Abstract
Nutrient transport, polymerization and expression of genetic information in cellular compartments are hallmarks of all life today, and must have appeared at some point during the origin and early evolution of life. Because the first cellular life lacked membrane transport systems based on highly evolved proteins, they presumably depended on simpler processes of nutrient uptake. Using a system consisting of an RNA polymerase and DNA template entrapped in submicrometre-sized lipid vesicles (liposomes), we found that the liposome membrane could be made sufficiently permeable to allow access of ionized substrate molecules as large as nucleoside triphosphates (NTPs) to the enzyme. The encapsulated polymerase transcribed the template-specific base sequences of the DNA to the RNA that was synthesized. These experiments demonstrate that units of genetic information can be associated with a functional catalyst in a single compartment, and that transcription of gene-sized DNA fragments can be achieved by relying solely on passive diffusion to supply NTPs substrates.
Keywords: encapsulated transcription, liposomes, nutrient uptake, origin of life, passive diffusion, protocell
1. Introduction
A fundamental property of life is polymer synthesis in a confined space, using free energy and nutrients available in the environment (Deamer et al. 1994; Walde et al. 1994; Deamer 1997; Tawfik & Griffiths 1998; Szostak et al. 2001). In contemporary cellular life, two polymers—nucleic acids and proteins—are central to this process. In order to have the capacity for evolution, the two polymers must necessarily be linked through an information transfer process that allows genetic changes to be translated into altered phenotypes and then transmitted to ensuing generations. The origin of cellular life presumably occurred by self-assembly of organic compounds on the prebiotic Earth into encapsulated molecular systems capable of catalysed polymer synthesis. Although it is unknown whether nucleic acids and proteins were components of the first living systems, analogous polymers must have been synthesized by an yet unknown pathway, which were capable of the linked interactions that permit genetic variation and evolutionary selection of expressed phenotypes.
Living cells are defined by lipid bilayer membranes, and liposomes have been explored previously as model protocells (Deamer & Oro 1980; Lazcano 1994a,b; Luisi et al. 1999; Szostak et al. 2001; Hanczyc & Szostak 2004). Investigations of enzymatic reactions within the boundaries of vesicles or liposomes have been performed in a variety of molecular systems (Chakrabarti et al. 1994; Walde et al. 1994; Oberholzer et al. 1995, 1999; Nomura et al. 2003; Ishikawa et al. 2004; Noireaux & Libchaber 2004; Chen et al. 2005). Two approaches to the design of these molecular systems have generally been followed. The first approach uses vesicular systems with diameters up to tens of micrometres to study coupled transcription and translation (Nomura et al. 2003; Ishikawa et al. 2004; Noireaux & Libchaber 2004) or to encapsulate DNA/histone complexes (Nomura et al. 2001). In the second approach, smaller vesicles in the submicrometre range were investigated to study reactions such as random RNA polymerization (Walde et al. 1994), DNA amplification (PCR; Oberholzer et al. 1995) and ribozyme cleavage (Chen et al. 2005). Although the size of primitive protocells is unknown, it seems probable that simple genetic/catalytic machinery could have fitted into a volume smaller than that of contemporary bacteria. A large membrane-defined volume, being relatively fragile, might have been disadvantageous in a natural environment, which is prone to sudden changes in pH, temperature or ionic strength. Furthermore, self-assembly of protocells was likely to have been a simple process. For example, in one plausible scenario, it is proposed that dried films of mixed lipid and polymers were rehydrated, producing large vesicles that would then be fragmented into smaller compartments (Hanczyc & Szostak 2004).
Typical lipid bilayer systems have a significant limitation with respect to primitive cellular life, which is their relative impermeability to polar or ionic solutes required as substrates. To circumvent this limitation, in previous investigations, ionic substrates were typically encapsulated simultaneously with the enzyme machinery, ensuring an adequate substrate supply at least during the early stages of an encapsulated polymerization reaction (Oberholzer et al. 1995; Nomura et al. 2003; Ishikawa et al. 2004). In contemporary cells, the permeability barrier is essential for maintaining ionic concentration gradients that drive many bioenergetic processes. For this reason, complex systems of transport proteins that facilitate the movement of ions, nutrients and metabolites across the barrier are incorporated in biological membranes (Aidley & Stanfield 1996). The first forms of cellular life presumably lacked such systems and instead relied on simpler mechanisms, such as passive diffusion across their boundaries to accumulate nutrients from the environment (Deamer 1997).
In order to provide a laboratory model of a protocell, we have developed a liposome system consisting of dimyristoylphosphatidylcholine (DMPC) vesicles with an encapsulated RNA transcription system composed of an enzyme, T7 RNA polymerase, its DNA template and magnesium ions. These vesicles have an average diameter in the submicrometre range. Substrate molecules for the transcription, nucleoside triphosphates (NTPs), are present in the external medium and must cross the bilayer barrier by passive diffusion to become available to the enzymatic system. In other words, the substrate molecules must diffuse through transient defects produced by disturbances in the lipid packing order (Paula et al. 1996). The frequency of such defects significantly increases at the lipid phase transition temperature (Kanehisa & Tsong 1978; Mouritsen et al. 1995), an additional experimental variable. We also know that the bilayers of DMPC vesicles display a selective permeability barrier that permits permeation of monomers while retaining nucleotide dimers and higher oligomers (Monnard & Deamer 2001).
This system allowed us to address the following questions:
How readily can macromolecules such as a polymerase enzyme and its template be captured within the volume of a single lipid vesicle with an average diameter in the submicrometre range?
Can ionized substrates, such as NTPs, be made available to the polymerase at a rate sufficient to permit RNA synthesis?
Does the microenvironment of a lipid vesicle affect the fidelity with which an encapsulated T7 polymerase transcribes a nucleotide sequence from a DNA template to RNA? In other words, how small can a compartment be without interfering with the polymerase activity?
2. Material and methods
Reagents were purchased from the following sources: phospholipids from Avanti Polar Lipids (Alabaster, AL); bovine serum albumin (BSA), inorganic pyrophosphatase (iPPase) and sodium cholate from Sigma Chemical Co. (St Louis, MO); T7 RNA polymerase was purchased as in vitro transcription kits (MAXIscript and Megascript) from Ambion (Austin, TX), or expressed in bacteria from a plasmid, and enzyme stock solutions with a typical activity of 90–100 U μl−1 were prepared. RNase-free DNase I, proteinase K and plasmid template, pTRI-XEF, were obtained from Ambion, Inc. (Austin, TX) and the plasmid d56-33, encoding for a 248 nucleotide(nt)-long RNA fragment containing 70 nt RNA region flanked by hammerhead and hepatitis delta virus (HDV) ribozymes (Ferré-D'Amaré et al. 1998), was a kind gift of Dr A. Ferré-d'Amaré, Fred Hutchinson Cancer Research Center, Seattle, WA.
