Abstract
The genome of Arabidopsis (Arabidopsis thaliana) contains five sequences with high similarity to FLAVONOL SYNTHASE1 (AtFLS1), a previously characterized flavonol synthase gene that plays a central role in flavonoid metabolism. This apparent redundancy suggests the possibility that Arabidopsis uses multiple isoforms of FLS with different substrate specificities to mediate the production of the flavonols, quercetin and kaempferol, in a tissue-specific and inducible manner. However, biochemical and genetic analysis of the six AtFLS sequences indicates that, although several of the members are expressed, only AtFLS1 encodes a catalytically competent protein. AtFLS1 also appears to be the only member of this group that influences flavonoid levels and the root gravitropic response in seedlings under nonstressed conditions. This study showed that the other expressed AtFLS sequences have tissue- and cell type-specific promoter activities that overlap with those of AtFLS1 and encode proteins that interact with other flavonoid enzymes in yeast two-hybrid assays. Thus, it is possible that these “pseudogenes” have alternative, noncatalytic functions that have not yet been uncovered.
Flavonoids are well-known plant natural products that have a wide array of physiological functions in plants, while also contributing significant health-promoting properties to plant foods. Many of the roles in plants, including UV protection, regulation of auxin transport, modulation of flower color, and signaling, have been attributed to a subclass of flavonoids known as flavonols, which are among the most abundant flavonoids (Bohm et al., 1998; Harborne and Williams, 2000). These same compounds have also been identified with the antioxidant, antiproliferative, antiangiogenic, and neuropharmacological properties of flavonoids (Lee et al., 2005; Kim et al., 2006; Kim and Lee, 2007). Although concerns have been raised about the potential deleterious effects of high levels of dietary or supplemental flavonols, these appear to be largely unfounded (Havsteen, 2002; Okamoto, 2005). As a result, understanding the synthesis of flavonols is of particular interest from the perspective of metabolic engineering, as illustrated by recent efforts to up-regulate flavonol biosynthesis in tomato (Lycopersicon esculentum) fruit (Schijlen et al., 2006) and rice (Oryza sativa; Reddy et al., 2007) and to overproduce flavonols in Escherichia coli (Leonard et al., 2006; Katsuyama et al., 2007).
Most plants synthesize derivatives of one or more of the three major flavonols, quercetin, kaempferol, and myricetin, which differ by only a single hydroxyl group on the flavonoid B ring and yet can specify quite different biological activities. The ratio of these flavonols varies substantially among different tissues and can be altered in response to environmental cues (Winkel-Shirley, 2002). For example, UV-B light has been shown to specifically induce the accumulation of quercetin derivatives in Petunia, which have a higher antioxidant potential and therefore are deemed more effective sunscreens than other flavonols (Ryan et al., 2002). Quercetin has also been shown to be most effective at inhibiting the auxin efflux carrier (Jacobs and Rubery, 1988), and quercetin and kaempferol exhibit different spatial and temporal distribution patterns in Arabidopsis (Arabidopsis thaliana) roots that are consistent with roles in controlling auxin movement (Peer et al., 2001, 2004). Interestingly, quercetin is also frequently identified as a primary bioactive compound in medicinal and food plants (Havsteen, 2002; Kim et al., 2006; Nichenametla et al., 2006).
The synthesis of flavonol aglycones has long been attributed to a single enzyme, flavonol synthase (FLS), which competes with several other enzymes for dihydroflavonol substrates. Among these are flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′,5′-hydroxylase, which mediate the addition of hydroxyl groups to the B ring of flavanones, flavones, dihydroflavonols, and flavonols (Hagmann et al., 1983; Kaltenbach et al., 1999), and dihydroflavonol reductase (DFR), which drives flux away from flavonols into anthocyanin and proanthocyanidin biosynthesis (Davies et al., 2003). More recently, anthocyanidin synthase (ANS) has been shown to use both dihydroflavonols and leucoanthocyanidins in vitro for the synthesis of flavonols, the latter suggesting an alternative route to quercetin using a substrate normally associated with anthocyanin and proanthocyanidin biosynthesis (Turnbull et al., 2004; Wellmann et al., 2006; Lillo et al., 2008). Some of the competition for common substrates appears to be mediated by the differential expression of genes required for upstream (flavonol) versus downstream (anthocyanin and proanthocyanidin) pathways (Pelletier et al., 1997; Mehrtens et al., 2005). Yet, how these enzymes cooperate to control the metabolic balance among the branch pathways of flavonoid biosynthesis, possibly through participation in one or more enzyme complexes, remains to be fully determined. In fact, efforts to use enzymes such as FLS and ANS to engineer altered flavonoid profiles have had consistently unpredictable outcomes (Schijlen et al., 2006; Wellmann et al., 2006; Reddy et al., 2007).
Flavonoid biosynthesis in Arabidopsis is relatively simple compared with that in many other higher plants, involving the production of only three major classes of compounds: flavonols, anthocyanins, and proanthocyanidins. With only one apparent exception, the enzymes of the central flavonoid pathway, including chalcone synthase (CHS), chalcone isomerase (CHI), DFR, flavanone 3-hydroxylase (F3H), F3′H, ANS, and anthocyanidin reductase, are encoded by single genes. The exception is FLS, for which we have identified six homologs in the Arabidopsis genome. This raises the possibility that gene duplication has led to a group of differentially regulated genes encoding isoforms with varying substrate specificities, facilitating the synthesis of different flavonols to meet the dynamic physiological needs of the plant. Here, we describe an effort to test this hypothesis by examining the expression patterns and biochemical characteristics of the six Arabidopsis FLS isoforms as well as the impact of knockout mutations on phenotypes associated with flavonoid metabolism. The results of these experiments provide new insights into the mechanisms controlling flavonol accumulation in vivo.
RESULTS
Identification of a FLS Gene Family in Arabidopsis
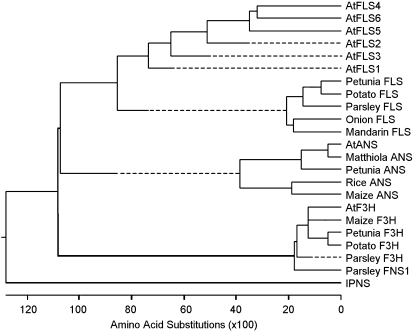
The first Arabidopsis gene with high homology to FLS genes from other plant species, AtFLS1 (At5g08640), was originally identified in the EST database a number of years ago (Pelletier et al., 1997). Analysis of flavonols in an En-induced mutant line and activity assays with recombinant protein confirmed that the gene encoded a protein with FLS activity (Wisman et al., 1998; Prescott et al., 2002). Five additional sequences with high homology to FLS genes were subsequently uncovered during sequencing of the Arabidopsis genome, which we have designated AtFLS2 (At5g63580), AtFLS3 (At5g63590), AtFLS4 (At5g63595), AtFLS5 (At5g63600), and AtFLS6 (At5g43935; Arabidopsis Genome Initiative, 2000). These sequences cluster more closely with FLS genes from other plants than with other plant flavonoid 2-oxoglutarate-dependent dioxygenases (2-ODDs), both at the nucleotide (data not shown) and predicted amino acid (Fig. 1) levels. The six genes are all located on chromosome 5, with AtFLS2, -3, -4, and -5 arranged in a 7.5-kb tandem array (Fig. 2). The four clustered genes are no more closely related to each other than to the other two genes, with AtFLS2 the most distantly related at the nucleotide level (48%–51% identity) and the others exhibiting 62% to 73% identity. This suggests that the duplications leading to the amplification of this gene family, including the AtFLS2 to -5 tandem array, are ancient events.
Figure 1.
Phylogeny of the AtFLS isoforms and other dioxygenases of the flavonoid pathway based on predicted amino acid sequences. IPNS, Isopenicillin N synthase.
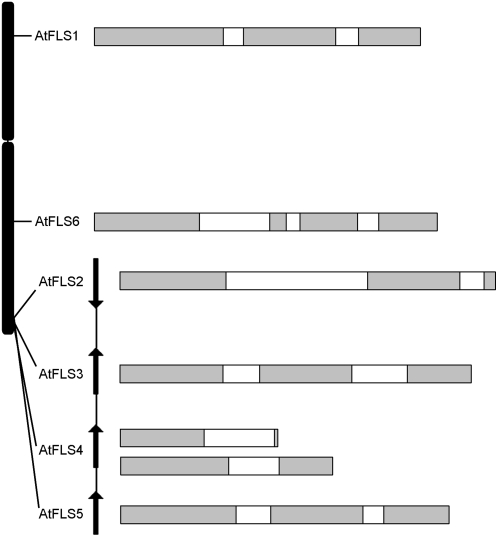
Figure 2.
Arrangement of the AtFLS genes in the Arabidopsis genome. All six genes are located on chromosome 5. AtFLS2 to -5 are clustered in a 7.5-kb region. AtFLS1, -3, and -5 appear to constitute full-length or nearly full-length coding sequences, while the AtFLS2, -4, and -6 coding regions are truncated, with the AtFLS4 gene giving rise to multiple forms, apparently due to alternative/aberrant splicing. Exons are shown in gray, and introns are shown in white.
