Abstract
A comprehensive knowledge of mechanisms regulating nitrogen (N) use efficiency is required to reduce excessive input of N fertilizers while maintaining acceptable crop yields under limited N supply. Studying plant species that are naturally adapted to low N conditions could facilitate the identification of novel regulatory genes conferring better N use efficiency. Here, we show that Thellungiella halophila, a halophytic relative of Arabidopsis (Arabidopsis thaliana), grows better than Arabidopsis under moderate (1 mm nitrate) and severe (0.4 mm nitrate) N-limiting conditions. Thellungiella exhibited a lower carbon to N ratio than Arabidopsis under N limitation, which was due to Thellungiella plants possessing higher N content, total amino acids, total soluble protein, and lower starch content compared with Arabidopsis. Furthermore, Thellungiella had higher amounts of several metabolites, such as soluble sugars and organic acids, under N-sufficient conditions (4 mm nitrate). Nitrate reductase activity and NR2 gene expression in Thellungiella displayed less of a reduction in response to N limitation than in Arabidopsis. Thellungiella shoot GS1 expression was more induced by low N than in Arabidopsis, while in roots, Thellungiella GS2 expression was maintained under N limitation but was decreased in Arabidopsis. Up-regulation of NRT2.1 and NRT3.1 expression was higher and repression of NRT1.1 was lower in Thellungiella roots under N-limiting conditions compared with Arabidopsis. Differential transporter gene expression was correlated with higher nitrate influx in Thellungiella at low 15NO3− supply. Taken together, our results suggest that Thellungiella is tolerant to N-limited conditions and could act as a model system to unravel the mechanisms for low N tolerance.
Nitrogen (N) is an essential macronutrient required in large quantities by plants to achieve optimal growth and development (Marschner, 1995). As such, N is a major limiting factor for crop growth (Diaz et al., 2006; Lea and Azevedo, 2006). During the past five decades, global N fertilizer applications have increased by 20-fold, with the present rate at approximately 1011 kg per annum (Glass, 2003). However, crop plants are able to utilize only 30% to 40% of this applied N (Raun and Johnson, 1999); the remaining N is lost by leaching, denitrification, volatilization, soil erosion, and microbial consumption. All of these factors increase crop production costs as well as N pollution in the environment (Good et al., 2004). To overcome these problems, there is an urgent need to improve the N use efficiency of crop plants by (1) increasing N uptake efficiency, (2) increasing N utilization efficiency, and (3) enhancing the adaptability of crops to N-limited conditions. In recent years, considerable efforts have been made to identify different genes involved in N transport and metabolism (Lea and Azevedo, 2006; Hirel et al., 2007). However, little is known about the molecular mechanisms regulating the adaptability of plants to N limitation (Peng et al., 2007b).
Several physiological and biochemical changes occur in plants as adaptive responses to N limitation, including an increase in N uptake by high-affinity transporters, remobilization of N from older to younger leaves and reproductive parts, retardation of growth and photosynthesis, and increased anthocyanin accumulation (Bongue-Bartelsman and Philips, 1995; Ono et al., 1996; Chalker-Scott, 1999; Ding et al., 2005; Diaz et al., 2006; Peng et al., 2007a, 2007b). In Arabidopsis (Arabidopsis thaliana), expression of nitrate transporter genes involved in nitrate uptake (NRT1.1, NRT1.2, NRT2.1, and NRT2.2) is induced or repressed depending upon nitrate availability (Lejay et al., 1999; Zhuo et al., 1999; Cerezo et al., 2001; Okamoto et al., 2003; Orsel et al., 2004; Krouk et al., 2006; Li et al., 2007). In addition, Autophagy proteins are thought to play an important role in the recycling of N and other nutrients in Arabidopsis under nutrient-limited conditions (Doelling et al., 2002). Recently, the NLA (for NITROGEN LIMITATION ADAPTATION) gene was identified as a positive regulator of Arabidopsis adaptation to low N (Peng et al., 2007b). Notably, most of these genes have been identified in Arabidopsis, whose growth is retarded under limited N availability (Martin et al., 2002; Bi et al., 2007; Peng et al., 2007a, 2007b). A complementary approach would be to unravel the molecular mechanisms in plant species that are naturally adapted to N limitation. However, it has not been easy to work with plant species that survive well under low N because of their genetic complexity and the lack of molecular and genetic tools for these plants. For example, modern maize (Zea mays) hybrids and wheat (Triticum aestivum) cultivars can grow better and produce higher yields than older lines under limited N conditions (McCullough et al., 1994; Ortiz-Monasterio et al., 1997; Ding et al., 2005). However, little is known about the molecular mechanisms leading to higher N use efficiency in these newer maize hybrids and wheat cultivars. Information on components of higher N uptake in any plant species would be useful for improving N uptake in crop plants (Hirel et al., 2007). Thus, there is a need for a model plant that is tolerant to N stress conditions that would facilitate investigation of the molecular mechanisms regulating adaptation to low N and that may confer an improved N use efficiency.
Thellungiella halophila (also know as Thellungiella salsuginea) has emerged as a new model plant for the molecular elucidation of abiotic stress tolerance (Inan et al., 2004; Amtmann et al., 2005; Kant et al., 2006). Thellungiella shares similar morphology and sequence identity with Arabidopsis, thus allowing for the utilization of Arabidopsis genetic information to investigate Thellungiella responses to stress (Inan et al., 2004; Taji et al., 2004; Wong et al., 2005). Several ecotypes of Thellungiella have been collected from Central Asia and North America, indicating the genetic diversity of this plant species (http://www.Thellungiella.org/). Until now, research on two ecotypes has been reported. The Shandong ecotype is native to the high-salinity coastal areas of the Shandong province in northeast China (Inan et al., 2004). The Yukon ecotype was isolated from saline meadows in the Yukon Territories, Canada, a subarctic and semiarid region (Wong et al., 2005, 2006). Both Shandong and Yukon ecotypes originate from extreme climatic conditions and have been reported to be tolerant to salinity, drought, and/or cold stresses (Inan et al., 2004; Kant et al., 2006; Volkov and Amtmann, 2006; Wong et al., 2006; Griffith et al., 2007). Interestingly, analysis of soil samples from the region where the Yukon ecotype grows wild shows a very low N (<1 mm) status of the soil (E. Weretilnyk, unpublished data). This is in contrast to the typical N concentration in agricultural soils, which ranges from 1 to 10 mm (Crawford and Forde, 2002). The naturally low N growth conditions of Thellungiella in the wild led us to hypothesize that Thellungiella could act as an Arabidopsis relative model system for investigating the molecular mechanisms of low N tolerance.
Here, physiological, biochemical, and molecular experiments have been conducted on both Shandong and Yukon ecotypes under varying N levels as well as a comparative genetic analysis with Arabidopsis. Based on these analyses, we confirm that Thellungiella has a higher tolerance than Arabidopsis to low external N supply. This is accompanied by a variety of changes in the expression of key genes involved in nitrate uptake and assimilation and a change in altered metabolic status.
RESULTS
Thellungiella Exhibits Better Growth Than Arabidopsis under Limiting N Conditions
Thellungiella and Arabidopsis plants were grown in a perlite and soil mixture to allow easy harvesting of roots. This medium was favored over a hydroponics system because root physiology can differ in hydroponics compared with soil-grown plants (Gibeaut et al., 1997). Moreover, a soil-based medium is closer to natural growth conditions. Among the two inorganic forms of N taken up by plants (i.e. nitrate and ammonium), nitrate is the main N source in most aerated agricultural soils (Crawford and Forde, 2002). Therefore, we used nitrate-N for all experiments.