NTPs, deoxynucleoside triphosphate (dNTPs) and RNase ONE were purchased from Promega (Madison, WI); the Decade marker from Ambion (Austin, TX); Superscript II RNase H− reverse transcriptase from Life Technologies (Gaithersburg, MD); RNeasy Mini spin columns from Qiagen, Inc. (Santa Clarita, CA); Bio-Gel A 15m (mesh 200–400), Micro Bio-spin 6 and 30 chromatography columns from BioRad (Hercules, CA); [α-33P]UTP and [α-33P]dATP from Amersham Life Science, Inc. (Arlington Heights, IL). Primers were prepared on an ABI 392 DNA/RNA synthesizer (Applied Biosystems) using standard phosphoramidite chemistry.
(a) Liposome encapsulation of T7 RNA polymerase and its templates
Encapsulation of the T7 RNA polymerase and its template was carried out according to a dehydration/rehydration method (Deamer & Barchfeld 1982) which can use small volumes and, at the same time, incorporates evaporation and wetting cycles characteristic of fluctuating terrestrial environments. In brief, a 20 mg DMPC film (29.5 μmole) was swollen in a Tris–HCl buffer containing MgCl2, β-mercaptoethanol and BSA. The suspension was then vortexed and heated at 55°C for 10 min to obtain a complete dispersion of the lipid film as multilamellar vesicles. Simultaneously, 250–350 U T7 RNA polymerase and 100 U iPPase were incubated at 37°C with 3.0×10−11 mole template (concentration during the encapsulation procedure, 0.16 μM). Both solutions were then thoroughly mixed (the final volume was 184.4 μl of 40 mM Tris (pH 8.0), 10 mM MgCl2, 10 μM β-mercaptoethanol and 50 μg ml−1 BSA). At this stage, 10 μCi [α-33P]UTP was added.
The resulting suspension was dehydrated for 2–3 h at 30°C under a constant N2 flow. The protein/DNA/DMPC film was carefully rehydrated, first using a moistened cloth plug at 37°C for 20 min to provide 100% relative humidity, then with RNase-free water to obtain a final lipid concentration of 160 mM. The suspensions were diluted to 120 mM DMPC, and eventually extruded 10 times through polycarbonate films (LiposoFast, Avestin, Inc.) of various pore sizes.
(b) Inactivation of the non-entrapped T7 RNA polymerase
In order to inhibit external RNA synthesis, a double digestion was performed that was designed to hydrolyse both the polymerase enzyme and any nucleic acid present in the medium external to the liposomes. To 100 μl suspension (containing 120 mM DMPC, 150 U T7 RNA polymerase and 7.5 μg template), 120 U RNase-free DNase I and 400 U exonuclease III were added. The resulting suspension was incubated for 1 h at 37°C. A second digestion with proteinase K (150 U) was then performed for 1 h at 37°C. In control experiments, in which the same amounts of T7 RNA polymerase and template were externally added to preformed, extruded liposomes, these quantities of hydrolytic enzymes were shown to inhibit RNA synthesis almost completely. Even after a 3 h incubation at 37°C with 2 mM NTP, only trace amounts of RNA product could be detected by the standard methods used here.
To determine template encapsulation efficiency, the procedure described previously was carried out in the absence of T7 RNA polymerase. Non-entrapped DNA plasmids were digested with DNase I/exonuclease III, and separated by spin column chromatography (Bio-Gel A 15m) for the 200 and 400 nm extruded liposomes, and by centrifugation for the 800 nm and unextruded liposomes (Monnard et al. 1997). The DNA encapsulation efficiency was monitored by UV absorption (260 nm) after solubilization of the liposomes with cholate (molar ratio detergent to lipid was 1.8).
(c) Liposome size after preparation of the reaction mixtures
Liposome suspensions extruded with 200 and 400 nm pore filters and diluted 100–200-fold (0.25–0.5 mM DMPC) were determined by dynamic light scattering (DLS; Precision Detectors, MA) to determine liposome size.
(d) RNA polymerization within the DMPC liposomes
Following digestion with DNase I/exonuclease III and proteinase K to remove external DNA and polymerase, aliquots (71.2 μl) of the liposome preparations (approx. 56 mM DMPC) were mixed with 50 μl NTPs mix (25 mM each, sodium salt, pH 8), giving a final reaction mixture containing approximately 33 mM DMPC and 10.3 mM of each NTP. Alternatively, 50 μl NTP mixture (32.3 mM ATP, CTP, GTP, and 0.24 mM UTP) was added to the liposomes in experiments with entrapped [α-33P]UTP.
These suspensions were incubated with temperature cycling (5 min at 23.3°C and 1 min at 37°C) in a common thermocycler. Aliquots were withdrawn at varying times up to 147 temperature cycles and analysed for the presence of RNA.
(e) RNA purification of pTRI-XEF transcripts
Incubated samples were immediately heated at 50°C for 10 min to inactivate entrapped T7 RNA polymerase. The liposomes were then solubilized with cholate (1.5 molar ratio cholate to DMPC) to allow DNase I/exonuclease III digestion of the DNA template (200 and 1000 U ml−1) for 1 h at 37°C. Purification of the RNA product was then performed with a Centricon YM-3 (Amicon).
(f) RNA purification of d56-33 products
The samples were first incubated at 55°C for 1 h to enhance the activity of the HDV ribozyme. The liposomes were solubilized, protein and lipid were extracted with phenol: chloroform (1 : 1) and the RNA was precipitated in NaOAc/EtOH at −20°C. Precipitates were resuspended in 20 μl Tris–EDTA (TE) buffer.
(g) Detection of RNA product
[α-33P or α-32P]-Labelled purified RNA (one-quarter to half of a reaction mixture) was fractionated on polyacrylamide gel (6–20%) and visualized by X-ray film autoradiography or exposed on image plates (Molecular Dynamics) and analysed using ImageQuant (Molecular Dynamics). Alternatively, the unlabelled RNA fragments were reverse transcribed according to the following procedure. The 20 μl batches contained 10 μl purified RNA, about 50 pmoles primer, 50 mM Tris–HCl (pH 8.3), 75 mM KCl, 1 mM DTT, 3 mM MgCl2, 1 mM of each dNTP with 3–5 μCi [α-33P]dATP and 200 U Superscript II RNase H− reverse transcriptase. The samples were first incubated at 42°C for 5 min, then at 37°C for 1 h. The solutions were loaded on a micro bio-spin 6 chromatography column to separate cDNA from non-incorporated [α-33P]dATP. [α-33P]-labelled cDNA was fractionated on polyacrylamide gel (6%) and visualized by X-ray film autoradiography.
