular biology of the cell. [Google Scholar]
- Andrianantoandro E, Basu S, Karig D.K, Weiss R. Synthetic biology: new engineering rules for an emerging discipline. Mol. Syst. Biol. 2006;2:E1–E14. doi: 10.1038/msb4100073. doi:10.1038/msb4100073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman P.A, Luisi P.L, Lang J. Autocatalytic self-replicating micelles as models for prebiotic structures. Nature. 1992;357:57–59. doi:10.1038/357057a0 [Google Scholar]
- Bartel D, Unrau P. Constructing an RNA world. Trends Cell Biol. 1999;9:M9–M13. doi:10.1016/S0962-8924(99)01669-4 [PubMed] [Google Scholar]
- Benner S.A, Sismour A.M. Synthetic biology. Nat. Rev. Genet. 2005;6:533–543. doi: 10.1038/nrg1637. doi:10.1038/nrg1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berclaz N, Blöchinger E, Müller M, Luisi P.L. Growth and transformation of vesicles studied by ferritin labeling and cryotransmission electron microscopy. J. Phys. Chem. B. 2001a;105:1056–1064. doi:10.1021/jp001298i [Google Scholar]
- Berclaz N, Blöchinger E, Müller M, Luisi P.L. Matrix effect of vesicle formation as investigated by cryotransmission electron microscopy. J. Phys. Chem. B. 2001b;105:1065–1071. doi:10.1021/jp002151u [Google Scholar]
- Božič B, Svetina S. A relationship between membrane properties forms the basis of a selectivity mechanism for vesicle self-reproduction. Eur. Biophys. J. 2004;33:565–571. doi: 10.1007/s00249-004-0404-5. doi:10.1007/s00249-004-0404-5 [DOI] [PubMed] [Google Scholar]
- Bray D. Protein molecules as computational elements in living cells. Nature. 1995;376:307–312. doi: 10.1038/376307a0. doi:10.1038/376307a0 [DOI] [PubMed] [Google Scholar]
- Brent R. A partnership between biology and engineering. Nat. Biotech. 2004;22:1211–1214. doi: 10.1038/nbt1004-1211. doi:10.1038/nbt1004-1211 [DOI] [PubMed] [Google Scholar]
- Browning S.T, Shuler M.L. Towards the development of a minimal cell model by generalization of a model of Escherichia coli: use of dimensionless rate parameters. Biotechnol. Bioeng. 2001;76:187–192. doi: 10.1002/bit.10007. doi:10.1002/bit.10007 [DOI] [PubMed] [Google Scholar]
- Chen J.C, Kim A.S. Brownian dynamics, molecular dynamics, and Monte Carlo modeling of colloidal systems. Adv. Coll. Int. Sci. 2004;112:159–173. doi: 10.1016/j.cis.2004.10.001. doi:10.1016/j.cis.2004.10.001 [DOI] [PubMed] [Google Scholar]
- Chen I.A, Szostak J.W. A kinetic study of the growth of fatty acid vesicles. Biophys. J. 2004;87:988–998. doi: 10.1529/biophysj.104.039875. doi:10.1529/biophysj.104.039875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.A, Roberts R.W, Szostak J.W. The emergence of competition between model protocells. Science. 2004;305:1474–1476. doi: 10.1126/science.1100757. doi:10.1126/science.1100757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronhjort M.B, Bloomberg C. Cluster compartmentalization may provide resistance to parasites for catalytic networks. Physica D. 1997;101:289–298. doi:10.1016/S0167-2789(97)87469-6 [Google Scholar]
- Deamer D. A giant step toward artificial life? Trends Biotech. 2005;23:336–338. doi: 10.1016/j.tibtech.2005.05.008. doi:10.1016/j.tibtech.2005.05.008 [DOI] [PubMed] [Google Scholar]
- Deamer D, Dworkin J, Sandford S, Bernstein M, Allamandola L. The first cell membranes. Astrobiology. 2002;2:371–381. doi: 10.1089/153110702762470482. doi:10.1089/153110702762470482 [DOI] [PubMed] [Google Scholar]