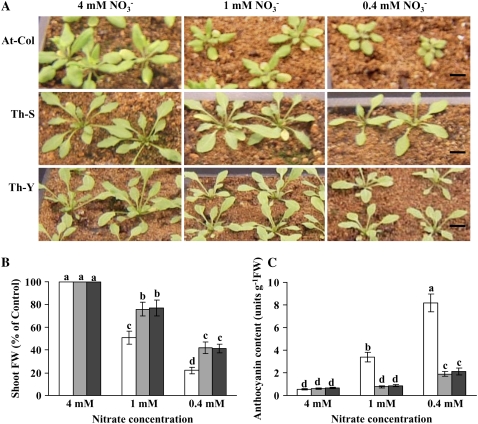
Figure 1A shows the growth of Arabidopsis and Thellungiella at three levels of nitrate: 4 mm nitrate (control; N sufficient), 1 mm nitrate (mild N limitation), and 0.4 mm nitrate (severe N limitation). Arabidopsis displayed a greater reduction in fresh weight than Thellungiella in N-limiting conditions. Under mild N limitation, both Thellungiella ecotypes showed an approximately 22% decrease in fresh weight compared with the control, while Arabidopsis showed a 48% drop in fresh weight (Fig. 1B). Under severe N limitation, Thellungiella and Arabidopsis displayed 53% and 75% reductions in fresh weight, respectively. Thellungiella plants also exhibited less of a reduction in leaf number under N-limiting conditions than Arabidopsis (data not shown). Furthermore, Arabidopsis could not complete its life cycle under severe N limitation, whereas Thellungiella plants were still able to produce viable seeds (data not shown).
Figure 1.
Effects of different nitrate levels on the growth of Arabidopsis Columbia ecotype (AT-Col) and Thellungiella Shandong (Th-S) and Yukon (Th-Y) ecotypes (A; bars = 1 cm), shoot fresh weight (B), and anthocyanin content (C). The plants were continuously grown at three different N levels, 4 mm nitrate (sufficient N), 1 mm nitrate (mild limited N), and 0.4 mm nitrate (severe limited N), for 20 and 23 d for Arabidopsis and Thellungiella, respectively. Data are means ± sd (n = 3), and each replicate consisted of a pool of 10 to 15 plants. Bars with different letters indicate a significant difference at P < 0.05 (Fisher's protected lsd test). White bars, Arabidopsis; light gray bars, Thellungiella Shandong ecotype; dark gray bars, Thellungiella Yukon ecotype. FW, Fresh weight.
The appearance of anthocyanins is often used as a marker of stress. Figure 1C shows that under control conditions, both species contained low levels of anthocyanins. When plants were subjected to mild N limitation, anthocyanin levels rose 6-fold in Arabidopsis but remained at control levels in Thellungiella. Under severe N limitation, anthocyanin content in Arabidopsis increased to 15-fold that of control plants, whereas in Thellungiella, anthocyanin content only increased by 3-fold.
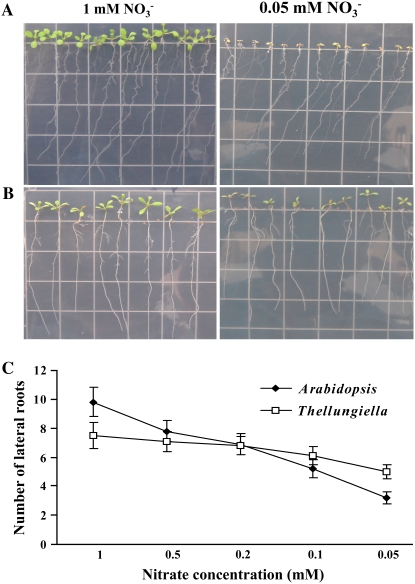
Since lateral root proliferation is reduced and shoots are stunted when seedlings are grown in medium containing a high Suc to nitrate ratio (Malamy and Ryan, 2001), we examined whether N limitation differentially affected root growth in the two species. Seedlings were grown on vertical plates containing nutrient agar supplemented with 2% Suc and nitrate levels ranging from 0.05 to 1 mm KNO3. Figure 2, A and B, shows that Arabidopsis and Thellungiella (Shandong) seedlings had green cotyledons and produced green true leaves when grown on 1 mm nitrate. Strikingly, Thellungiella (Shandong) was still able to produce green cotyledons on 0.05 mm nitrate, whereas Arabidopsis produced only small purple cotyledons. Arabidopsis and Thellungiella (Shandong) exhibited similar reductions in primary root length at 0.05 mm compared with 1 mm nitrate (Fig. 2, A and B). Furthermore, in both species, lateral root numbers declined with decreasing nitrate concentration (Fig. 2C). However, while Arabidopsis lateral root number was reduced 3-fold at 0.05 mm nitrate compared with the 1 mm nitrate treatment, Thellungiella lateral number declined by only 1.5-fold. The Yukon ecotype exhibited a similar root growth response to that seen for Shandong (data not shown).
Figure 2.
Effects of different nitrate levels on the growth and root architecture of Arabidopsis and Thellungiella seedlings. Arabidopsis (A) and Thellungiella Shandong ecotype (B) seedlings were grown on vertical plates (2% Suc, pH 5.8, and 0.8% agar) with different nitrate levels for 10 and 12 d, respectively. The number of lateral roots (C) visible was counted at the indicated nitrate levels. Data are means ± sd (n = 3), and each replicate consisted of a pool of 10 to 15 plants. Fisher's protected lsd test showed a significant difference (P < 0.05) at 1 and 0.05 mm nitrate between Arabidopsis and Thellungiella.
Thellungiella Displays Higher N Uptake and Assimilation Capacity Than Arabidopsis under N Limitation
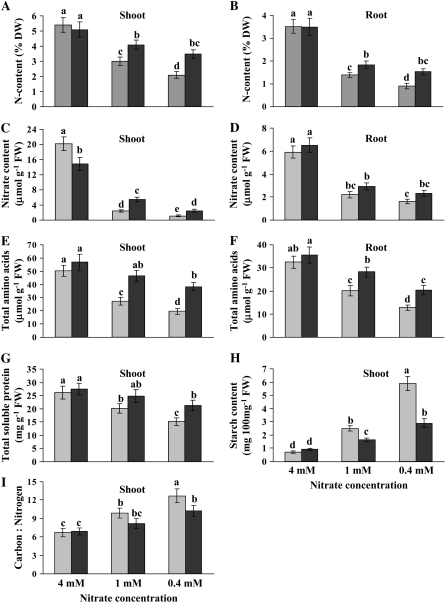
One possible explanation for the better growth of Thellungiella plants than Arabidopsis under low N is that Thellungiella has a higher capacity for N acquisition and/or assimilation. To test this possibility, we first analyzed total N content in shoots and roots. Since both Thellungiella ecotypes exhibited similar growth responses, only the data for the Shandong ecotype are presented hereafter. Under mild and severe N limitation, Arabidopsis exhibited 1.8- and 2.6-fold drops in total N content, respectively, whereas Thellungiella only showed 1.2- and 1.5-fold decreases, respectively (Fig. 3A). Similar results were obtained for roots, although the difference in total N content between Arabidopsis and Thellungiella was less than in shoots (Fig. 3B). The results showing greater N content in Thellungiella under N-limiting conditions were reflected in the total free nitrate content of shoots in the two species. For instance, whereas Arabidopsis displayed an 18-fold drop in free nitrate content under severe N limitation, Thellungiella only exhibited a 6-fold decrease (Fig. 3C). Similar results for free nitrate content were observed in roots under severe N limitation, again with a smaller difference between the two species than in shoots (Fig. 3D).
Figure 3.
Effects of different nitrate levels on N content in shoots (A) and roots (B), nitrate content in shoots (C) and roots (D), total amino acids in shoots (E) and roots (F), total soluble protein in shoots (G), starch content in shoots (H), and shoot carbon to N ratio (I) in Arabidopsis and Thellungiella plants. Experimental conditions were as described for Figure 1. Data are means ± sd (n = 3–5). Bars with different letters indicate a significant difference at P < 0.05 (Fisher's protected lsd test). Light gray bars, Arabidopsis; dark gray bars, Thellungiella Shandong ecotype. DW, Dry weight; FW, fresh weight.