(h) Quantification of the RNA product
RiboGreen RNA fluorescent staining was used to determine the RNA product of the entrapped reaction, and the fluorescence was measured with an SLM 8000C spectrofluorimeter (SLM Instruments, Inc.). The samples were prepared as described previously. However, to minimize the fluorescence due to DNA fragments, the samples (30 μl) after RNeasy purification were digested a second time with 12 U DNase I and 200 U exonuclease III, and the DNA fragments were separated on a P30 column. The assay was performed with half of the sample (total fluorescence). The second half was digested by RNase ONE (20 U). Fluorescence yields of the RNase ONE-digested sample, corresponding to DNA fragments, were subtracted from total fluorescence yields, and the corresponding amount of RNA was calculated from a standard curve prepared using ribosomal RNA.
3. Results and discussion
(a) Permeability measurements
Measurements of NTP permeation across DMPC liposomes have been previously published (Monnard & Deamer 2001). In brief, permeability of NTPs is only detectable under a minimum molar excess of NTPs to magnesium of 3.5. Assuming a concentration gradient of 1 mM, the calculated permeability coefficient (1.3×10−9 cm s−1) for pure DMPC liposomes at their phase transition temperature (23.3°C) is equivalent to approximately four ATP molecules per second entering a typical unilamellar liposome of 0.4 μm diameter. In contrast, the same liposomes at 37°C exhibited permeability about two orders of magnitude lower. Assuming that each liposome contains at most one enzyme molecule and several DNA templates, this rate would not provide saturating substrate concentrations with respect to the known turnover number of the T7 RNA polymerase, which is in the range of 200 per second (Chamberlin & Ryan 1985).
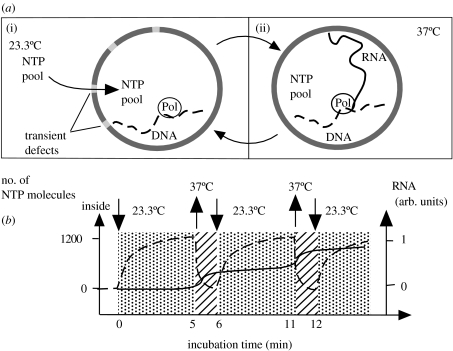
For this reason, a thermocycled incubation between 23.3 and 37°C was employed to increase the supply of nucleotides while still allowing a period of optimal enzyme activity at 37°C. We assume that the enzyme activity is significantly reduced at 23.3°C, because even at 30°C, the incorporation of NTPs is only 2 nt s−1 (Chamberlin & Ryan 1985). Using our values for the permeability coefficient of ATP, at most 1200 nucleotides have entered each liposome by the end of the 5 min incubation at 23.3°C (figure 1). During the incubation at 37°C, only a few nucleotides can cross the membrane, and the nucleotide supply within the liposomes is rapidly exhausted by the entrapped polymerase. In such a system, the T7 RNA polymerase has the advantage of being highly processive in the elongation phase of transcription, a feature that should help overcome occasional substrate exhaustion.
Figure 1.
Schematic of the T7 RNA polymerase liposomal system. (a(i)) NTPs diffuse across lipid membrane by passive diffusion across membrane defects at 23.3°C (phase transition temperature of DMPC bilayers). (a(ii)) At 37°C, whereas the diffusion of NTPs is impaired, the enzyme activity is at an optimum. (b) The internal concentration of NTPs (dashed line) and the RNA formation (solid line) following the temperature cycles: when the temperature is set at 23.3°C, NTPs accumulate in the internal medium; and when it is set at 37°C, the internal supply of monomers is rapidly depleted to transcribe the template to RNA.
(b) Polymerization reactions in extruded and unextruded liposomes
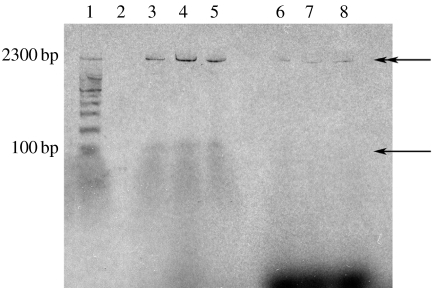
We used the dehydration/rehydration method (Deamer & Barchfeld 1982) to encapsulate the enzyme, the circular, non-linearized pTRI-EXF plasmid template and magnesium cofactor. When the internally produced RNA fragments were to be directly radiolabelled, [α-33P or α-32P]-labelled UTP was also encapsulated. This relatively gentle method also reduced loss of enzyme activity due to denaturation. The non-entrapped template and enzyme were enzymatically digested, and NTPs added to the external medium before DMPC liposome suspensions were incubated for an increasing number of temperature cycles (figure 1) using a thermocycler. After several cycles, transcription products could be detected on a 6% polyacrylamide gel (figure 2, lanes 3–5). RNAs shorter than 100 nt (single-head arrow), presumably products of aborted transcription initiation and elongation, are present as a smeared band. Other long polymers whose lengths are difficult to properly evaluate on the gel even at low polyacrylamide concentrations are also visible (double-head arrow). A relatively long product was surprising, because control reactions run in the external medium of liposomes yielded RNA polymers as long as 3900–4000 nt (data not shown), when the circular pTRI-EXF plasmid was used as the template in aqueous solutions. Both product bands were tested for RNase sensitivity and completely hydrolysed when treated with RNase ONE (9 U in 10 μl solution containing half of the RNA product), confirming that RNA had been produced.
Figure 2.
Gel electrophoresis of radioactively labelled RNA products. T7 RNA polymerase, its template and [α-33P]UTP were entrapped in DMPC liposomes which were subsequently extruded through filters of 400 nm pore size. Lane 1 contains size marker, lanes 2–5 show the product formation within liposomes after 0, 25, 53 and 90 temperature cycles. Lanes 6–8 show the residual reactions outside 400 nm extruded liposomes after digestion with DNase I/exonuclease III and proteinase K: after 0.5, 1.5 and 2.5 h incubation at 37°C. In all lanes, the same reaction volume was loaded for a direct comparison.
