Abstract
Many regard metabolism as one of the central phenomena (or criteria) of life. Yet, the earliest infrabiological systems may have been devoid of metabolism: such systems would have been extreme heterotrophs. We do not know what level of complexity is attainable for chemical systems without enzymatic aid. Lack of template-instructed enzymatic catalysis may put a ceiling on complexity owing to inevitable spontaneous decay and wear and tear of chemodynamical machines. Views on the origin of metabolism critically depend on the assumptions concerning the sites of synthesis and consumption of organic compounds. If these sites are different, non-enzymatic origin of autotrophy is excluded. Whether autotrophy is secondary or not, it seems that protocell boundaries may have become more selective with time, concurrent with the enzymatization of the metabolic network. Primary heterotrophy and autotrophy imply pathway innovation and retention, respectively. The idea of metabolism–membrane coevolution leads to a scenario of progressive sequestration of the emerging living system from its exterior milieu. Comparative data on current protein enzymes may shed some light on such a primeval process by analogy, since two main ideas about enzymatization (the retroevolution and the patchwork scenarios) may not necessarily be mutually exclusive and the earliest enzymatic system may have used ribozymes rather than proteins.
Keywords: metabolism, coevolution, infrabiological systems, retroevolution, patchwork evolution, enzymatization
1. Introduction
Metabolism is considered to be one of the central phenomena of life. Following a very long tradition, Gánti (1971, 2003a,b) considers metabolism to be one of the criteria of life. His definition is the following:
By metabolism we understand the active or passive entrance of material and energy into the system which transforms them by chemical processes into its own internal constituents. Waste products are also produced so that the chemical reactions result in a regulated and controlled increase of the inner constituents as well as in the energy supply of the system. The waste products finally leave the system, either actively or passively. (Gánti 2003a, p. 76).
There are some assumptions hidden in this definition. First, ‘entry into the system’ presupposes that there is a boundary for the system. Second, there is a distinction between inner constituents and external material. Third, some quantity of the inner constituents increases as a result of metabolism. Fourth, this increase is a regulated and controlled process. In agreement with other life criteria, it is obvious that this regulatory effect is due at least to some of the inner constituents. If so, metabolism is autocatalytic.
This paper follows Gánti's concept of metabolism and investigates its further logical development, as well as its application to some problems of abiogenesis. Section 2 discusses why autocatalysis is necessary not only for growth and replication but also for self-maintenance, and how this relates to the tribology of chemical systems. Section 3 analyses the autocatalytic nature of intermediary metabolism and whether it holds even for heterotrophs. Section 4 explains the progressive sequestration (metabolism–membrane coevolution) and how it applies in the cases of primary heterotrophy and autotrophy. Section 5 describes the evaluation of the relevance of comparative work on contemporary enzymes. Finally, §6 provides a short discussion.
2. Decay and tribology of chemodynamical machines
Gánti has also formulated a family of models related to the concept of a minimal living system. Those models (including that of the chemoton, comprising three subsystems: one for metabolism; one for template replication; and the other for compartment formation; Gánti 1978) are not only abstract, but also idealized. This is not a drawback, but a merit. However, one way of developing such theoretical constructions is to drop the idealizations one after the other. Thus, an inevitable increase in complexity of the model system ensues.
One of the idealizations important for our present purpose is the total neglect of side reactions. In the model, one can set the concentrations of the raw and waste materials such that the system ‘stops’ and waits indefinitely until the concentration of the raw materials increases again. Obviously, this is not the case for real chemical machineries. Following the terminology of Bauer (1967), living systems are basically chemodynamical machines (note that the original work of Ervin Bauer about the inequilibrium of living systems was published in Russian in 1935). Now, all machines suffer from decay as well as from wear and tear. I make a distinction between these two effects in relation to the activity of the system: the former occurs in inactive state as well (e.g. the spontaneous degradation of nucleic acids or proteins), whereas the latter is a loss of internal material due to the activity of the system (such as photorespiration in plants). One could say that the latter is due to ‘chemical friction’ between the chemical parts (Szathmáry 1989).