- Discher D.E, Eisenberg A. Polymer vesicles. Science. 2002;297:967–973. doi: 10.1126/science.1074972. doi:10.1126/science.1074972 [DOI] [PubMed] [Google Scholar]
- Du Q, Liu C, Wang X. Simulating the deformation of vesicle membranes under elastic bending energy in three dimensions. J. Comput. Phys. 2006;212:757–777. doi:10.1016/j.jcp.2005.07.020 [Google Scholar]
- Dyson F. Cambridge University Press; Cambridge, UK: 1999. Origins of life. [Google Scholar]
- Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. doi:10.1038/nature04342 [DOI] [PubMed] [Google Scholar]
- Felermann H, Solé R.V. Minimal model of self-replicating nanocells: a physically embodied information-free scenario. Phil. Trans. R. Soc. B. 2007;362:1803–1811. doi: 10.1098/rstb.2007.2072. doi:10.1098/rstb.2007.2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández P, Solé R.V. The role of computation in complex regulatory networks. In: Koonin E, editor. Scale-free networks and genome biology. Landes Bioscience; Berlin, Germany: 2003. pp. 206–225. [Google Scholar]
- Fernando C, Di Paolo E. The chemoton: a model for the origin of long RNA templates. In: Pollack J, Bedau M, Husbands P, Ikegami T, Watson R, editors. Artificial Life IX: Proc. Ninth Int. Conf. on the Simulation and Synthesis of Life. MIT Press; Cambridge, MA: 2004. pp. 1–8. [Google Scholar]
- Fischer A, Franco A, Oberholzer T. Giant vesicles as microreactors for enzymatic mRNA synthesis. ChemBioChem. 2002;3:409–417. doi: 10.1002/1439-7633(20020503)3:5<409::AID-CBIC409>3.0.CO;2-P. doi:10.1002/1439-7633(20020503)3:5<409::AID-CBIC409>3.0.CO;2-P [DOI] [PubMed] [Google Scholar]
- Flamm C, Endler L, Müller S, Widder S, Schuster P. A minimal and self-consistent in silico cell model based on macromolecular interactions. Phil. Trans. R. Soc. B. 2007;362:1831–1839. doi: 10.1098/rstb.2007.2075. doi:10.1098/rstb.2007.2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanari J.F, Santos M, Száthmary E. Coexistence and error propagation in pre-biotic vesicle models: a group selection approach. J. Theor. Biol. 2006;239:247–256. doi: 10.1016/j.jtbi.2005.08.039. doi:10.1016/j.jtbi.2005.08.039 [DOI] [PubMed] [Google Scholar]
- Françis P, Hakim V. Design of genetic networks with specified functions by evolution in silico. Proc. Natl Acad. Sci. USA. 2004;101:580–585. doi: 10.1073/pnas.0304532101. doi:10.1073/pnas.0304532101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldo´n T, Pereto´ J, Montero F, Gil R, Latorre A, Moya A. Structural analyses of a hypothetical minimal metabolism. Phil. Trans. R. Soc. B. 2007;362:1751–1762. doi: 10.1098/rstb.2007.2067. doi:10.1098/rstb.2007.2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gánti T. Organization of chemical reactions into dividing and metabolizing units: the chemotons. Biosystems. 1975;7:189–195. doi: 10.1016/0303-2647(75)90038-6. doi:10.1016/0303-2647(75)90057-X [DOI] [PubMed] [Google Scholar]
- Gánti T. Oxford University Press; Oxford, UK: 2003. principles of life. [Google Scholar]
- Gil R, Silva F, Pereto J, Moya A. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 2004;68:518–537. doi: 10.1128/MMBR.68.3.518-537.2004. doi:10.1128/MMBR.68.3.518-537.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanczyc M, Szostak J. Replicating vesicles as models of primitive cell growth and division. Curr. Opin. Chem. Biol. 2004;8:660–664. doi: 10.1016/j.cbpa.2004.10.002. doi:10.1016/j.cbpa.2004.10.002 [DOI] [PubMed] [Google Scholar]
