Abstract
The signal transduction of the phytohormone cytokinin is mediated by a multistep histidine-to-aspartate phosphorelay system. One component of this system are B-type response regulators, transcription factors mediating at least part of the response to cytokinin. In planta functional analysis of this family is hampered by the high level of functional redundancy of its 11 members. We generated a dominant repressor version of the Arabidopsis (Arabidopsis thaliana) response regulator ARR1 (ARR1-SRDX) using chimeric repressor silencing technology in order to study the extent of the contribution of B-type response regulators to cytokinin activities. In a protoplast test system, ARR1-SRDX suppressed ARR6:β-glucuronidase reporter gene activation by different B-type ARRs. 35S:ARR1-SRDX transgenic Arabidopsis plants showed phenotypic changes reminiscent of plants with a reduced cytokinin status, such as a strongly reduced leaf size, an enhanced root system, and larger seeds. Several bioassays showed that 35S:ARR1-SRDX plants have an increased resistance toward cytokinin. The rapid induction of a large part of the cytokinin response genes was dampened. The transcript levels of more than 500 genes were more than 2.5-fold reduced in 35S:ARR1-SRDX transgenic seedlings, suggesting a broad function of B-type ARRs. Collectively, the suppression of pleiotropic cytokinin activities by a dominant repressor version of a B-type ARR indicates that this protein family is involved in mediating most, if not all, of the cytokinin activities in Arabidopsis. In addition, a role for B-type ARRs in mediating cross talk with other pathways is supported by the resistance of 35S:ARR1-SRDX seeds to phytochrome B-mediated inhibition of germination by far-red light. This study demonstrates the usefulness of chimeric repressor silencing technology to overcome redundancy in transcription factor families for functional studies.
The plant hormone cytokinin is involved in many developmental processes and plays a critical role in numerous physiological responses to changes in the environment (Mok and Mok, 2001). In recent years, significant progress has been made toward the understanding of how the cytokinin signal is perceived and transduced (Heyl and Schmülling, 2003; Kakimoto, 2003; Mizuno, 2004; Heyl et al., 2006; Hwang and Sakakibara, 2006; Müller and Sheen, 2007). In the current model, which has been developed mainly in Arabidopsis (Arabidopsis thaliana), the hormone is perceived by membrane-bound hybrid His-kinase receptors, which autophosphorylate upon binding of the hormone ligand. After transphosphorylation within the receptor, the phosphate group is transferred to His phosphotransfer proteins, which subsequently locate to the nucleus, where they activate B-type response regulators (ARRs) via phosphorylation. These transcription factors regulate the transcription of their target genes, one group of which codes for A-type response regulators. A negative feedback on the cytokinin signaling pathway was shown to be mediated by members of this protein class (Hwang and Sheen, 2001; To et al., 2004).
Arabidopsis encodes 11 B-type ARRs. They possess, in addition to the response regulator domain, a MYB-class DNA-binding domain (Riechmann et al., 2000). The DNA-binding domains share a high degree of sequence conservation, in particular in the nine amino acids that were identified in ARR10 to be most likely in direct contact with the DNA (Hosoda et al., 2002). Several B-type ARRs have been shown to bind to the same or very similar sequence motifs (Sakai et al., 2000; Hosoda et al., 2002; Imamura et al., 2003). B-type ARRs regulate the transcription of their target genes in response to cytokinin treatment (Sakai et al., 2000; Hwang and Sheen, 2001; Lohrmann et al., 2001; Imamura et al., 2003). Reverse transcription-PCR, microarray, and promoter-GUS fusion experiments have demonstrated that the members of the B-type ARR family have large and overlapping expression domains (Mason et al., 2004; Tajima et al., 2004; Zimmermann et al., 2004; summarized by Heyl et al., 2006). The analysis of B-type ARR mutants has revealed their involvement in cytokinin signaling but also a high degree of functional redundancy (Sakai et al., 2001; Horák et al., 2003; Hass et al., 2004; Mason et al., 2005). Loss-of-function mutants of single B-type ARR genes (ARR1, ARR2, ARR10, ARR11, and ARR18) caused no strong phenotypic alterations. However, different cytokinin response assays showed increasing cytokinin resistance for higher order mutants (Mason et al., 2005; Yokoyama et al., 2007). Surprisingly, besides quantitative changes of the root system and a defect of vascular development in the arr1 arr10 arr12 triple mutant (Mason et al., 2005; Yokoyama et al., 2007), no strong morphological alterations were described. This indicates that the degree of redundancy among B-type ARRs is very high and/or that other transcriptional regulators compensate for the loss of B-type ARRs, such as the cytokinin response factors of the ethylene response factor (ERF) transcription factor family (Rashotte et al., 2006).
In order to explore the functions of B-type response regulators, we took advantage of the chimeric repressor silencing technology (CRES-T; Hiratsu et al., 2003). This technology has been developed to study the consequences of silencing of the target genes of single transcription factors and has also been used to overcome the experimental limitations caused by functional redundancy of transcription factor families. In 2001, Ohta and colleagues mapped a repression motif of transcriptional repressors of the class II ERFs (Ohta et al., 2001). This ERF-associated amphiphilic repression motif or variations of it are found in numerous plant transcriptional repressors (Ohta et al., 2001; Tiwari et al., 2004; Kazan, 2006). Further improvement resulted in the SRDX motif (Hiratsu et al., 2003). Fusion of this motif to transcriptional activators converts them into dominant repressors, even in the presence of the original activator domain (Hiratsu et al., 2003). Interestingly, these dominant repressors may repress not only the transcription of their own target genes but also the expression of target genes of other members of their respective gene families and thus overcome functional redundancy (Hiratsu et al., 2003; Mitsuda et al., 2007).
In this study, the SRDX motif was fused to the C terminal end of ARR1 and expressed under the control of a 35S promoter in wild-type Arabidopsis plants. The resulting transgenic plants showed a cytokinin deficiency phenotype, including reduced shoot and enhanced root growth, altered reproductive organs, and increased resistance to cytokinin. Molecular analysis revealed the attenuation of early transcriptional responses to cytokinin. The implications for the function of this class of transcription factors are discussed.
RESULTS
ARR1-SRDX Suppresses the Cytokinin-Dependent Induction of a Cytokinin Primary Response Gene in Planta
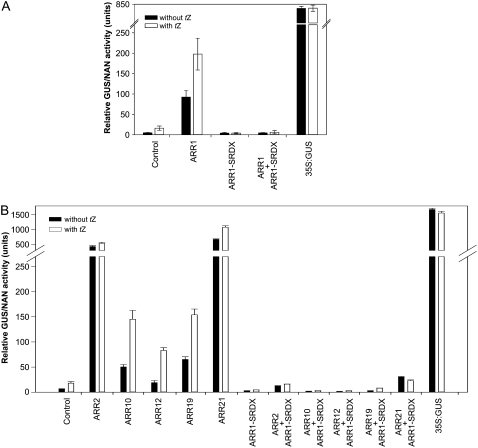
A dominant repressor for the B-type ARRs was generated by introducing a 36-bp DNA sequence (5′-CTGGATCTGGATCTGGAACTGCGCCTGGGCTTTGCG-3′) encoding the SRDX domain (LDLDLELRLGFA; Hiratsu et al., 2003) replacing the last 20 nucleotides 5′ of the ARR1 stop codon (Supplemental Fig. S1). ARR1 was chosen as it is the best characterized B-type response regulator in Arabidopsis (Sakai et al., 2000, 2001, Mason et al., 2004, 2005; Taniguchi et al., 2007). The in planta effect of ARR1-SRDX was first examined in a protoplast transactivation assay (Ehlert et al., 2006) using a 1,000-bp promoter fragment upstream of the translational start of ARR6, a primary cytokinin response gene (Hwang and Sheen, 2001), fused to the GUS reporter gene. The addition of cytokinin resulted in a more than 3-fold induction of the reporter gene expression compared with the noninduced condition (Fig. 1A). Cotransfection with ARR1 under the control of a 35S promoter led to a strong increase in GUS activity even in the absence of cytokinin. The addition of cytokinin caused a further 2-fold increase of the GUS activity, indicating that ARR1 mediates the cytokinin response in this assay. In contrast, the expression of ARR1-SRDX under the control of a 35S promoter effectively suppressed the cytokinin induction of GUS expression. Furthermore, ARR1-SRDX completely abolished the ARR1-caused expression of the reporter gene in the absence and presence of cytokinin (Fig. 1A). The expression of the GUS reporter gene under the control of a 35S promoter was not cytokinin inducible. In the next step, we tested in the same assay system whether other B-type ARRs activate the transcription of the ARR6:GUS reporter and whether this activation could be suppressed by ARR1-SRDX. For these experiments, ARR2 was chosen as this is the B-type ARR most closely related to ARR1. ARR10 and ARR12 were tested as well, as both have been shown to be at least partly functionally redundant to ARR1 (Yokoyama et al., 2007; Ishida et al., 2008). Furthermore, ARR19 and ARR21 were included in the analysis as representatives of the two pairs of more distantly related B-type ARRs. All of these B-type ARRs increased ARR6:GUS expression without cytokinin to a different extent (Fig. 1B). The addition of cytokinin in most cases increased the reporter gene activity further. The coexpression of ARR1-SRDX effectively repressed reporter gene activation by all of the B-type ARRs in the absence and presence of cytokinin (Fig. 1B). These results clearly demonstrate the potential of ARR1-SRDX as a dominant repressor of the primary transcriptional response to cytokinin in planta.
Figure 1.