Tribology (Zum Gahr 1985) is the engineering science dealing with the wear and tear of machines. One of its central observations is that it is very hard to analyse such phenomena without the knowledge of the structure and functioning of the machine. One could say that the only valid generalization is that the machines will wear out. By the same token, I have formulated the concept of a ‘tribology of chemodynamical machines’ (Szathmáry 1989), applicable not only to living but also to other relevant systems (chemical automata). As for conventional machines, it also holds for chemodynamical machines that their tribology can be made quantitative only by knowing the organization of the component processes, the structure of the chemical ‘gadgets’, etc. Let us consider the following ‘didactic’ equation:
| (2.1) |
where x is the ‘chemomass’ of the chemical machine in question; s is the concentration of raw materials; and k and d are the rate constants for autocatalytic growth and spontaneous decay, respectively. Clearly, if s<d/k, the system decays. If s is too small, then living systems can maintain themselves by adaptations that lead to a decrease in d, such as desiccation and hibernation. It is also clear that unless S can spontaneously be converted to X, self-maintenance requires autocatalysis, matter and energy. This is well known in microbiology where there is a special term for ‘maintenance metabolism’ (e.g. Dauner et al. 2001) in the balance equations.
The special problem of side reactions for the origin of life is that with rudimentary catalysis, k is bound to be much smaller than that for contemporary living systems. There are of course many more side reactions than organized reactions. We know from living systems that the problem is solvable with sufficiently high k, but we are much less confident about prebiotic systems. What degree of chemical complexity can non-enzymatic chemodynamical machines have? I think we do not know the answer. This worry projects into considerations of the primitive ancestry of autotrophy.
3. Autocatalytic nature of metabolism
Gánti (1971, 1978) has called attention to the fact that at the heart of metabolism in many organisms, there is an autocatalytic core of small metabolites, such as the Calvin cycle in plants. The reductive citric acid cycle in some bacteria has a similar nature. Wächtershäuser (1988, 1992) thought that an archaic version of the latter could be at the origin of biological metabolism and that under the right conditions it could have functioned without enzymes. Gánti (2003a,b) thinks that the formose reaction played the role of the primordial metabolic network in the earliest living systems. However, there is no decisive support at present for either of these ideas.
It is more relevant for us to look at the autocatalytic nature of metabolism, also important for Gánti. Immediately, one can raise the question about the nature of metabolism in heterotrophs, where there is no apparent autocatalytic core that would be similar to the Calvin or the reductive citric acid cycle. It is clear that the macromolecular set of cells is autocatalytic, because enzymes acting on metabolism are necessary for making more enzymes, but this is a different issue. The question to formulate is as follows: could one kick-start metabolism just from the macromolecules, membranes or raw materials? Of course, different cells may be different in this regard: neither the set of raw materials that can be taken up through the membrane nor the metabolic networks involved are conserved in evolution. This defines an empirical research programme. I make only one relevant observation in this regard. Gil et al. (2004) have defined the gene content of a hypothetical minimal cell. Although they assume that this cell can take up palmitate, serine, d-glucose, adenine, guanine, uracil, folic acid, nicotinamide, riboflavin, thiamine, methionine, pyridoxal, pantothenic acid and cysteine, at least phosphoenolpyruvate (PEP) should be present inside the system to get started, without which even the first step of glycolysis cannot proceed. A comparative analysis of the metabolism of reduced cells would be welcome.
The issue of an autocatalytic metabolism differs from other, and later, ideas of reflexively autocatalytic sets. Dyson (1985) and Kauffman (1986), for example, consider autocatalytic protein networks in the sense that members of the set would catalyse the formation of other members of the same set, so that in the end the formation of each member is catalysed by at least one member in the set. Such imaginary sets are utterly different from small molecular autocatalytic networks, such as the formose reaction. In the latter, the elementary reactions are stoichiometric transformations rather than catalytic actions. Only the cycle as a whole acts as a catalyst of the net reaction from food source to by-products; in the case of small molecular autocatalytic cycles, the latter include the inner compounds produced in excess.
4. Build-up of primordial metabolism
How did early evolution shape metabolism? The question must be subdivided into smaller parts. Was there a significant phase of non-enzymatic metabolism? Were the earliest cells heterotrophic or autotrophic? All combinations are possible: one can imagine, for example, an early non-enzymatic heterotrophic or autotrophic metabolism. Some claims have been made about the occurrence of essentially contemporary biosynthetic pathways in Miller-type experiments, for example, for purine synthesis. These claims have little, if any, empirical support (Lazcano & Miller 1999), at least if we stay in the realm of demonstrated reactions in the prebiotic soup framework.