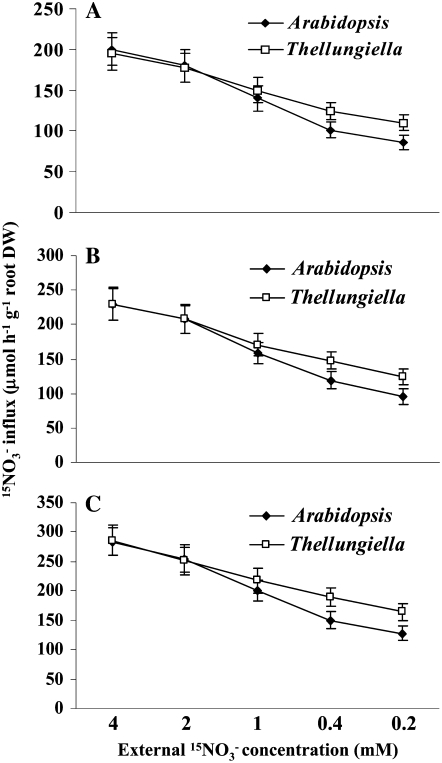
Thellungiella plants had higher total N and free nitrate contents than Arabidopsis specifically under limited N availability, which could be ascribed to the higher nitrate uptake capacity of Thellungiella. To address this possibility, 15NO3− influx in roots was compared between Thellungiella and Arabidopsis (Fig. 4). The plants were first grown on vertical agar plates for 10 to 12 d at 4, 1, and 0.4 mm nitrate and then incubated with 0.2, 0.4, 1, 2, or 4 mm 15NO3− for 5 min. Figure 4 shows that overall 15NO3− influx of both species increased as the levels of low N stress at which the plants were grown rose. For instance, roots from plants grown at 4 mm nitrate exhibited a 15NO3− influx of approximately 200 μmol h−1 g−1 root dry weight when incubated at 4 mm 15NO3− (Fig. 4A), whereas plants grown under severe N limitation displayed a 15NO3− influx of approximately 300 μmol h−1 g−1 root dry weight (Fig. 4C). For plants grown at all levels of nitrate, Thellungiella roots had a similar 15NO3− influx to Arabidopsis when 15NO3− availability was in the higher range (up to 1 mm 15NO3−). However, Thellungiella roots had significantly higher 15NO3− influx than Arabidopsis when 15NO3− availability was in the lower range (0.4 and 0.2 mm 15NO3−), suggesting that nitrate uptake in Thellungiella is constitutively higher than in Arabidopsis at low levels of NO3− in the growth medium.
Figure 4.
15NO3− influx in Arabidopsis and Thellungiella roots. Seedlings were grown on vertical plates with nutrient solution containing 4 mm (A), 1 mm (B), and 0.4 mm (C) nitrate for 10 and 12 d, respectively, and then transferred to nutrient solution containing the indicated concentrations of 15NO3− (99% atom excess 15N) for 5 min. Data are means ± sd (n = 5), and each replicate consisted of a pool of 15 to 20 plants. Fisher's protected lsd test showed a significant difference (P < 0.05) at 0.4 and 0.2 mm 15NO3− between Arabidopsis and Thellungiella.
N assimilation requires the use of large amounts of organic acids from photosynthesis to incorporate inorganic N into amino acids. N deficiency alters the source-sink balance of plants, with a lower percentage of carbon skeletons used for N assimilation meaning that more of this carbon is diverted to starch production. Therefore, increased starch accumulation and decreased nitrogenous metabolite levels occur in plants supplied with lower N (Paul and Driscoll, 1997). In Arabidopsis shoots, amino acid levels were reduced by 46% and 61% under mild and severe N limitation, respectively, compared with control N levels, whereas only 15% and 32% drops occurred in Thellungiella shoots (Fig. 3E). A similar trend was observed in roots, although roots had overall lower amino acid levels than shoots (Fig. 3F). Thellungiella also displayed higher shoot total soluble protein content under severe N limitation than Arabidopsis (Fig. 3G). These results suggest that under N-limiting conditions, Thellungiella has a higher capacity for nitrate assimilation than Arabidopsis. Figure 3H shows that under N-limiting conditions, particularly severe N limitation, Arabidopsis accumulated up to twice the amount of starch than in Thellungiella, suggesting that in Arabidopsis more carbon skeletons are diverted to storage products such as starch instead of to nitrate assimilation. The trend of higher nitrogenous metabolites and lower starch levels under limited N in Thellungiella was reflected in a lower carbon to N ratio, where the increase in the carbon to N ratio under N limitation was lower in Thellungiella shoots compared with Arabidopsis (Fig. 3I).
To further investigate differences in the carbohydrate and N metabolism of the two species, we examined their metabolic profiles. Table I shows selected metabolites of the polar fraction. Strikingly, Thellungiella accumulated 288%, 203%, and 60% more malate than Arabidopsis under N-sufficient, mild N-limiting, and severe N-limiting conditions, respectively. Malate is the immediate precursor of oxaloacetate, which provides carbon skeletons for amino acid synthesis (Siedow and Day, 2000). Malate itself can be produced from both fumarate in the citric acid cycle and from Glc and Fru via phosphoenolpyruvate in the cytosol (Dennis and Blakeley, 2000). Our metabolite analysis showed a large reduction in fumarate in Thellungiella compared with Arabidopsis under N-sufficient and mild N-limiting conditions, although under severe N limitation fumarate was undetectable in both species. Nevertheless, these results are consistent with a reduction in the fumarate pool to produce malate. Thellungiella also exhibited increased accumulation of Glc and Fru, products of Suc breakdown. This observation could indicate the use of Glc and Fru in the production of malate, possibly for anaplerotic carbon reactions. Overall, these findings suggest a greater diversion of carbohydrates to nitrate assimilation in Thellungiella than in Arabidopsis and agree with our results showing greater accumulation of starch in Arabidopsis (Fig. 3H).
Table I.
Metabolite analysis in Arabidopsis and Thellungiella shoots at different nitrate levels
Data are mean values expressed as percentages of the internal standard, ribitol. Shoots of seedlings grown on plates for the root study were used for metabolite analysis. Data are means ± sd (n = 3). ND, Not detectable.
| Metabolite | Arabidopsis
|
Thellungiella
|
||||
|---|---|---|---|---|---|---|
| 1 mm | 0.2 mm | 0.05 mm | 1 mm | 0.2 mm | 0.05 mm | |
| Citric acid | 6.5 ± 0.9 | 12.1 ± 1.7 | 10.5 ± 1.2 | 9.1 ± 1.1 | 17.2 ± 2.1 | 12.9 ± 1.6 |
| Fru | 11.3 ± 1.2 | 20.2 ± 2.8 | 29.0 ± 3.2 | 23.1 ± 2.1 | 55.0 ± 6 | 70.0 ± 7.2 |
| Fumaric acid | 24.5 ± 2.5 | 11.3 ± 1.8 | ND | 1.2 ± 0.2 | 0.1 ± 0.02 | ND |
| Glc | 20.2 ± 1.8 | 68.4 ± 8.2 | 125 ± 14 | 39.2 ± 3.5 | 87.2 ± 10 | 147 ± 15 |
| Gly | 2.9 ± 0.4 | 2.1 ± 0.3 | 1.7 ± 0.3 | 3.5 ± 0.5 | 3.1 ± 0.5 | 2.6 ± 0.3 |
| Malate | 25.3 ± 3.3 | 53.4 ± 6.5 | 42.4 ± 5.2 | 98.3 ± 11 | 162 ± 16 | 68.2 ± 8.1 |
| Myoinositol | 2.7 ± 0.4 | 4.8 ± 0.4 | 6.1 ± 0.8 | 2.2 ± 0.4 | 11.2 ± 1.4 | 7.5 ± 0.9 |
| Pro | 3.2 ± 0.5 | 2.5 ± 0.3 | 2.1 ± 0.3 | 4.6 ± 0.6 | 3.6 ± 0.5 | 2.9 ± 0.4 |
| Suc | 84.2 ± 9 | 169 ± 19 | 203 ± 25 | 116 ± 11 | 142 ± 16 | 155 ± 17 |
Higher N Assimilation in Thellungiella Is Mediated by Maintenance of N Assimilation Enzyme Activity
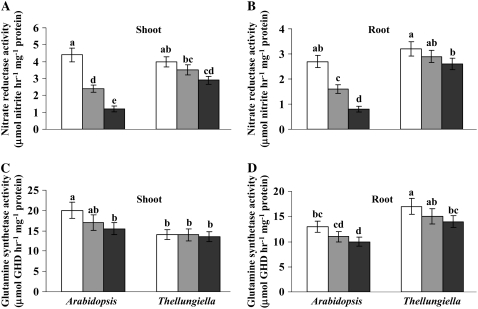
Higher assimilation of N in Thellungiella might imply differences in the activity of N assimilation enzymes between the two species. To test this hypothesis, the activities of nitrate reductase (NR) and Gln synthetase (GS), via which N is assimilated into amino acids (Crawford, 1995; Stitt, 1999), were measured. Figure 5, A and B, shows that in Arabidopsis shoots and roots, NR activity decreased by approximately 3.7- and 3.4-fold, respectively, under severe N limitation compared with control conditions. In contrast, NR activity in Thellungiella shoots and roots was maintained at all levels of nitrate supply. On the other hand, there was little difference in the response of GS activity to N limitation between the two species (Fig. 5, C and D). However, there are several isoforms of GS in plants that have nonoverlapping functions (Edwards et al., 1990; Coruzzi and Last, 2000), and measurement of total GS activity is likely to mask any isoform-specific differences.