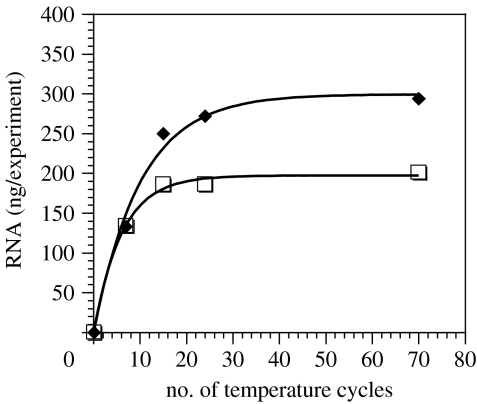
The amount of RNA synthesized was assayed using RiboGreen (Molecular Probes, Inc.), a dye that intercalates between the bases. Typically, the fluorescence yields increased with increasing incubation cycles as shown in figure 3, reaching 294 ng RNA per reaction volume for 400 nm extruded liposomes. The fluorescence yield obtained in samples after 0 cycle always surpassed the background, but was not affected by addition of RNase ONE, which indicated that DNA fragments were still present in the samples. In control experiments with RNA, RNase ONE treatment was sufficient to reduce the fluorescence yield of 10 μg ml−1 ribosomal RNA to background values. We infer from the data in figure 3 that the polymerization reaction proceeded rapidly during the early temperature cycles, and then slowed after approximately 15–20 further cycles, probably due to the progressive inactivation of the encapsulated enzymes.
Figure 3.
Accumulation of RNA within liposomes over time. RNA polymerization within 400 nm (filled diamonds) and 800 nm (open squares) extruded liposomes was assayed by fluorescence (RiboGreen assay) at various times during 70 temperature cycles.
(c) Vesicle dimensions and RNA synthesis
We further investigated transcription as a function of liposome size. When formed by dehydration/rehydration, liposomes tend to be multilamellar and their size varies from 100 nm to 10 μm in diameter. To prepare a more homogeneous size distribution and establish the influence of compartment size on the transcription, liposomes of various sizes were prepared by extrusion through 800, 400 and 200 nm polycarbonate filters sequentially. Each extrusion series consisted of four suspensions prepared from the same starting liposome batch with encapsulated template and enzyme. Although the transcription reaction occurred in all liposome preparations that were tested, the 400 and 800 nm extruded liposomes always yielded more products (table 1). The RNA yields varied by as much as a factor of 2 between extrusion series, probably owing to day-to-day variations in the encapsulation efficiency for the enzyme and template molecules. However, we consistently observed the same RNA yield dependence on liposome size (400 nm≥800 nm≫unextruded≈200 nm extruded liposomes).
Table 1.
DNA-template encapsulation yields and amount of RNA (RiboGreen assay) produced within DMPC liposomes after 70 temperature cycles as function pore size of the filters used for the extrusion
| pore size of filters used for extrusion (nm) | encapsulation efficiency (% of initial DNA template)a | RNA amount present within liposomes (ng per experiment)b | estimated RNA amount (μg per ml of internal volume)c |
|---|---|---|---|
| 200 | 5.3 | 99 | 7.6 |
| 400 | 8 | 294 | 8.4 |
| 800 | 10.5 | 201 | d,e |
| unextruded | 11 | 101 | d,e |
These values are somewhat lower than previously published results (Shew & Deamer 1985), probably due to the fact that not only DNA but also two proteins (T7 RNA polymerase and BSA) and several other components of the transcription machinery were simultaneously encapsulated.
The data shown represent the average of at least three separated measurements. The data were obtained using a standard reaction composition and the same total reaction volume.
The values were calculated assuming a monodisperse size distribution of unilamellar liposomes with an average size of 130 and 195 nm for 200 and 400 nm extruded liposomes (DLS data), a head group area of 0.589 nm2 and a bilayer thickness of 4.45 nm (Kucerka et al. 2004). This assumption is biased as liposome extrusion never achieves a perfect size distribution or unilamellarity (Monnard et al. 1999).
The diameter ranges obtained for these liposomes cannot be reported owing to the polydisperse character of the size distribution.
The calculation of a theoretical internal volume for this liposome size is too imprecise, even for estimation purposes.
In general, these observations are consistent with the known physical characteristics of extruded and unextruded liposomes, as well as the DLS measurements: 200 nm extruded liposomes had an average diameter between 120 and 140 nm; and 400 nm extruded ones between 170 and 220 nm. Extruded liposomes tend to be largely unilamellar or oligolamellar, and the ratio of surface area to internal volume increases as the diameter decreases. This ratio determines the amount of substrate diffusion into the liposome interior during a given time-interval. On the other hand, when liposome suspensions are extruded with filters of pore size smaller than 400 nm, the encapsulation efficiency decreases significantly (table 1; Monnard et al. 1997). It follows that the amount of enzyme and template molecules co-encapsulated decreases with the vesicle diameter and, consequently, the yield of RNA for a given incubation would be reduced in 200 nm extruded vesicles.
The encapsulation efficiency of unextruded liposomes was approximately twice that of 200 nm extruded liposomes (table 1). However, higher content of DNA and enzyme did not result in higher yields of RNA. Reduced yields would result if a substantial fraction of enzyme and template was trapped between multiple layers of membrane, and therefore had less access to substrate. It is also possible that templates and enzymes were more often encapsulated in different compartments of multilamellar vesicles, thereby preventing their association. At the same time, the permeation of substrates across multiple bilayer membranes should decrease their availability to encapsulated enzymes. A slightly higher multilamellarity of larger vesicles may also explain the difference observed between 400 and 800 nm extruded species.
To address the possibility that RNA synthesis was occurring outside the vesicles, we carried out the following experiments. The complete digestion of non-entrapped enzyme and template molecules was checked by externally adding 2 mM of each NTP except 0.24 mM UTP and 10 μCi [α-33P]UTP. After an incubation of up to 3 h at 37°C, only traces of product (figure 2, lanes 6–8) could be detected on a 6% polyacrylamide gel, whereas an undigested enzyme mixture outside DMPC liposomes produced a large amount of RNA (data not shown). Note also that a 90-cycle incubation (figure 2, lane 5) was only equivalent to 1.5 h incubation under optimal conditions. Controls conducted with 200 nm and unextruded liposomes showed no product bands.
We also investigated any residual external RNA polymerization in each liposome size using RiboGreen assay. The fluorescence yields were so low that only data for 400 and 800 nm extruded liposomes could be obtained. After 120 min incubation at 37°C with NTPs, only 15 ng RNA (the largest value obtained during three independent measurements) were detected, equivalent to 5% of the total RNA detected within 400 nm extruded liposomes after 70 temperature cycles.