35S:ARR1-SRDX represses the transactivation capacity of B-type ARRs in a protoplast transactivation assay. A, 35S:ARR1-SRDX represses the activation of an ARR6:GUS reporter gene by ARR1. B, 35S:ARR1-SRDX represses the activation of an ARR6:GUS reporter gene by several B-type ARRs. The activation of the ARR6:GUS reporter gene was measured without and 16 h after the addition of 500 nm t-zeatin (tZ). The ARR6:GUS reporter construct and a 35S:GUS construct without any effector plasmid were used as controls. Protoplasts were cotransfected with the ARR6:GUS reporter and an effector plasmid expressing ARR1, ARR2, ARR10, ARR12, ARR19, ARR21, or ARR1-SRDX or two effector plasmids expressing the respective B-type ARR and ARR1-SRDX. Variations in transformation efficiencies were normalized using a 35S:NAN reporter construct. The mean values and sd of four independent transfection assays were calculated and shown as relative GUS/NAN activity units.
Transgenic Arabidopsis Plants Expressing a Dominant ARR1-SRDX Repressor
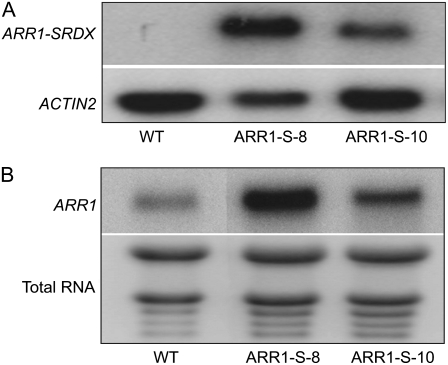
Next, we established transgenic lines by transforming the 35S:ARR1-SRDX gene into wild-type Columbia (Col-0) plants. Because B-type ARR expression was detected in virtually all tissues investigated (Heyl et al., 2006) and we aimed to study the effects of the repression of as many B-type ARRs as possible (not just ARR1 in the ARR1 expression domain), we purposefully chose the 35S promoter to drive gene expression. The 35S promoter has been previously used successfully in combination with CRES-T (Koyama et al., 2007). More than 10 independent transgenic lines with a similar phenotype, but distinct from the wild type, were recovered (Supplemental Fig. S2). Two of those lines, ARR1-S-8 and ARR1-S-10, which showed a characteristic phenotype with different expressivity, were characterized in more detail. Experiments were carried out with homozygote and phenotypically uniform progeny. Northern-blot analysis using a SRDX-specific probe showed the expression of the ARR1-SRDX transcript in both lines (Fig. 2A). The steady-state transcript level was higher in ARR1-S-8 compared with ARR1-S-10 (Fig. 2A). In order to check whether the transcript level of the ARR1 gene itself was altered in the transgenic lines, we compared its level with the wild type using a probe that specifically recognizes the wild-type ARR1 but not ARR1-SRDX transcripts. This analysis revealed an increased ARR1 transcript level in both 35S:ARR1-SRDX transgenic lines, indicating a feedback regulation of ARR1 (Fig. 2B). This increase in transcript level was specific for ARR1 and did not affect four other B-type ARR genes, which were detectable in our microarray experiments (see below) and showed similar transcript levels in the transgenic lines and the wild type.
Figure 2.
Expression of the 35S:ARR1-SRDX gene fusion and of the ARR1 gene. A, Total RNA samples were isolated from 3-week-old plants, and the transcript was detected by northern-blot hybridization using a probe specific for the ARR1-SRDX gene fusion. The ACTIN2 transcript was used as a loading control. WT, Wild-type Col-0; ARR1-S-8 and ARR1-S-10 are independent lines of 35S:ARR1-SRDX transgenic plants. B, Analysis of ARR1 transcript levels. Total RNA was isolated from 5-d-old wild-type and 35S:ARR1-SRDX transgenic seedlings. Northern blots were hybridized with a probe specific for that C-terminal part of ARR1 that had been deleted during the construction of the 35S:ARR1-SRDX gene and thus hybridizes only with the native ARR1 and not with the ARR1-SRDX fusion transcript. Total RNA was used as a loading control.
35S:ARR1-SRDX Transgenic Plants Display a Pleiotropic Shoot Phenotype
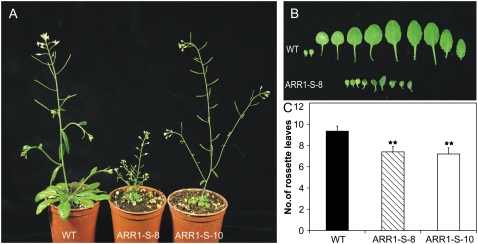
Transgenic plants expressing the 35S:ARR1-SRDX construct displayed a strong pleiotropic shoot phenotype, with a higher expressivity in line ARR1-S-8 than in line ARR1-S-10 (Fig. 3A). Plants were generally smaller, and after flowering numerous individuals showed to a varying degree enhanced branching of the shoot. The leaves of the dominant repressors were strongly reduced in both size and number compared with wild-type plants. In the stronger expressing line ARR1-S-8, the true leaves were only about the size of the cotyledons in the wild type (Fig. 3B). Twenty days after germination (DAG), the wild-type plant had on average nine rosette leaves, while the ARR1-SRDX-expressing plants had only seven leaves at that time point (Fig. 3C).
Figure 3.
Shoot phenotype of 35S:ARR1-SRDX transgenic plants. A, Shoot phenotype of transgenic 35S:ARR1-SRDX plants at 35 DAG compared with the wild type. B, Leaf phenotypes of a plant of line ARR1-S-8 and of a wild-type plant at 20 DAG. The leaves on the left are the cotyledons, followed by the true leaves in order of their formation. C, 35S:ARR1-SRDX transgenic plants have formed fewer leaves than the wild type at the onset of flowering. The total number of rosette leaves was counted at 19 DAG. Error bars represent sd (n ≥ 20). Pairwise Student's t test was used to compare values with the wild-type. **, 0.001 < P < 0.01. ARR1-S-8 and ARR1-S-10 are two independent lines of 35S:ARR1-SRDX transgenic plants. [See online article for color version of this figure.]
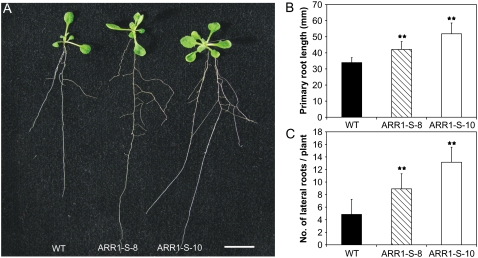
35S:ARR1-SRDX Plants Develop an Enlarged Root System
In contrast to the reduced shoot size, 35S:ARR1-SRDX transgenic plants showed a generally enhanced root system compared with the wild type (Fig. 4A). While the ARR1-S-8 plants had only a slightly longer primary root than the wild type at 10 DAG, the primary root of line ARR1-S-10 was more than 30% longer (34.9 ± 3.1 mm in the wild type compared with 51.9 ± 6.6 mm in line ARR1-S-10; Fig. 4B). The difference between the transgenic lines and the wild type was also marked in the number of lateral roots. At 10 DAG, plants of line ARR1-S-8 had developed about twice as many lateral roots as the wild type and line ARR1-S-10 had about three times more (13.2 ± 1.9 lateral roots compared with 4.8 ± 1.1 lateral roots in the wild type; Fig. 4C).
Figure 4.
Root phenotype of 35S:ARR1-SRDX transgenic plants. A, Transgenic plants show longer and more branched roots than wild-type plants. Seedlings were grown vertically on MS medium plates. The photograph was taken at 10 DAG. Bar = 1 cm. B, 35S:ARR1-SRDX transgenic plants produce longer primary roots than wild-type plants. The length of the primary root was determined at 10 DAG. C, 35S:ARR1-SRDX transgenic plants produce more lateral roots compared with the wild type. The number of emerged lateral roots was determined at 10 DAG. Results shown in B and C represent means from at least three independent replicates for each line. Error bars represent sd (n ≥ 15). Pairwise Student's t test was used to compare values with the wild type. **, 0.001 < P < 0.01. ARR1-S-8 and ARR1-S-10 are two independent lines of 35S:ARR1-SRDX transgenic plants. [See online article for color version of this figure.]
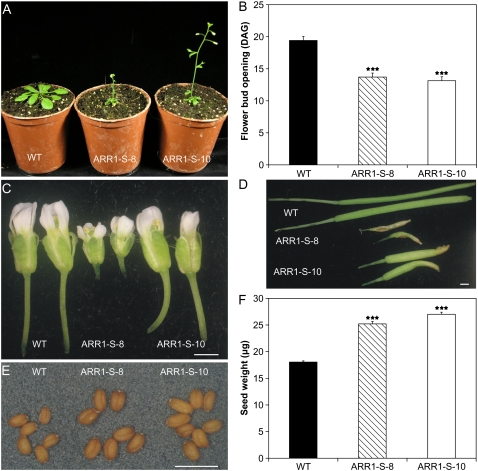
Reproductive Development of 35S:ARR1-SRDX Plants Is Altered
The 35S:ARR1-SRDX transgenic plants flowered earlier than wild-type plants (Fig. 5A). In the wild type, the first flower buds opened at 19 DAG. In contrast, both transgenic lines flowered at around 14 DAG (Fig. 5B). In the 35S:ARR1-SRDX transgenic plants, all reproductive organs were reduced in size. This was more pronounced in line ARR1-S-8, in which the flowers were only half the size of the wild-type flowers (Fig. 5C). The smaller flowers of the 35S:ARR1-SRDX plants gave rise to smaller siliques. The length of the siliques of line ARR1-S-8 was about 30% of the length of the wild-type siliques. The shape differed also from the wild type, as it was twisted and crooked (Fig. 5D). The phenotype of line ARR1-S-10 was weaker, as the reduction of the silique size was not as dramatic as in line ARR1-S-8, and the shape resembled more that of the wild type (Fig. 5D). While the number of seeds obtained by selfing was considerably lower in the 35S:ARR1-SRDX plants (data not shown), the seeds themselves showed increased size (Fig. 5E). Their weight was 40% and 50% higher in the lines ARR1-S-8 and ARR1-S-10, respectively (Fig. 5F).