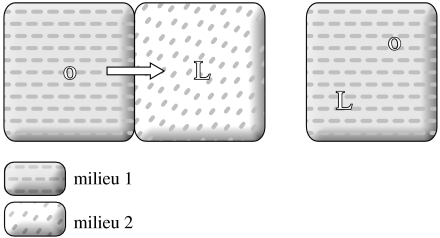
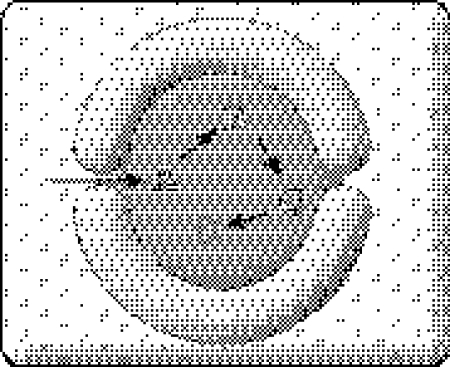
One must realize that the autotrophy/heterotrophy issue is closely related to the one about the geological setting of life's origin. The first relevant question is whether the synthesis of organics and their consumption occurred in the same milieu or not (figure 1). According to the classic Miller scenario, the synthesis of organics took place in the atmosphere, and they were consumed in the liquid phase. This implies that the earliest systems were heterotrophic in the manner Oparin (1961) imagined them to be (originally in 1924). In contrast, if organics were synthesized and consumed in the same milieu, then there is room for autotrophy. Clearly, Wächtershäuser's pyrite-pulled scenario is a case in point. However, as Lazcano & Miller (1999) observed, if protocells consume the organics that leave the surface where first synthesized, they should be considered heterotrophic. An Oparin–Haldane–Miller-type scenario rules out a non-enzymatic origin of metabolism in toto, since for most organic compounds it holds that they would have been created somewhere else (the same holds for cometary delivery). This further implies that the majority of metabolic reactions have been invented by the evolving enzymes. (In this paper, I assume that they were similar to ribozymes, although this does not crucially affect the logic of the argument.) Invention does not mean that enzymes would catalyse impossible reactions. It just means that in the given milieu, the rate of spontaneous reactions would be modest or small. Of course, it is expected that among the slow reactions, enzymes would still choose those with relatively higher rate. In contrast, in the case of primitive autotrophy, the enzymes would have presumably been grafted on top of the network that had been largely in place already.
Figure 1.
Organic synthesis (O) and primitive life-like (L) system may or may not be found in the same milieu on the early earth.
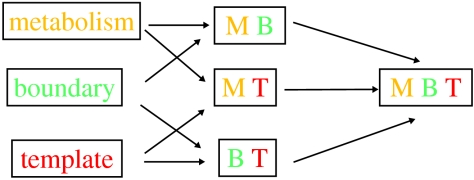
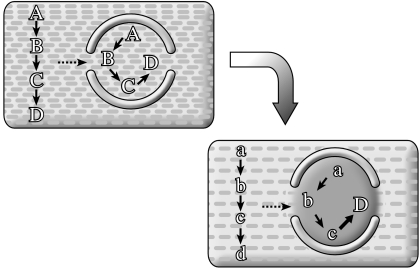
Next, I investigate subsequent evolution of the network. At this point, we must consider another crucial aspect: what was the systemic context of metabolism's evolution? By this I mean the following. I find it hard to believe that a living chemical system, such as described in the chemoton model (Gánti 2003), could have sprung into existence out of some chemical mayhem. I follow the view (e.g. Maynard Smith & Szathmáry 1995) that there was a protracted phase of evolution of natural selection of what I call infrabiological systems (figure 2; see Fernando et al. 2005; Szathmáry 2005). Naturally, the picture of the evolution of metabolism depends on what kind of infrabiological system serves as context.
Figure 2.
Elementary combinatorics of infrabiological systems (Fernando et al. 2005).
Interestingly, the earliest instantiation of the chemoton did not have a boundary: it was a metabolic network coupled with template replication (Gánti 1971). Such a system could not work without some spatial confinement. Adsorption to a surface is a poor man's form of compartment formation. Such a metabolism–template system could develop enzymatic functionality, due to the beneficial selective effect of population structure on the surface (Czárán & Szathmáry 2000; Szabó et al. 2002). This is also the view how Wächtershäuser (1988) imagines evolution before membranes.