AtFLS1, -3, and -5 appear to encode full-length proteins and all contain two introns at identical positions, corresponding to two of the five intron sites that are conserved among plant 2-ODD genes (Prescott and John, 1996). The AtFLS2 gene contains a large second intron, and the predicted coding sequence encodes a truncated protein lacking key C-terminal residues required for Fe2+ coordination (H220, D222, and H276 in AtFLS1) and α-ketoglutarate binding (R286 and S288 in AtFLS1; Lukacin and Britsch, 1997; Wilmouth et al., 2002). The situation is more complex for AtFLS4 and AtFLS6, both of which are predicted to contain an additional intron relative to the other four AtFLS genes in what is otherwise the second exon (The Arabidopsis Information Resource [TAIR] 7.0 genome sequence, released April 23, 2007; Swarbreck et al., 2008). To date, no full-length cDNA sequences have been reported for AtFLS4. Of the four AtFLS4 EST sequences available in GenBank (Alexandrov et al., 2006) and the RIKEN Genomic Sciences Center (Seki et al., 2004), only one spans the region containing the predicted additional intron, and these sequences are not spliced out, severely truncating the coding region. As described in further detail below, reverse transcription (RT)-PCR analysis of ecotype Landsberg erecta (Ler) roots identified multiple transcripts for AtFLS4 that apparently arise from a complex differential splicing scheme. Sequence analysis of four of these cDNAs showed erroneous splicing at the 3′ ends of exons 1 and 2, resulting in premature stop codons (Fig. 2); all also retained the additional predicted intron sequences. In the case of AtFLS6, no cDNA or EST sequences have been reported to date, and efforts to amplify transcripts from root RNA by RT-PCR were unsuccessful (data not shown). If this gene is expressed at all, the transcript is likely to be processed in a manner similar to that used for AtFLS4. Thus, it appears that AtFLS2, -4, and -6 are pseudogenes that are unlikely to contribute to flavonol synthase activity in Arabidopsis.
AtFLS Gene Expression Patterns
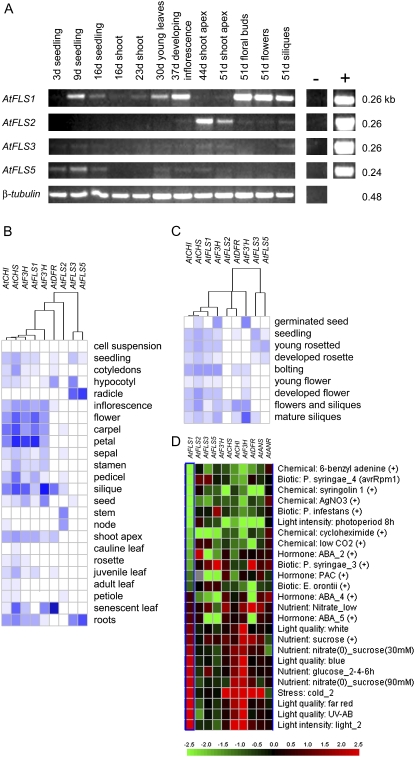
To test the possibility that the AtFLS genes have acquired differential patterns of expression, transcript abundance and promoter activities were examined over the course of Arabidopsis plant growth and development. Plants growing in soil under a 16-h photoperiod were sampled at regular intervals over a 7-week period. Semiquantitative RT-PCR was used to compare the abundance of the transcripts in whole seedlings and in plant organs known to accumulate high levels of flavonols (Shirley et al., 1995). AtFLS1 displayed the broadest pattern of expression (Fig. 3A). The highest AtFLS1 transcript levels were detected during the reproductive stage, in the developing inflorescence, floral buds, flowers, and siliques. Lower, but still substantial, levels were detected in the roots and shoots of young seedlings and in leaves of later vegetative stages. This pattern is consistent with the publicly available microarray data for different stages of Arabidopsis development (Genevestigator [Fig. 3, B and C] and AtGenExpress [data not shown]). Interestingly, the AtFLS2 pseudogene appears to be expressed at high levels in the shoot apex and lower stem, tissues in which AtFLS1 transcripts were not detected, and at low levels in flowers and siliques (Fig. 3A). These patterns are also reflected in the microarray data (Fig. 3, B and C). AtFLS5 appeared to be expressed at much lower levels, with transcripts detected primarily in seedling roots, while AtFLS3 expression was undetectable or extremely low in all samples examined; these findings are again consistent with the microarray data (Fig. 3, B and C). Expression of the AtFLS4 and -6 pseudogenes was not examined, as these genes appear to have little, if any, expression based on the EST databases; they are also not represented on either the 8 K or 22 K array used to generate the data compiled in Genevestigator.
Figure 3.
Analysis of AtFLS gene expression. A, Semiquantitative RT-PCR analysis of plants and plant tissues at various developmental stages. Positive controls (+) contained cDNA clones for each gene, and negative controls (−) contained no template. RNA samples from whole seedlings and shoots of 16-d-old plants were analyzed and used to infer gene expression in roots. B to D, Data from the public microarray databases for the AtFLS genes and other select flavonoid genes obtained using Genevestigator (Zimmermann et al., 2004).
It is interesting that the microarray data indicate that expression of AtFLS1, but not AtFLS2, -3, or -5, parallels that of the other “early” flavonoid genes during development and in the response to light and several other external cues (Fig. 3, B–D). This is also reflected in the ATTED-II database, where AtFLS1 expression has a 0.83 to 0.84 correlation score with other “early” flavonoid genes, while AtFLS3 and AtFLS5 are correlated with each other (score of 0.70) but not with any other flavonoid genes (Obayashi et al., 2007).
Developmental gene expression patterns were further investigated by analyzing transgenic plants containing AtFLS1, -2, -3, and -5 promoter sequences fused to the GUS gene. AtFLS1, -3, and -5 were expressed in the root-shoot transition zone of 3-d-old seedlings and along the length of the roots at 9 d (Fig. 4, A–C and G–I). In 9-d-old seedlings, AtFLS3 promoter activity was strongest in the vascular bundle, while the AtFLS5 promoter was active from the vascular bundle up to, but not including, the epidermis, although it was not possible to resolve staining differences between the endodermis and cortex. Compared with AtFLS3 and -5 in 9-d-old roots, AtFLS1 expression appeared more sporadically, with no consistent expression pattern emerging in roots at this stage of development. AtFLS1 and -3 root expression decreased in later vegetative stages, but AtFLS5 was sporadically detected in various positions of older roots (data not shown). All three isoforms showed expression in initiating lateral roots, especially in young plants (Fig. 4, J–L).
Figure 4.
Developmental, organ-specific, and parasite-induced expression of the AtFLS genes. Promoter-GUS fusions were analyzed in multiple independent transgenic lines by histochemical staining with 5-bromo-4-chloro-3-indolyl-β-d-glucuronide. Staining was observed primarily in 3-d-old seedlings (A–C), 9-d-old seedlings (D–I), initiating lateral roots in plants of various ages (J–L), trichomes on 30-d-old plants (M–O), and reproductive structures of 51-d-old plants (P–U). Arrows identify the root-shoot transition zone in A to C and G to I and lateral roots in J to L. Unlike the AtCHS promoter (V), expression of the AtFLS1 to -5 promoters was not induced by infection with the plant parasite O. aegyptiaca (W–AA).
In vegetative shoots, AtFLS1 promoter activity was consistently detected in young leaves, appearing in the upper epidermal tissues and especially concentrated in the youngest initiating leaves (near the shoot apical meristem), including the trichomes (Fig. 4M). The AtFLS3 and -5 promoters were also active in young leaves, but they were limited to trichomes for AtFLS3 and to the meristem for AtFLS5 (Fig. 4, N and O). While AtFLS1 expression was visible in leaf tissues for all transgenic lines that we investigated, this was not the case for AtFLS3 and -5, in which expression was limited to a few lines each, consistent with the overall lower gene expression levels for these isoforms as assessed by RT-PCR. High levels of AtFLS1 promoter activity were also detected in reproductive tissues, specifically in petals, stamens (filament and anther), carpels (stigma), and siliques (pedicel/valve junction), and sporadically through the perianth of young bud clusters and mature flowers (Fig. 4, P–R), consistent with the results of RT-PCR analysis. No AtFLS3 or AtFLS5 promoter activity was detected in any of these tissues. However, this is the one stage at which the AtFLS2 promoter was observed, with the highest GUS activity occurring in the shoot apex at the base of the inflorescence bolt and in the pedicel/valve junction (Fig. 4, S–U), consistent with the RT-PCR experiments and the Genevestigator microarray database.
A hallmark of flavonoid genes such as CHS, CHI, and DFR is that their expression is strongly induced by a variety of environmental factors, including both biotic and abiotic factors that cause mechanical damage to the plant (McKhann and Hirsch, 1994; Djordjevic et al., 1997; Reymond et al., 2000; Richard et al., 2000; Peters and Constabel, 2002; Pang et al., 2005). One example is the induction of CHS gene expression in diverse plant species by Orobanche aegyptiaca, a plant parasite that forms a physical connection with host roots and activates a variety of wound- and jasmonic acid-inducible genes (Griffitts et al., 2004; J.H. Westwood, unpublished data). To test whether the AtFLS genes were also induced by infection with this parasite, the promoter-GUS plants were grown for 3 weeks in a semihydroponic system and then infected with O. aegyptiaca as described by Westwood (2000). Unlike the AtCHS promoter, which was strongly induced upon invasion of the O. aegyptiaca haustorium, the AtFLS1 to -5 promoters did not exhibit any detectable activity in this assay (Fig. 4, V–AA). The AtFLS1 to -5 promoters were also not induced when roots were accidentally damaged during handling, unlike the CHS promoter, which showed strong activation at sites of breakage (data not shown). A similar lack of wound inducibility of FLS genes relative to other flavonoid genes was recently reported in Populus and was suggested to reflect the lack of participation of FLS in the synthesis of condensed tannin defense molecules (Tsai et al., 2006). Therefore, although AtFLS1 is coordinately expressed with other flavonoid genes during development (Pelletier et al., 1997; Fig. 3), it is also subject to distinct regulation in response to environmental factors.