- Hanczyc M.M, Fujikawa S.M, Szostak J. Experimental models of primitive cellular compartments: encapsulation, growth, and division. Science. 2003;302:618–622. doi: 10.1126/science.1089904. doi:10.1126/science.1089904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson C.A, Peterson S.N, Gill S.R, Cline R.T, White O, Fraser C.M, Smith S.O, Venter J.C. Global transposon mutagenesis and a minimal mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. doi:10.1126/science.286.5447.2165 [DOI] [PubMed] [Google Scholar]
- Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu. Rev. Genomics Hum. Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. doi:10.1146/annurev.genom.2.1.343 [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Sato K, Shima Y, Urabe I, Yomo T. Expression of a cascading genetic network within liposomes. FEBS Lett. 2004;576:387–390. doi: 10.1016/j.febslet.2004.09.046. doi:10.1016/j.febslet.2004.09.046 [DOI] [PubMed] [Google Scholar]
- Jülicher F. The morphology of vesicles of higher topological genus: conformal degeneracy and conformal modes. J. Phys. II France. 1996;6:1797–1824. doi:10.1051/jp2:1996161 [Google Scholar]
- Jung H.T, Coldren B, Zasadzinski J.A, Iampietro D.J, Kaler E.W. The origins of stability of spontaneous vesicles. Proc. Natl Acad. Sci. USA. 2001;98:1353–1357. doi: 10.1073/pnas.041420998. doi:10.1073/pnas.041420998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko K. On recursive production and evolvabilty of cells: catalytic reaction network approach. Adv. Chem. Phys. 2005;130:543–598. [Google Scholar]
- Kobayashi H, Kaern M, Araki M, Chung K, Gardner T.S, Cantor C.R, Collins J.J. Programmable cells: interfacing natural and engineered gene networks. Proc. Natl Acad. Sci. USA. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. doi:10.1073/pnas.0402940101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V. How many genes can make a cell: the minimal-gene-set concept. Annu. Rev. Genomics Hum. Genet. 2000;1:99–116. doi: 10.1146/annurev.genom.1.1.99. doi:10.1146/annurev.genom.1.1.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V. Comparative genomic, minimal-gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 2003;1:127–136. doi: 10.1038/nrmicro751. doi:10.1038/nrmicro751 [DOI] [PubMed] [Google Scholar]
- Luisi P.L, Walde P, Oberholzer T. Lipid vesicles as possible intermediates in the origin of life. Curr. Opin. Coll. Int. Sci. 1999;4:33–39. doi:10.1016/S1359-0294(99)00012-6 [Google Scholar]
- Luisi P.L, Oberholzer T, Lazcano A. The notion of a DNA minimal cell: a general discourse and some guidelines for an experimental approach. Helvetica Chim. Acta. 2002;85:1759–1777. doi:10.1002/1522-2675(200206)85:6<1759::AID-HLCA1759>3.0.CO;2-7 [Google Scholar]
- Luisi P.L, Ferri F, Stano P. Approaches to semi-synthetic minimal cells: a review. Naturwissenschaften. 2006;93:1–13. doi: 10.1007/s00114-005-0056-z. doi:10.1007/s00114-005-0056-z [DOI] [PubMed] [Google Scholar]
- Lyubartsev A.P. Multiscale modeling of lipids and lipid bilayers. Eur. Biophys. 2005;35:53–61. doi: 10.1007/s00249-005-0005-y. doi:10.1007/s00249-005-0005-y [DOI] [PubMed] [Google Scholar]
- Macía J, Solé R.V. Protocell self-reproduction in a spatially explicit metabolism-vesicle system. J. Theor. Biol. 2007a;245:400–410. doi: 10.1016/j.jtbi.2006.10.021. [DOI] [PubMed] [Google Scholar]