These results support the conclusion that template-directed RNA polymerization can occur within DMPC liposomes, using externally added NTPs as substrate. Therefore, temperature cycling, a compromise between the optimal permeability of the DMPC bilayers at a phase transition temperature of 23.3°C (Monnard & Deamer 2001) and the T7 RNA polymerase optimal activity at 37°C (Chamberlin & Ryan 1985), seemed to support an efficient entrapped transcription.
(d) Fidelity of encapsulated RNA transcription
The encapsulated transcription of a DNA plasmid sequence would be successful if it can be demonstrated that the transcribed RNA sequence corresponds to that of the template. To probe the RNA product from the pTRI-XEF transcription, reverse transcription (RT) was carried out in separate experiments using two 20-mer primers (P1, nucleotide 280–299, and P2, nucleotide 1001–1020 downstream from the transcription start). A satisfactory result would be that both P1 and P2 could bind to their respective complementary sequences in the RNA and initiate RT. This would demonstrate the transcription of the DNA template at two different sequence locations. Ideally, one might expect the synthesis of two different cDNAs with lengths of 299 or 1020 nt.
Control experiments were also carried out in which the transcription and RT reactions with primer P2 were run outside liposomes. These controls produced relatively large amounts of radioactively labelled cDNA ranging from 100 to more than 1000 bases in length with no distinct bands that could be related to the length of the expected template sequences.
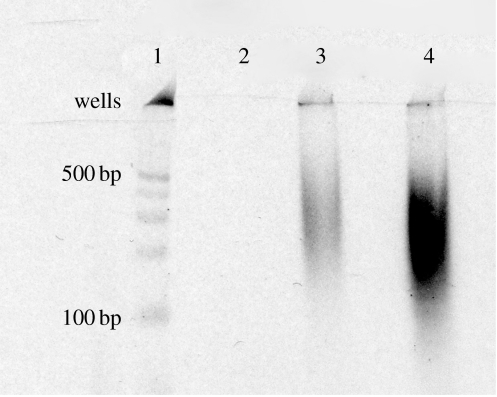
Only products of transcriptions encapsulated in 400 and 800 nm extruded liposomes could be reverse transcribed. These reactions yielded smeared bands of cDNAs of length shorter than those observed in the RT of RNA transcribed outside liposomes. As the number of temperature cycles increased, so did the amount of cDNA present in the smeared band (figure 4). Similar results were obtained with primer P1. In the absence of RNA (either omitted, digested by RNase ONE or after 0 cycle; figure 4) or with a primer that lacked a sequence complementary to the RNA (P2 with two centred mismatches), no cDNA was produced.
Figure 4.
RT of the RNA products transcribed from the pTRI-EXF plasmid in 400 nm extruded liposomes. Lane 1 contains the DNA marker ladder. Lanes 2–4 show RT products of liposome-encapsulated reactions after 0, 15 and 25 temperature cycles.
It follows that both primers P1 and P2 were annealing to complementary sequences on the RNA and initiating cDNA synthesis, which satisfied our expectation that transcription of RNA using a DNA template could be catalysed by encapsulated polymerases. However, the RT did not yield products of defined lengths. The absence of specific cDNA bands could be related to degradation specific to pTRI-EXF products, the formation of complex aggregates (as suggested by the migration patterns on both polyacrylamide and agarose gels) or RNA tertiary structures that may not have been fully denatured at the RT temperature (42°C). On the other hand, RNA fragments transcribed from a circular template within liposomes could also be concatemeric, so that a range of cDNA products should then be expected.
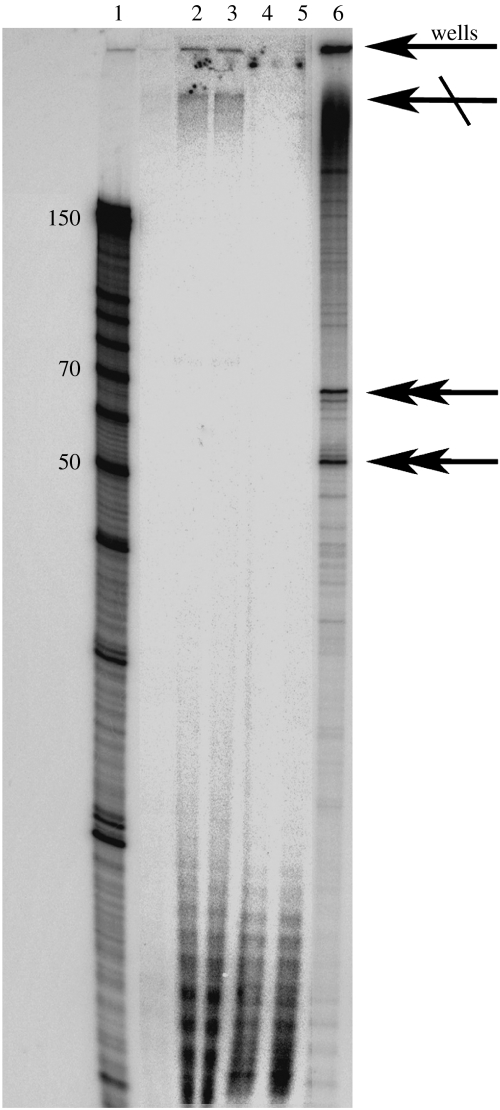
Successful transcription could also be demonstrated by short products of defined length obtained both inside and outside liposomes. To produce a short RNA, we used the plasmid d56-33 that encodes for a 248 nt long RNA construct comprising a hammerhead ribozyme (53 nt) and an HDV ribozyme flanking a 70 nt region (Ferré-D'Amaré et al. 1998). This plasmid was not linearized. Thus, the transcription did not terminate at the end of this 248 nt insert, but rather proceeded in a run-on fashion (figure 5, lane 6, the single-head and crossed arrows, the top band—single-head arrow—probably represents RNA product aggregates in the wells). In figure 5, lane 6, the ribozyme self-cleavage products, the two bands of 70 and 53 nt, are also readily observed (two double-head arrows). With this construct, the HDV ribozyme cannot be resolved because it cleaves its 5′ end and the transcription extends beyond the 3′ end. Comparison of the lanes 2–5 with lane 6 established that long RNAs (run-on transcripts, crossed arrows; product aggregates in the wells, single-head arrow) were present in the encapsulated reactions, but absent in the control reactions (lanes 4 and 5). It follows that digestion of the non-encapsulated template and enzymes prevented the production of full-length RNA transcript, and we concluded that the full-length RNAs in lanes 2 and 3 were synthesized inside the vesicular reactors. It was also possible to infer from the bands in the 20-mer region that in reaction mixtures 4 and 5, the T7 RNA polymerase was still active but lacked the full-length templates. Template digestion by both nucleases was apparently more efficient than the protein digestion by the proteinase K, which is prone to self-digestion.