In Vitro Enzyme Activity of AtFLS1, -3, and -5
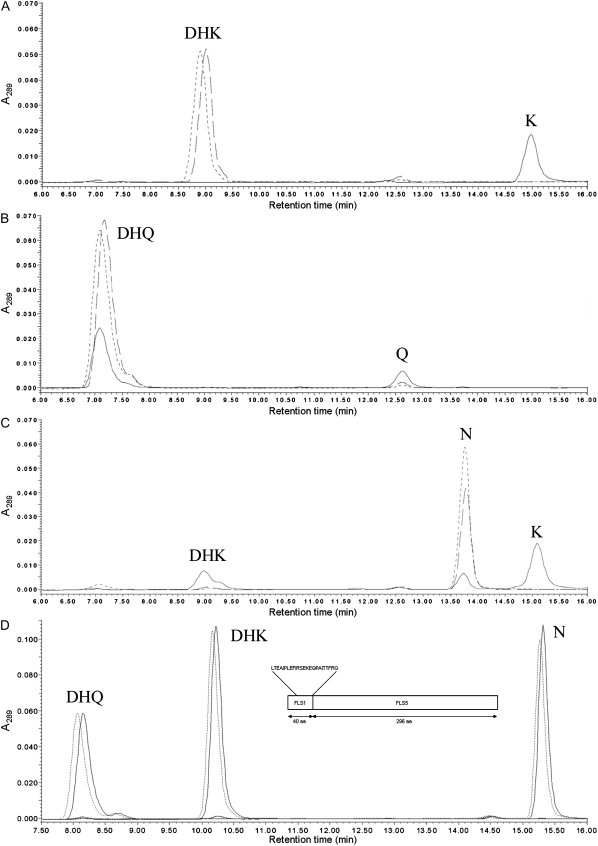
In Arabidopsis, as in other plant species, the relative levels of quercetin and kaempferol vary substantially depending on the tissue and cell type (Peer et al., 2001; Tohge et al., 2005; Kerhoas et al., 2006; Stracke et al., 2007). To test the possibility that differential expression of AtFLS isoforms with distinct substrate specificities could determine the relative ratios of these two flavonols, AtFLS1, -3, and -5 enzymes were produced in E. coli as thioredoxin fusion proteins and assayed using a variety of substrates. Consistent with previous reports, AtFLS1 was very effective at converting dihydrokaempferol (DHK) to kaempferol (Fig. 5A; Wisman et al., 1998; Lukacin et al., 2003), while only a portion of the supplied dihydroquercetin (DHQ) was converted to quercetin by this enzyme under the same conditions (Fig. 5B; Turnbull et al., 2004). In addition, a portion of naringenin, normally the substrate for F3H, was converted to DHK by AtFLS1, some of which was subsequently converted to kaempferol (Fig. 5C; Prescott et al., 2002). Thus, AtFLS1 exhibited a clear preference for DHK in these assays, while, surprisingly, DHQ was used less well than even naringenin. However, neither AtFLS3 nor AtFLS5 appeared to have enzyme activity with any of the substrates under a variety of conditions that included variations in pH, temperature, enzyme and substrate concentration, and enzyme enrichment and cleavage procedures (Fig. 5, A–C; data not shown).
Figure 5.
AtFLS1, AtFLS3, and AtFLS5 enzyme activity. Recombinant AtFLS1 (solid line), AtFLS3 (dashed line), and AtFLS5 (dotted line) proteins were assayed with the substrates DHK (A), DHQ (B), and naringenin (C). HPLC scans extracted at 289 nm are shown, with peaks labeled as DHK, DHQ, K (kaempferol), Q (quercetin), and N (naringenin). D, Analysis of the AtFLS1/AtFLS5 chimera. HPLC scans of assays of the AtFLS1/AtFLS5 chimera (solid line) and a thioredoxin negative control (dotted line) with DHQ, DHK, and N or without substrate are shown. The inset shows the structure of the AtFLS1/AtFLS5 chimera formed from the 40 N-terminal amino acids of AtFLS1 and the 296 C-terminal amino acids of AtFLS5. Introduced amino acids are shown above the structure.
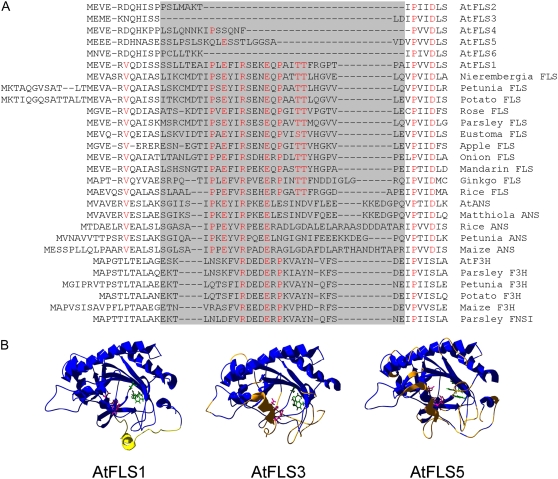
Close inspection of the primary sequences of the AtFLS proteins identified a region spanning approximately 30 amino acids that is present in AtFLS1 and all other plant flavonoid dioxygenases but that is altered or absent in AtFLS2 to -6 (Fig. 6A). Included in this region are Arg and Glu residues (Arg-25 and Glu-29 in AtFLS1) that are invariant in all other plant dioxygenases as well as numerous other residues that are strictly conserved among the flavonoid 2-ODD enzymes, FLS, F3H, ANS, and FNS1. To analyze this region on a structural level, homology models were constructed based on the crystal structure of Arabidopsis ANS (Protein Data Bank identifier 1GP4, Wilmouth et al., 2002), with which AtFLS1, -3, and -5 exhibit 37.8%, 33.9%, and 31.4% amino acid identity, respectively (Fig. 6B). The root mean square deviation (RMSD) values for the homology models of AtFLS3 and AtFLS5 compared with AtFLS1 were 1.23 and 1.48 Å, respectively, indicating that the structures are quite similar overall, including the architecture of the jellyroll core. However, there appeared to be substantial differences in the positions of several key active site residues among these proteins. In particular, the Fe2+-coordinating residue Asp-222 differed by 3.28 Å between AtFLS1 and -3, while the adjacent His-220 varied by 2.55 Å between AtFLS1 and -5. The largest apparent differences are near the N terminus and in the largely unstructured C terminus. The conserved 30 amino acids that are altered or absent in the AtFLS2 to -6 proteins constitute a region in the AtFLS1 model that contains a seven-residue α-helix (residues 26–31) near the mouth of the jellyroll motif and is otherwise largely unstructured. The absolutely conserved Arg-25 is adjacent to this helix, while the conserved Glu-29 is in the center of the helix. This structural element appears to be missing in AtFLS3 and AtFLS5. An additional N-terminal α-helix in the homology model (AtFLS1 residues 5–8) is also absent in AtFLS3 and is present, but appears to be positioned differently, in AtFLS5.
Figure 6.
Structural analysis of the AtFLS1, -3, and -5 proteins. A, N-terminal sequence alignment showing the highly conserved region that is altered or missing in AtFLS2 to -6 (highlighted in gray), including residues that are strictly conserved in the various enzyme subclasses (shown in red). B, Homology models generated based on the crystal structure of AtANS (At4g22880) are shown looking into the core of the jellyroll motif. The predicted Fe2+-coordinating residues are shown in pink, and the α-ketoglutarate binding residues are shown in green. The yellow region in AtFLS1 identifies the N-terminal fragment missing in all other AtFLS isoforms. Regions colored orange in AtFLS3 and -5 are those with RMSD values greater than 2.75 Å relative to AtFLS1.
Further evidence for the functional importance of this N-terminal region comes from analysis of expression constructs derived from AtFLS1 clone EST 153O10T7, which lacks the coding sequences for the 21 N-terminal amino acids (Pelletier et al., 1999). The truncation completely eliminates the first α-helix and the first seven residues in an unstructured region of the conserved 30-amino acid fragment. Protein produced from this construct had no activity with any of the tested substrates when assayed under the same conditions as the full-length AtFLS1 (data not shown).
To test the possibility that the N-terminal region of AtFLS1 could restore the activity of the inactive AtFLS isoforms, a chimeric construct was generated in which the N-terminal 30 amino acids of AtFLS5 were replaced with the first 40 amino acids of AtFLS1 (Fig. 5D). However, the chimeric protein also had no detectable activity with any of the tested substrates. This indicates that the 21 N-terminal amino acids of AtFLS1 are required for activity in that enzyme but are not sufficient to restore activity to AtFLS5. This suggests that the structural integrity of the remaining AtFLS5 gene product underwent further decay following loss of the critical N-terminal residues.
Two-Hybrid Analysis of Interactions with Other Flavonoid Enzymes
The possibility that the FLS proteins may serve nonenzymatic roles as part of a flavonoid biosynthetic metabolon was investigated by yeast two-hybrid analysis of potential interactions of AtFLS1, -3, and -5 with AtCHS, AtCHI, AtF3H, and AtDFR. The proteins were analyzed in all possible pairwise combinations, fused to either the activation domain or the binding domain of GAL4 (Chevray and Nathans, 1992; Kohalmi et al., 1998). The observed interactions are summarized in Table I. AtFLS1, -3, and -5 interacted with AtCHS when they were fused to the GAL4 activation domain but not when fused to the bait domain. AtFLS1 also interacted with AtF3H and AtDFR in both configurations. The only other positive result was for AtFLS5 fused to the GAL4 bait domain with AtDFR. These findings are reminiscent of those reported previously for AtCHS, AtCHI, and AtDFR (Burbulis and Winkel-Shirley, 1999) and suggest that AtFLS1 may function as yet another component of a flavonoid multienzyme complex. Moreover, although AtFLS3 and AtFLS5 do not have measurable enzyme activity, these proteins appear to have retained the ability to interact physically with other members of the central flavonoid biosynthetic pathway and could conceivably play structural and/or regulatory roles in flavonoid metabolism.