- Macía J, Solé R.V. Synthetic Turing protocells: vesicle self-reproduction through symmetry-breaking instabilities. Phil. Trans. R. Soc. B. 2007b;362:1821–1829. doi: 10.1098/rstb.2007.2074. doi:10.1098/rstb.2007.2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavelli F, Luisi L. Autopoietic self-reproducing vesicles: a simplified kinetic model. J. Phys. Chem. 1996;100:16 600–16 607. doi:10.1021/jp960524e [Google Scholar]
- Mavelli F, Ruiz-Merazo K. Stochastic simulations of minimal self-reproducing cellular systems. Phil. Trans. R. Soc. B. 2007;362:1789–1802. doi: 10.1098/rstb.2007.2071. doi:10.1098/rstb.2007.2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer B, Köhler G, Rasmussen S. Simulation and dynamics of entropy-driven, molecular self-assembly processes. Phys. Rev. E. 1997;55:4489–4499. doi:10.1103/PhysRevE.55.4489 [Google Scholar]
- McDaniel R, Weiss R. Advances in synthetic biology: on the path from prototypes to applications. Curr. Opin. Biotech. 2005;16:476–483. doi: 10.1016/j.copbio.2005.07.002. doi:10.1016/j.copbio.2005.07.002 [DOI] [PubMed] [Google Scholar]
- Monnard P, Deamer D. Nutrient uptake by protocells: a liposome model system. Orig. Life Evol. Biosph. 2001;31:147–155. doi: 10.1023/a:1006769503968. doi:10.1023/A:1006769503968 [DOI] [PubMed] [Google Scholar]
- Monnard P, Deamer D. Membrane self-assembly processes: steps toward the first cellular life. Anat. Rec. 2002;268:196–207. doi: 10.1002/ar.10154. doi:10.1002/ar.10154 [DOI] [PubMed] [Google Scholar]
- Munteanu A, Attolini C.S.-O, Rasmussen S, Ziock H, Solé R.V. Generic Darwinian selection in catalytic protocell assemblies. Phil. Trans. R. Soc. B. 2007;362:1847–1855. doi: 10.1098/rstb.2007.2077. doi:10.1098/rstb.2007.2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.D. Springer; Berlin, Germany: 1989. Mathematical biology. [Google Scholar]
- Mushegian A.R, Koonin E.V. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl Acad. Sci. USA. 1996;93:10 268–10 273. doi: 10.1073/pnas.93.19.10268. doi:10.1073/pnas.93.19.10268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M, Rasmussen S, Mayer B, Whitten D.G. Constructive molecular dynamics lattice gases: 3-D molecular self-assembly. In: Moore D.G.C, editor. New constructions in cellular automata. Oxford University Press; Oxford, NY: 2003. pp. 275–290. [Google Scholar]
- Noguchi H, Takasu M. Adhesion of nanoparticles to vesicles: a Brownian dynamics simulation. Biophys. J. 2002;83:299–308. doi: 10.1016/S0006-3495(02)75170-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noireaux V, Libchaber A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl Acad. Sci. USA. 2004;101:17 669–17 674. doi: 10.1073/pnas.0408236101. doi:10.1073/pnas.0408236101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noireaux V, Bar-Ziv R, Godefroy J, Salman H, Libchaber A. Toward an artificial cell based on gene expression in vesicles. Phys. Biol. 2005;2:1–8. doi: 10.1088/1478-3975/2/3/P01. doi:10.1088/1478-3975/2/3/P01 [DOI] [PubMed] [Google Scholar]
- Nomura S.M, Tsumoto K, Hamada T, Akiyoshi K, Nakatani Y, Yoshikawa K. Gene expression within cell-sized lipid vesicles. ChemBioChem. 2003;4:1172–1175. doi: 10.1002/cbic.200300630. doi:10.1002/cbic.200300630 [DOI] [PubMed] [Google Scholar]