Figure 5.
Reproductive development of 35S:ARR1-SRDX transgenic plants. A, Flower induction occurs earlier in 35S:ARR1-SRDX transgenic plants compared with wild-type plants. The plants were grown under long-day conditions and photographed at 19 DAG. B, Quantitative analysis of the early-flowering phenotype of 35S:ARR1-SRDX transgenic plants. The graph shows the time (DAG) of flower bud opening in transgenic plants compared with the wild type. C, Flower morphology of 35S:ARR1-SRDX transgenic plants compared with the wild type. D, Siliques of 35S:ARR1-SRDX transgenic plants compared with wild-type plants. E, Seeds of 35S:ARR1-SRDX transgenic lines compared with the wild type. F, The seeds of 35S:ARR1-SRDX transgenic plants have increased weight. Seed weight was determined by weighing 10 pools of 200 seeds for each line. Error bars represent sd. Bars in C to E = 1.0 mm. Pairwise Student's t test was used to compare values with the wild type. ***, P < 0.001. ARR1-S-8 and ARR1-S-10 are two independent lines of 35S:ARR1-SRDX transgenic plants. [See online article for color version of this figure.]
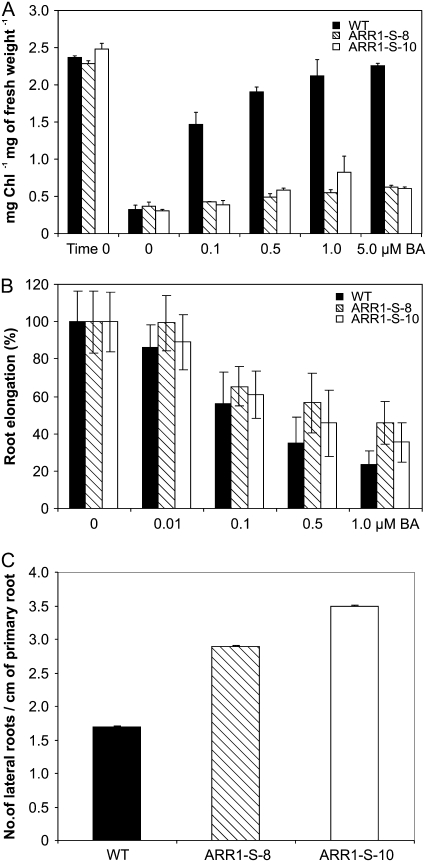
The Cytokinin Response of 35S:ARR1-SRDX Plants Is Altered in Shoots and Roots
As the phenotype of the 35S:ARR1-SRDX plants is reminiscent of plants with reduced cytokinin signaling (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006), we carried out several cytokinin sensitivity assays.
Cytokinin is known to delay the onset of leaf senescence and to increase the chlorophyll retention in detached leaves incubated in the dark (Richmond and Lang, 1957; Riefler et al., 2006). Detached wild-type leaves kept in the dark for 10 d lost more than 80% of their chlorophyll compared with fresh leaves (Fig. 6A). The addition of increasing amounts of cytokinin reversed this effect. In wild-type leaves, 0.1 μm 6-benzyladenine (BA) strongly increased chlorophyll retention, and at 5 μm BA the chlorophyll level of dark-incubated detached leaves was similar to that in fresh leaves. In contrast, while in both 35S:ARR1-SRDX lines the loss of chlorophyll under dark conditions was similar to that in the wild type, the addition of cytokinin to the medium caused only a minor increase in chlorophyll retention, even at the highest concentration (Fig. 6A). This indicates that the cytokinin sensitivity in the leaves had been almost completely lost.
Figure 6.
35S:ARR1-SRDX plants are less sensitive to cytokinin. A, Chlorophyll retention in dark-incubated leaves by cytokinin. Fully expanded leaves were excised from 24-d-old plants and floated for 10 d in the dark on water supplemented with various concentrations of cytokinin before the chlorophyll concentration was determined. Time 0 indicates the chlorophyll concentration at the beginning of the experiment. B, Cytokinin sensitivity of primary root elongation. Root elongation was measured for each line between 4 and 9 DAG. The root elongation of each line is expressed as a percentage of its DMSO control. The original data for the transgenic lines were significantly different from the wild-type data, with 0.001 < P < 0.01 as calculated by pairwise Student's t test. C, Cytokinin sensitivity of lateral root formation. The number of lateral roots was determined following growth on vertical plates for 9 DAG. The number of lateral roots between the wild type and the transgenic lines was significantly different (P < 0.01). Error bars represent sd (n ≥ 15). ARR1-S-8 and ARR1-S-10 are two independent lines of 35S:ARR1-SRDX transgenic plants.
To investigate the cytokinin response in roots, seedlings of the wild type and the 35S:ARR1-SRDX lines were grown on medium containing increasing amounts of cytokinin. On control medium without cytokinin, the transgenic seedlings developed a longer primary root compared with the wild type (Fig. 4A). The relative difference in root length increased with increasing cytokinin concentrations (Fig. 6B). Seedlings of line ARR1-S-8 displayed a higher resistance to cytokinin than line ARR1-S-10. A 50% inhibition of root elongation was achieved by an approximately 5-fold higher cytokinin concentration. However, the sensitivity to cytokinin, while being clearly reduced, was not completely eliminated in either ARR1-SRDX line (Fig. 6B). Subsequently, we tested whether lateral root formation was altered in the presence of the hormone. In all seedlings, the addition of cytokinin to the medium led to a dramatic decrease in the number of lateral roots (Fig. 6C). But whereas the lateral root formation of the wild type was almost totally repressed at 0.01 μm BA, lateral root formation of both 35S:ARR1-SRDX lines was only halved at this cytokinin concentration. At 0.1 μm BA, the wild type was unable to produce lateral roots, while the dominant repressor lines still showed a significant number (Fig. 6C).
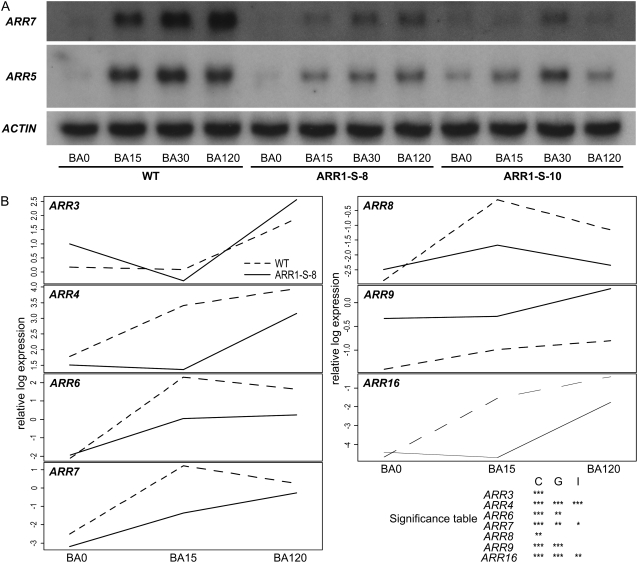
35S:ARR1-SRDX Plants Show a Reduced Induction of Cytokinin Response Genes
The phenotypic changes and cytokinin bioassays described above report altered long-term responses to the hormone. To analyze whether rapid cytokinin responses were also altered in the 35S:ARR1-SRDX transgenic lines, the expression of two known primary cytokinin response genes was tested. The transcript levels of ARR5 and ARR7, which both encode A-type ARRs, are direct targets of the B-type ARRs and rapidly induced by cytokinin (D'Agostino et al., 2000; Romanov et al., 2002; To et al., 2004). Both genes were rapidly and strongly induced in wild-type seedlings following the application of cytokinin. This induction was strongly diminished in both transgenic lines (Fig. 7A).
Figure 7.
35S:ARR1-SRDX gene expression dampens the early cytokinin response. A, Northern-blot analysis of ARR5 and ARR7 transcript levels. Total RNA was isolated from 5-d-old wild-type and 35S:ARR1-SRDX seedlings before treatment and after 15, 30, and 120 min of treatment with 5 μm BA. Northern blots were hybridized with a probe specific for the cytokinin response genes ARR5 and ARR7. The ACTIN2 gene was used as a loading control. B, Interaction plots of A-type ARR genes. Interaction plots show the relative normalized expression values for all seven A-type ARR genes present on the CATMA V2.4 array plotted against the time points of cytokinin treatment: BA0 (control), BA15 (15 min of BA treatment), and BA120 (120 min of BA treatment). The table in the lower right corner shows the significance of differences in gene expression. C (cytokinin effect) represents the responsiveness of the respective gene to cytokinin treatment. G (genotype effect) denotes whether the average expression of the respective gene in line ARR1-S-8 differs significantly from that in the wild type. The cytokinin response of genes with a significant I (interaction effect) is dependent on the genotype. The significance codes are based on the uncorrected P value for each gene. Symbols are as follows: no symbol, P > 0.05; *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001.
Transcriptome Analysis Reveals Complex Changes in 35S:ARR1-SRDX Plants
Next, we investigated the changes occurring at 15 and 120 min following cytokinin treatment of 5-d-old seedlings in the transcriptome of the wild type and line ARR1-S-8 using the CATMA V2.4 array. Seedlings of both genotypes show only minor morphological differences at this stage. In all experiments, we regarded genes in which the expression was changed by ≥2.5-fold, the false discovery rate (FDR)-corrected P value (q value) was ≤0.05, and that had 50% or more spots above background in the higher expressed condition as significantly regulated. In total, 658 genes were found with a lower steady-state mRNA level and 505 genes were found with a higher steady-state mRNA level in seedlings of line ARR1-S-8 compared with the wild type (Table I; Supplemental Table S1).
Table I.