Figure 5.
Effects of different nitrate levels on enzyme activity of NR in shoots (A) and roots (B) and of GS in shoots (C) and roots (D) of Arabidopsis and Thellungiella. Experimental conditions were as described for Figure 1. Data are means ± sd (n = 3). Bars with different letters indicate a significant difference at P < 0.05 (Fisher's protected lsd test). White bars, 4 mm nitrate; light gray bars, 1 mm nitrate; dark gray bars, 0.4 mm nitrate. For each part, left panel bars represent Arabidopsis and right panel bars represent Thellungiella Shandong ecotype.
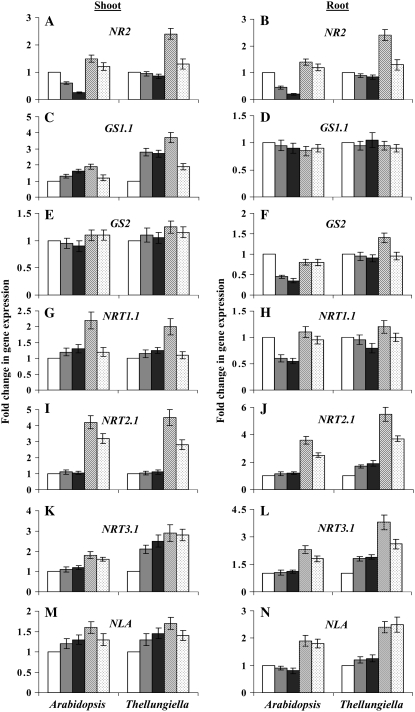
To further investigate differences in nitrate assimilation enzymes, Arabidopsis cDNA sequences were used to prepare primers for real-time PCR analysis of shoot and root expression of Arabidopsis and Thellungiella genes encoding NR and GS isoforms. PCR products amplified by the gene-specific primers were sequenced, and the Arabidopsis and Thellungiella orthologs were shown to have 80% to 100% sequence identity (Supplemental Table S1) and to produce single product-specific peaks from dissociation curves (data not shown). Arabidopsis has two NR genes, NR1 (Nia1) and NR2 (Nia2), with the latter gene being responsible for 90% of the total NR activity in Arabidopsis (Yu et al., 1998; Wilkinson and Crawford, 1991). Figure 6, A and B, shows that Arabidopsis NR2 expression in both shoots and roots decreased with greater N limitation, whereas Thellungiella maintained NR2 gene expression at all levels of nitrate supply. These results correlate well with our analysis of NR enzyme activity in the two species (Fig. 5, A and B).
Figure 6.
Effects of different nitrate levels on the relative expression of various genes in shoots and roots of Arabidopsis and Thellungiella. Experimental conditions were as described for Figure 1, with two additional nitrate treatments: induction, switching plants 2 h before harvesting from 1 to 4 mm nitrate; reduction, switching plants 2 h before harvesting from 4 to 1 mm nitrate. Relative transcript levels were determined by real-time PCR according to Livak and Schmittgen (2001) using UBQ10 as an internal control. The gene expression for each nitrate treatment was normalized to the control level (4 mm nitrate), which was assigned a value of 1. Data are means ± sd (n = 3) and are representative of similar results from three independent experiments. Data shown are as follows: shoot (A) and root (B) NR2; shoot (C) and root (D) GS1.1; shoot (E) and root (F) GS2; shoot (G) and root (H) NRT1.1; shoot (I) and root (J) NRT2.1; shoot (K) and root (L) NRT3.1; and shoot (M) and root (N) NLA. White bars, 4 mm nitrate; light gray bars, 1 mm nitrate; dark gray bars, 0.4 mm nitrate; hatched bars, nitrate induction; dotted bars, nitrate reduction. For each part, left panel bars represent Arabidopsis and right panel bars represent Thellungiella Shandong ecotype.
The GS enzyme activity is encoded by (1) cytosolic GS1 (Gln-1), which comprises five isoforms, GS1.1 to GS1.5, and is predominantly expressed in roots; and (2) chloroplastic GS2 (Gln-2), which is targeted to both chloroplasts and mitochondria and is expressed more strongly in shoots (Cren and Hirel, 1999; Taira et al., 2004). Shoot GS1.1 expression in both species increased under severe N-limiting conditions. However, while Arabidopsis GS1.1 rose by only 1.5-fold, Thellungiella GS1.1 increased almost 3-fold in response to N-limiting conditions (Fig. 6C). Moreover, when plants grown under mild N limitation were switched to 4 mm nitrate for 2 h (N induction), Thellungiella shoot GS1.1 expression exhibited a further increase in expression, whereas in Arabidopsis little change in GS1.1 expression was observed. No differences were found in root GS1.1 expression between the two species (Fig. 6D). GS1.3 and GS1.4 expression showed similar results to GS1.1 expression in shoots and roots of both species (data not shown). GS2 expression displayed no difference between Arabidopsis and Thellungiella in shoots (Fig. 6E). In roots, however, Arabidopsis GS2 expression decreased under N-limiting conditions, whereas it was maintained in Thellungiella (Fig. 6F). The GS expression analysis suggests that there may be isoform-specific differences in GS activity between Arabidopsis and Thellungiella that is masked when measuring total GS activity. However, for transcript copy numbers of GS1 and GS2 under control N-sufficient conditions, we observed contrasting differences between the two species (Table II). In shoots, Thellungiella had lower transcript copies of GS1.1 and GS2 than Arabidopsis, but in roots, Thellungiella had a higher number of transcripts of these genes compared with Arabidopsis. This trend correlates with tissue-specific GS enzyme activity, which was higher in Arabidopsis shoots than roots, whereas the opposite was observed in Thellungiella, with higher GS activity in roots than shoots (Fig. 5, C and D).
Table II.
