Abstract
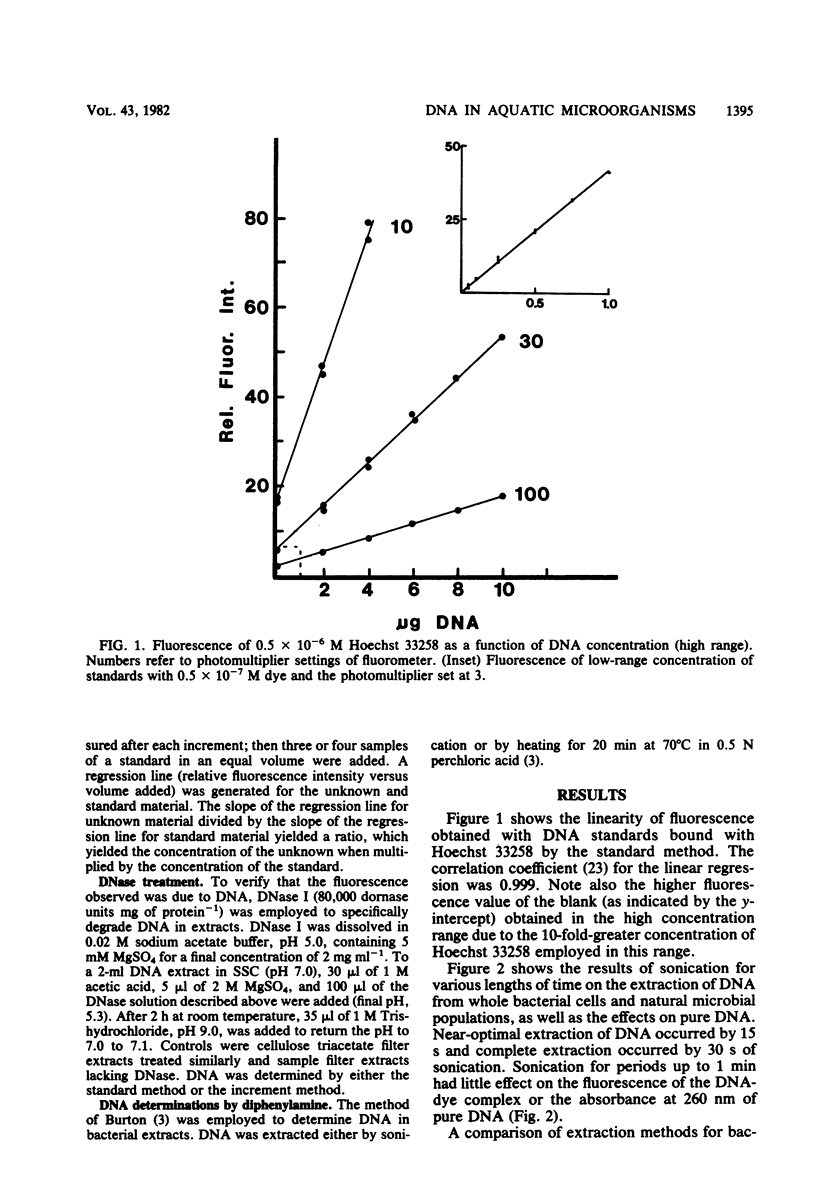
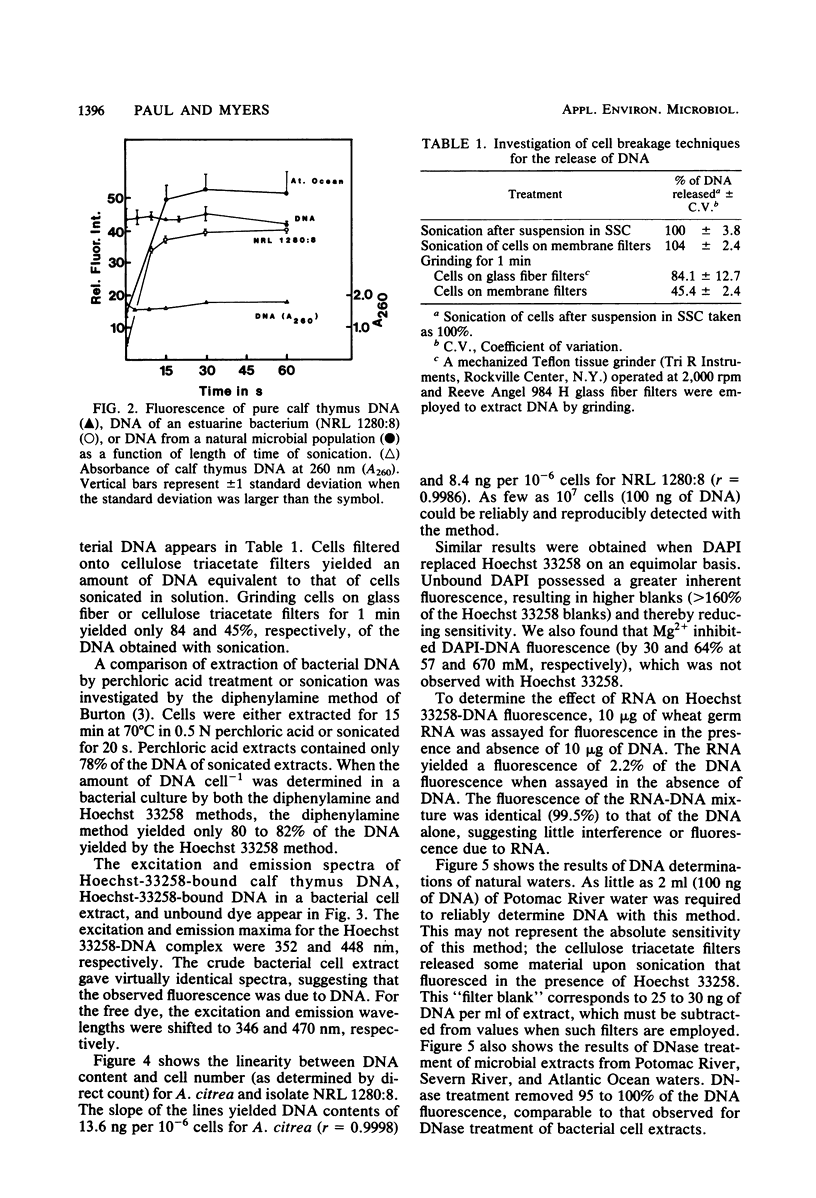
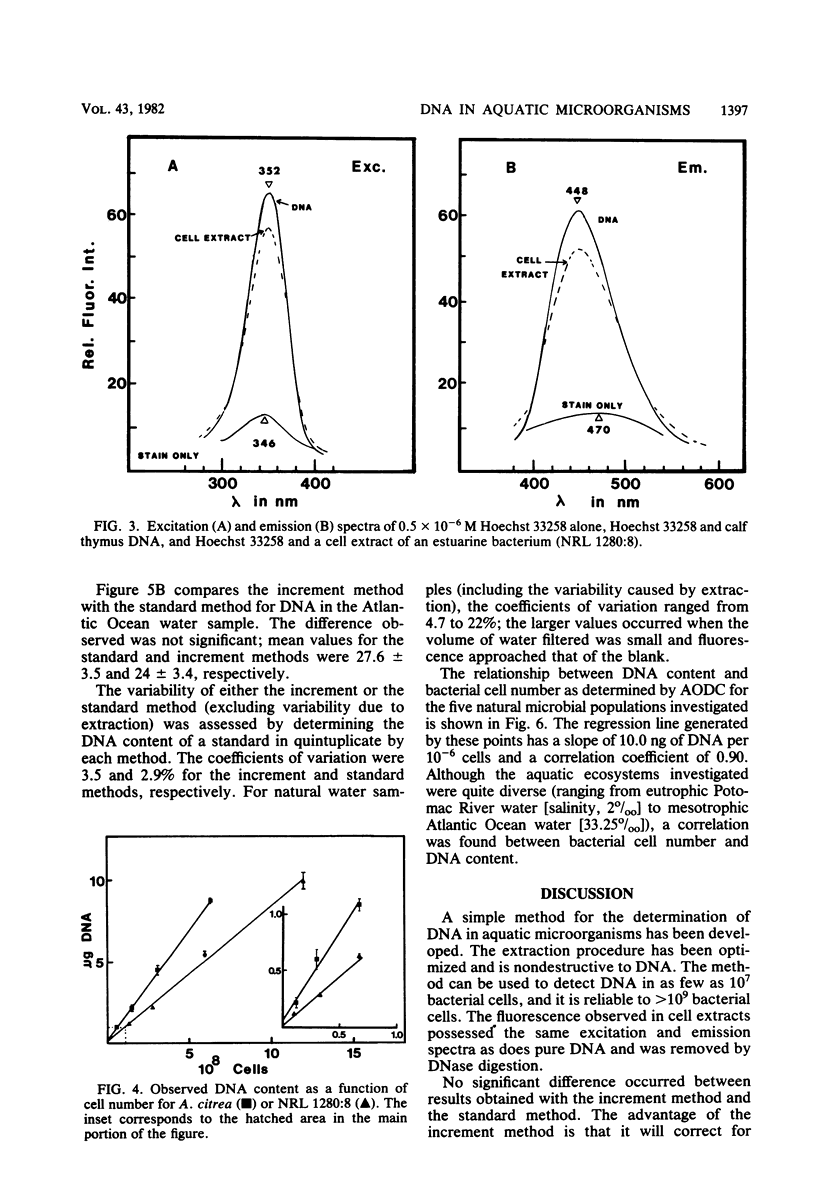
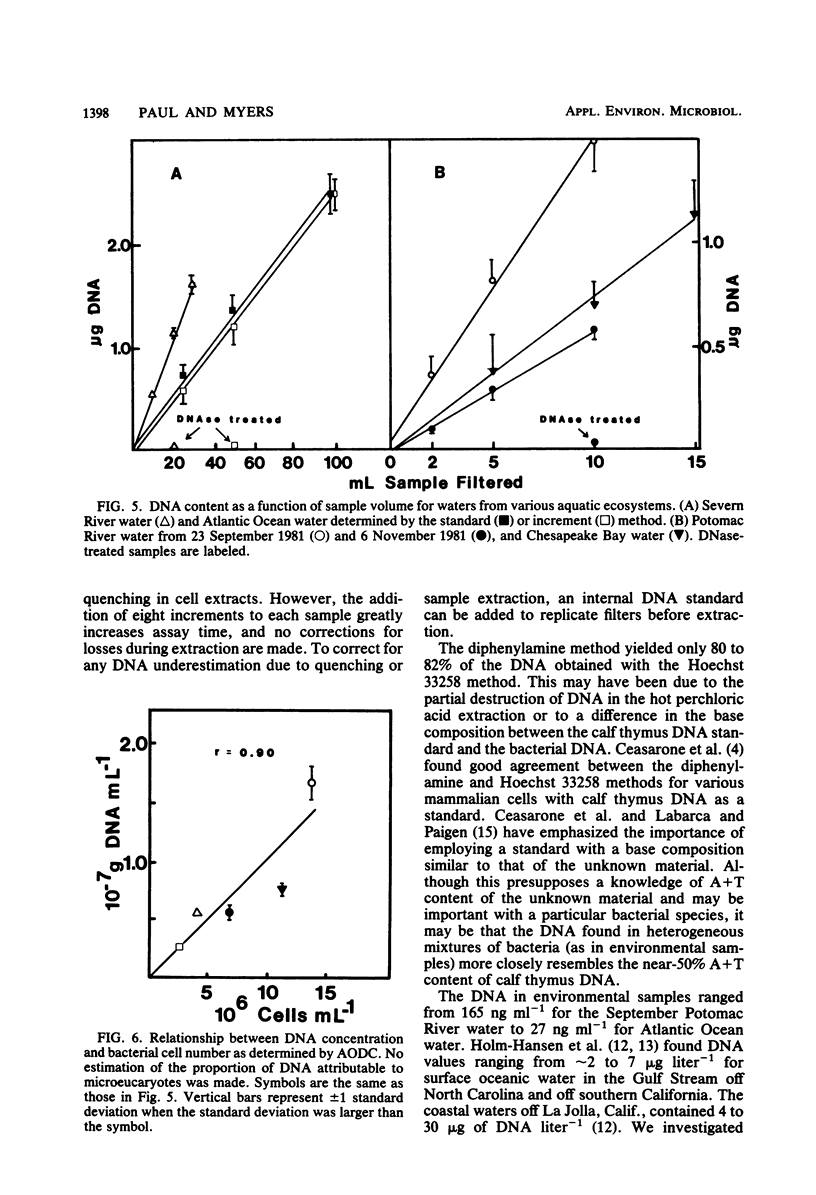
A method for the determination of microbial DNA in aquatic environments by the use of Hoechst 33258 has been developed. With unsophisticated instrumentation and simple extraction procedures, it is possible to detect from 0.05 to 10 μg of DNA in bacterial cultures or natural water samples. The method is specific for DNA; DNase I treatment of extracts of natural microbial populations removed 95 to 100% of the observed fluorescence. DNA content ranged from 165 ng ml−1 for relatively eutrophic Potomac River water to 27 ng ml−1 for coastal Atlantic Ocean water and was correlated to an acridine orange direct count (r = 0.90).