Examples of down-regulated and up-regulated genes in 35S:ARR1-SRDX transgenic seedlings
Steady-state mRNA levels of seedlings of the wild type and line ARR1-S-8 were analyzed at 5 DAG. The data are sorted according to the ratio. The rank in the table of all significantly regulated genes (>2.5-fold difference, q < 0.05) is given. The q value is the FDR-corrected P value and was calculated as described (Benjamini and Hochberg, 1995).
| Rank | CATMA Identifier | Expression in Col-0 | Expression in ARR1-S-8 | Ratio | q Value | Arabidopsis Genome Initiative Code | Description |
|---|---|---|---|---|---|---|---|
| Down-regulated genes | |||||||
| 3 | CATMA1b35155 | 14.72 | 0.20 | 0.01 | 7.29E-08 | AT1G37130 | Nitrate reductase 2 (NR2) |
| 21 | CATMA3a01035 | 0.48 | 0.03 | 0.07 | 3.38E-09 | AT3G02050 | Potassium transporter (KUP3) |
| 24 | CATMA1a10175 | 14.59 | 1.03 | 0.07 | 1.18E-12 | AT1G11260 | Glucose transporter (STP1) |
| 26 | CATMA4a37435 | 1.45 | 0.12 | 0.08 | 4.80E-10 | AT4G35790 | Phospholipase Dδ/PLDδ (PLDDELTA) |
| 28 | CATMA5a22070 | 40.19 | 3.50 | 0.09 | 7.36E-11 | AT5G24470 | Pseudo response regulator 5 (APRR5) |
| 35 | CATMA1b13933 | 0.75 | 0.07 | 0.09 | 1.16E-11 | AT1G14920 | GA response modulator (GAI; RGA2) |
| 41 | CATMA4a27720 | 3.68 | 0.37 | 0.10 | 5.33E-10 | AT4G26200 | 1-Amino-cyclopropane-1-carboxylate synthase 7 (ACS7) |
| 86 | CATMA2a00615 | 2.28 | 0.34 | 0.15 | 1.43E-03 | AT2G01570 | GA response modulator (RGA1) |
| 103 | CATMA5a57125 | 2.68 | 0.43 | 0.16 | 2.11E-13 | AT5G61520 | Hexose transporter, putative |
| 127 | CATMA3a19545 | 6.01 | 1.04 | 0.17 | 2.71E-10 | AT3G19930 | Sugar transport protein (STP4) |
| 139 | CATMA5a53340 | 3.15 | 0.56 | 0.18 | 1.95E-14 | AT5G57630 | CBL-interacting protein kinase 21, putative (CIPK21) |
| 152 | CATMA2a36595 | 4.21 | 0.79 | 0.19 | 1.04E-13 | AT2G38290 | Ammonium transporter 2 (AMT2) |
| 155 | CATMA1a09640 | 1.47 | 0.28 | 0.19 | 1.16E-05 | AT1G10760 | Starch excess protein (SEX1) |
| 170 | CATMA3b13000 | 0.45 | 0.09 | 0.20 | 3.67E-05 | AT1g13790 | β-Fructosidase (BFRUCT1) |
| 200 | CATMA2a42185 | 1.65 | 0.36 | 0.22 | 7.04E-11 | AT2G43790 | Mitogen-activated protein kinase, putative (MPK6) |
| 202 | CATMA1a59165 | 11.17 | 2.47 | 0.22 | 3.27E-10 | AT1G69850 | Nitrate transporter (NTL1) |
| 247 | CATMA4a39020 | 3.69 | 0.91 | 0.25 | 2.08E-08 | AT4G37450 | Lys-rich arabinogalactan protein (AGP18) |
| 333 | CATMA3a14960 | 0.34 | 0.10 | 0.28 | 3.07E-09 | AT3G15540 | Indoleacetic acid-induced protein 19 (IAA19) |
| 338 | CATMA5a39960 | 1.07 | 0.31 | 0.28 | 4.04E-10 | AT5G42210 | Ethylene response factor subfamily B-1 (ATERF-9) |
| 343 | CATMA3a47160 | 2.85 | 0.82 | 0.29 | 5.31E-03 | AT3G54220 | Scarecrow transcription factor, putative |
| 396 | CATMA5a61005 | 2.59 | 0.80 | 0.31 | 5.92E-09 | AT5G65670 | Indoleacetic acid-inducible protein 9 (IAA9) |
| 398 | CATMA1a24230 | 2.80 | 0.86 | 0.31 | 4.17E-02 | AT1G25540 | Phytochrome and flowering time regulatory protein (PFT1) |
| 412 | CATMA3a53395 | 0.30 | 0.09 | 0.31 | 5.95E-15 | AT3G60390 | Homeobox-Leu zipper protein 3 (HAT3) |
| 456 | CATMA3b54606 | 68.70 | 22.74 | 0.33 | 1.27E-06 | AT3G61470 | Chlorophyll a/b-binding protein (LHCA2) |
| 459 | CATMA3a39720 | 6.48 | 2.15 | 0.33 | 1.06E-09 | AT3G46640 | Phytoclock 1 (PCL1) |
| 470 | CATMA5a55870 | 0.62 | 0.21 | 0.34 | 1.07E-05 | AT5G60120 | AP2 domain-containing transcription factor, putative |
| 551 | CATMA1a32530 | 2.19 | 0.81 | 0.37 | 9.91E-06 | AT1G34210 | Somatic embryogenesis receptor-like kinase 2 (SERK2) |
| 562 | CATMA1a08395 | 16.80 | 6.28 | 0.37 | 1.01E-10 | AT1G09530 | Phytochrome-interacting factor 3 (PIF3) |
| 573 | CATMA4a31370 | 0.15 | 0.05 | 0.38 | 4.11E-09 | AT4G29740 | Cytokinin oxidase family protein (CKX4) |
| 619 | CATMA1a61230 | 3.11 | 1.21 | 0.39 | 1.87E-09 | AT1G72010 | TCP family transcription factor, putative (PCF2) |
| 621 | CATMA5a01890 | 1.57 | 0.61 | 0.39 | 1.08E-11 | AT5G02810 | Pseudo response regulator 7 (APRR7) |
| Up-regulated genes | |||||||
| 2 | CATMA3a22775 | 0.09 | 14.47 | 161.47 | 3.53E-14 | AT3G22840 | Chlorophyll a/b-binding family protein/early light-induced protein (ELIP) |
| 9 | CATMA5a07985 | 0.34 | 10.52 | 31.12 | 1.62E-16 | AT5G08640 | Flavonol synthase 1 (FLS1) |
| 11 | CATMA1a54343 | 0.05 | 0.95 | 20.43 | 1.50E-11 | AT1G65060 | Isoform of 4-coumarate:CoA ligase (4CL3) |
| 30 | CATMA2a25985 | 0.22 | 2.44 | 11.00 | 7.43E-11 | AT2G27550 | Centroradialis protein, putative (CEN) |
| 46 | CATMA1a54085 | 1.34 | 11.66 | 8.67 | 1.38E-12 | AT1G64780 | Ammonium transporter 1, member 2 (AMT1.2) |
| 68 | CATMA5a12150 | 19.01 | 135.85 | 7.14 | 1.58E-14 | AT5G13930 | Chalcone synthase/naringenin-chalcone synthase |
| 88 | CATMA3a21086 | 0.15 | 0.84 | 5.74 | 1.66E-05 | AT3G21240 | Encodes an isoform of 4-coumarate:CoA ligase (4CL2) |
| 99 | CATMA5a22550 | 3.30 | 17.66 | 5.36 | 1.83E-13 | AT5G24850 | Cryptochrome dash (CRYD) |
| 106 | CATMA3a48130 | 1.93 | 9.94 | 5.14 | 2.24E-14 | AT3G55120 | Chalcone-flavanone isomerase/chalcone isomerase (CHI) |
| 114 | CATMA2a31150 | 3.33 | 16.23 | 4.88 | 6.43E-08 | AT2G32950 | COP1 regulatory protein/FUSCA protein (FUS1) |
| 134 | CATMA3a24230 | 0.11 | 0.53 | 4.62 | 1.68E-03 | AT3G24290 | Ammonium transporter, putative |
| 138 | CATMA2a44715 | 4.58 | 20.90 | 4.57 | 1.99E-07 | AT2G46340 | Phytochrome A suppressor SPA1 (SPA1) |
| 145 | CATMA5a04585 | 1.18 | 5.30 | 4.47 | 2.22E-06 | AT5G05410 | DREB subfamily A-2 of ERF/AP2 transcription factor family (DREB2A) |
| 226 | CATMA3a43674 | 0.16 | 0.56 | 3.46 | 3.14E-04 | AT3G50630 | Kip-related protein 2 (KRP2)/cyclin-dependent kinase inhibitor 2 (ICK2) |
| 235 | CATMA2a26350 | 0.82 | 2.78 | 3.40 | 6.71E-07 | AT2G27960 | Cyclin-dependent kinase/CDK (CKS1) |
| 252 | CATMA3a16940 | 5.06 | 16.65 | 3.29 | 1.27E-07 | AT3G17510 | CBL-interacting protein kinase 1 (CIPK1) |
| 400 | CATMA1a64750 | 1.28 | 3.56 | 2.77 | 5.40E-07 | AT1G75410 | BEL1-like homeodomain 3 protein (BLH3) |
| 402 | CATMA4a17105 | 0.17 | 0.47 | 2.77 | 5.25E-03 | AT4G16280 | Flowering time control protein/FCAγ (FCA) |
| 485 | CATMA5a17440 | 0.32 | 0.81 | 2.54 | 8.27E-05 | AT5G19040 | Adenylate isopentenyltransferase 5/cytokinin synthase (IPT5) |
First we analyzed whether the expression of known ARR1 target genes was altered. Eleven of the 23 ARR1 target genes described by Taniguchi et al. (2007) were also on our array and could be detected. All of these were also induced in the wild type under our conditions, 10 showed a diminished and/or delayed induction in the 35S:ARR1-SRDX plants, and seven showed a lower basal expression level in the 35S:ARR1-SRDX plants compared with wild-type plants (Supplemental Table S2). Together, this shows a significant repressive influence of ARR1-SRDX at least on ARR1 target gene expression. Target genes of other B-type ARRs could not be analyzed as they are mostly not known.