Transcript copy numbers of genes in shoots and roots of Arabidopsis and Thellungiella under control (4 mm nitrate) conditions
The transcript copy numbers were quantified by relating the real-time PCR signal for each gene to a standard curve. Data are means ± sd (n = 3).
| Gene | Arabidopsis
|
Thellungiella
|
||
|---|---|---|---|---|
| Shoot | Root | Shoot | Root | |
| NRT1.1 | 14.7 ± 2.2 | 7.4 ± 1.4 | 7.2 ± 1.2 | 15.4 ± 2.9 |
| NRT2.1 | 1.9 ± 0.4 | 85 ± 12 | 1.5 ± 0.4 | 84 ± 14 |
| NRT3.1 | 7 ± 1.2 | 78 ± 12 | 11 ± 1.2 | 128 ± 20 |
| CLCa | 32 ± 6 | 37 ± 5 | 29 ± 4 | 47 ± 6 |
| NLA | 7 ± 1 | 50 ± 8 | 15 ± 2 | 81 ± 15 |
| NR2 | 35 ± 5 | 11 ± 2 | 32 ± 6 | 14 ± 3 |
| GS1.1 | 23 ± 4 | 59 ± 8 | 10 ± 1.5 | 116 ± 18 |
| GS2 | 117 ± 18 | 7 ± 1.1 | 84 ± 15 | 15 ± 2.2 |
Comparative Analysis of Expression of Nitrate Transporter and Other Related Genes in Thellungiella and Arabidopsis
In Arabidopsis, there are two types of nitrate transporters: NRT1 and NRT2, with 53 NRT1 and seven NRT2 genes identified (Miller et al., 2007; Tsay et al., 2007). However, two of the NRT1 genes, NRT1.1 (CHL1) and NRT1.2, and two NRT2 genes, NRT2.1 and NRT2.2, are known to be involved in nitrate uptake from external medium. NRT2 proteins are high-affinity nitrate transporters, while most of the NRT1 family members are low-affinity nitrate transporters, except NRT1.1, which is a dual-affinity nitrate transporter (Wang et al., 1998; Liu et al., 1999; Miller et al., 2007; Tsay et al., 2007). Among NRT1 and NRT2 nitrate transporters, NRT1.1 and NRT2.1 genes are well characterized and known to play a vital role in nitrate uptake (Okamoto et al., 2003; Li et al., 2007). Expression of NRT1.1 increased slightly under low N in both Thellungiella and Arabidopsis shoots. In roots, NRT1.1 expression was reduced under low N, although this reduction was lower in Thellungiella than in Arabidopsis (Fig. 6, G and H). Analysis of transcript copy number of NRT1.1 under control conditions demonstrated contrasting tissue-specific differences between the two species (Table II). Arabidopsis shoots had 2-fold higher NRT1.1 transcript levels than roots, while the reverse was observed in Thellungiella, with roots showing higher transcript levels than shoots.
Expression of the high-affinity nitrate transport gene NRT2.1 remained virtually unchanged in shoots of both species under different stable nitrate levels, while nitrate induction (1 mm to 4 mm nitrate) and reduction (4 mm to 1 mm nitrate) treatments led to up-regulation of NRT2.1 expression (Fig. 6, I and J). This suggests that a swift change in N status (by either increased or decreased nitrate levels) leads to increased accumulation of NRT2.1 transcripts. In Arabidopsis roots, expression of NRT2.1 was unchanged under stable nitrate levels, whereas nitrate induction and reduction treatments led to approximately 4- and 2.5-fold up-regulation compared with control, respectively. However, in Thellungiella roots, the up-regulation of NRT2.1 at 0.4 mm nitrate was approximately 2-fold, and nitrate induction and reduction led to up-regulation by approximately 6- and 3.5-fold over control, respectively.
NRT2.1 needs to be coexpressed with another protein, NRT3.1 (NAR2), to mediate nitrate transport activity. Although NRT3.1 itself has no known transport activity, it is an essential component of high-affinity nitrate transport (Okamoto et al., 2006; Orsel et al., 2006; Miller et al., 2007). In this study, expression of NRT3.1 in Arabidopsis tissues was not changed by stable low nitrate levels. However, its expression increased by approximately 2-fold with nitrate induction treatment. In Thellungiella shoots and roots, expression of NRT3.1 increased by 2.5- and 1.9-fold, respectively, when comparing severe N limitation with control. Nitrate induction treatment in Thellungiella tissues led to up-regulation of NRT3.1 expression by approximately 4-fold (Fig. 6, K and L). In addition, Thellungiella shoots and roots had significantly higher basal NRT3.1 transcript copy numbers than in Arabidopsis (Table II).
The anion channel nitrate/proton antiporter CLCa mediates nitrate accumulation in plant vacuoles (De Angeli et al., 2006). However, no differences in CLCa expression were observed between Thellungiella and Arabidopsis: CLCa expression was reduced in shoots and roots of both species under limited N (data not shown).
Plants have developed adaptive responses to certain levels of N limitation. Recently, NLA, a gene essential for these responses, was identified (Peng et al., 2007b). Expression of NLA was up-regulated in shoots of Thellungiella and Arabidopsis grown under limited N (Fig. 6O). However, in Arabidopsis roots, its expression was slightly repressed under low nitrate, whereas in Thellungiella roots, NLA expression was slightly induced by low nitrate treatment (Fig. 6P). The largest difference was that Thellungiella had 2- and 1.6-fold higher basal transcript copy numbers than Arabidopsis in shoots and roots, respectively (Table II), possibly indicating that Thellungiella has a higher constitutive adaptive response to cope with N-limiting conditions.
DISCUSSION
In this study, we report that the halophytic Arabidopsis relative Thellungiella is also tolerant to low N stress. Growth reduction due to N limitation was much less in Thellungiella than in Arabidopsis (Figs. 1, A and B, and 2), and under severe N limitation, only Thellungiella completed its life cycle (data not shown). Moreover, analysis of anthocyanin content, a useful marker of abiotic stress in plants (Bongue-Bartelsman and Philips, 1995; Chalker-Scott, 1999; Peng et al., 2007b), suggested that Arabidopsis was far more sensitive to even mild N limitation than Thellungiella (Fig. 1C). A low N supply generally leads to a reduction in photosynthesis, lower root growth, suppression of lateral root initiation, early leaf senescence, and an increase in the carbon to N ratio in plants (Paul and Driscoll, 1997; Malamy and Ryan, 2001; Martin et al., 2002; Malamy, 2005; Wingler et al., 2006; Zhang et al., 2007). However, suppression of lateral roots was comparatively lower in Thellungiella than in Arabidopsis when grown on high Suc to nitrate medium (Fig. 2C), and Thellungiella also displayed a lower increase in carbon to N ratio than was observed in Arabidopsis under N limitation (Fig. 3I). This N stress-tolerant phenotype of Thellungiella is consistent with the prevailing conditions in its native habitat, where soil N levels are low (E. Weretilnyk, unpublished data). Furthermore, the high salt levels in these soils could exacerbate N-limiting conditions, since Na+ ions can compete with nutrients for root uptake sites (Sagi et al., 1997).
We were able to demonstrate that under N-limiting conditions, Thellungiella contained higher free nitrate levels (Fig. 3, C and D), which could be ascribed to higher nitrate uptake (Fig. 4). To some extent, the higher nitrate uptake capacity of Thellungiella could be correlated with differences in the expression of nitrate transporter genes between Thellungiella and Arabidopsis. For instance, NRT1.1 expression was repressed to a lesser degree in Thellungiella roots than in Arabidopsis roots by N limitation (Fig. 6H), while up-regulation of NRT2.1 expression by low nitrate and nitrate induction treatment was higher in Thellungiella roots than in Arabidopsis (Fig. 6J). Up-regulation of NRT3.1 by N limitation was also higher in Thellungiella shoots and roots than in Arabidopsis (Fig. 6, K and L). Furthermore, NRT3.1 was expressed at a constitutively higher level in Thellungiella compared with Arabidopsis under N-sufficient conditions (Table II). It is not clear at this stage how these complex differences in nitrate transporter expression function to increase the nitrate uptake capacity of Thellungiella under N limitation, and it is important to note that there is significant posttranslational regulation of nitrate transporters (Miller et al., 2007; Wirth et al., 2007). Indeed, NRT3.1 itself has not been shown to display nitrate transporter activity; rather, it modulates the activity of other transporters, such as NRT1.1 and NRT2.1 (Okamoto et al., 2006; Orsel et al., 2006; Miller et al., 2007).