Szostak et al. (2001) conceived a system composed of one membrane and two ribozymes only, where one ribozyme would help build the membrane and the other would serve as replicase for both ribozymes. This system has no metabolism in Gánti's sense, although it consumes matter and energy. There is nothing a priori impossible about such a system: of course, it is an ultimate heterotroph, which according to Gánti would not be qualified as living. Yet, it could evolve into a living cell. The path I am investigating in detail also assumes compartment as a starting point, with the difference that it could be either autotrophic or heterotrophic. The former would mean that there is a non-enzymatic metabolic network inside. How does the network get enzymatized?
Enzymatization is a process whereby an expanding set of genetically specified enzymes evolve by natural selection to catalyse more and more reactions of a metabolic network. This means that these enzymes (and their templates: in the case of ribozymes, one strand acts as the enzyme and the complementary strand serves as the gene) must come from somewhere. How does the presence of enzymes relate to the autotrophy/heterotrophy problem? If one considers macromolecular synthesis as a ‘set’, then, of course, all macromolecular replicators are heterotrophs since they are agents feeding on raw materials produced by some other set. This is not the way how I am using the term heterotrophy in this paper. The crucial point is an inside/outside distinction, as aptly emphasized by Varela et al. (1974) and Varela (1979): living systems generate and maintain their own boundary. Heterotrophy means that the system cannot work without organic compounds coming from outside. Note that logically this can apply to an infrabiological system.
As many have realized (reviewed by Fernando et al. 2005), permeability of the membrane is a crucial issue of early evolution: the cells die if they exhaust their internal package of food and there is no way to replenish the resources. Self-suffocation is by necessity a dead end. Let us consider the heterotrophic and the autotrophic cases in turn.
In the case of heterotrophy, practically all raw materials must be taken up in directly consumable form, and not much non-enzymatic ‘metabolism’ can take place for the reasons discussed before. Therefore, one would start with a membrane that would be permeable to everything, except the membrane constituents and the templates (maybe acting as ribozymes). In the case of autotrophy, curiously, the easiest start is the same: since organic synthesis and consumption are allowed to occur in the same milieu, very selective compartmentation may not make sense. However, some could argue that this may not hold.
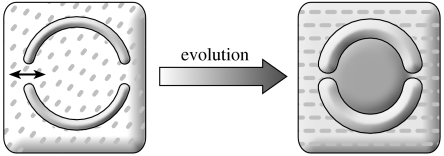
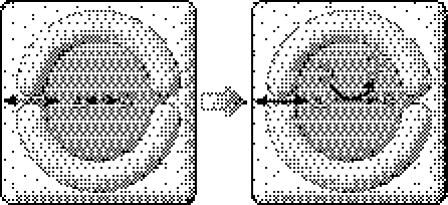
Imagine the formose reaction and associated pathways forming something similar to the prebiotic chemoton (Gánti 2003b). This hypothetical protocell lives on a minimal diet, so the membrane could allow the passage of very simple compounds (such as formaldehyde) only. The snag is that ions have been forgotten. It is a good question to ask whether ions are necessary in principle. A reformulation of this question is as follows: given the abstract scheme of the chemoton, what real chemicals could realize it? Chemistry is not advanced enough to answer such a question. We still do not know much about how chemicals behave under conditions radically different from those prevailing on Earth. It is suffice to say that most ribozymes are metalloenzymes (Doudna & Lorsch 2005); hence, in each generation, metal ions must be replenished so that they are not diluted. In the case of a very selective membrane, their transport must be solved by some mechanism, one that is presumably evolved rather than provided by chance. This is the true reason why in both scenarios I start with a membrane that allows the passage of small molecules and ions freely. Thus, evolution must proceed through progressive sequestration from the environment (figure 3). This process entails three different components: (i) increasing distinction of inside from outside, (ii) build up of enzymatic pathways, and (iii) evolution of membrane structure and permeability. This also implies the coevolution of membrane and metabolism.
Figure 3.
Progressive coevolution of compartmentation and metabolism. Initially, the membrane may have permitted free passage of all small molecules and ions; hence, the only difference between exterior and interior milieus could have been the presence of macromolecules (not shown) in the latter. Membranes may have become much more selective (indicated by thicker boundary and narrower channels) in the course of evolution, resulting in a progressive differentiation between exterior and interior milieus.