We then studied broader cytokinin-induced changes in the transcript abundance, which should reveal whether there is an overall dampening of the cytokinin response or whether specific changes could be observed. In 35S:ARR1-SRDX transgenic seedlings, the total number of genes up-regulated by cytokinin treatment was about half the number observed in the wild type (Supplemental Table S1). The induction of the 10 strongest cytokinin-regulated genes in the wild type was reduced in line ARR1-S-8 to 6% and 30% after 15 and 120 min, respectively (Supplemental Table S1). Comparison of other subsets of up-regulated genes between the wild type and 35S:ARR1-SRDX transgenic seedlings yielded similar results. For example, the response to cytokinin of the majority of the seven A-type ARR genes that are represented in the CATMA V2.4 array was dampened and/or delayed (Fig. 7B), which is consistent with the result of the northern-blot analysis (Fig. 7A). However, the response curves show individual differences. For example, for ARR4 and ARR16, the early increase of transcript abundance seen in the wild type is completely missing in the 35S:ARR1-SRDX transgenic seedlings. Other genes, like ARR6 and ARR7, show a weaker response at the early time point (Fig. 7B). In total, 73% of the genes induced by cytokinin in the wild type were not induced in line ARR1-S-8.
We then checked whether genes that are down-regulated by cytokinin would show enhanced repression in 35S:ARR1-SRDX seedlings. Analysis of a set of 56 early or late cytokinin down-regulated genes (Brenner et al., 2005) revealed only five examples that show a stronger down-regulation following cytokinin treatment in the 35S:ARR1-SRDX seedlings compared with the wild type. In addition, this concerned only genes that were down-regulated at 2 h after cytokinin treatment but none of the genes that were rapidly (15 min) down-regulated. This result argues against the possibility that ARR1-SRDX enhances a putative repressor function of ARR1.
Analysis of the classes of genes and individual genes that are constitutively down-regulated in 35S:ARR1-SRDX plants should yield indications about the direct or indirect regulatory functions of B-type ARRs. There are numerous genes in line ARR1-S-8 that showed only one-tenth or even less of their wild-type transcript level, indicating that they are directly or indirectly regulated by B-type ARRs. Prominent examples are the nitrate reductase gene NR2 and the ammonium transporter gene AMT2, the nitrate transporter gene NTL1, two sugar transporter genes (STP1 and STP4), and the genes encoding the repressors of GA responses, RGA1 and RGA2 (Table I; Supplemental Table S1).
Among the individual genes that were very strongly up-regulated in ARR1-S-8 is a gene encoding an ELIP (for early light-induced protein) of the chlorophyll a/b-binding protein family and the cyclin-dependent kinase inhibitor gene ICK2. Among the cytokinin-related genes, IPT5 was 2.5-fold up-regulated (Table I; Supplemental Table S1).
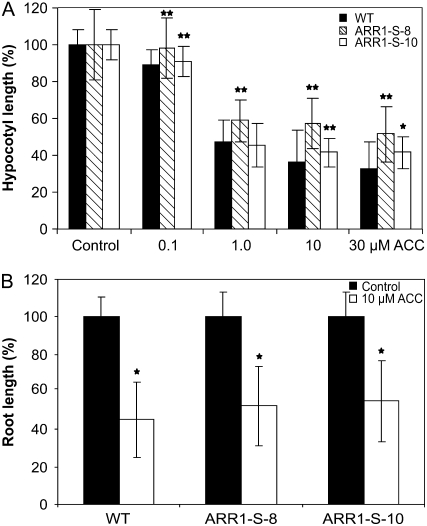
The Ethylene Response of 35S:ARR1-SRDX Seedlings
The large number of differentially expressed genes in the 35S:ARR1-SRDX transgenic seedlings indicates the possibility that this family of transcription factors might not have a function solely in cytokinin signaling but also in other pathways. An involvement of the B-type ARR2 in ethylene signaling has been discussed (Hass et al., 2004; Mason et al., 2005). To test the possible involvement of B-type ARRs in mediating the ethylene response, the growth response of 35S:ARR1-SRDX seedlings on medium containing the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) was compared with that in wild-type seedlings. The hypocotyl lengths of wild-type and 35S:ARR1-SRDX seedlings decreased with increasing concentrations of ACC in the medium (Fig. 8A). In contrast to the wild type, in which hypocotyl length decreased more with further increasing ACC concentrations, the 35S:ARR1-SRDX seedlings showed resistance to ACC concentrations greater than 1 μm, as their hypocotyl length did not decrease further (Fig. 8A). However, root shortening of wild-type and 35S:ARR1-SRDX seedlings was similar at low and high ethylene concentrations (Fig. 8B). Also, the bending of the apical hook did not differ between the genotypes (data not shown).
Figure 8.
Hypocotyl and root elongation of 35S:ARR1-SRDX transgenic seedlings in response to ethylene. A, Hypocotyl elongation of 35S:ARR1-SRDX transgenic seedlings on ACC-containing medium compared with the wild type. Transgenic seedlings show resistance at higher concentrations of ACC. Seedlings were placed on vertical MS medium plates containing 0.1, 1.0, 10, or 30 μm ACC. Hypocotyl lengths were measured after 4 d of dark incubation. B, Primary root elongation of 35S:ARR1-SRDX transgenic plants on ACC-containing medium. Root elongation was inhibited to a similar extent in both wild-type and transgenic seedlings. Seedlings were grown on medium containing 10 μm ACC for 4 d in the dark. More than 15 seedlings in two replicates were analyzed for each genotype. Error bars represent sd (n ≥ 15). Pairwise Student's t test was used to compare values with the wild type. *, 0.01 < P < 0.05; **, 0.001 < P < 0.01. ARR1-S-8 and ARR1-S-10 are two independent lines of 35S:ARR1-SRDX transgenic plants.
Germination of 35S:ARR1-SRDX Seeds Is Resistant to Inhibition by Far-Red Light
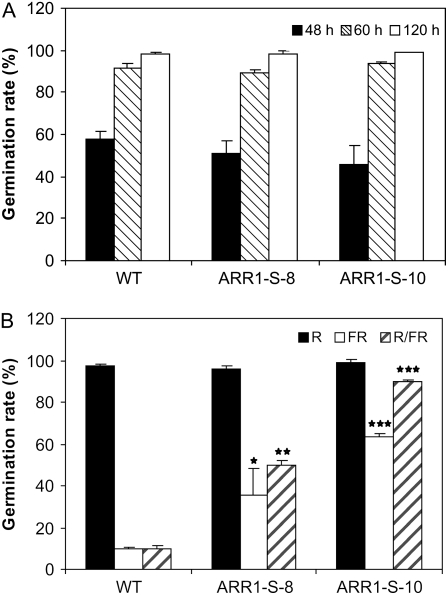
Several experiments have indicated an involvement of the two-component signaling system in red light signaling (Sweere et al., 2001; To et al., 2004; Riefler et al., 2006, Mira-Rodado et al., 2007). We used a seed germination assay to explore the possible role of B-type ARRs in response to red light. Far-red light inhibits seed germination, a response that is, under our experimental conditions, mediated by phytochrome B and reversible by red light (Shinomura, 1997; Sullivan and Deng, 2003). The germination behavior of 35S:ARR1-SRDX transgenic seeds and wild-type seeds in white light was similar. At 48 and 60 h after transfer from cold treatment to room temperature, a similar percentage of the seeds of all genotypes had germinated and eventually reached about 100%, indicating that there was no marked difference in the timing or capability of germination (Fig. 9A). Similarly, about 100% of the seeds germinated after a white light pulse or a red light pulse of 5 min followed by a dark period of 7 d (Fig. 9B). However, when wild-type seeds were exposed to a 5-min far-red light pulse prior to dark incubation, only around 10% of the seeds germinated (Fig. 9B). In contrast, 35% and 60% of the seeds of lines ARR1-S-8 and ARR1-S-10, respectively, germinated following this treatment. A similar difference between the wild type and the transgenic lines was seen when the far-red light treatment was preceded by a 5-min red light pulse. Then, the germination rate of the transgenic lines reached about 80%, while it remained around 10% for wild-type seeds (Fig. 9B).
Figure 9.
Germination behavior of 35S:ARR1-SRDX seedlings under different light conditions. A, Germination rate after treatment by a white light pulse (5 min, 30 μE) and subsequent incubation in the dark. B, Germination rate after red (R), far-red (FR), or R/FR treatment. Seedlings were treated for 5 min with the indicated light and kept subsequently in the dark. Germination frequency was determined from batches of approximately 100 seeds after 7 d of dark incubation (n = 2). Error bars represent sd. Pairwise Student's t test was used to compare values with the wild type. No symbol, P > 0.05; *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001. ARR1-S-8 and ARR1-S-10 are two independent lines of 35S:ARR1-SRDX transgenic plants.
DISCUSSION
Application of the CRES-T Results in a Dominant Negative Effect and Thus Allows the Investigation of B-Type ARR Functions
Previous studies have shown the involvement of several members of the B-type ARRs in mediating cytokinin signaling (Imamura et al., 2003; Tajima et al., 2004; Mason et al., 2005; Yokoyama et al., 2007). However, because of a high degree of functional redundancy, the extent of their contribution was unclear. Here, we have shown that a dominant repressor form of a B-type ARR, ARR1-SRDX, causes a strong cytokinin deficiency phenotype, which is most likely due to the concerted repression of the activities of ARR1 and related transcription factors. This suggests that the activity of B-type ARRs is involved in most, if not all, of the cytokinin-regulated processes.