Thellungiella plants not only exhibited increased nitrate uptake under N-limiting conditions compared with Arabidopsis, they also displayed higher levels of nitrate assimilation. In general, N-stressed plants respond by reducing the synthesis of nitrogenous compounds (Paul and Driscoll, 1997; Peng et al., 2007b), and indeed, a reduction in amino acid and soluble protein content was observed in both Arabidopsis and Thellungiella under N limitation (Fig. 3, E–G). However, Thellungiella was able to maintain substantially higher levels of amino acids and soluble protein than Arabidopsis. To support this increased level of nitrate assimilation under N limitation, Thellungiella was able to maintain NR activity at virtually control levels, whereas NR activity was drastically reduced in N-stressed Arabidopsis (Fig. 5, A and B). The pattern of NR2 gene expression closely followed that of NR activity in both species (Fig. 6, A and B), suggesting that control of NR activity under N limitation is exerted at the level of NR2 transcript accumulation. The results for GS activity and gene expression were more complex. Although no difference in total GS activity was observed between Arabidopsis and Thellungiella (Fig. 5, C and D), there were differences in GS isoform-specific gene expression between the two species (Fig. 6, C–F). Thellungiella shoot GS1.1, GS1.3, and GS1.4 expression was induced to a greater extent by N limitation (Fig. 6C; data not shown), and the basal level of Thellungiella root GS1.1 transcript accumulation was higher, than in Arabidopsis (Table II). Under N limitation, Thellungiella root GS2 expression was maintained at control levels, whereas it decreased in Arabidopsis (Fig. 6F). Moreover, basal levels of root GS2 transcript accumulation were also higher in Thellungiella (Table II). As with nitrate transporter gene expression, it is not clear how these isoform-specific and tissue-specific differences in GS expression contribute to higher nitrate assimilation in Thellungiella under N limitation. However, the fact that there is a little or no species-specific difference in shoot GS2 expression suggests that it may be important to maintain plastidic GS2 activity even in N-stressed Arabidopsis, in which there is a decrease in nitrate assimilation. Since protein breakdown occurs under N stress (Coruzzi, 2003; Bi et al., 2007), plastidic GS2 activity could be vital for the reassimilation of ammonia in the GS/GOGAT cycle. On the other hand, the increased induction of expression of shoot cytosolic GS1 isoforms by N limitation in Thellungiella would be important in maintaining primary nitrate assimilation.
The assimilation of nitrate into amino acids not only depends upon a supply of N but also upon the availability of carbon skeletons. The fact that Arabidopsis exhibited reduced nitrate uptake and assimilation compared with Thellungiella suggests a reduced demand for carbon skeletons for the generation of amino acids. This reduced demand was reflected in the use of carbon to produce the higher starch levels and the concomitant increase in carbon to N ratio observed in Arabidopsis (Fig. 3, H and I). Conversely, for Thellungiella to maintain higher levels of nitrate assimilation under N limitation, carbon skeletons would have to be supplied. Our metabolite analysis suggested that these carbon skeletons are supplied via a large increase in the malate pool in Thellungiella (Table I), presumably allowing an increase in the generation of oxaloacetate for amino acid synthesis (Coruzzi and Last, 2000; Siedow and Day, 2000). On the other hand, continuous export of oxaloacetate for nitrate assimilation could eventually lead to a deficiency in citric acid intermediates such as the reduced accumulation of fumarate (Table I). However, malate can also be produced in the cytosol from Glc and Fru via phosphoenolpyruvate and can be transported back into the mitochondria, providing anaplerotic replenishment of citric acid intermediates (Dennis and Blakeley, 2000; Siedow and Day, 2000). Both Glc and Fru displayed greater accumulation in Thellungiella than in Arabidopsis, suggesting that these two sugars may also be involved in the increased production of malate observed in Thellungiella. Glc and Fru are derived from Suc, yet Suc accumulation did not differ greatly between the two species (Table I). This suggests that in order to maintain Suc pools while at the same time accumulating more Glc and Fru, Thellungiella would need to generate more Suc than Arabidopsis via photosynthesis. The possibility of a higher level of photosynthesis in Thellungiella under N limitation is supported by (1) a higher level of growth of Thellungiella (Fig. 1); (2) a higher leaf number in Thellungiella (data not shown); and (3) the ability of Thellungiella to maintain green leaves and cotyledons while Arabidopsis produced cotyledons that bleached, indicating low N-mediated damage to the photosynthetic machinery (Fig. 2).
In conclusion, our physiological, biochemical, and gene expression analyses suggest that Thellungiella is better adapted to efficiently acquire and utilize nitrate under N-limiting conditions. Thellungiella and Arabidopsis are closely related, and it is likely that they share many of the same mechanisms and associated genes involved in N acquisition and assimilation. Thus, subtle variations in gene regulation (transcriptional, posttranscriptional, and posttranslational) may at least partly explain the differences in the ability of Thellungiella and Arabidopsis to withstand low N stress. Such a hypothesis has been put forward to explain the differing salt tolerance of the two species (Zhu, 2001), and evidence supporting this hypothesis was recently published (Kant et al., 2006). Our results showing differential expression of nitrate transporter and assimilation genes further support this contention.
Until now, Thellungiella has been suggested as an Arabidopsis relative model system for investigating natural tolerance to salt stress (Inan et al., 2004), since it has all of the attributes of a model plant system, including short life cycle, relatively small genome, copious seed production, and ease of transformation with foreign genes. We propose that Thellungiella would be equally useful for studying the mechanisms of natural low N tolerance. The ongoing sequencing of the Thellungiella genome, production of bacterial artificial chromosome and cDNA libraries, and generation of EST and T-DNA insertion collections will further enhance the power of the Thellungiella system for identifying key components governing N utilization under low N conditions.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Thellungiella halophila (Shandong and Yukon ecotypes) and Arabidopsis (Arabidopsis thaliana Columbia ecotype) were used for all experiments. Seeds were mixed with 0.12% (w/v) agarose and stratified at 4°C in the dark for 6, 2, and 4 d for Shandong, Yukon, and Columbia ecotypes, respectively. Our preliminary work has shown that Shandong ecotype requires 6 d of stratification for uniform germination, and for Yukon ecotype 2 d of stratification is optimum. Seeds were sown in a 1:1 mixture of perlite and nutrient-free LB2 soil (Sun Gro Horticulture Canada; http://www.sungro.com/) in small pots, and fine vermiculite was sprinkled on the soil to prevent algae growth. This soil mixture is nutrient free, allows easy flow through of nutrient solution to prevent the buildup of nutrients, and also permits easy harvesting of roots. The plants were grown in controlled growth chambers at 16 h of light/8 h of dark, 23°C day/18°C night, light intensity of 150 μE m−2 s−1, and 65% relative humidity. The nutrient solution contained 2 mm NaH2PO4 (pH 6.0), 1.5 mm MgSO4, 1.5 mm CaCl2, 100 μm Fe-EDTA, 50 μm H3BO3, 12 μm MnCl2, 2 μm ZnSO4, 1 μm CuSO4, and 0.2 μm Na2MoO4. We conducted preliminary experiments to determine the optimum and limiting N conditions for Thellungiella and Arabidopsis. Plants were grown at different N levels ranging from 0.2 to 10 mm nitrate. At 4 mm and above, growth was similar for both plant species, while below 0.4 mm nitrate, neither species was able to complete its life cycle (data not shown). Therefore, three nitrate levels were chosen: N sufficient (4 mm KNO3−), mild N limitation (1 mm KNO3−), and severe N limitation (0.4 mm KNO3−). The potassium level was balanced with KCl to maintain 4 mm potassium. For gene expression studies, two more treatments were added. Two hours before harvesting, (1) the nitrate concentration of a subsample of plants treated with 1 mm nitrate was raised to 4 mm (N induction), or (2) the nitrate concentration of a subsample of plants treated with 4 mm nitrate was reduced to 1 mm (N reduction).
Since the initial growth of Thellungiella is slower than that of Arabidopsis (Taji et al., 2004; Gong et al., 2005; Kant et al., 2006), plants were harvested at different times (i.e. 20 d after germination for Arabidopsis and 23 d after germination for Thellungiella). This ensured a similar physiological stage and comparable plant size for both species. Shoots and roots were harvested separately, frozen in liquid N, and stored at −80°C until use. To analyze the effect of nitrate levels on root architecture, sterilized seeds were grown on vertical square plates (2% Suc, pH 5.8, and 0.8% agar). To visualize the early effects on roots, nutrient concentrations in plate medium were kept lower (Remans et al., 2006) than in soil experiments: 0.7 mm NaH2PO4, 0.5 mm MgSO4, 0.5 mm CaCl2, with N levels of 1, 0.5, 0.2, 0.1, and 0.05 mm KNO3, and 1 mm potassium maintained with KCl. The micronutrient composition and growth conditions were similar to those in the soil experiments mentioned above. Measurements for root length and lateral root number were taken at 10 and 12 d after germination for Arabidopsis and Thellungiella, respectively. The shoots were separated and stored at −80°C.