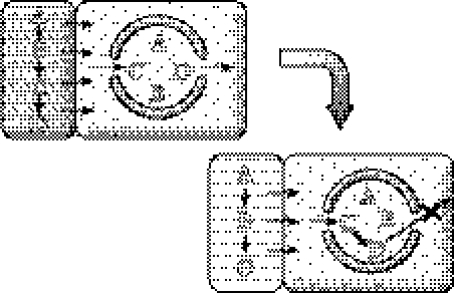
As discussed previously, a heterotrophic start implies pathway innovation: all enzymatic steps cannot be identical to their non-enzymatic counterparts in a different environment (figure 4). If compound D is the most complex, it is likely to be depleted first through growth of the system. Thus, any enzyme catalysing the C→D reaction would be selected for. At the same time, free passage of D becomes undesired: it would just leak into the external milieu. Thus, any ‘genetic’ change in the membrane, resulting in the selective retention of D, would also be selected for. Figure 5 depicts the ‘final’ stage of metabolic innovation; note that depending on the nature of A, this could be either a heterotroph or an autotroph. In addition, enzymes evolve in an order that is reverse to what would be now regarded as ‘forward’: thus, this scenario is consonant with the view of Horowitz (1945) of the evolution of pathways (see below).
Figure 4.
Evolution of heterotrophic metabolism (pathway innovation). Capital letters stand for metabolites. Thin arrows indicate spontaneous chemical reactions; the only thick black arrow (on the right) stands for a catalysed reaction. X indicates that free passage of D is hindered as a result of evolution of membrane permeability. The monomolecular nature of the pathway is only a didactic simplification, as in all remaining figures.
Figure 5.
The ‘end state’ of evolution. Only A can pass through the membrane and all reactions are enzymatized.
Primitive autotrophy evolves through pathway retention (figure 6). Also, in this case, the inside and the outside will become progressively different (partly owing to the activity of existing cells: e.g. in the case how blue-green algae have changed the biota by the mass production of oxygen, a poisonous gas). This implies that the conditions will slowly change and the organic yield in the exterior milieu will decrease, hence autotrophy is progressively relegated to the interior milieu. If, again, D is the most complex compound, then it is probable that the last reaction (C→D) will become enzymatized first, because this change implies the simplest associated evolutionary change in the membrane.
Figure 6.
Evolution of autotrophic metabolism (pathway retention). Initially, the same spontaneous reactions occur everywhere. Change in the environment decreases the yield of reactions in the exterior milieu, implying smaller concentrations (indicated by small letters). Enzymatization of reactions inside the vesicle is thus favoured by natural selection (concentration of D is reset inside).
There are come corollaries to such a picture. For example, some evolution of irreversibility is expected (figure 7). Consider the reversible reaction A←→C. Suppose that this reaction is already enzymatized and that C cannot leak out. But due to reversibility, it can leak out via A. Hence, if, say, due to a fluctuation the concentration of A is reduced in the environment, this reaction will flow in the unfavourable backward direction, depleting valuable C. Imagine, however, that this reaction gets coupled with the irreversible reaction B→D (such as ATP→ADP+Pi), where neither B nor D is permeable; this confers a marked selective advantage on the compartment because C cannot leak out any longer.
Figure 7.
The advantage of transforming a reversible reaction into an irreversible one. See text for details.
I conclude that enzymatization is likely to proceed in a backward direction and it is bound to coevolve with membrane permeability (membrane content and transport mechanisms). Note that we already have an artificially selected RNA that can facilitate diffusion (a passive RNA transporter of tryptophan: Janas et al. 2004).
5. What can be inferred from comtemporary enzyme homologies?
As mentioned previously, the oldest idea of enzymatization is the idea of retroevolution (Horowitz 1945). Suppose that the cell uses D for replication, and this compound is depleted, but C is abundantly present. Then, there will be selection for an enzyme catalysing the C→D reaction. The same evolutionary sequence is now repeated for C, which then triggers enzymatization of reaction B→C. Ultimately, one has a fully enzymatized pathway.
This scenario contrasts with that advocated by Jensen (1976), the so-called patchwork model. According to this idea, enzymes are recruited from anywhere in the network on the basis of similarity of the catalysed reaction mechanisms. A corollary to this is that enzymes are (or can be) less specific in the beginning, to be replaced later by more specific and efficient enzymes. This emerging division of labour (Maynard Smith & Szathmáry 1995) can pay off in terms of cell fitness, as shown in theory (Kacser & Beeby 1984) and simulation (Beeby & Kacser 1990).