The results of the protoplast transactivation assay clearly demonstrate the dominant negative effect of the ARR1-SRDX protein on ARR1-dependent activation of a reporter gene as well as its induction by cytokinin (Fig. 1A). This dominant negative activity also suppressed the activity of all other B-type ARRs tested (Fig. 1B), further validating the chosen approach. The transcript level of the resident ARR1 gene was even enhanced in the 35S:ARR1-SRDX transgenic plants, indicating that the observed changes of gene expression and plant morphology were not due to cosuppression. The phenotypic changes described here were not observed in 35S:ARR1-overexpressing plants (Sakai et al., 2001; our unpublished data), making it unlikely that nonspecific negative effects such as transcriptional squelching (Kornberg, 2005) contribute to the phenotype. Importantly, the phenotypic changes occur in tissues expressing B-type ARR genes. Binding sites of several B-type ARRs are similar but not identical (Imamura et al., 2003). It is likely, therefore, that ARR1-SRDX suppresses the activity of most if not all B-type ARRs, which is supported by the results of the protoplast transactivation assay (Fig. 1B). However, we cannot completely exclude that part of the changes in phenotype and gene regulations caused by ARR1-SRDX are off-target effects due to interference with other related transcription factors. Candidates are the MYB domain-containing pseudo response regulators APRR2 and APRR4, which show in their DNA binding domains similarity to B-type ARRs, but nothing is known about the function of these APRRs (Makino et al., 2000). Recent experiments have indicated that the DNA motif necessary for ARR1 binding is actually longer than previously reported (Taniguchi et al., 2007), making it less likely that the effect of the ARR1-SRDX dominant repressor spreads far beyond the B-type ARRs. One also has to be aware that individual B-type ARRs are likely to have different functions and that some of the phenotypes seen in the 35S:ARR1-SRDX transgenic plants might be due to the repression of specific ARRs or combinations thereof and do not indicate functions for all members of this protein family. However, taken together, the phenotypic characteristics of the 35S:ARR1-SRDX plants clearly demonstrate that the CRES-T is useful to study larger transcription factor families and can be applied to study other regulatory pathways involving transcriptional regulators hampered by redundancy (Koyama et al., 2007).
The Morphological Phenotype of 35S:ARR1-SRDX Plants Is Consistent with a Broad Function of B-Type ARRs in Cytokinin Signaling
The phenotype of the 35S:ARR1-SRDX transgenic plants resembled in various aspects the phenotype of cytokinin receptor triple mutants and cytokinin-deficient plants (Werner et al., 2003; Yang et al., 2003; Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006; Supplemental Fig. S3). As in those mutants, the shoot growth and in particular leaf size were strongly reduced, while seed size and the root system were enlarged. An exception to the similarities between 35S:ARR1-SRDX plants and plants with a reduced cytokinin status is the early-flowering phenotype, which contrasts with the retarded flowering reported for triple receptor mutants and cytokinin-deficient plants (Werner et al., 2003; Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006). Earlier flowering in 35S:ARR1-SRDX plants was developmentally regulated, as fewer leaves had formed at the onset of flowering. This indicates that this function may be cytokinin-independent, as cytokinin deficiency causes only minor changes in leaf number at the onset of flower formation (Werner et al., 2003). A cytokinin-independent role in flowering has been suggested for the rice B-type response regulator gene Edh1 (Doi et al., 2004). Interestingly, arr2 mutant plants also flower earlier than wild-type plants, indicating a negative regulatory function of ARR2 in the induction of flowering (Hass et al., 2004). It could be that the early-flowering phenotype of the 35S:ARR1-SRDX plants is at least partially due to the suppression of ARR2 function, which was demonstrated in the protoplast transactivation assay (Fig. 1B).
35S:ARR1-SRDX Plants Show Specific Changes of Cytokinin Sensitivity
A decreased cytokinin sensitivity of the 35S:ARR1-SRDX transgenic plants was detected in several bioassays. 35S:ARR1-SRDX plants showed complete resistance to cytokinin in the chlorophyll retention assay, which is similar to the phenotype of ahk2 ahk3 receptor mutants (Riefler et al., 2006). This indicates that the cytokinin activities in regulating chlorophyll retention are entirely dependent on B-type ARRs. In particular, ARR2, the closest homolog of ARR1, has been shown to play a role in mediating cytokinin-dependent chlorophyll retention (Kim et al., 2006) and thus may be suppressed in leaves of 35S:ARR1-SRDX plants.
The reduction of cytokinin sensitivity was less pronounced in roots of 35S:ARR1-SRDX plants. One possibility to explain this distinction is that different sets of B-type ARRs act in shoots and roots and that the B-type ARRs in the roots are less affected by the dominant repressor activity of ARR1-SRDX. Alternatively, the affected B-type ARRs could play antagonistic roles in the root. The latter possibility is supported by the observation that the inclusion of an arr2 knockout allele in arr mutant combinations causes a reduction in the strength of the root phenotype, indicating that ARR2 might have an antagonistic function to the other B-type ARRs in regulating root elongation and root branching (Mason et al., 2005). Therefore, reduced activity of ARR2 in the dominant repression plants could explain the somewhat weaker root phenotype in the 35S:ARR1-SRDX transgenic plants.
Early Cytokinin Responses Are Attenuated in 35S:ARR1-SRDX Seedlings
The expression and inducibility of A-type ARR genes and other early cytokinin response genes, which have been shown to be direct target genes of B-type ARRs in the past (Hwang and Sheen, 2001; Taniguchi et al., 2007), were dampened in 35S:ARR1-SRDX transgenic seedlings (Fig. 7; Supplemental Table S1). This clearly shows that the ARR1-SRDX protein suppresses the activation of direct target genes of endogenous B-type ARRs. Interestingly, the response of the majority of the response genes was generally reduced, but each to a different extent. These differences may reflect the different contributions of ARR1 and related B-type ARRs to the cytokinin-dependent regulation of the respective genes. Furthermore, recent work has shown that the transcription of A-type ARRs is also regulated via a B-type ARR-independent pathway. The target genes of the cytokinin response factors of the ERF family of transcription factors partially overlap with those of the B-type ARRs (Rashotte et al., 2006).
We have identified large sets of genes that show significant changes in their transcript levels in association with the cytokinin deficiency syndrome of the 35S:ARR1-SRDX plants. The function of the deregulated genes can be linked, in many cases, to known functions of cytokinin in plant physiology and development, such as nitrogen metabolism (Sakakibara et al., 2006) and GA action (Brenner et al., 2005; Jasinski et al., 2005). The fact that the transcript levels of genes related to these processes (e.g. NR2, AMT2, NTL1, RGA1, and RGA2; Table I) change in plants with repressed B-type ARR functions confirms that cytokinin plays an important role in the control of these processes and that the B-type ARRs are at least partly involved in this regulation. This data set will be an important source to identify and study processes downstream of B-type ARRs.
B-Type ARRs Are Possible Nodes for the Integration of Different Signaling Pathways
As developmental programs are subject to fine-tuning by different factors, the integration of different signaling pathways is necessary. Evidence for a function for B-type ARRs in cross talk came from the red light experiments. Red light is a key regulator of seed germination, and its effect is mediated by phytochromes (Shinomura, 1997; Sullivan and Deng, 2003). Red light stimulates seed germination, while far-red light is inhibitory. The response under our light regime is mediated by phytochrome B, and the reversibility of red light-stimulated germination by far-red light is a hallmark of its activity (Shinomura, 1997). This reversibility was almost completely lost in 35S:ARR1-SRDX transgenic seeds (Fig. 9B). This result is consistent with the previously described reduced far-red light sensitivity of cytokinin receptor mutant seeds (Riefler et al., 2006) and seeds of cytokinin-deficient plants (data not shown). A plausible explanation is that cytokinin in an unknown way alters the Pfr-Pr ratio of phytochrome B (e.g. by stabilization of the active Pfr form). This may be mediated directly by downstream factors such as A-type ARRs, as was proposed for red light-dependent regulation of hypocotyl elongation (Sweere et al., 2001; Mira-Rodado et al., 2007). The dominant repressor effect of ARR1-SRDX could block the cytokinin signal involved in regulating this process. Alternatively, B-type ARRs may be involved in regulating red light responses independent of cytokinin (e.g. by acting downstream of phytochromes). In any case, there is now considerable evidence that the cytokinin status has a strong influence on the reaction to red light.
CONCLUSION
Using the CRES-T, we generated a dominant repressor of B-type ARR function and demonstrated that this approach is useful to overcome functional redundancy in this transcription factor family. Overall, we show that B-type ARRs are required for normal growth and development of the shoot and root, where they exert positive and negative regulatory functions. Comparisons with the phenotypes of cytokinin receptor mutants and cytokinin-deficient plants clearly show that the B-type ARRs, collectively, are involved in regulating most cytokinin-dependent processes in Arabidopsis. 35S:ARR1-SRDX transgenic plants and the data sets of deregulated genes will be a valuable tool for further investigation of the function of this class of transcription factors, in particular of their role in mediating the cross talk of the two-component signaling system with other pathways.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Plants of the Col-0 accession of Arabidopsis (Arabidopsis thaliana) were used as the wild type. The plants were grown in the greenhouse on soil at 22°C under long-day conditions (16 h of light/8 h of dark). For in vitro experiments, seeds were surface sterilized with saturated calcium hypochlorate solution. After sowing, they were kept at 4°C for 3 d in the dark and then exposed to white light (approximately 75 μE). Seedlings were grown at 22°C on medium containing 1× Murashige and Skoog (MS) salts, 3% Suc, 0.05% MES, and 0.9% agar (Merck) unless specified otherwise. For ethylene and red light experiments, Suc was omitted from the medium. For the flowering phenotype, the total number of rosette leaves was counted upon flower bud initiation.