15NO3− Uptake
Measurements of 15NO3− influx in roots were performed according to Munos et al. (2004) with minor modifications. Plants were grown on vertical agar plates (1% Suc, pH 5.8, and 0.8% agar) with 4, 1, and 0.4 mm nitrate and other nutrient concentrations the same as in the soil experiments. Ten- and 12-d-old seedlings of Arabidopsis and Thellungiella, respectively, were transferred to 0.1 mm CaSO4 solution for 1 min, and then to nutrient solution containing 0.2, 0.4, 1, 2, and 4 mm 15NO3− (99% atom excess 15N) for 5 min, and finally to 0.1 mm CaSO4 for 1 min. Roots were separated from shoots and dried at 70°C for 48 h. Influx of 15NO3− was calculated from the 15N content of the roots, as analyzed using an isotope mass spectrometer (PDZ Europa).
Biochemical Analyses
Frozen shoot and root tissue was used for the following biochemical assays. Nitrate content was analyzed by colorimetric assay according to Cataldo et al. (1975). Percentages of total N and total carbon in the dried tissues were measured by the Micro-Dumas combustion analysis method using a Carlo Erba NA1500 C/N analyzer (Carlo Erba Strumentazione). Total amino acids were extracted successively with 80%, 50%, and 0% ethanol in 10 mm HEPES-KOH (pH 7.4), supernatants were pooled, and total amino acids were assayed according to Rosen (1957). Total soluble protein was extracted with buffer containing 100 mm HEPES-KOH, pH 7.5, and 0.1% Triton X-100 and assayed using the Bio-Rad Protein Assay Kit (http://www.biorad.com). Starch was extracted according to Delatte et al. (2005) and quantified using a commercial total starch assay kit (Megazyme; http://www.megazyme.com). Relative anthocyanin content was analyzed based on Neff and Chory (1998).
NR and GS Enzyme Activity
NR activity was measured according to Yu et al. (1998) with slight modifications. In brief, the frozen tissue was homogenized in extraction buffer containing 100 mm K2PO4 (pH 7.5), 1 mm EDTA, 5 mm dithiothreitol, 0.5 mm phenylmethylsulfonyl fluoride, 1 μm Na2MoO4, 5 μm FAD, and a protease inhibitor tablet (Roche Diagnostics). Extracts were centrifuged at 10,000g for 10 min. An aliquot of supernatant was incubated with reaction buffer (50 mm K2PO4, pH 7.5, 40 mm KNO3, and 0.2 mm NADH) for 30 min at room temperature. The reaction was terminated by adding 1% sulfanilamide in 3 n HCl. N-Naphthylethylenediamine dihydrochloride (0.02%) was added, and NO2− content was measured spectrophotometrically at A540.
GS activity was determined by transferase reaction, which measures the formation of γ-glutamyl hydroxymate, as described by Shapiro and Stadtman (1970). Tissue was homogenized in extraction buffer (100 mm Tris, pH 7.5, 10 mm MgCl2, 10% glycerol, 5 mm dithiothreitol, 0.05% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and a protease inhibitor tablet). Extracts were centrifuged at 10,000g for 10 min, and supernatant was incubated with reaction buffer (100 mm HEPES, pH 7.0, 60 mm l-Gln, 45 mm potassium arsenate, 5 mm MnCl2, 1 mm ADP-Na, and 125 mm hydroxylamine-HCl, with the final pH adjusted to 7.2) for 30 min at 30°C. The reaction was stopped by adding stop solution containing 5% FeCl3 and 4% trichloroacetic acid in 0.5 n HCl, and γ-glutamyl hydroxymate formation was measured spectrophotometrically at A540.
Metabolite Analysis
Shoots (50 mg fresh weight) from seedlings employed in the root study were used for metabolite analysis essentially according to Fiehn et al. (2000). Ribitol was added as an internal standard to the samples during extraction, and extracts were separated into polar (methanol/water) and apolar/lipid (chloroform) phases. Polar fractions were vacuum dried and derivatized. The derivatized samples were diluted 10-fold with hexane, and then 1 μL was injected into the splitless injection port of a TRACE DSQ gas chromatography-mass spectrometry system (Thermo Finnigan). Chromatography was performed using a 30-m × 0.25-mm Rtx-5MS column (Chromatographic Specialties). For each run, the gas chromatograph oven was set at 50°C initial temperature, which was held for 2.5 min, increased at 7.5°C min−1 to 70°C, and finally increased at 5°C min−1 to 310°C, at which it was held for 6 min. The injection temperature was 230°C, and the ion source was kept at 200°C. Mass spectra were recorded at three scans per second with an m/z scanning range of 50 to 650. The data analysis was performed using the automated mass spectral deconvolution and identification system (AMDIS) software (http://chemdata.nist.gov/mass-spc/amdis).
Expression Analysis by Quantitative Real-Time PCR
Total RNA was isolated from shoot and root tissue using Trizol reagent (Invitrogen). To eliminate any residual genomic DNA, total RNA was treated with RQ1 ribonuclease-free DNase (Promega). The first-strand cDNA was synthesized from total RNA using the Reverse Transcription System kit (Promega). Since Thellungiella ESTs exhibit up to 95% identity with Arabidopsis cDNA sequences (Taji et al., 2004; Wang et al., 2004; Wong et al., 2005), the Arabidopsis cDNA sequences from GenBank (http://www.ncbi.nlm.nih.gov/) were used to design primers for real-time PCR measurement of gene expression in both species (Kant et al., 2006). Primer Express 2.0 software (Applied Biosystems) was used to design the primers. The PCR products of Thellungiella for each orthologous gene were examined for a similar size as amplified from the respective Arabidopsis gene primers and sequenced to ensure that they encoded the expected gene product. Primer sequences for each gene are given in Supplemental Table S2. Real-time PCR was performed according to Kant et al. (2006). Relative quantification values for each target gene were calculated by the 2−ΔΔCT method (Livak and Schmittgen, 2001) using UBIQUITIN10 (UBQ10) and/or GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE A SUBUNIT as an internal reference gene to compare data from different PCR runs or cDNA samples. To ensure the validity of the 2−ΔΔCT method, 2-fold serial dilutions of cDNA from control plants of Thellungiella and Arabidopsis were used to create standard curves, and the amplification efficiencies of the target and reference genes were shown to be approximately equal (Livak and Schmittgen, 2001). Expression of most of the genes was also confirmed by semiquantitative reverse transcription-PCR using UBQ10 as an internal control, and similar results were obtained as for real-time PCR (data not shown). The PCR conditions and specific primers for semiquantitative reverse transcription-PCR are available upon request.
Real-time PCR analysis gives relative changes in gene expression, with control treatment normalized to a value of 1. Hence, comparison of basal expression levels of genes between two species under the control treatment (4 mm nitrate) requires absolute quantification. To compare basal transcript levels, quantification of absolute transcript copy number was performed according to Kant et al. (2007). Real-time PCR products for each gene were gel purified (QIAEX II Gel Extraction Kit; Qiagen) and quantified with a NanoDrop spectrophotometer (ND-1000; NanoDrop Technologies). To determine the transcript copies per nanogram of PCR product, we first calculated the molecular mass of the PCR product, then converted to number of picomoles per nanogram, and further converted to number of transcripts per nanogram; the calculations were based on formulae given in the PCR Application Manual (third edition, online version; Roche Molecular Biochemicals). Ten-fold serial dilutions of each PCR product with known transcript copies per nanogram were used to create standard curves. The slope and intercept of the standard curve and the threshold cycle (CT) of each gene were used to calculate transcript copy number for each respective gene.