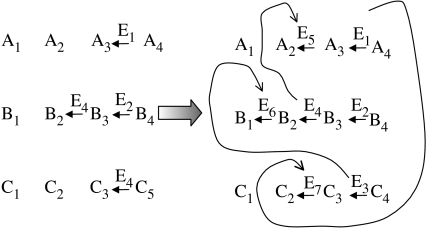
The two scenarios make different predictions, although both hinge on duplication and divergence of genes for enzymes. Retroevolution predicts that enzymes belonging to the same metabolic pathways should be homologous. The patchwork model, in agreement with its name, predicts that homologous enzymes should be scattered all over the network. Comparative evidence (such as Light & Kraulis 2004) is consonant with the patchwork model, and is taken to be against the retroevolution scenario. There are two serious objections against this conclusion, however. First, if one takes the view that enzymatic pathways were evolving in parallel, then retroevolution in the historical sense is compatible with enzyme recruitment as envisaged by the patchwork model. Each pathway could retroevolve while recruiting enzymes from anywhere in the network (i.e. from other pathways: figure 8). Second, comparative analysis of protein enzymes may say next to nothing about primitive pathway development. If the RNA world (Gilbert 1986) was metabolically complex (Benner et al. 1989), then the primary pathways would have become fixed in that world (Lazcano & Miller 1999). Thus, primary enzymatization by ribozymes may have been overwritten by a secondary wave of enzymatization by proteins. If this is true, the pattern inferred from proteins is at best analogous, rather than homologous, to the primary pattern. To make things worse, the conditions of primary and secondary enzymatization are different. Secondary enzymatization affects a network that is already enzymatic, thus the costs and benefits may be different from those of primary enzymatization. This is a serious methodological challenge, which also upgrades the value of insights one can gain from modelling.
Figure 8.
Simultaneous coevolution of metabolic pathways allows for reconciliation between retroevolution and the patchwork model. Straight arrows stand for enzymatized reactions; curved arrows indicate evolutionary homology (enzyme recruitment). Note that each pathway retroevolves.
6. Discussion
There are some additional modes of metabolic evolution. For example, Granick (1957) imagined a forward evolution of metabolic pathways, which would be the direct opposite of the retroevolution scenario. Such extension of core metabolism is of course possible. But as Lazcano & Miller (1999) rightly observed, a necessary precondition for this evolutionary mode is that each novel compound of the emerging pathway should be useful for the system, at least during the critical phase of fixation of the corresponding enzyme. Granick himself thought that such a process could have been the origin and evolution of photosynthesis.
A ‘semi-enzymatic’ contribution to the emerging pathways cannot be ruled out either. A novel enzyme catalysing a particular reaction distorts the dynamics of the network. Some by-products will now become by-products of this new enzymatic step; hence, some side reactions will be speeded up, albeit without direct selection. Some products of these reactions may turn out to be useful, so such side reactions may also later become enzymatized (Lazcano & Miller 1999). This goes in the direction of forward evolution.
Recently, Anet (2004) presented a pessimistic appraisal of the metabolism-first approaches. Whereas several of his observations hold, it must be said that at the moment, there is no unequivocal support for either this or the rival replication-first hypothesis. In the present paper, I have assumed a stage when some replicators are present that directly or indirectly can exert catalytic activity. Otherwise, my scenario is open for various metabolic scenarios. Even if metabolism came first, surely enzymatic metabolism did not. I am interested in the logic of enzymatization of whatever metabolism prevails.
The qualitative scenario outlined in this paper about progressive sequestration and metabolism–membrane coevolution should be illustrated by a quantitative model in the future. Hence, I call attention to an attempt (Pfeiffer et al. 2005) which (in modified form) is a probable ingredient of an appropriate approach. The authors build primarily on the patchwork (Jensen 1976) and the division of enzymatic labour by duplication (Kacser & Beeby 1984) models, but they also investigate the role of group transfer reactions in the emerging network. The structure of the evolved networks is rather similar to known ones in biochemistry, in that the network is nearly scale-free and hubs (similar to ATP) appear. It will be interesting to see what kind of networks appear in more detailed simulations. As Light et al. (2005) have shown, ‘preferential attachment’ seems to have worked in protein-catalysed metabolism of Escherichia coli: one explanation being that enzymes when they arise through duplication and divergence, at least, partly inherit their substrates. (Curiously, this logic favours the idea that homologous enzymes have very similar or identical substrates, but this is in contrast with the patchwork model where the reaction mechanism plays a stronger role.) Enzyme involved in nucleotide, amino acid and energy metabolism seems to be both ancient and highly connected.