Figure 5.
Transcription of short RNAs (17% PAGE gel). Lane 1, decade marker containing RNA molecular weight markers of 150 (full-length marker) and shorter RNAs of 100, 90, 80, 70, 60, 50, 40, 30, 20 and 10 nt. Lanes 2 and 3, two independent transcription reactions of plasmid d56-33 in the liposomes; lanes 4 and 5, two independent control reactions of the inactivation of the non-entrapped transcription system; lane 6, positive control: transcription performed outside liposomes. The single-head arrows indicate transcription products (single-head arrow, well aggregate; crossed arrow, run-on transcription) and the double-head products of the ribozyme indicate self-cleavage.
The absence of 53 and 70 nt products in the internalized transcription (figure 5, lanes 2 and 3) may be due to the low concentration of free magnesium. Magnesium (II) is required as a cofactor for the activity of both the hammerhead and HDV ribozymes at low ionic strength (Murray et al. 1998). In control experiments (data not shown), it was established that doubling the lipid concentration significantly reduced the activity of the hammerhead ribozyme, which could be re-established by doubling the magnesium concentration. Although phosphatidylcholine bilayers are often considered neutral owing to the zwitterionic characters of their constituents, the interactions between charged head groups and metal ions should not be neglected.
From the results obtained with both transcription protocols, we conclude that RNA strands containing specific sequences were in fact produced, as verified by annealing of two randomly selected primers to the RNA product of pTRI-XEF followed by initiation of RT. The length of the RNA fragments obtained in the encapsulated transcription of plasmid d56-33 also supported this conclusion.
Finally, as the P2 sequence is found only between 1001 and 1020 bp downstream of the T7 promoter in the template DNA, the T7 RNA polymerase transcribed, probably with considerable fidelity, a template sequence of at least 1020 bp. Achieving efficient transcription of a functional ribozyme RNA will require further optimization of the liposomal systems.
4. Conclusions
Our current understanding of self-assembly processes in contemporary cellular life leads to a variety of hypothetical scenarios about how life can begin on a planetary surface (Morowitz 1992). Given the presence of liquid water, there is little doubt that mixtures of organic compounds present on the prebiotic Earth would become organized into more complex systems by self-assembly. Since polymer synthesis defines growth in all living systems today, a molecular self-assembling system capable of such reactions is well on its way towards the living state. Therefore, the most probable next step towards life would take place when a few self-assembling systems happened to contain a particular set of molecules that allowed capture of energy and nutrients from the environment to be used for polymer synthesis. Significantly, if one of the growing systems contained molecules that could be used as templates to direct further growth, a second polymeric molecule could be synthesized, which is a replica of the first molecule. Passing the information content of one molecule to a second molecule would represent the origin of replication, an essential feature of the definition of life, and understanding encapsulated replication processes is the goal of the research presented here.
(a) Lipid bilayers and membrane transport of nutrients
Standard liposome preparations are able to capture macromolecules such as enzymes and nucleic acids (table 1), but their bilayers are relatively impermeable to polar and ionic solutes. Because growth and reproduction require the efficient transport of nutrients across the cell membrane, contemporary cells employ complex protein assemblies to catalyse the transport process. Before such proteins had evolved, what mechanism was available to transport the nutrients required for cell growth? Our research is based on the assumption that early membranes were more permeable than those of contemporary cells, and thereby ionic substances, probably coordinated by ions of the opposite charge, would have crossed the membrane barrier by passive diffusion. The composition of early membranes remains an unresolved issue. However, both modern lipids and simpler amphiphilic molecules, including fatty acids, can form reasonably stable vesicles composed of bilayer membranes (Hargreaves & Deamer 1978; Walde 1994). Their permeability is strongly dependent on amphiphile chain length (Paula et al. 1996) and can also be enhanced by artificially increasing bilayer packing defects. These properties have permitted systems of encapsulated catalysts and nucleic acids to be used as models of primitive cell-like structures (Deamer 1997; Luisi et al. 1999; Szostak et al. 2001).
In this report, we described a model system of genetic transcription in a primitive cell, in which T7 RNA polymerase is encapsulated in liposomes together with a DNA template. To mimic nutrient uptake by a primitive cell, substrate molecules in the form of NTPs were added to the external medium, and they entered the liposome-encapsulated volume by passive diffusion through transient defects in the bilayers. Carrying out a T7 RNA polymerase-mediated transcription within the aqueous compartments of DMPC liposomes requires that four anionic nucleotides diffuse across the membranes at a rate sufficient to supply T7 RNA polymerase, an enzyme with a relatively high turnover of approximately 200 nt s−1 at 37°C (Chamberlin & Ryan 1985), a temperature at which nucleotide permeability is markedly decreased. A thermocycled incubation is the simplest solution under laboratory conditions in which vesicles composed of a synthetic phospholipid allowed a sufficient substrate permeation rate as well as efficient enzymatic activity. Even though early cellular life presumably used mixtures of amphiphilic compounds with permeability properties that did not require temperature cycling, achieving a sustained substrate supply across DMPC bilayer membranes—presumably less permeable than those of primitive cells—clearly demonstrates that passive diffusion could have been a plausible uptake mechanism for a primitive cell.
(b) Progress towards a model protocell
In earlier experiments using vesicular compartments of submicrometre diameter and substrate supply by permeation across lipid bilayers, an RNA polymerase called polynucleotide phosphorylase was encapsulated either by a simulated tide pool cycle (Shew & Deamer 1985) in liposomes composed of DMPC (Chakrabarti et al. 1994) or in oleic acid vesicles prepared by pH adjustment (Walde et al. 1994). This enzyme does not depend on a template to synthesize RNA. Instead, it can use nucleotide diphosphates such as ADP as both an energy source and a monomer to be incorporated into a random RNA, which was synthesized even if a protease was present in the external medium. In other words, the lipid bilayer protected the polymerase in the vesicle interior.