- Oberholzer T, Luisi P.L. The use of liposomes for constructing cell models. J. Biol. Phys. 2002;28:733–744. doi: 10.1023/A:1021267512805. doi:10.1023/A:1021267512805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer T, Wick R, Luisi P.L, Biebricher C.K. RNA enzymatic replication in self-reproducing vesicles: an approach to a minimal cell. Biochem. Biophys. Res. Commun. 1995;207:250–257. doi: 10.1006/bbrc.1995.1180. doi:10.1006/bbrc.1995.1180 [DOI] [PubMed] [Google Scholar]
- Oberholzer T, Nierhaus K.H, Luisi P.L. Protein expression in liposomes. Biochem. Biophys. Res. Commun. 1999;261:238–241. doi: 10.1006/bbrc.1999.0404. doi:10.1006/bbrc.1999.0404 [DOI] [PubMed] [Google Scholar]
- Oparin A.I, Gladilin K.L. Evolution of self-assembly of protobionts. Biosystems. 1980;12:133–145. doi: 10.1016/0303-2647(80)90011-8. doi:10.1016/0303-2647(80)90011-8 [DOI] [PubMed] [Google Scholar]
- Pantazatos D.P, MacDonald R.C. Directly observed membrane fusion between oppositely charged phospholipid bilayers. J. Membr. Biol. 1999;170:27–38. doi: 10.1007/s002329900535. doi:10.1007/s002329900535 [DOI] [PubMed] [Google Scholar]
- Paul N, Joyce G.F. Minimal self-replicating systems. Curr. Opin. Chem. Biol. 2004;8:634–639. doi: 10.1016/j.cbpa.2004.09.005. doi:10.1016/j.cbpa.2004.09.005 [DOI] [PubMed] [Google Scholar]
- Piotto S, Mavelli F. Monte Carlo simulations of vesicles and fluid membranes transformations. Orig. Life Evol. Biosph. 2004;34:225–235. doi: 10.1023/b:orig.0000009842.58634.a0. doi:10.1023/B:ORIG.0000009842.58634.a0 [DOI] [PubMed] [Google Scholar]
- Pohorille A, Deamer D. Artificial cells: prospects for biotechnology. Trends Biotech. 2002;20:123–128. doi: 10.1016/s0167-7799(02)01909-1. doi:10.1016/S0167-7799(02)01909-1 [DOI] [PubMed] [Google Scholar]
- Pohorille A, New M.H. Models of protocellular structures, functions and evolution. In: Celniker L.M, Tran Thanh Van J, editors. Proc. XII Rencontres de Blois “Frontiers of Life”. The Gioi Publishers; Hanoi, Vietnam: 2001. pp. 37–42. [Google Scholar]
- Pohorille A, Chipot C, New M.H, Wilson M.A. Molecular modeling of protocellular functions. In: Hunter L, Klein T.E, editors. Pacific symposium on biocomputing. World Scientific; Singapore: 1996. pp. 550–569. [PubMed] [Google Scholar]
- Pohorille P, Schweighofer K, Wilson M.A. The origin and evolution of early membrane channels. Astrobiology. 2005;5:1–17. doi: 10.1089/ast.2005.5.1. doi:10.1089/ast.2005.5.1 [DOI] [PubMed] [Google Scholar]
- Rashevsky N. Mathematical biophysics: physico-mathematical foundations of biology. vol. 1. Dover Publications; New York, NY: 1960. [Google Scholar]
- Rasmussen S, Chen L, Nilsson M, Abe S. Bridging nonliving and living matter. Artif. Life. 2003;9:269–316. doi: 10.1162/106454603322392479. doi:10.1162/106454603322392479 [DOI] [PubMed] [Google Scholar]
- Restrepo H.F, Mange D, Sipper M. A self-replicating universal Turing machine: from von Neumann's dream to new embryonic circuits. In: Bedau M.A, McCaskill J.S, Packard N.H, Rasmussen S, editors. Artificial Life VII: Proc. Seventh Int. Conf. on Artificial Life. The MIT Press; Cambridge, MA: 2000. pp. 3–12. [Google Scholar]
- Richard P, Pitard B, Rigaud J.-L. ATP synthesis by the F0F1-ATPase from the thermophilic Bacillus PS3 co-reconstituted with bacteriorhodopsin into liposomes. J. Biol. Chem. 1995;270:21 571–21 578. doi: 10.1074/jbc.270.37.21571. doi:10.1074/jbc.270.5.2010 [DOI] [PubMed] [Google Scholar]