Table I.
Yeast two-hybrid analysis of interactions between AtFLS1, -3, and -5 and other flavonoid enzymes
AtFLS fused to the activation domain/AtFLS fused to the binding domain.
ND, Not determined.
AtFLS1 to -6 in Planta Gene Function
To further investigate the possibility that AtFLS genes play unanticipated roles in flavonoid biosynthesis in planta, knockout lines were identified for each of the genes in the SALK and GABI-KAT T-DNA collections (Alonso et al., 2003; Rosso et al., 2003). Homozygous lines were obtained in the Columbia (Col) background for AtFLS2 (GABI 429B10), AtFLS3 (SALK_050041), AtFLS4 (SALK_002309), AtFLS5 (GABI 317E12), and AtFLS6 (SALK_003879) as described in “Materials and Methods.” The only knockout candidate for AtFLS1, SALK_076420, was found to be embryo lethal in the homozygous state (data not shown). However, this insertion lies in the intergenic region shared by AtFLS1 and a divergently transcribed gene (At5g08630) that encodes a DDT domain-containing protein of unknown function. Two other T-DNA insertions, in the coding region of At5g08630 (SALK_004358 and SALK_039219), were also homozygous lethal (data not shown), indicating that this phenotype in the SALK_076420 line was due to disruption of the adjacent gene, not AtFLS1. A line homozygous for an insertion in the 5′ untranslated region of AtFLS1, AJ588535, was subsequently recovered in the ecotype Wassilewskija (Ws) background from the INRA collection (Ortega et al., 2002).
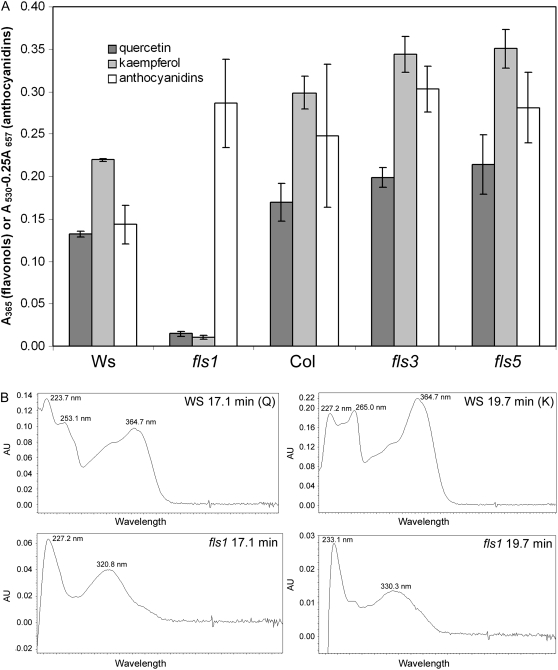
The T-DNA lines were then used to explore the contributions of the AtFLS1 to -6 genes to flavonoid biosynthesis. Extracts were prepared from whole seedlings and from flowers, in which these genes were found to be expressed at high levels in the experiments described above. The insertion in AtFLS1 resulted in a substantial reduction in peaks with retention times corresponding to those of quercetin and kaempferol, and these peaks had different UV-visible light absorption spectra than did those from wild-type Ws and the authentic standards (Fig. 7; supplemental material). This suggested that these compounds in fls1 were sinapate esters, not flavonols, similar to what is observed in Arabidopsis CHS and F3H null mutants (Li et al., 1993; Owens et al., 2008; Fig. 7B). To examine this possibility further, seedling extracts were analyzed by liquid chromatography-mass spectrometry (LC-MS); surprisingly, small quantities of both quercetin and kaempferol were detected in fls1 (Supplemental Fig. S2). The fls1 plants also exhibited a much more intense red coloration of the hypocotyl and cotyledons during germination and at the base of the stalk of mature plants compared with the wild type (data not shown). Analysis of anthocyanidin levels in seedlings showed that fls1 seedlings accumulated approximately twice as much of these pigments per gram dry weight as the wild-type Ws counterpart (Fig. 7A). This apparent diversion of flux into neighboring branch pathways is similar to what has been reported for other flavonoid mutants, such as banyuls, which is deficient in ANS (Devic et al., 1999). In contrast, neither the fls3 and fls5 lines nor any of the other FLS mutant lines exhibited a detectable effect on flavonol or anthocyanidin accumulation, either in flowers or seedlings (Fig. 7A; supplemental material; data not shown). This suggests that only AtFLS1 contributes to flavonol synthesis in Arabidopsis.
Figure 7.
Effects of fls1, fls3, and fls5 mutations on flavonol and anthocyanidin levels in 4-d-old seedlings. A, Quercetin and kaempferol levels were quantified by HPLC by extracting chromatograms at 365 nm and integrating peaks corresponding to authentic standards. Anthocyanidin levels were determined spectrophotometrically. B, Representative UV-visible light spectra for peaks in Ws and fls1 with retention times corresponding to quercetin (Q) and kaempferol (K) standards.
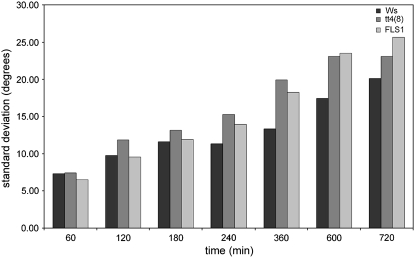
The fls1 mutant also provides a new genetic tool for exploring the role of flavonols in root gravitropism. Extensive work over the past several years with Arabidopsis tt4 mutants has provided strong support for a model in which flavonoids, and flavonols in particular, function to slow auxin transport in specific cell files in order to cause root curvature (Brown et al., 2001; Buer and Muday, 2004; Lewis et al., 2007; Peer and Murphy, 2007). To provide further support for the specific role of flavonols, as opposed to other flavonoids, in this process, the gravity response of fls1 roots was compared with that of tt4(8), an allele in the Ws background, and the wild-type Ws. Surprisingly, neither tt4(8) nor fls1 exhibited a substantial difference in the response of roots to gravity relative to the Ws wild type (data not shown). This could reflect differences in the ecotype that was used (Ws versus Col) compared with previously published experiments. However, both fls1 and tt4(8) showed a distinct difference from Ws in the amount of variation in the response among seedlings (Fig. 8). This suggests that, although the overall response is similar in Ws, with or without flavonols present, the precision of the response is much higher in the presence of these compounds. The similarity between fls1 and tt4(8) provides further evidence that it is flavonols, and not other flavonoids, that mediate root bending in response to gravity.
Figure 8.
Effects of the fls1 and tt4(8) mutations on root gravitropism. Murashige and Skoog/Suc/agar plates containing 4-d-old wild-type and mutant seedlings were rotated 90°, and root angles were measured relative to the original direction of growth. Bar graph shows the sd in bending angle for the tt4(8) and fls1 mutants versus the corresponding Ws wild type.
DISCUSSION
Gene families are common features of the genomes of complex organisms, including plants (Jander and Barth, 2007). Still, the finding that the Arabidopsis genome contains six sequences with high homology to FLS was surprising in that all other flavonoid enzymes in this species appear to be encoded by single-copy genes (Winkel, 2006). The presence of multiple AtFLS genes suggested the possibility of differentially expressed isoforms with different substrate specificities. This could explain the different relative levels of kaempferol and quercetin that are present in various tissues and under different environmental conditions in Arabidopsis (Peer et al., 2001; Ryan et al., 2002; Tohge et al., 2005; Kerhoas et al., 2006; Lea et al., 2007; Stracke et al., 2007). It was also consistent with the report by Wisman et al. (1998) that En-induced disruption of the AtFLS1 gene abolished quercetin accumulation but had no effect on kaempferol levels, suggesting that another source of FLS activity was present.
Therefore, we carried out a thorough biochemical and genetic analysis of the six predicted AtFLS genes. Unlike the situation for the putative Arabidopsis cinnamyl alcohol dehydrogenase multigene family, in which the products of six genes had high cinnamyl alcohol dehydrogenase activity and three had low activity (and eight additional genes were simply misannotated; Kim et al., 2004), in this case only one of six genes was found to encode a catalytically competent protein. The products of the other five genes encode products that appear to lack critical functional residues, as a result of premature stop codons (AtFLS2 and -6), alternative splicing and missplicing (AtFLS4), or loss of a small region near the 5′ end of the gene that may have resulted in further functional degeneration of the downstream sequences (AtFLS3 and -5). Thus, the theory that Arabidopsis uses different FLS genes to mediate differential synthesis of quercetin and kaempferol in different tissue or cell types appears to be incorrect. One possibility is that the FLS activity of the ANS enzyme may contribute to the differential accumulation of kaempferol and quercetin, as suggested by Lillo et al. (2008). Differential expression of the F3′H enzyme could also mediate these ratios, as illustrated by the large increases in kaempferol levels observed in Petunia flowers expressing an antisense construct for F3′H (Lewis et al., 2006). In addition, recent work on the PAP1 and PFG1-3 R2R3-MYB factors indicates that this variation is regulated, at least in part, at the level of gene expression, with a network of many different transcription factors interacting with the various flavonoid gene promoters to orchestrate the differential biosynthesis of flavonoid products (Tohge et al., 2005; Stracke et al., 2007).
This network of transcriptional control also explains how AtCHS and AtFLS1 may be coordinately regulated during development but differentially expressed in response to parasitization by Orobanche. Even though flavonoids are not required for the parasitization process, in that the CHS mutant tt4(2YY6) is just as efficiently parasitized as the wild type, CHS may still contribute to the localized production of flavonoids as part of the plant stress response system, as parasitized tt4 plants accumulated a lower root mass than wild-type controls (Westwood, 2000). The lack of induction of the AtFLS1 promoter by the parasite suggests that this does not involve FLS activity, which is surprising since flavonols are known to have potent free radical-scavenging activity (Braca et al., 2003).