The next obvious step towards a laboratory model of a protocell is to encapsulate a system capable of not only simple random polymerization but also transcription which will be required for an experimental, evolving molecular system (Wilson & Szostak 1999). The results reported here demonstrate that template-directed synthesis of RNA is possible within submicrometre-sized DMPC liposomes, using entrapped T7 RNA polymerase, plasmid template and four substrates added to the external medium. The fact that we could use a template-dependent enzyme with a relatively high turnover rate also demonstrates that compartmentalization in small vesicles was not necessarily an obstacle to the development of complex metabolic systems. Furthermore, the relative stability of simple lipid bilayers, as demonstrated by the lack of product release, is an essential prerequisite for a protocell which would then be able to gradually increase molecular complexity in the interior volume, while releasing metabolic end products. The efficient transcription of a DNA plasmid represents the first step in expressing compartmentalized genetic information. The transcript length obtained in the system described here allows us to consider transcription of sequence lengths having biological relevance. For instance, a 1020 nt transcribed fragment is promising when compared to the average size of natural ribozymes (50–2000 nt) or to the length of artificial catalytic RNA.
In the future, it should be possible to build on this progress by capturing both a reverse transcriptase and the T7 RNA polymerase in liposomes. This represents a significant hurdle: seven molecules (two functional enzymes, two primers and one template) must be present in the same liposome and eight different substrates (four ribonucleotide triphosphates and four deoxyribonucleotide triphosphates) must cross the membrane barrier at a rate sufficient to supply the encapsulated enzymes with substrates. A functional system with entrapped transcription and RT could allow for evolutionary selection of a ribozyme (Wright & Joyce 1997), but in a confined environment with all the advantages of a compartmented selection (Griffiths & Tawfik 2000; Monnard 2005). In a compartmentalized selection, the general scheme would call for the individual encapsulation of a single DNA template with all the necessary enzymatic machinery in each compartment, much like in living cells. During synthesis, more than one copy of the active molecule can be obtained for each template and remain co-localized within the same compartment. Intact compartments with an encapsulated active biopolymer can be recovered individually at the end of the selection. Thus, such a system could provide an individual transcription of each template instead of bulk transcription, if only one DNA strand is encapsulated per compartment. It could permit an easier isolation of active RNA species because each compartment with a detected RNA activity would contain both multiple copies of the active RNA and the corresponding template.
Additional investigations of encapsulated replicating catalytic systems, similar to the one presented here, will help us better understand how self-assembled molecular systems, protocells and artificial cells, can grow, reproduce and evolve. Directly modelling the protocellular systems present some 3.5 Gyr ago on the early Earth will further require the use of plausible catalysts, such as RNA polymerase ribozymes (that have not been discovered yet), to simulate more accurately the emergence of the earliest forms of microbial life.
Acknowledgments
This research was supported by NASA grants NAG5-4665 and SC-00-35 to D.W.D. We thank J. W. Szostak for his support.
Footnotes
One contribution of 13 to a Theme Issue ‘Towards the artificial cell’.
References
- Aidley D.J, Stanfield P.R. Cambridge University Press; New York, NY: 1996. Ion channels. Molecules in action. [Google Scholar]
- Chakrabarti A.C, Breaker R.R, Joyce G.F, Deamer D.W. Production of RNA by polymerase protein encapsulated within phospholipid vesicles. J. Mol. Evol. 1994;39:555–559. doi: 10.1007/BF00160400. doi:10.1007/BF00160400 [DOI] [PubMed] [Google Scholar]
- Chamberlin M, Ryan T. Bacteriophage DNA-dependent RNA polymerases. In: Boyer P.D, editor. The enzymes. Academic Press; New York, NY: 1985. pp. 87–108. [Google Scholar]
- Chen I, Salehi-Ashtiani K, Szostak J.W. RNA catalysis in model protocell vesicles. J. Am. Chem. Soc. 2005;127:13 213–13 219. doi: 10.1021/ja051784p. doi:10.1021/ja051784p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer D.W. The first living systems: a bioenergetic perspective. Microbiol. Mol. Biol. Rev. 1997;61:230–261. doi: 10.1128/mmbr.61.2.239-261.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer D.W, Barchfeld G.L. Encapsulation of macromolecules by lipid vesicles under simulated prebiotic conditions. J. Mol. Evol. 1982;18:203–206. doi: 10.1007/BF01733047. doi:10.1007/BF01733047 [DOI] [PubMed] [Google Scholar]
- Deamer D.W, Oro J. Role of lipids in prebiotic structures. BioSystems. 1980;12:167–175. doi: 10.1016/0303-2647(80)90014-3. doi:10.1016/0303-2647(80)90014-3 [DOI] [PubMed] [Google Scholar]
- Deamer D.W, Mahon E.H, Bosco G. Self-assembly and function of primitive membrane structures. In: Bengtson S, editor. Early life on Earth. Columbia University Press; New York, NY; Chichester, UK: 1994. pp. 107–123. [Google Scholar]
- Ferré-D'Amaré A, Zhou K, Doudna J.A. A general module for RNA crystallization. J. Mol. Biol. 1998;279:621–631. doi: 10.1006/jmbi.1998.1789. [DOI] [PubMed] [Google Scholar]
- Griffiths A.D, Tawfik D.S. Man-made enzymes—from design to in vitro compartmentalisation. Curr. Opin. Biotechnol. 2000;11:338–353. doi: 10.1016/s0958-1669(00)00109-9. doi:10.1016/S0958-1669(00)00109-9 [DOI] [PubMed] [Google Scholar]
- Hanczyc M.M, Szostak J.W. Replicating vesicles as models of primitive cell growth and division. Curr. Opin. Chem. Biol. 2004;8:660–664. doi: 10.1016/j.cbpa.2004.10.002. doi:10.1016/j.cbpa.2004.10.002 [DOI] [PubMed] [Google Scholar]