A computer model investigating progressive sequestration should account for the evolution of membrane permeability and also investigate maybe non-trivial consequences of primary heterotrophy versus autotrophy.
Acknowledgments
This work was supported by the Hungarian Scientific Research Fund (OTKA T047245), the National Office for Research and Technology (NAP 2005/KCKHA005) and the COST D27 action (Prebiotic chemistry and early evolution). I am grateful to two anonymous referees for their very helpful comments. Figures have kindly been drawn by Sa´ndor Breznay.
Footnotes
One contribution of 13 to a Theme Issue ‘Towards the artificial cell’.
References
- Anet F.A.L. The place of metabolism in the origin of life. Curr. Opin. Chem. Biol. 2004;8:654–659. doi: 10.1016/j.cbpa.2004.10.005. doi:10.1016/j.cbpa.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Bauer E. Akadémiai Kiadó; Budapest, Hungary: 1967. Theoretical biology. [Google Scholar]
- Beeby R, Kacser H. Metabolic constraints in evolution. In: Maynard Smith J, Vida G, editors. Organizational constraints on the dynamics of evolution. Manchester University Press; Manchester, UK: 1990. pp. 1–22. [Google Scholar]
- Benner S.A, Ellington A.D, Tauer A. Modern metabolism as a palimpsest of the RNA world. Proc. Natl Acad. Sci USA. 1989;86:7054–7058. doi: 10.1073/pnas.86.18.7054. doi:10.1073/pnas.86.18.7054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czárán T, Szathmáry E. Coexistence of replicators in prebiotic evolution. In: Dieckmann U, Law R, Metz J.A.J, editors. The geometry of ecological interactions: simplifying spatial complexity. IIASA and Cambridge University Press; Cambridge, UK: 2000. pp. 116–134. [Google Scholar]
- Dauner M, Storni T, Sauer U. Bacillus subtilis metabolism and energetics in carbon-limited and excess-carbon chemostat culture. J. Bacteriol. 2001;183:7308–7317. doi: 10.1128/JB.183.24.7308-7317.2001. doi:10.1128/JB.183.24.7308-7317.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J.A, Lorsch J.R. Ribozyme catalysis: not different, just worse. Nat. Struct. Mol. Biol. 2005;12:395–402. doi: 10.1038/nsmb932. doi:10.1038/nsmb932 [DOI] [PubMed] [Google Scholar]
- Dyson F.J. Cambridge University Press; Cambridge, UK: 1985. Origins of life. [Google Scholar]
- Fernando C, Santos M, Szathmáry E. Evolutionary potential and requirements for minimal protocells. Top. Curr. Chem. 2005;259:167–211. [Google Scholar]
- Gánti T. Gondolat; Budapest, Hungary: 1971. Az élet princípiuma (The principle of life) [Google Scholar]
- Gánti T. Gondolat; Budapest, Hungary: 1978. Az élet princípiuma (The principle of life) [Google Scholar]
- Gánti T. Oxford University Press; Oxford, UK: 2003a. The principles of life. [Google Scholar]
- Gánti T. Kluwer; Boston, MA: 2003b. Chemoton theory I-II. [Google Scholar]
- Gil R, Silva F.J, Peretó J, Moya A. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 2004;68:518–537. doi: 10.1128/MMBR.68.3.518-537.2004. doi:10.1128/MMBR.68.3.518-537.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. The RNA world. Nature. 1986;319:618. doi:10.1038/319618a0 [Google Scholar]
- Granick S. Speculations on the origins of evolution of photosynthesis. Ann. N. Y. Acad. Sci. 1957;69:292–308. doi: 10.1111/j.1749-6632.1957.tb49665.x. doi:10.1111/j.1749-6632.1957.tb49665.x [DOI] [PubMed] [Google Scholar]