It also remains to be explained how the fls1 T-DNA knockout line produced small quantities of quercetin and kaempferol at the seedling stage (Fig. 7; Supplemental Fig. S2) while fls1 En mutants accumulated quercetin, both in UV-treated mature plants and in seeds (Wisman et al., 1998; Routaboul et al., 2006). Although it appears that none of the other AtFLS genes contribute FLS activity, it is possible that AtANS is able to do so (Turnbull et al., 2004; Lillo et al., 2008). Like AtFLS1, AtANS can produce flavonols at high efficiency in vitro from naringenin, DHK, and DHQ. The fact that AtANS is not able to fully substitute for AtFLS1 in vivo suggests that the intracellular organization and/or localization of the flavonoid pathway could restrict the access of ANS to these intermediates. Interestingly, AtANS also produces quercetin via an alternative route, from its “natural” substrate, leucocyanidin (Turnbull et al., 2000, 2004). Because quercetin is the preferred product of this reaction in vitro, it has been suggested that the production of cyanidin glycosides involves channeling of the flav-2-en-3,4-diol intermediate directly from ANS to a flavonoid glycosyltransferase (Nakajima et al., 2001; Turnbull et al., 2003). This channel may be sufficiently “leaky” to allow some accumulation of quercetin, which is uncovered in the fls1 mutant lines.
If the AtFLS2 to -6 genes do not contribute to flavonol biosynthesis, then what drove the duplication of these genes at two sites far removed from AtFLS1 in the Arabidopsis genome? Perhaps part of the explanation has to do with the fact that AtFLS1 is located in a 1-Mb region exhibiting the second highest level of evidence of recent positive selection; this region of the genome may thus have limited potential for diversification and the evolution of new gene function (Clark et al., 2007). Does the fact that the AtFLS2 to -6 genes have apparently been maintained over substantial evolutionary time indicate that they once had, or still retain, functional importance? AtFLS4 and -6 appear to be fully quiescent, nonfunctional pseudogenes. However, AtFLS2, -3, and -5 are still expressed in patterns that partially overlap with that of AtFLS1. Yeast two-hybrid assays suggest that AtFLS3 and -5 could compete with AtFLS1 for interactions with other proteins, perhaps during the assembly and/or localization of the flavonoid enzyme complex.
The possibility also remains that all four expressed FLS genes have other as yet unknown functions. We recently reported that CHS and CHI are localized not just at the endoplasmic reticulum but also in the nucleus (Saslowsky et al., 2005), and this now also appears to be the case for an (iso)flavone malonyltransferase from Medicago truncatula (Yu et al., 2008). Therefore, these proteins may have “moonlighting” functions, similar to a growing list of enzymes in plants and other organisms with functions independent of their catalytic activities (Moore, 2004; Sriram et al., 2005). It should also be noted that the Arabidopsis genome also contains distant relatives of CHS, CHI, F3H, and DFR, although with much less similarity than for the FLS gene family (14%–44% amino acid identity, and one exception, at 63%, for CHI; Supplemental Table S3; TAIR 7.0 genome sequence, released April 23, 2007; Swarbreck et al., 2008). The phenotypes of mutations in CHS, CHI, F3H, and DFR (tt4, tt5, tt6, and tt3, respectively) indicate that the distant relatives are unlikely to contribute directly to flavonoid biosynthesis. Therefore, it appears that the gene family model described for FLS, with one catalytically active member and several pseudogenes, may also apply to other flavonoid genes, particularly in the case of CHI. There is also a growing awareness that the “promiscuity” of metabolic enzymes such as ANS, as well as flavonoid glycosyltransferases (Lim et al., 2004) and O-methyltransferases (Deavours et al., 2006), is more the rule than the exception (Taglieber et al., 2007). These “alternative” functions of otherwise well-characterized proteins may represent new paradigms that must be taken into account in efforts to develop framework models of cellular metabolism.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown in 7.5- × 5.5- × 5.5-cm pots containing Sunshine Mix 1 soil (Sungro Horticulture Processing) in a climate-controlled incubator (I-66LLVL; Percival Scientific) with a 16-/8-h photoperiod, 45 μmol m−2 s−1 fluorescent light, and 20°C constant temperature. The soil was amended with Osmocote controlled-release fertilizer (Scotts) or weekly fertilizing with 0.015% (w/v) Miracle-Gro 15-30-15 (Scotts). Under these conditions, inflorescence development was prominent at 6 weeks after planting. Seedlings for the analysis of flavonol and anthocyanidin content were grown on the surface of Murashige and Skoog/Suc/agar plates under continuous light as described previously (Saslowsky and Winkel-Shirley, 2001).
Sequence Analysis of the AtFLS Gene Family
Sequences for the six members of the AtFLS gene family in Col (AtFLS1, accession no. At5g08640; AtFLS2, At5g63580; AtFLS3, At5g63590; AtFLS4, At5g63595; AtFLS5, At5g63600; AtFLS6, At5g43935) were obtained from TAIR and analyzed using Lasergene (DNAStar). Gene maps were prepared by comparing Col and Ler sequences using published ESTs (TAIR) and cloned Ler cDNAs (see below).
Construction of AtFLS Promoter-GUS Reporter Gene Fusions, Arabidopsis Transformation, and Histochemical Localization of GUS Activity
Intergenic regions upstream of the start codon of each AtFLS isoform (1,002 bp for AtFLS1, 1,109 bp for AtFLS2, 768 bp for AtFLS3, 645 bp for AtFLS4, and 1,374 bp for AtFLS5) were amplified from Ler genomic DNA by PCR using Elongase (Invitrogen) or Taq polymerase, incorporating a SphI site in the forward primers and HindIII in the reverse primers (Supplemental Table S1). The fragments were first cloned into pBluescript KS+ (Stratagene) and sequences were confirmed prior to subcloning into the BamHI/HindIII sites in the binary vector pBI121 (Clontech), replacing the cauliflower mosaic virus 35S promoter. AtFLS:pGUS fusion constructs and positive and negative controls (pBI121 and pBI101 vectors, respectively) were introduced into Agrobacterium tumefaciens (GV3101) and then used to transform Ler plants by the floral dip method (Clough and Bent, 1998). Ten independent transgenic T1 lines were selected for each construct and control vector on solid Murashige and Skoog growth medium with 0.005% (w/v) kanamycin. Transgenic lines were confirmed by PCR using the forward primers (Supplemental Table S1) with a reverse primer complementary to the 5′ end of the GUS gene, 5′-ACTTTGCCGTAATGAGTG-3′.
To test for the expression of promoter-GUS constructs, T2 plants were grown on soil (six plants per pot) for 3, 9, 16, 23, 30, 37, 44, and 51 d as described above or in a semihydroponic system for infection with Orobanche aegyptiaca as described by Griffitts et al. (2004). GUS activity was assayed using a histochemical procedure modified from Sieburth and Meyerowitz (1997). Plants were submerged in 90% (v/v) 4°C acetone for 15 min and rinsed in water, and soil particles were removed from roots using forceps. Plants were then blotted on tissue paper, placed into microcentrifuge tubes, covered with staining solution [50 mm phosphate buffer, pH 7.2, 0.5 mm K3Fe(CN)6, 0.5 mm K4Fe(CN)6, and 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (cyclohexylammonium salt; Gold Biotechnology)], vacuum infiltrated three times for 30 s at 5 to 10 Torr, and then incubated overnight at 37°C. Chlorophyll was removed with subsequent rinses of 15%, 30%, 50%, 75%, 80%, and 100% (v/v) ethanol. GUS-stained plants were then transferred to water in petri dishes (16 d or older plants) or onto glass microscope slides and photographed using a digital camera system (3CCD; MIT) mounted on a dissecting microscope (Stemi SVII Apo; Zeiss).
Determination of AtFLS Gene Expression by RT-PCR
Tissues from two independent biological replicates of representative developmental stages were flash frozen in liquid nitrogen and stored at −80°C. Total RNA was extracted using the RNeasy Plant Mini Kit with optional DNase treatment (Qiagen). cDNA was prepared from 5 μg of total RNA in a 33-μL final volume using the NotI-d(T)18 primer and other standard components included with the First-Strand cDNA Synthesis Kit (Amersham Biosciences). The resulting cDNA served as a template for PCR amplification of either 0.3 kb of each AtFLS isoform or 0.5 kb of β-tubulin (At5g62690; Chen et al., 2003) as a control using the primers listed in Supplemental Table S1. RT-PCRs contained 20 pmol of each primer, 2 mm deoxynucleoside triphosphate, 0.5 unit of Taq polymerase (New England Biolabs), and template cDNA (either 2.5 μL of cDNA for AtFLS reactions or 0.5 μL of cDNA for β-tubulin reactions). Reactions used the following parameters: 94°C for 2 min, and 26 cycles of 94°C for 30 s, 60°C (AtFLS1, AtFLS2, and β-tubulin) or 56°C (AtFLS3 and AtFLS5) for 30 s, and 72°C for 1 min. cDNA clones for AtFLS1, AtFLS2, AtFLS3, and AtFLS5 in pBluescript KS+ (described below) were used as a positive control for cDNA amplification at 56°C and 60°C (50 ng of vector per reaction). These constructs were also used to confirm the specificity of the primers. A second independent (biological) replicate of this experiment was performed and produced similar results.