- Hargreaves W.R, Deamer D.W. Liposomes from ionic, single-chain amphiphiles. Biochemistry. 1978;17:3759–3768. doi: 10.1021/bi00611a014. doi:10.1021/bi00611a014 [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Sato K, Shima Y, Urabe I, Yomo T. Expression of a cascading genetic network within liposomes. FEBS Lett. 2004;576:387–390. doi: 10.1016/j.febslet.2004.09.046. doi:10.1016/j.febslet.2004.09.046 [DOI] [PubMed] [Google Scholar]
- Kanehisa M.I, Tsong T.Y. Cluster model of lipid phase transition with application to passive permeation of molecules and structures relaxations in lipid bilayers. J. Am. Chem. Soc. 1978;100:424–432. doi:10.1021/ja00470a011 [Google Scholar]
- Kucerka N, Kiselev M.A, Balgavy P. Determination of bilayer thickness and lipid surface area in unilamellar dimyristoylphsophatidylcholine vesicles from small-angle neutron scattering curves: a comparison of evaluatrion methods. Eur. Biophys. J. 2004;33:328–344. doi: 10.1007/s00249-003-0349-0. [DOI] [PubMed] [Google Scholar]
- Lazcano A. The transition from nonliving too living. In: Bengtson S, editor. Early life on Earth. Columbia University Press; New York, NY; Chichester, UK: 1994a. pp. 60–69. [Google Scholar]
- Lazcano A. The RNA world, its predecessors, and its descendants. In: Bengtson S, editor. Early life on Earth. Columbia University Press; New York, NY; Chichester, UK: 1994b. pp. 70–80. [Google Scholar]
- Luisi P.L, Walde P, Oberholzer T. Lipid vesicles as possible intermediates in the origin of life. Curr. Opin. Colloid Interface Sci. 1999;4:33–38. doi:10.1016/S1359-0294(99)00012-6 [Google Scholar]
- Monnard P.-A. Catalysis in abiotic structured media: an approach to selective synthesis of biopolymers. Cell. Mol. Life Sci. 2005;62:520–534. doi: 10.1007/s00018-004-4342-2. doi:10.1007/s00018-004-4342-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnard P.-A, Deamer D.W. Nutrient uptake by protocells: a liposome model system. Orig. Life Evol. Biosph. 2001;31:147–155. doi: 10.1023/a:1006769503968. doi:10.1023/A:1006769503968 [DOI] [PubMed] [Google Scholar]
- Monnard P.-A, Oberholzer T, Luisi P.L. Encapsulation of polynucleotides in liposomes. Biochim. Biophys. Acta. 1997;1329:39–50. doi: 10.1016/s0005-2736(97)00066-7. doi:10.1016/S0005-2736(97)00066-7 [DOI] [PubMed] [Google Scholar]
- Monnard P.-A, Berclaz N, Conde-Frieboes K, Oberholzer T. Decreased solute entrapment in POPC liposomes prepared by freeze/thaw in the presence of physiological amounts of monovalent salts. Langmuir. 1999;15:7504–7509. doi:10.1021/la990068w [Google Scholar]
- Morowitz H.J. Yale University Press; New Haven, CO; London, UK: 1992. Beginnings of cellular life. Metabolism recapitulates biogenesis. [Google Scholar]
- Mouritsen O.G, Jorgenseb K, Honger T. Permeability of lipid bilayers near the phase transition. In: Disalvo E.A, Simon S.A, editors. Permeability and stability of lipid bilayers. CRC Press; Boca Raton, FL: 1995. pp. 137–160. [Google Scholar]
- Murray J.B, Seyhan A.A, Walter N.G, Burke J.M, Scott W.G. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. doi:10.1016/S1074-5521(98)90116-8 [DOI] [PubMed] [Google Scholar]
- Noireaux V, Libchaber A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl Acad. Sci. USA. 2004;101:17 669–17 674. doi: 10.1073/pnas.0408236101. doi:10.1073/pnas.0408236101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura S.-I.M, Yoshikawa Y, Yoshikawa K, Tsumoto K, Dannemuller O, Chasserot-Golaz S, Ourisson G, Nakatani Y. Towards proto-cells: “Primitive” lipid vesicles encapsulating giant DNA and its histone complex. Chembiochem. 2001;2:457–459. doi: 10.1002/1439-7633(20010601)2:6<457::AID-CBIC457>3.0.CO;2-F. doi:10.1002/1439-7633(20010601)2:6<457::AID-CBIC457>3.0.CO;2-F [DOI] [PubMed] [Google Scholar]
- Nomura S.-I.M, Tsumoto K, Hamada T, Akiyoshi K, Nakatani Y, Yoshikawa K. Gene expression within cell-sized lipid vesicles. Chembiochem. 2003;4:1172–1175. doi: 10.1002/cbic.200300630. doi:10.1002/cbic.200300630 [DOI] [PubMed] [Google Scholar]
- Oberholzer T, Albrizio M, Luisi P.L. Polymerase chain reaction in liposomes. Chem. Biol. 1995;2:677–682. doi: 10.1016/1074-5521(95)90031-4. doi:10.1016/1074-5521(95)90031-4 [DOI] [PubMed] [Google Scholar]
- Oberholzer T, Meyer E, Amato I, Lustig A, Monnard P.-A. Enzymatic reactions in liposomes using the detergent-induced liposome loading method. Biochim. Biophys. Acta. 1999;1416:57–68. doi: 10.1016/s0005-2736(98)00210-7. doi:10.1016/S0005-2736(98)00210-7 [DOI] [PubMed] [Google Scholar]
- Paula S, Volkov A.G, Van Hoek A.N, Haines T.H, Deamer D.W. Permeation of proton, potassium ions, and small polar molecules through phospholipid bilayers as a function of membrane thickness. Biophys. J. 1996;70:339–348. doi: 10.1016/S0006-3495(96)79575-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shew R.L, Deamer D.W. A novel method for encapsulation of macromolecules in liposomes. Biochim. Biophys. Acta. 1985;816:1–8. doi: 10.1016/0005-2736(85)90386-4. doi:10.1016/0005-2736(85)90386-4 [DOI] [PubMed] [Google Scholar]
- Szostak J.W, Bartel D.P, Luisi P.L. Synthesizing life. Nature. 2001;409:387–390. doi: 10.1038/35053176. doi:10.1038/35053176 [DOI] [PubMed] [Google Scholar]
- Tawfik D.S, Griffiths A.D. Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 1998;16:652–656. doi: 10.1038/nbt0798-652. doi:10.1038/nbt0798-652 [DOI] [PubMed] [Google Scholar]
- Walde P. Self-reproducing vesicles. In: Fleischaker G.F, Colonna S, Luisi P.L, editors. Self-production of supramolecular structures. From synthetic structures to model of minimal living systems. Kluwer Academic Publishers; Dordrecht, The Netherlands: 1994. pp. 209–216. [Google Scholar]
- Walde P, Goto A, Monnard P.-A, Wessicken M, Luisi P.L. Oparin's reaction revisited: enzymatic synthesis of poly(adenyl acid) in micelles and self-reproducing vesicles. J. Am. Chem. Soc. 1994;116:7541–7547. doi:10.1021/ja00096a010 [Google Scholar]
- Wilson C, Szostak J.W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. doi:10.1146/annurev.biochem.68.1.611 [DOI] [PubMed] [Google Scholar]
- Wright M.C, Joyce G.F. Continuous in vitro evolution of catalytic function. Science. 1997;276:614–617. doi: 10.1126/science.276.5312.614. doi:10.1126/science.276.5312.614 [DOI] [PubMed] [Google Scholar]