Gene Cloning and Transformation
The protein-coding region of the ARR1 gene was amplified by PCR using a cDNA library from Arabidopsis C24 (Minet et al., 1992) and inserted in the plasmid pDONR201 (Invitrogen), yielding pDONR201-ARR1 (Dortay et al., 2006). The DNA fragment coding for the SRDX peptide (LDLDLELRLGFA) was synthesized with a TAA stop codon and a BsrGI restriction site at the 3′ end and an in-frame HhaI site at the 5′ end. The ARR1 gene was isolated from the plasmid pDONR201-ARR1 by restriction digestion with BsrGI, and the resulting fragment was further digested with HhaI. The DNA fragments were ligated and recloned into pDONR201. The resulting ARR1-SRDX gene was shuttled into the vector pB2GW7 (Karimi et al., 2002) for overexpression under the control of the 35S promoter. This construct was transformed using Agrobacterium tumefaciens-mediated transformation into Arabidopsis plants by the floral dip method (Clough and Bent, 1998).
For the protoplast assay, the reporter plasmid was generated by amplifying the 1,000-bp fragment directly upstream of the ARR6 gene (for primers used, see Supplemental Table S3). The resulting PCR product was digested with HindIII and XbaI and ligated into the pBT10-GUS vector (Sprenger-Haussels and Weisshaar, 2000). The B-type ARR overexpression constructs were generated by shuttling the respective genes from the pDONR vector into the pB2GW7 vector (Karimi et al., 2002) for expression under the control of the 35S promoter. All B-type ARR genes were cloned as cDNAs, except for ARR2, which was a genomic clone (for primers used, see Supplemental Table S3).
Protoplast Transformation and GUS Assays
Protoplast isolation and transformation were carried out according to the method described by Hwang and Sheen (2001). For isolation of mesophyll protoplasts, 4- to 5-week-old rosette leaves were used. Transformation of protoplasts was mediated by 40% polyethylene glycol solution. For cytokinin treatment, protoplasts were incubated overnight with 500 nm trans-zeatin. For the transactivation assays, 9 μg of the ARR6:GUS reporter plasmid and 14 μg of each effector plasmid carrying 35S:ARR1 or other B-type ARRs and 35S:ARR1-SRDX were used. For normalization, 3 μg of a plasmid harboring the 35S:NAN gene (Kirby and Kavanagh, 2002) was added. Both GUS and NAN enzyme assays were performed according to Kirby and Kavanagh (2002). The ratios of GUS and NAN activities were calculated as relative GUS/NAN activity units.
Gene Expression Analysis by Northern Blot
Total RNA was extracted from 3-week-old soil-grown plants (Fig. 2) or 5-d-old seedlings grown in 0.5× MS liquid medium (Fig. 7A). Seedlings were treated with 5 μm BA and incubated at 0, 15, 30, and 120 min prior to harvesting. Preparation of total RNA and northern-blot analysis were performed as described (Brenner et al., 2005). For northern-blot analysis, 20 μg of RNA was separated on a denaturing agarose-formaldehyde gel (1.2%) containing 10% of 10× MOPS and 3% of 37% formaldehyde, transferred to a Hybond-N+ nylon membrane (Amersham), and after fixation hybridized with radioactive [α-32P]dCTP-labeled DNA. Hybridization was performed at 68°C in a phosphate buffer containing 7% SDS and 1% bovine serum albumin. Washing was done with 2× SSC and 0.2× SSC, 0.1% SDS at 65°C. As a control for loading, blots were reprobed with an ACTIN2 probe. The different probes were generated by amplifying the respective cDNAs (for primers used, see Supplemental Table S3).
Root Growth Assay
Arabidopsis seeds were grown on vertical MS medium plates containing different concentrations of BA ranging from 0.01 to 1.0 μm. Dimethyl sulfoxide (DMSO; 0.1%) was included as a vehicle control. The primary root lengths were determined at 10 DAG (Fig. 4), between days 4 and 9 after germination (Fig. 6), or after 4 d of growth in the dark (Fig. 8). Photographs were taken with a digital camera (Nikon Coolpix 8800), and root lengths were determined using the Scion Image program version Beta 4.0.3 (www.scioncorp.com). The number of lateral roots emerging from the epidermis of the primary root was counted with a microscope at 10 DAG. The experiments were performed using three independent replicates and at least 15 seedlings in each replicate.
Chlorophyll Retention Assay
This assay was performed as described previously (Riefler et al., 2006). Either the sixth or the seventh leaf was detached from 24-d-old in vitro grown seedlings and floated on distilled water supplemented with 0, 0.01, 0.1, 0.5, 1.0, or 5.0 μm BA in 0.1% DMSO in small petri dishes for 10 d at room temperature in the dark. Three replicates of each genotype, each consisting of five leaves, were prepared and two chlorophyll measurements were taken per plate. Chlorophyll was extracted with methanol for 24 h in the dark. The amount of chlorophyll was measured with a spectrophotometer and normalized to fresh weight, and the chlorophyll content was calculated as described (Porra et al., 1989).
Ethylene Response Assay
Seeds were surface sterilized and placed on vertical plates containing sugar-free MS medium. Different concentrations of ACC (0.1, 1.0, 10, or 30 μM) were included in the medium. The seeds were grown in the dark at room temperature. After 4 d, the hypocotyl lengths were measured using the Scion Image program version Beta 4.0.3 (www.scioncorp.com). The experiment was done in two replicates, each with more than 15 seedlings. The root elongation assay was performed in the same way, but using only one ACC concentration (10 μm). The experiment was done with 15 seedlings per genotype in two replicates.
Determination of Seed Size and Weight
Determination of seed size and seed weight was carried out as described by Riefler et al. (2006) and Werner et al. (2003), respectively. Seed size of the wild type and ARR1-SRDX overexpressors was determined by measuring the length and width of 60 seeds harvested from two different plants. The volume was estimated by calculating with the formula for a spheroid (volume = 4/3 × π × length × width × depth). Biomass of the seed was weighed using a fine balance LE244S (Sartorius). The weight of one seed was calculated from the weight of pools of 200 seeds. The sample size for each genotype was 10.
Seed Germination Assay
Seeds of wild-type and 35S:ARR1-SRDX plants used for germination experiments had been harvested and stored under the same conditions. Seed batches of approximately 100 seeds were sown on petri dishes on four layers of filter paper and moistened with 5 mL of distilled water. Seeds were vernalized for 3 d at 4°C in the dark, followed by 6 h at 35°C to alleviate the seeds of hypersensitivity to red light (Fankhauser and Casal, 2004), and for 30 min at 25°C immediately prior to the irradiation. For the different light treatments, seeds were exposed for 5 min to white light (30 μE), red light (660 nm, 6 μE), or far-red light (724 nm, 15 μE) or to a sequential treatment of 5 min of red light followed by 5 min of far-red light. Seeds were stored at room temperature in a dark place for 7 d. Radicle protrusion was employed as the criterion for germination.
Microarray Experiments
For microarray experiments, surface-sterilized seedlings of the wild type and line ARR1-S-8 were grown in sterile 0.5× MS liquid medium (1 g L−1 Suc and 0.5 g L−1 MES, pH 5.7) in petri dishes and cultured under the conditions described above. At 5 DAG, the seedlings were treated with either 5 μm BA dissolved in 1.13 mm KCl for 15 or 120 min (BA15 and BA120, respectively) or with 1.13 mm KCl solution for 120 min as a control (BA0). Timing of the treatment was such that the plants were harvested within an interval of 6 to 8 h after the onset of light. The content of two petri dishes was pooled to yield one sample of biological material. Two biological samples were harvested for each experimental condition. RNA was extracted as described previously (Brenner et al., 2005). To reduce the number of false positives due to nonspecific changes in gene expression, we hybridized RNA samples extracted from uninduced wild-type seedlings (BA0) as a reference sample onto an additional set of microarrays. Genes with a significant expression change between this reference sample and the experimental Col-0 BA0 sample were excluded from the genotype comparison.
Preparation of the labeled samples for microarray hybridization was performed following the linear amplification protocol (Puskas et al., 2002) with modifications. A first-strand reaction was performed starting with 5 μg of total RNA using SuperScript II reverse transcriptase (Invitrogen). The whole product was used for second-strand synthesis with Escherichia coli DNA polymerase, E. coli DNA ligase, and E. coli RNase H (Invitrogen). After purification with the QiaQuick PCR purification kit (Qiagen), the whole second-strand product was used for in vitro transcription with the MEGAScript High Yield Transcription Kit (Ambion/Applera). The resulting copy RNA was purified with RNeasy mini columns (Qiagen). Five micrograms of the purified copy RNA was used for labeling with the SuperScript II reverse transcriptase (Invitrogen), 2 μg of random nonamers as primers, and 1.5 nmol Cy5-dCTP supplemented with a mixture of 2 nmol dCTP and 10 nmol dDTP. The labeled cDNA was purified with the help of QiaQuick PCR purification columns.