AtFLS Cloning, Expression, and Activity Assays
The AtFLS1 coding region was amplified by PCR from cDNA generated using the iScript cDNA Synthesis Kit (Bio-Rad) and RNA isolated with the RNeasy Plant Mini Kit (Qiagen) from 15-d-old Arabidopsis Ler roots. The AtFLS2 coding region was amplified by PCR from Arabidopsis Col EST clone SQ202h01 (accession no. AV564339). AtFLS3 and AtFLS5 were amplified by RT-PCR utilizing RNA isolated from 4-d-old Arabidopsis Ler seedlings using the method described by Pelletier and Shirley (1996). All reactions used primers that incorporated EcoRI and XhoI sites (Supplemental Table S1) to allow cloning into the corresponding sites in pET32a (Novagen).
An AtFLS1/AtFLS5 chimeric construct was generated by amplifying a 925-bp fragment from the SphI site in pET32a through the first 120 bp in AtFLS1 using the primers shown in Supplemental Table S1. The product was used to replace the corresponding fragment in pET32a-AtFLS5. The integrity of all clones was confirmed by DNA sequencing. It should be noted that although the AtFLS1, -3, and -5 sequences were derived from the Ler ecotype, identical products are encoded by the corresponding genes in Col.
The pET-FLS constructs were used to transform BL21(DE3) pLysS cells and produce recombinant protein essentially as described by Pelletier et al. (1999). Expression was induced by the addition of isopropylthio-β-galactoside to 1 mm final concentration and incubation for 4 h at room temperature and 250 rpm. Similar levels of expression were observed for all of the constructs, as assessed by SDS-PAGE. Cells were harvested by centrifugation at 7,400g and 4°C for 10 min and stored at −80°C. Frozen cells were resuspended in 3 mL of 0.2 m Gly (pH 8.5) and lysed by sonication on ice. The resulting cell slurry was centrifuged at 16,170g and 4°C for 40 min, and the supernatant was used as the source of crude enzyme in activity assays.
FLS Activity Assays
The FLS assay was based on the method of Britsch and Grisebach (1986). Each 100-μL reaction contained 10 mm α-ketoglutaric acid (disodium salt), 10 mm ascorbic acid, 0.25 mm ferrous sulfate, 50 mm Gly (pH 8.5), and 0.1 mm substrate. All flavonoid compounds were dissolved in 80% HPLC-grade methanol at a starting concentration of 10 mm. The ferrous sulfate solution was prepared in 50 mm HEPES, pH 7.5, containing 10 mm ascorbic acid to inhibit the oxidation of Fe2+. All other assay components were suspended in 50 mm HEPES, pH 7.5. The solutions were degassed under vacuum for 10 min, equilibrated under N2 for 5 min, and again degassed under vacuum for 10 min immediately before use.
Activity assays were performed at 25°C for up to 60 min using crude extracts containing similar amounts of each recombinant protein (approximately 3.5–100 μg, depending on the experiment, as assessed by comparison with a dilution series of bovine serum albumin on a Coomassie Blue-stained SDS-PAGE gel). Reactions were initiated by the addition of substrate and terminated by extraction with ethyl acetate (1:1, v/v), performed by adding 200 μL of ethyl acetate and mixing well for 1 min. Solvent layers were separated by centrifugation at 13,000 rpm for 5 min. A 100-μL aliquot of the organic layer was then reextracted with another 200 μL of ethyl acetate and 200 μL of the organic layer combined with the initial 100 μL of extract (R. Lukacin, personal communication). The solvent was evaporated in a SpeedVac under low heat. Dried samples were reconstituted in 50 μL of 80% methanol, mixed for 5 min, and spun at 13,000 rpm.
Supernatants were analyzed by HPLC using a Waters system with a 2996 photodiode array and Millenium 3.2 or Empower 2 software. Samples were kept at 4°C prior to analysis. A 20-μL aliquot was injected and fractionated at room temperature as described by Pelletier and Shirley (1996), except that the absorbance was monitored from 200 to 600 nm. The resulting data were analyzed by extracting a single wavelength chromatogram at 289 nm; an unidentified peak that coelutes with DHQ was subtracted from all of the samples.
Protein Structure Modeling
Homology models were generated for AtFLS1, AtFLS3, and AtFLS5 based on the crystal structure of Arabidopsis ANS (Wilmouth et al., 2002). The sequence of each protein was aligned with ANS, and five models were generated using MODELLER6 essentially as described by Dana et al. (2006). These five structures were then combined by coordinated averaging with the first structure used as the reference, and overlay was on the backbone to generate a single structure. The resulting average structure was subjected to 500 steps of steepest descent minimization using the Sander module of AMBER7. The structure was solvated, and the net charge of the system was brought to zero by the addition of Na+ atoms using LeaP. Equilibration was performed on the water and counter ions by molecular dynamics at constant volume for 100 ps. The solvent and counter ions as well as the entire system were subjected to 500 steps of steepest descent minimization. All molecular dynamics calculations were performed using the AMBER94 force field with a time step of 2 fs and coordinates collected every 1 ps. Molecular dynamics consisted of an 80-ps heating phase to raise the temperature from 0 to 300 K, a 100-ps constant volume equilibration, and a 1-ns constant pressure phase. All calculations were performed using up to eight processors on Virginia Tech's Laboratory for Advanced Scientific Computing and Applications Linux cluster (Anantham). Final models were generated by coordinated averaging from the last 100 ps of dynamics simulation and minimization data. The solvent and Na+ ion coordinates were removed from the analyzed files using the Vi text editor to improve the visualization of the model. Models were visualized and analyzed using DeepView/Swiss-Pdb Viewer v3.7 sp5 and rendered with POV-ray v3.5. Structural comparisons were performed by aligning the isoform homology models using the DeepView iterative magic fit function and calculating the corresponding RMSD values.
Yeast Two-Hybrid Analysis
Coding regions for AtFLS1, AtFLS3, AtFLS5, and AtDFR were amplified from the pET32a constructs described above and for AtF3H from a pBluescript KS+ construct (Pelletier and Shirley, 1996) using the primers listed in Supplemental Table S1. Each PCR product was then digested with SalI (AtFLS1, AtFLS3, and AtDFR), XhoI (AtFLS5), or PstI (AtF3H) and NotI and then inserted into the corresponding sites in the yeast two-hybrid vectors pBI880 and pBI881 (Kohalmi et al., 1998). Plasmids were transformed into Escherichia coli DH10B cells by electroporation. The sequence integrity of all clones was confirmed by sequencing. HF7c yeast cells (Feilotter et al., 1994) were transformed simultaneously with bait and prey constructs essentially as described by Kohalmi et al. (1998). Several independent colonies from each transformation were used to inoculate −Leu−Trp broth and then cultured on −Leu−Trp−His solid medium at 30°C.
Characterization of T-DNA Knockout Lines
Lines segregating for T-DNA insertions in the AtFLS1, -2, -3, -4, and -6 genes were obtained from the SALK and INRA collections; homozygous T-DNA knockout lines were obtained for AtFLS2 and AtFLS5 from GABI-KAT. These included INRA AJ588535 (insertion in the 5′ untranslated region of AtFLS1), SALK_076420 (AtFLS1 promoter), GABI 429B10 (second intron of AtFLS2), SALK_050041 (third exon of AtFLS3), SALK_002309 (third exon of AtFLS4), GABI 317E12 (first intron of AtFLS5), and SALK_003879 (third intron of AtFLS6). Homozygous lines were identified/confirmed by PCR analysis using slight modifications of the method of Edwards et al. (1991) to extract genomic DNA from one large leaf from each plant. In the first method, extraction was in 750 μL of 50 mm Tris, pH 8, and 10 mm EDTA, pH 8. Following incubation at 65°C for 10 min, 200 μL of 5 m KOAc was added and the sample was incubated on ice for 20 min. The sample was then centrifuged at 13,000 rpm for 10 min, the supernatant was mixed with 750 μL of isopropanol and spun at 13,000 rpm for 10 min, and the pellet was rinsed twice in cold 80% ethanol. The pellet was then resuspended in 1 mm Tris, pH 7.5, and 0.1 mm EDTA for 15 min at 37°C. In the second method, extraction was in 350 μL of 200 mm Tris, pH 7.5, and 25 mm EDTA, pH 7.5. The samples were incubated at 65°C for 10 min and centrifuged at 13,000 rpm for 10 min, and the supernatant was mixed with an equivalent volume of isopropanol followed by 5 min of incubation at room temperature. The DNA was pelleted by centrifugation at 13,000 rpm for 10 min and then resuspended overnight in 100 μL of distilled, deionized water. PCR was performed using 1 to 2 μL of each sample with the primers and annealing temperatures given in Supplemental Table S2 in a total volume of 10 to 20 μL. PCR products were analyzed by agarose gel electrophoresis.
Anthocyanidin and Flavonol Assays
Four-day-old seedlings were collected in preweighed 2-mL cryotubes (Corning) containing two 3-mm-diameter stainless steel balls, type 316 (Small Parts). Tissue was then flash frozen in liquid nitrogen and freeze dried for 36 to 48 h in a lyophilizer in the same tubes. For HPLC analysis of flavonols, 50 μL of 1% acetic acid in 80% methanol was added per milligram of tissue dry weight. Samples were ground by agitation for 3 min in a 5-G paint mixer (IDEX) and then clarified by centrifugation at 13,000 rpm and 4°C for 15 min. The samples were then hydrolyzed by the addition of an equal volume of 2 n HCl, followed by incubation at 70°C for 40 min. An equal volume of 100% methanol was added to prevent the precipitation of aglycones. Samples were again centrifuged at 13,000 rpm and 4°C for 15 min and then analyzed by HPLC as described above for the FLS activity assays, except that chromatograms were extracted at 365 nm. For spectrophotometric analysis of anthocyanidins, 30 μL of 1% HCl in methanol was added per milligram of tissue dry weight. Samples were ground and clarified as above, except that centrifugation was at room temperature. The supernatant was mixed with two-thirds volume of distilled, deionized water and then back extracted with an equivalent volume of chloroform to remove chlorophyll. Samples were centrifuged at 13,000 rpm for 10 min, and the upper, aqueous phase was mixed with two volumes of 60% extraction buffer and 40% water. Absorbance at 530 and 657 nm was used to determine the relative levels of anthocyanidins in these samples, as described by Mancinelli and Schwartz (1984). Three independent biological replicates were analyzed for each genotype.