- Roux A, Capello G, Cartaud J, Prost J, Goud B, Bassereau P. A minimal system allowing tubulation using molecular motors pulling on giant liposomes. Proc. Natl Acad. Sci. USA. 2002;99:5394–5399. doi: 10.1073/pnas.082107299. doi:10.1073/pnas.082107299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savageau M.A. Design principles of elementary gene circuits: elements, methods and examples. Chaos. 2001;11:142–159. doi: 10.1063/1.1349892. doi:10.1063/1.1349892 [DOI] [PubMed] [Google Scholar]
- Segré D, Ben-Eli D, Deamer D, lancet D. The lipid world. Orig. Life Evol. Biosph. 2001;31:119–145. doi: 10.1023/a:1006746807104. [DOI] [PubMed] [Google Scholar]
- Seifert U, Berndl K, Lipowsky R. Shape transformations of vesicles: phase diagram for spontaneous-curvature and bilayer-coupling models. Phys. Rev. A. 1991;44:1182–1202. doi: 10.1103/physreva.44.1182. doi:10.1103/PhysRevA.44.1182 [DOI] [PubMed] [Google Scholar]
- Serra, R., Carletti, T. & Poli, I. In press. Synchronization phenomena in surface-reaction models of protocells, Artif. Life [DOI] [PubMed]
- Shapiro E, Benenson Y. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. doi:10.1038/nature04396 [DOI] [PubMed] [Google Scholar]
- Shapiro E, Benenson Y. Bringing DNA computers to life. Sci. Am. 2006;294:44–51. [PubMed] [Google Scholar]
- Slepchenko B.M, Schaff J.C, Carson J.H, Loew L.M. Computational cell biology: spatiotemporal simulation of cellular events. Annu. Rev. Biomol. Struct. 2002;31:423–441. doi: 10.1146/annurev.biophys.31.101101.140930. doi:10.1146/annurev.biophys.31.101101.140930 [DOI] [PubMed] [Google Scholar]
- Smith E, Morowitz H.J. Universality in intermediary metabolism. Proc. Natl Acad. Sci. USA. 2004;101:13 168–13 173. doi: 10.1073/pnas.0404922101. doi:10.1073/pnas.0404922101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solé R.V, Macía J, Fellermann H, Munteanu A, Sardanyés J, Valverde S. Models of protocell replication. In: Steen R, editor. Protocells: bridging living and non-living matter. MIT Press; Cambridge, MA: 2006. [Google Scholar]
- Sprinzak D, Elowitz M.B. Reconstruction of genetic circuits. Nature. 2005;438:443–448. doi: 10.1038/nature04335. doi:10.1038/nature04335 [DOI] [PubMed] [Google Scholar]
- Svetina S, Žekš B. Membrane bending energy and shape determination of phospholipid vesicles and red blood cells. Eur. Biophys. J. 1989;17:101–111. doi: 10.1007/BF00257107. doi:10.1007/BF00257107 [DOI] [PubMed] [Google Scholar]
- Szabó P, Scheuring I, Czárán T, Szathmáry E. In silico simulations reveal that replicators with limited dispersal evolve towards higher efficiency. Nature. 2002;420:340–343. doi: 10.1038/nature01187. [DOI] [PubMed] [Google Scholar]
- Szathmáry E, Maynard-Smith J. From replicators to reproducers: the first major transitions leading to life. J. Theor. Biol. 1997;187:555–571. doi: 10.1006/jtbi.1996.0389. doi:10.1006/jtbi.1996.0389 [DOI] [PubMed] [Google Scholar]
- Száthmary E, Santos M, Fernando C. Evolutionary potential and requirements for minimal protocells. In: Walde P, editor. Topics in current chemistry. Prebiotic chemistry. Springer; Berlin, Germany: 2005. pp. 167–211. [Google Scholar]
- Szostak J.W, Bartel D.P, Luisi P.L. Synthesizing life. Nature. 2001;409:387–390. doi: 10.1038/35053176. doi:10.1038/35053176 [DOI] [PubMed] [Google Scholar]