We followed the hybridization strategy used in the European Compendium of Arabidopsis Gene Expression project (http://www.cagecompendium.org), employing a Cy3-labeled mixture of oligonucleotides complementary to the secondary primers for gene-specific tag amplification as a common reference sample and Cy5 labeling for the biological samples. Forty picomoles of incorporated Cy5 and 1 pmol Cy3-labeled oligonucleotide reference were dissolved in a mixture of 25% Microarray Hybridization Buffer (Amersham), 50% formamide, and 33.3 ng μL−1 poly(dT35) DNA, applied onto CATMA V2.4 microarrays (ArrayExpress accession no. A-MEXP-274; http://www.ebi.ac.uk/aerep), covered with a coverslip, and incubated in hybridization chambers (Scienion) in a water bath at 42°C for 15 h. The arrays were washed once with 1× SSC, 0.2% SDS for 10 min at 55°C, twice with 0.1× SSC, 0.2% SDS for 10 min at 55°C, and once with 0.1× SSC at 37°C and finally dipped five times into distilled water at room temperature. After drying with an air stream, the microarrays were scanned with a Fuji FLA-5000 scanner at 5-μm resolution and photomultiplier tube gain settings of 93 and 83 for Cy5 and Cy3, respectively. The images were saved in TIFF format. Each biological sample was hybridized onto two microarrays, resulting in a total of four hybridizations per experimental condition. The raw data were submitted to the ArrayExpress database at the European Bioinformatics Institute (www.ebi.ac.uk/aerep) and normalized with a preprocessing pipeline (Allemeersch, 2006) written in R/Bioconductor (Gentleman et al., 2004; RDC Team, 2005). After normalization, general linear models were fitted for each gene (Kerr et al., 2000; Wolfinger et al., 2001). The resulting data were used to generate the interaction plots as well as a table of all expression values, ratios, and P values. FDR-corrected P values (q values) were also included. This table was exported into Microsoft Access, the filtering functions of which were used to identify significantly regulated genes and intersections between groups of genes. A gene was called significantly regulated if it had a q value of ≤0.05, a fold change of ≥2.5, and at least half of the spots above background.
Accession Numbers
The Arabidopsis Genome Initiative locus identifier for ARR1 is At3g16857. The raw data from the microarray experiment are accessible through the ArrayExpress database at the European Bioinformatics Institute (www.ebi.ac.uk/aerep).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Schematic representation of the ARR1-SRDX fusion protein.
Supplemental Figure S2. Phenotype of different 35S:ARR1-SRDX lines.
Supplemental Figure S3. Phenotype of the 35S:ARR1-SRDX transgenic plants compared with a triple cytokinin receptor mutant and a cytokinin-deficient plant.
Supplemental Table S1. Lists of all significantly regulated genes in the wild type and line ARR1-S-8.
Supplemental Table S2. Regulation of ARR1 target genes in 35S:ARR1-SRDX transgenic seedlings.
Supplemental Table S3. List of the primers used in this study.
Supplementary Material
Acknowledgments
We thank Cordula Braatz for skillful technical assistance and Klaus Harter and Virtudes Mira-Rodado for the use of and technical help with the red light chambers. We are grateful to Andrea Ehlert and Wolfgang Dröge-Laser for support with the protoplast assays. We thank also Yves Moreau for help in bioinformatics and Takeshi Mizuno for sending plasmids harboring cDNAs of ARR12, ARR19, and ARR21.
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. Schm 814/20–2) in the frame of the Arabidopsis Functional Genomics Network.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Thomas Schmülling (tschmue@zedat.fu-berlin.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Allemeersch J (2006) Statistical analysis of microarray data: applications in platform comparison, compendium data, and array CGH. PhD thesis. Katholieke Universiteit, Leuven, The Netherlands
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57 289–300 [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T (2005) Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44 314–333 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- D'Agostino IB, Deruere J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortay H, Mehnert N, Bürkle L, Schmülling T, Heyl A (2006) Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J 273 4631–4644 [DOI] [PubMed] [Google Scholar]
- Ehlert A, Weltmeier F, Wang X, Mayer CS, Smeekens S, Vicente-Carbajosa J, Dröge-Laser W (2006) Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J 46 890–900 [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Casal JJ (2004) Phenotypic characterization of a photomorphogenic mutant. Plant J 39 747–760 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass C, Lohrmann J, Albrecht V, Sweere U, Hummel F, Yoo SD, Hwang I, Zhu T, Schäfer E, Kudla J, et al (2004) The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J 23 3290–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A, Schmülling T (2003) Cytokinin signal perception and transduction. Curr Opin Plant Biol 6 480–488 [DOI] [PubMed] [Google Scholar]
- Heyl A, Werner T, Schmülling T (2006) Cytokinin metabolism and signal transduction. In P Hedden, SG Thomas, eds, Plant Hormone Signaling: Annual Plant Reviews, Vol 24. Blackwell Publishing, Oxford, pp 93–123
- Higuchi M, Pischke MS, Mahönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34 733–739 [DOI] [PubMed] [Google Scholar]
- Horák J, Brzobohatý B, Lexa M (2003) Molecular and physiological characterisation of an insertion mutant in the ARR21 putative response regulator gene from Arabidopsis thaliana. Plant Biol 5 245–254 [Google Scholar]
- Hosoda K, Imamura A, Katoh E, Hatta T, Tachiki M, Yamada H, Mizuno T, Yamazaki T (2002) Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14 2015–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sakakibara H (2006) Cytokinin biosynthesis and perception. Planta 126 528–538 [Google Scholar]
- Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413 383–389 [DOI] [PubMed] [Google Scholar]
- Imamura A, Kiba T, Tajima Y, Yamashino T, Mizuno T (2003) In vivo and in vitro characterization of the ARR11 response regulator implicated in the His-to-Asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol 44 122–131 [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Yokoyama A, Mizuno T (2008) Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol 49 47–57 [DOI] [PubMed] [Google Scholar]
- Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M (2005) KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15 1560–1565 [DOI] [PubMed] [Google Scholar]
- Kakimoto T (2003) Perception and signal transduction of cytokinins. Annu Rev Plant Biol 54 605–627 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7 193–195 [DOI] [PubMed] [Google Scholar]
- Kazan K (2006) Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci 11 109–112 [DOI] [PubMed] [Google Scholar]
- Kerr MK, Martin M, Churchill GA (2000) Analysis of variance for gene expression microarray data. J Comput Biol 7 819–837 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 103 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby J, Kavanagh TA (2002) NAN fusions: a synthetic sialidase reporter gene as a sensitive and versatile partner for GUS. Plant J 32 391–400 [DOI] [PubMed] [Google Scholar]
- Kornberg RD (2005) Mediator and the mechanism of transcriptional activation. Trends Biochem Sci 30 235–239 [DOI] [PubMed] [Google Scholar]
- Koyama T, Furutani M, Tasaka M, Ohme-Takagi M (2007) TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrmann J, Sweere U, Zabaleta E, Bäurle I, Keitel C, Kozma BL, Brennicke A, Schäfer E, Kudla J, Harter K (2001) The response regulator ARR2: a pollen-specific transcription factor involved in the expression of nuclear genes for components of mitochondrial complex I in Arabidopsis. Mol Genet Genomics 265 2–13 [DOI] [PubMed] [Google Scholar]
- Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Ueguchi C, Sugiyama T, Mizuno T (2000) Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol 41 791–803 [DOI] [PubMed] [Google Scholar]
- Mason MG, Li J, Mathews DE, Kieber JJ, Schaller GE (2004) Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol 135 927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J 2 417–422 [DOI] [PubMed] [Google Scholar]
- Mira-Rodado V, Sweere U, Grefen C, Kunkel T, Fejes E, Nagy F, Schäfer E, Harter K (2007) Functional cross-talk between two-component and phytochrome B signal transduction in Arabidopsis. J Exp Bot 58 2595–2607 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T (2004) Plant response regulators implicated in signal transduction and circadian rhythm. Curr Opin Plant Biol 7 499–505 [DOI] [PubMed] [Google Scholar]
- Mok DW, Mok MC (2001) Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol 52 89–118 [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J (2007) Advances in cytokinin signaling. Science 318 68–69 [DOI] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann E (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b with four different solvents: verification of the concentrations of chlorophyll standards by atomic adsorption spectroscopy. Biochim Biophys Acta 975 384–394 [Google Scholar]
- Puskas LG, Zvara A, Hackler L Jr, Van Hummelen P (2002) RNA amplification results in reproducible microarray data with slight ratio bias. Biotechniques 32 1330–1340 [DOI] [PubMed] [Google Scholar]
- Rashotte AM, Mason MG, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ (2006) A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc Natl Acad Sci USA 103 11081–11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RDC Team (2005) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna
- Richmond AE, Lang A (1957) Effect of kinetin on protein content and survival of detached Xanthium leaves. Science 125 650–65113421662 [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290 2105–2110 [DOI] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov GA, Kieber JJ, Schmülling T (2002) A rapid cytokinin response assay in Arabidopsis indicates a role for phospholipase D in cytokinin signalling. FEBS Lett 515 39–43 [DOI] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Oka A (2000) Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24 703–711 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294 1519–1521 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Takei K, Hirose N (2006) Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci 11 440–448 [DOI] [PubMed] [Google Scholar]
- Shinomura T (1997) Phytochrome regulation of seed germination. J Plant Res 110 151–161 [DOI] [PubMed] [Google Scholar]
- Sprenger-Haussels M, Weisshaar B (2000) Transactivation properties of parsley proline-rich bZIP transcription factors. Plant J 22 1–8 [DOI] [PubMed] [Google Scholar]
- Sullivan JA, Deng XW (2003) From seed to seed: the role of photoreceptors in Arabidopsis development. Dev Biol 26 289–297 [DOI] [PubMed] [Google Scholar]
- Sweere U, Eichenberg K, Lohrmann J, Mira-Rodado V, Baurle I, Kudla J, Nagy F, Schäfer E, Harter K (2001) Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 294 1108–1111 [DOI] [PubMed] [Google Scholar]
- Tajima Y, Imamura A, Kiba T, Amano Y, Yamashino T, Mizuno T (2004) Comparative studies on the type-B response regulators revealing their distinctive properties in the His-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol 45 28–39 [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A (2007) ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol 48 263–277 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruere J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, van Onckelen H, Schmülling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS (2001) Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol 8 625–637 [DOI] [PubMed] [Google Scholar]
- Yang SH, Yu H, Goh CJ (2003) Functional characterisation of a cytokinin oxidase gene DSCKX1 in Dendrobium orchid. Plant Mol Biol 51 237–248 [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Yamashino T, Amano Y, Tajima Y, Imamura A, Sakakibara H, Mizuno T (2007) Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol 48 84–96 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.