Gravitropism Assays
Seedlings were grown on the surface of Murashige and Skoog/2% Suc/agar plates under continuous light at 23°C for 4 d. Plates were rotated 90° relative to the initial growth orientation and placed at room temperature under normal ambient light conditions. Seedlings were photographed every 30 min for the first 5 h, then every 60 min for another 5 h; a final photograph was taken at 12 h. Changes in the angle of root tips relative to the original orientation were measured using Photoshop and analyzed using Microsoft Excel.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effects on flavonol accumulation of T-DNA insertions in the AtFLS genes.
Supplemental Figure S2. LC-MS analysis of flavonols in fls1.
Supplemental Table S1. Primers used in cloning and RT-PCR.
Supplemental Table S2. Primers and annealing temperatures used to identify/confirm homozygous FLS knockout lines.
Supplemental Table S3. Arabidopsis flavonoid gene homologs.
Supplemental Materials and Methods S1. LC-MS analysis of plant extracts.
Supplementary Material
Acknowledgments
We thank David Lally of the PREP program and Cheryl Weidow's Fall 2006 Biotechnology Class at Louisa High School (Jessica Agee, Thomas Baker, Randy Fisher, Rosa Lee Harkrader, Andrew Harris, Fletcher Jones, Ashley Love, Whitney Perkins, Kelsey Mawyer, Ryan Minnick, Rachel Musser, Jesse Sestito, Eric Stone, Dakota Willis, and Samantha Woolfolk), who first pointed out the enhanced pigmentation phenotype of fls1 plants. We also thank Chris Dana for constructing the AtF3H two-hybrid constructs used in this work and for help with the molecular modeling work. We thank Kim Harich for expert assistance with LC-MS analysis. We are grateful to the Arabidopsis Biological Resource Center for providing a number of essential clones and plant lines and to INRA and GABI-KAT for additional T-DNA knockout lines. We thank Hernán Mauricio at Pennsylvania State University for the pBI101 plasmid and Isabelle Debeaujon at the INRA-Versailles for providing the tt4(8) allele in Ws.
This work was supported by grants from the U.S. Department of Agriculture (grant no. 0189385) and the National Science Foundation (grant nos. MCB–0131010, MCB–0445878, and DGE–0523658).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Brenda S.J. Winkel (winkel@vt.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alexandrov NN, Troukhan ME, Brover VV, Tatarinova T, Flavell RB, Feldmann KA (2006) Features of Arabidopsis genes and genome discovered using full-length cDNAs. Plant Mol Biol 60 69–85 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Bohm H, Boeing H, Hempel J, Raab B, Kroke A (1998) Flavonols, flavones and anthocyanins as native antioxidants of food and their possible role in the prevention of chronic diseases. Z Ernahrungswiss 37 147–163 [DOI] [PubMed] [Google Scholar]
- Braca A, Fico G, Morelli I, De Simone F, Tome F, De Tommasi N (2003) Antioxidant and free radical scavenging activity of flavonol glycosides from different Aconitum species. J Ethnopharmacol 86 63–67 [DOI] [PubMed] [Google Scholar]
- Britsch L, Grisebach H (1986) Purification and characterization of (2S)-flavanone 3-hydroxylase from Petunia hybrida. Eur J Biochem 156 569–577 [DOI] [PubMed] [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol 126 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Muday GK (2004) The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16 1191–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulis IE, Winkel-Shirley B (1999) Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc Natl Acad Sci USA 96 12929–12934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, D'Auria JC, Tholl D, Ross JR, Gershenzon J, Noel JP, Pichersky E (2003) An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J 36 577–588 [DOI] [PubMed] [Google Scholar]
- Chevray PM, Nathans D (1992) Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci USA 89 5789–5793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al (2007) Common sequence polymorphisms shaping genetic diversity in Arabidopsis. Science 317 338–342 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Dana CD, Bevan DR, Winkel BSJ (2006) Molecular modeling of the effects of mutant alleles on chalcone synthase protein structure. J Mol Model 12 905–914 [DOI] [PubMed] [Google Scholar]
- Davies KM, Schwinn KE, Deroles SC, Manson DG, Lewis DH, Bloor SJ, Bradley JM (2003) Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica 131 259–268 [Google Scholar]
- Deavours BE, Liu CJ, Naoumkina MA, Tang YH, Farag MA, Sumner LW, Noel JP, Dixon RA (2006) Functional analysis of members of the isoflavone and isoflavanone O-methyltransferase enzyme families from the model legume Medicago truncatula. Plant Mol Biol 62 715–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devic M, Guilleminot J, Debeaujon I, Bechtold N, Bensaude E, Koornneef M, Pelletier G, Delseny M (1999) The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J 19 387–398 [DOI] [PubMed] [Google Scholar]
- Djordjevic MA, Mathesius U, Arioli T, Weinman JJ, Gartner E (1997) Chalcone synthase gene expression in transgenic subterranean clover correlates with localised accumulation of flavonoids. Aust J Plant Physiol 24 119–132 [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feilotter HE, Hannon GJ, Ruddell CJ, Beach D (1994) Construction of an improved host strain for two hybrid screening. Nucleic Acids Res 22 1502–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffitts AA, Cramer CL, Westwood JH (2004) Host gene expression in response to Egyptian broomrape (Orobanche aegyptiaca). Weed Sci 52 697–703 [Google Scholar]
- Hagmann ML, Heller W, Grisebach H (1983) Induction and characterization of a microsomal flavonoid 3′-hydroxylase from parsley cell cultures. Eur J Biochem 134 547–554 [DOI] [PubMed] [Google Scholar]
- Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55 481–504 [DOI] [PubMed] [Google Scholar]
- Havsteen BH (2002) The biochemistry and medical significance of the flavonoids. Pharmacol Ther 96 67–202 [DOI] [PubMed] [Google Scholar]
- Jacobs M, Rubery PH (1988) Naturally occurring auxin transport regulators. Science 241 346–349 [DOI] [PubMed] [Google Scholar]
- Jander G, Barth C (2007) Tandem gene arrays: a challenge for functional genomics. Trends Plant Sci 12 203–210 [DOI] [PubMed] [Google Scholar]
- Kaltenbach M, Schroder G, Schmelzer E, Lutz V, Schroder J (1999) Flavonoid hydroxylase from Catharanthus roseus: cDNA, heterologous expression, enzyme properties and cell-type specific expression in plants. Plant J 19 183–193 [DOI] [PubMed] [Google Scholar]
- Katsuyama Y, Funa N, Miyahisa I, Horinouchi S (2007) Synthesis of unnatural flavonoids and stilbenes by exploiting the plant biosynthetic pathway in Escherichia coli. Chem Biol 14 613–621 [DOI] [PubMed] [Google Scholar]
- Kerhoas L, Aouak D, Cingoz A, Routaboul JM, Lepiniec L, Einhorn J, Birlirakis N (2006) Structural characterization of the major flavonoid glycosides from Arabidopsis seeds. J Agric Food Chem 54 6603–6612 [DOI] [PubMed] [Google Scholar]
- Kim JD, Liu L, Guo W, Meydani M (2006) Chemical structure of flavonols in relation to modulation of angiogenesis and immune-endothelial cell adhesion. J Nutr Biochem 17 165–176 [DOI] [PubMed] [Google Scholar]
- Kim SJ, Kim MR, Bedgar DL, Moinuddin SG, Cardenas CL, Davin LB, Kang C, Lewis NG (2004) Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc Natl Acad Sci USA 101 1455–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Lee YJ (2007) TRAIL apoptosis is enhanced by quercetin through Akt dephosphorylation. J Cell Biochem 100 998–1009 [DOI] [PubMed] [Google Scholar]
- Kohalmi SE, Reader LJV, Samach A, Nowak J, Haughn GW, Crosby WL (1998) Identification and characterization of protein interactions using the yeast 2-hybrid system. In SB Gelvin, RA Schiperoort, eds, Plant Molecular Biology Manual M1. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–30
- Lea US, Slimestad R, Smedvig P, Lillo C (2007) Nitrogen deficiency enhances expression of specific MYB and bHLH transcription factors and accumulation of end products in the flavonoid pathway. Planta 225 1245–1253 [DOI] [PubMed] [Google Scholar]
- Lee BH, Jeong SM, Lee JH, Kim JH, Yoon IS, Lee JH, Choi SH, Lee SM, Chang CG, Kim HC, et al (2005) Quercetin inhibits the 5-hydroxytryptamine type 3 receptor-mediated ion current by interacting with pre-transmembrane domain I. Mol Cells 20 69–73 [PubMed] [Google Scholar]
- Leonard E, Yan Y, Koffas MA (2006) Functional expression of a P450 flavonoid hydroxylase for the biosynthesis of plant-specific hydroxylated flavonols in Escherichia coli. Metab Eng 8 172–181 [DOI] [PubMed] [Google Scholar]
- Lewis D, Bradley M, Bloor S, Swinny E, Deroles S, Winefield C, Davies K (2006) Altering expression of the flavonoid 3′-hydroxylase gene modified flavonol ratios and pollen germination in transgenic Mitchell petunia plants. Funct Plant Biol 33 1141–1152 [DOI] [PubMed] [Google Scholar]