- Horowitz N.H. On the evolution of biochemical synthesis. Proc. Natl Acad. Sci. USA. 1945;31:153–157. doi: 10.1073/pnas.31.6.153. doi:10.1073/pnas.31.6.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas T, Janas T, Yarus M. A membrane transporter for tryptophan composed of RNA. RNA. 2004;10:1541–1549. doi: 10.1261/rna.7112704. doi:10.1261/rna.7112704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.A. Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. doi:10.1146/annurev.mi.30.100176.002205 [DOI] [PubMed] [Google Scholar]
- Kacser H, Beeby R. Evolution of catalytic proteins or on the origin of enzyme species by means of natural selection. J. Mol. Evol. 1984;20:38–51. doi: 10.1007/BF02101984. doi:10.1007/BF02101984 [DOI] [PubMed] [Google Scholar]
- Kauffman S.A. Autocatalytic sets of proteins. J. Theor. Biol. 1986;119:1–24. doi: 10.1016/s0022-5193(86)80047-9. doi:10.1016/S0022-5193(86)80047-9 [DOI] [PubMed] [Google Scholar]
- Lazcano A, Miller S.L. On the origin of metabolic pathways. J. Mol. Evol. 1999;49:424–431. doi:10.1007/PL00006565 [PubMed] [Google Scholar]
- Light S, Kraulis P. Network analyis of metabolic enzyme evolution in Escherichia coli. BMC Bioinformatics. 2004;5:15. doi: 10.1186/1471-2105-5-15. http://www.biomedcentral.com/1471-2105/5/15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light S, Kraulis P, Elofsson A. Preferntial attachment in the evolution of metabolic networks. BMC Genomics. 2005;6:159. doi: 10.1186/1471-2164-6-159. doi:10.1186/1471-2164-6-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Szathmáry E. Freeman & Co; Oxford, UK: 1995. The major transitions in evolution. [Google Scholar]
- Oparin A.I. Academic Press; New York, NY: 1961. Life, its nature, origin and development. [Google Scholar]
- Pfeiffer T, Soyer O.S, Bonhoeffer S. The evolution of connectivity in metabolic networks. PLoS Biol. 2005;3:e228. doi: 10.1371/journal.pbio.0030228. doi:10.1371/journal.pbio.0030228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó P, Scheuring I, Czárán T, Szathmáry E. In silico simulations reveal that replicators with limited dispersal evolve towards higher efficiency and fidelity. Nature. 2002;420:360–363. doi: 10.1038/nature01187. [DOI] [PubMed] [Google Scholar]
- Szathmáry E. A romlás virágai (Flowers of doom) In: Koch S, editor. A tökéletlenség és korlátosság dicsérete (In praise of imperfection and limitedness) Gondolat; Budapest, Hungary: 1989. pp. 215–274. [Google Scholar]
- Szathmáry E. Life: in search of the simplest cell. Nature. 2005;433:469–470. doi: 10.1038/433469a. doi:10.1038/433469a [DOI] [PubMed] [Google Scholar]
- Szostak J.W, Bartel D.P, Luisi P.L. Synthesizing life. Nature. 2001;409:387–390. doi: 10.1038/35053176. doi:10.1038/35053176 [DOI] [PubMed] [Google Scholar]
- Varela F.G. North-Holland; New York, NY: 1979. Principles of biological autonomy. [Google Scholar]
- Varela F.G, Maturana H.R, Uribe R. Autopoiesis: the organization of living systems, its characterization and a model. BioSystems. 1974;5:187–196. doi: 10.1016/0303-2647(74)90031-8. doi:10.1016/0303-2647(74)90031-8 [DOI] [PubMed] [Google Scholar]
- Wächtershäuser G. Before enzymes and templates: theory of surface metabolism. Microbiol. Rev. 1988;52:452–484. doi: 10.1128/mr.52.4.452-484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wächtershäuser G. Groundworks for an evolutionary biochemistry: the iron–sulphur world. Prog. Biophys. Mol. Biol. 1992;58:85–201. doi: 10.1016/0079-6107(92)90022-x. doi:10.1016/0079-6107(92)90022-X [DOI] [PubMed] [Google Scholar]
- Zum Gahr K.H. Tribologie: Reibung—Verschleiss—Schmierung. Naturwissenschaften. 1985;72:260–267. doi: 10.1007/BF00448687. doi:10.1007/BF00448687 [DOI] [PubMed] [Google Scholar]