- Takakura K, Sugawara T. Membrane dynamics of a myelin-like giant membrane: multilamellar vesicle applicable to a self-reproducing system. Langmuir. 2004;20:3832–3834. doi: 10.1021/la049738a. doi:10.1021/la049738a [DOI] [PubMed] [Google Scholar]
- Takakura K, Toyota T, Sugawara T. A novel system of self-reproducing giant vesicles. J. Am. Chem. Soc. 2003;125:8134–8140. doi: 10.1021/ja029379a. doi:10.1021/ja029379a [DOI] [PubMed] [Google Scholar]
- Tsumoto K, Nomura S.M, Nakatani Y, Yoshikawa K. Giant liposome as a biochemical reactor: transcription of DNA and transportation by laser tweezers. Langmuir. 2001;17:7225–7228. doi:10.1021/la010887s [Google Scholar]
- Veronese A, Berclaz N, Luisi P.L. Photoinduced formation of bilayer vesicles. J. Phys. Chem. B. 1998;102:7078–7080. doi:10.1021/jp981017v [Google Scholar]
- von Kiedrowski G. Minimal replicator theory I: parabolic versus exponential growth. Bioorg. Chem. Front. 1993;3:113–146. [Google Scholar]
- von Neumann J. University of Illinois Press; Urbana, IL: 1966. Theory of self-reproducing automata. [Google Scholar]
- Walde P, Goto A, Monnard P.-A, Wessicken M, Luisi P.L. Oparin's reactions revisited: enzymatic synthesis of poly (adenylic acid) in micelles and self-reproducing vesicles. J. Am. Chem. Soc. 1994a;116:7541–7547. doi:10.1021/ja00096a010 [Google Scholar]
- Walde P, Wick R, Fresta M, Mangone A, Luisi P.L. Autopoietic self-reproduction of fatty acid vesicles. J. Am. Chem. Soc. 1994b;116:11 649–11 654. doi:10.1021/ja00105a004 [Google Scholar]
- Waugh R.E. Elastic energy of curvature-driven bump formation on red blood cell membrane. Biophys. J. 1996;70:1027–1035. doi: 10.1016/S0006-3495(96)79648-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R, Basu S, Hooshangi S, Kalmbach A, Karig D, Mehreja R, Netravali I. Genetic circuit building blocks for cellular computation, communications, and signal processing. Nat. Comput. 2003;2:47–84. doi:10.1023/A:1023307812034 [Google Scholar]
- Yokobayashi Y, Collins C.H, Leadbetter J.R, Weiss R, Arnold F.H. Directed evolution of a genetic circuit. Proc. Natl Acad. Sci. USA. 2002;99:16 587–16 591. doi: 10.1073/pnas.252535999. doi:10.1073/pnas.252535999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi Y, Collins C.H, Leadbetter J.R, Weiss R, Arnold F.H. Evolutionary design of genetic circuits and cell–cell communications. Adv. Complex Syst. 2003;6:37–45. doi:10.1142/S0219525903000700 [Google Scholar]
- You L, Cox R.S, III, Weiss R, Arnold F.H. Programmed population control by cell–cell communication and regulated killing. Nature. 2004;428:868–871. doi: 10.1038/nature02491. doi:10.1038/nature02491 [DOI] [PubMed] [Google Scholar]
- Zepik H.H, Blöchliger E, Luisi P.L. A chemical model of homeostasis. Angewandte Chemie. 2001;113:205–208. doi: 10.1002/1521-3773(20010105)40:1<199::AID-ANIE199>3.0.CO;2-H. doi:10.1002/1521-3757(20010105)113:1<205::AID-ANGE205>3.0.CO;2-A [DOI] [PubMed] [Google Scholar]
- Zhong-can O.-Y, Helfrich W. Instability and deformation of a spherical vesicle by pressure. Phys. Rev. Lett. 1987;59:2486–2488. doi: 10.1103/PhysRevLett.59.2486. doi:10.1103/PhysRevLett.59.2486 [DOI] [PubMed] [Google Scholar]
- Zintzaras E, Santos M, Szathmáry E. “Living” under the challenge of information decay: the stochastic corrector model vs. hypercycles. J. Theor. Biol. 2002;217:167–181. doi: 10.1006/jtbi.2002.3026. doi:10.1006/jtbi.2002.3026 [DOI] [PubMed] [Google Scholar]