- Lewis DR, Miller ND, Splitt BL, Wu G, Spalding EP (2007) Separating the roles of acropetal and basipetal auxin transport on gravitropism with mutations in two Arabidopsis multidrug resistance-like ABC transporter genes. Plant Cell 19 1838–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ou-Lee T-M, Raba R, Amundson RG, Last RL (1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillo C, Lea US, Ruoff P (2008) Nutrient depletion as a key factor for manipulating gene expression and product formation in different branches of the flavonoid pathway. Plant Cell Environ 31 587–601 [DOI] [PubMed] [Google Scholar]
- Lim EK, Ashford DA, Hou B, Jackson RG, Bowles DJ (2004) Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol Bioeng 87 623–631 [DOI] [PubMed] [Google Scholar]
- Lukacin R, Britsch L (1997) Identification of strictly conserved histidine and arginine residues as part of the active site in Petunia hybrida flavanone 3β-hydroxylase. Eur J Biochem 249 748–757 [DOI] [PubMed] [Google Scholar]
- Lukacin R, Wellmann F, Britsch L, Martens S, Matern U (2003) Flavonol synthase from Citrus unshiu is a bifunctional dioxygenase. Phytochemistry 62 287–292 [DOI] [PubMed] [Google Scholar]
- Mancinelli AL, Schwartz OM (1984) The photoregulation of anthocyanin synthesis. IX. The photosensitivity of the response in dark and light-grown tomato seedlings. Plant Cell Physiol 25 93–105 [Google Scholar]
- McKhann HI, Hirsch AM (1994) Isolation of chalcone synthase and chalcone isomerase cDNAs from alfalfa (Medicago sativa L.): highest transcript levels occur in young roots and root tips. Plant Mol Biol 24 767–777 [DOI] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BD (2004) Bifunctional and moonlighting enzymes: lighting the way to regulatory control. Trends Plant Sci 9 221–228 [DOI] [PubMed] [Google Scholar]
- Nakajima J, Tanaka Y, Yamazaki M, Saito K (2001) Reaction mechanism from leucoanthocyanidin to anthocyanidin 3-glucoside, a key reaction for coloring in anthocyanin biosynthesis. J Biol Chem 276 25797–25803 [DOI] [PubMed] [Google Scholar]
- Nichenametla SN, Taruscio TG, Barney DL, Exon JH (2006) A review of the effects and mechanisms of polyphenolics in cancer. Crit Rev Food Sci Nutr 46 161–183 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H (2007) ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res 35 D863–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T (2005) Safety of quercetin for clinical application. Review. Int J Mol Med 16 275–278 [PubMed] [Google Scholar]
- Ortega D, Raynal M, Laudie M, Llauro C, Cooke R, Devic M, Genestier S, Picard G, Abad P, Contard P, et al (2002) Flanking sequence tags in Arabidopsis T-DNA insertion lines: a pilot study. C R Biol 325 773–780 [DOI] [PubMed] [Google Scholar]
- Owens DK, Crosby KC, Runac J, Howard BA, Winkel BSJ (2008) Biochemical and genetic characterization of Arabidopsis flavanone 3β-hydroxylase. Plant Physiol Biochem (in press) [DOI] [PubMed]
- Pang YZ, Shen GA, Wu WS, Liu XF, Lin J, Tan F, Sun XF, Tang KX (2005) Characterization and expression of chalcone synthase gene from Ginkgo biloba. Plant Sci 168 1525–1531 [Google Scholar]
- Peer WA, Bandyopadhyay A, Blakeslee JJ, Makam SN, Chen RJ, Masson PH, Murphy AS (2004) Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis. Plant Cell 16 1898–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Brown DE, Tague BW, Muday GK, Taiz L, Murphy AS (2001) Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol 126 536–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Murphy AS (2007) Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci 12 556–563 [DOI] [PubMed] [Google Scholar]
- Pelletier MK, Burbulis IE, Shirley BW (1999) Disruption of specific flavonoid genes enhances the accumulation of flavonoid enzymes and endproducts in Arabidopsis seedlings. Plant Mol Biol 40 45–54 [DOI] [PubMed] [Google Scholar]
- Pelletier MK, Murrell J, Shirley BW (1997) Arabidopsis flavonol synthase and leucoanthocyanidin dioxygenase: further evidence for distinct regulation of “early” and “late” flavonoid biosynthetic genes. Plant Physiol 113 1437–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier MK, Shirley BW (1996) Analysis of flavanone 3-hydroxylase in Arabidopsis seedlings: coordinate regulation with chalcone synthase and chalcone isomerase. Plant Physiol 111 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters DJ, Constabel CP (2002) Molecular analysis of herbivore-induced condensed tannin synthesis: cloning and expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). Plant J 32 701–712 [DOI] [PubMed] [Google Scholar]
- Prescott AG, John P (1996) Dioxygenases: molecular structure and role in plant metabolism. Annu Rev Plant Physiol Plant Mol Biol 47 245–271 [DOI] [PubMed] [Google Scholar]
- Prescott AG, Stamford NP, Wheeler G, Firmin JL (2002) In vitro properties of a recombinant flavonol synthase from Arabidopsis. Phytochemistry 60 589–593 [DOI] [PubMed] [Google Scholar]
- Reddy AM, Reddy VS, Scheffler BE, Wienand U, Reddy AR (2007) Novel transgenic rice overexpressing anthocyanidin synthase accumulates a mixture of flavonoids leading to an increased antioxidant potential. Metab Eng 9 95–111 [DOI] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard S, Lapointe G, Rutledge RG, Seguin A (2000) Induction of chalcone synthase expression in white spruce by wounding and jasmonate. Plant Cell Physiol 41 982–987 [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53 247–259 [DOI] [PubMed] [Google Scholar]
- Routaboul JM, Kerhoas L, Debeaujon I, Pourcel L, Caboche M, Einhorn J, Lepiniec L (2006) Flavonoid diversity and biosynthesis in seed of Arabidopsis. Planta 224 96–107 [DOI] [PubMed] [Google Scholar]
- Ryan KG, Swinny EE, Markham KR, Winefield C (2002) Flavonoid gene expression and UV photoprotection in transgenic and mutant Petunia leaves. Phytochemistry 59 23–32 [DOI] [PubMed] [Google Scholar]
- Saslowsky D, Warek U, Winkel BSJ (2005) Nuclear localization of flavonoid metabolism in Arabidopsis. J Biol Chem 280 23735–23740 [DOI] [PubMed] [Google Scholar]
- Saslowsky D, Winkel-Shirley B (2001) Localization of flavonoid enzymes in Arabidopsis roots. Plant J 27 37–48 [DOI] [PubMed] [Google Scholar]
- Schijlen E, Ric de Vos CH, Jonker H, van den Broeck H, Molthoff J, van Tunen A, Martens S, Bovy A (2006) Pathway engineering for healthy phytochemicals leading to the production of novel flavonoids in tomato fruit. Plant Biotechnol J 4 433–444 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296 141–145 [DOI] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8 659–671 [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM (1997) Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram G, Martinez JA, McCabe ER, Liao JC, Dipple KM (2005) Single-gene disorders: what role could moonlighting enzymes play? Am J Hum Genet 76 911–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Barsch GHA, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis seedling. Plant J 50 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36 D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglieber A, Hoebenreich H, Carballeira JD, Mondiere R, Reetz MT (2007) Alternate-site enzyme promiscuity. Angew Chem Int Ed 46 8597–8600 [DOI] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42 218–235 [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Harding SA, Tschaplinski TJ, Lindroth RL, Yuan YN (2006) Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol 172 47–62 [DOI] [PubMed] [Google Scholar]
- Turnbull JJ, Nagle MJ, Seibel JF, Welford RWD, Grant GH, Schofield CJ (2003) The C-4 stereochemistry of leucocyanidin substrates for anthocyanidin synthase affects product selectivity. Bioorg Med Chem Lett 13 3853–3857 [DOI] [PubMed] [Google Scholar]
- Turnbull JJ, Nakajima JI, Welford RW, Yamazaki M, Saito K, Schofield CJ (2004) Mechanistic studies on three 2-oxoglutarate-dependent oxygenases of flavonoid biosynthesis. J Biol Chem 279 1206–1216 [DOI] [PubMed] [Google Scholar]
- Turnbull JJ, Sobey WJ, Aplin RT, Hassan A, Firmin JL, Schofield CJ, Prescott AG (2000) Are anthocyanidins the immediate precursors of anthocyanidin synthase? Chem Commun 24 2473–2474 [Google Scholar]
- Wellmann F, Griesser M, Schwab W, Martens S, Eisenreich W, Matern U, Lukacin R (2006) Anthocyanidin synthase from Gerbera hybrida catalyzes the conversion of (+)-catechin to cyanidin and a novel procyanidin. FEBS Lett 580 1642–1648 [DOI] [PubMed] [Google Scholar]
- Westwood JH (2000) Characterization of the Orobanche-Arabidopsis system for studying parasite-host interactions. Weed Sci 48 742–748 [Google Scholar]
- Wilmouth RC, Turnbull JJ, Welford RW, Clifton IJ, Prescott AG, Schofield CJ (2002) Structure and mechanism of anthocyanidin synthase from Arabidopsis. Structure 10 93–103 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5 218–223 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (2006) The biosynthesis of flavonoids. In E Grotewold, ed, The Science of Flavonoids. Springer Science & Business Media, New York, pp 71–95
- Wisman E, Hartmann U, Sagasser M, Baumann E, Palme K, Hahlbrock K, Saedler H, Weisshaar B (1998) Knock-out mutants from an En-1 mutagenized Arabidopsis population generate phenylpropanoid biosynthesis phenotypes. Proc Natl Acad Sci USA 95 12432–12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XH, Chen M-H, Liu CJ (2008) Nucleocytoplasmic-localized acyltransferases catalyze the malonylation of 7-O-glycosidic (iso)flavones in Medicago truncatula. Plant J (in press) [DOI] [PubMed]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.