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barth C. A., Willershausen B. S. Improvement of DNA quantitation following proteolytic extraction. Anal Biochem. 1978 Oct 1;90(1):167–173. doi: 10.1016/0003-2697(78)90020-9. [DOI] [PubMed] [Google Scholar]
- Brunk C. F., Jones K. C., James T. W. Assay for nanogram quantities of DNA in cellular homogenates. Anal Biochem. 1979 Jan 15;92(2):497–500. doi: 10.1016/0003-2697(79)90690-0. [DOI] [PubMed] [Google Scholar]
- Cesarone C. F., Bolognesi C., Santi L. Improved microfluorometric DNA determination in biological material using 33258 Hoechst. Anal Biochem. 1979 Nov 15;100(1):188–197. doi: 10.1016/0003-2697(79)90131-3. [DOI] [PubMed] [Google Scholar]
- Comings D. E. Mechanisms of chromosome banding. VIII. Hoechst 33258-DNA interaction. Chromosoma. 1975 Oct 14;52(3):229–243. doi: 10.1007/BF00332113. [DOI] [PubMed] [Google Scholar]
- Dalbow D. G., Bartuska B. M. Modification of the quantitative DNA assay using adriamycin. Anal Biochem. 1979 Jun;95(2):559–562. doi: 10.1016/0003-2697(79)90772-3. [DOI] [PubMed] [Google Scholar]
- Erickson L. C., Osieka R., Sharkey N. A., Kohn K. W. Measurement of DNA damage in unlabeled mammalian cells analyzed by alkaline elution and a fluorometric DNA assay. Anal Biochem. 1980 Jul 15;106(1):169–174. doi: 10.1016/0003-2697(80)90133-5. [DOI] [PubMed] [Google Scholar]
- Fiszer-Szafarz B., Szafarz D., Guevara de Murillo A. A general, fast, and sensitive micromethod for DNA determination application to rat and mouse liver, rat hepatoma, human leukocytes, chicken fibroblasts, and yeast cells. Anal Biochem. 1981 Jan 1;110(1):165–170. doi: 10.1016/0003-2697(81)90130-5. [DOI] [PubMed] [Google Scholar]
- Groyer A., Robel P. DNA measurement by mithramycin fluorescence in chromatin solubilized by heparin. Anal Biochem. 1980 Jul 15;106(1):262–268. doi: 10.1016/0003-2697(80)90146-3. [DOI] [PubMed] [Google Scholar]
- Hill B. T. The use of anti-tumour antibiotics for simple quantitative assays for DNA. Anal Biochem. 1976 Feb;70(2):635–638. doi: 10.1016/0003-2697(76)90493-0. [DOI] [PubMed] [Google Scholar]
- Hobbie J. E., Daley R. J., Jasper S. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977 May;33(5):1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KISSANE J. M., ROBINS E. The fluorometric measurement of deoxyribonucleic acid in animal tissues with special reference to the central nervous system. J Biol Chem. 1958 Jul;233(1):184–188. [PubMed] [Google Scholar]
- Labarca C., Paigen K. A simple, rapid, and sensitive DNA assay procedure. Anal Biochem. 1980 Mar 1;102(2):344–352. doi: 10.1016/0003-2697(80)90165-7. [DOI] [PubMed] [Google Scholar]
- Latt S. A., Stetten G. Spectral studies on 33258 Hoechst and related bisbenzimidazole dyes useful for fluorescent detection of deoxyribonucleic acid synthesis. J Histochem Cytochem. 1976 Jan;24(1):24–33. doi: 10.1177/24.1.943439. [DOI] [PubMed] [Google Scholar]
- Le Pecq J. B., Paoletti C. A new fluorometric method for RNA and DNA determination. Anal Biochem. 1966 Oct;17(1):100–107. doi: 10.1016/0003-2697(66)90012-1. [DOI] [PubMed] [Google Scholar]
- Markovits J., Roques B. P., Le Pecq J. B. Ethidium dimer: a new reagent for the fluorimetric determination of nucleic acids. Anal Biochem. 1979 Apr 15;94(2):259–264. doi: 10.1016/0003-2697(79)90357-9. [DOI] [PubMed] [Google Scholar]
- Munro H. N. The determination of nucleic acids. Methods Biochem Anal. 1966;14:113–176. doi: 10.1002/9780470110324.ch5. [DOI] [PubMed] [Google Scholar]
- Paul J. H. Use of hoechst dyes 33258 and 33342 for enumeration of attached and planktonic bacteria. Appl Environ Microbiol. 1982 Apr;43(4):939–944. doi: 10.1128/aem.43.4.939-944.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W. C. Simplified isolation and quantitation of cytoplasmic DNA from rat liver. Anal Biochem. 1980 Apr;103(2):413–418. doi: 10.1016/0003-2697(80)90632-6. [DOI] [PubMed] [Google Scholar]
- Weisblum B., Haenssler E. Fluorometric properties of the bibenzimidazole derivative Hoechst 33258, a fluorescent probe specific for AT concentration in chromosomal DNA. Chromosoma. 1974;46(3):255–260. doi: 10.1007/BF00284881. [DOI] [PubMed] [Google Scholar]
- Williams S. K., Sasaki A. W., Matthews M. A., Wagner R. C. Quantitative determination of deoxyribonucleic acid from cells collected on filters. Anal Biochem. 1980 Sep 1;107(1):17–20. doi: 10.1016/0003-2697(80)90485-6. [DOI] [PubMed] [Google Scholar]