Statistics
The results shown are representative of three independent experiments, and within each experiment treatments were replicated three times, unless otherwise stated. Data were statistically analyzed by Fisher's protected lsd test using SAS statistical software (SAS Institute).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Comparison of the partial sequences of genes in Arabidopsis and Thellungiella.
Supplemental Table S2. Primers used for real-time PCR analysis of gene expression.
Supplementary Material
Acknowledgments
We thank David Guevara and Jeff Dedrick for their help in gas chromatography-mass spectrometry data analysis.
This work was supported by the Natural Sciences and Engineering Research Council of Canada and the Ontario Research and Development Challenge Fund.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Steven J. Rothstein (rothstei@uoguelph.ca).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Amtmann A, Bohnert HJ, Bressan RA (2005) Abiotic stress and plant genome evolution: search for new models. Plant Physiol 138 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi YM, Wang RL, Zhu T, Rothstein SJ (2007) Global transcription profiling reveals differential responses to chronic nitrogen stress and putative nitrogen regulatory components in Arabidopsis. BMC Genomics 8 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongue-Bartelsman M, Philips DA (1995) Nitrogen stress regulates gene expression of enzymes in the flavonoid biosynthetic pathway of tomato. Plant Physiol Biochem 33 539–546 [Google Scholar]
- Cataldo DA, Haroon M, Schrader TE, Youngs VL (1975) Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid. Commun Soil Sci Plant Anal 6 71–80 [Google Scholar]
- Cerezo M, Tillard P, Filleur S, Munos S, Daniel-Vedele F, Gojon A (2001) Major alterations of the regulation of root NO3− uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol 127 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker-Scott L (1999) Environmental significance of anthocyanins in plant stress responses. Photochem Photobiol 70 1–9 [Google Scholar]
- Coruzzi GM (2003) Primary N-assimilation into amino acids in Arabidopsis. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi: 10.1199/tab.0010, www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Coruzzi GM, Last RL (2000) Amino acids. In RG Buchanan, W Gruissem, R Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 358–410
- Crawford NM (1995) Nitrate: nutrient and signal for plant growth. Plant Cell 7 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM, Forde BG (2002) Molecular and developmental biology of inorganic nitrogen nutrition. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi: 10.1199/tab.0011, www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Cren M, Hirel B (1999) Glutamine synthetase in higher plants: regulation of gene and protein expression from the organ to the cell. Plant Cell Physiol 40 1187–1193 [Google Scholar]
- De Angeli A, Monachello D, Ephritikhine G, Frachisse JM, Thomine S, Gambale F, Barbier-Brygoo H (2006) The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 442 939–942 [DOI] [PubMed] [Google Scholar]
- Delatte T, Trevisan M, Parker ML, Zeeman SC (2005) Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J 41 815–830 [DOI] [PubMed] [Google Scholar]
- Dennis DT, Blakeley SD (2000) Carbohydrate metabolism. In RG Buchanan, W Gruissem, R Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 630–675
- Diaz U, Saliba-Colombani V, Loudet O, Belluomo P, Moreau L, Daniel-Vedele F, Morot-Gaudry JF, Maselaux-Daubresse U (2006) Leaf yellowing and anthocyanin accumulation are two genetically independent strategies in response to nitrogen limitation in Arabidopsis thaliana. Plant Cell Physiol 47 74–83 [DOI] [PubMed] [Google Scholar]
- Ding L, Wang KJ, Jing GM, Biswas DK, Xu H, Li LF, Li YH (2005) Effects of nitrogen deficiency on photosynthetic traits of maize hybrids released in different years. Ann Bot (Lond) 96 925–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277 33105–33114 [DOI] [PubMed] [Google Scholar]
- Edwards JW, Walker EL, Coruzzi GM (1990) Cell-specific expression in transgenic plants reveals nonoverlapping roles for chloroplast and cytosolic glutamine synthetase. Proc Natl Acad Sci USA 87 3459–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L (2000) Metabolite profiling for plant functional genomics. Nat Biotechnol 18 1157–1161 [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Hulett J, Cramer GR, Seemann JR (1997) Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol 115 317–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass ADM (2003) Nitrogen use efficiency of crop plants: physiological constraints upon nitrate adsorption. Crit Rev Plant Sci 22 453–470 [Google Scholar]
- Gong Q, Li P, Ma S, Rupassara SI, Bohnert HJ (2005) Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J 44 826–839 [DOI] [PubMed] [Google Scholar]
- Good AG, Shrawat AK, Muench DG (2004) Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci 9 597–605 [DOI] [PubMed] [Google Scholar]
- Griffith M, Timonin M, Wong CEA, Gray GR, Akhter SR, Saldanha M, Rogers MA, Weretilnyk EA, Moffatt BA (2007) Thellungiella: an Arabidopsis-related model plant adapted to cold temperatures. Plant Cell Environ 30 529–538 [DOI] [PubMed] [Google Scholar]
- Hirel B, Gouis JL, Ney B, Gallais A (2007) The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. J Exp Bot 58 2369–2387 [DOI] [PubMed] [Google Scholar]
- Inan G, Zhang Q, Li P, Wang Z, Cao Z, Zhang H, Zhang C, Quist TM, Goodwin SM, Zhu J, et al (2004) Salt cress: a halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol 135 1718–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant P, Kant S, Gordon M, Shaked R, Barak S (2007) STRESS RESPONSE SUPPRESSOR1 and STRESS RESPONSE SUPPRESSOR2, two DEAD-box RNA helicases that attenuate Arabidopsis response to multiple abiotic stresses. Plant Physiol 145 814–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Kant P, Raveh E, Barak S (2006) Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T. halophila. Plant Cell Environ 29 1220–1234 [DOI] [PubMed] [Google Scholar]
- Krouk G, Tillard P, Gojon A (2006) Regulation of the high-affinity NO3− uptake system by NRT1.1-mediated NO3− demand signaling in Arabidopsis. Plant Physiol 142 1075–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea PJ, Azevedo RA (2006) Nitrogen use efficiency. 1. Uptake of nitrogen from the soil. Ann Appl Biol 149 243–247 [Google Scholar]
- Lejay L, Tillard P, Lepetit M, Olive FD, Filleur S, Daniel-Vedele F, Gojon A (1999) Molecular and functional regulation of two NO3− uptake systems by N- and C-status of Arabidopsis plants. Plant J 18 509–519 [DOI] [PubMed] [Google Scholar]
- Li W, Wang Y, Okamoto M, Crawford NM, Siddiqi MY, Glass ADM (2007) Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol 143 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Huang CY, Tsay YF (1999) CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Malamy JE (2005) Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 28 67–77 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Ryan KS (2001) Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol 127 899–909 [PMC free article] [PubMed] [Google Scholar]
- Marschner H (1995) Mineral Nutrition of Higher Plants, Ed 2. Academic Press, London
- Martin T, Oswald O, Graham IA (2002) Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol 128 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough DE, Girardin PH, Mihajlovic M, Aguilera A, Tollenaar M (1994) Influence of N supply on development and dry matter accumulation of an old and a new maize hybrid. Can J Plant Sci 74 471–477 [Google Scholar]
- Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM (2007) Nitrate transport and signalling. J Exp Bot 58 2297–2306 [DOI] [PubMed] [Google Scholar]
- Munos S, Cazettes C, Fizames C, Gaymard F, Tillard P, Lepetit M, Lejay L, Gojon A (2004) Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell 16 2433–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Kumar A, Li W, Wang Y, Siddiqi MY, Crawford NM, Glass ADM (2006) High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol 140 1036–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Vidmar JJ, Glass ADM (2003) Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: responses to nitrate provision. Plant Cell Physiol 44 304–317 [DOI] [PubMed] [Google Scholar]
- Ono K, Terashima I, Watanabe A (1996) Interaction between nitrogen deficit of a plant and nitrogen content in the old leaves. Plant Cell Physiol 37 1083–1089 [Google Scholar]
- Orsel M, Chopin F, Leleu O, Smith SJ, Krapp A, Daniel-Vedele F, Miller AJ (2006) Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis: physiology and protein-protein interaction. Plant Physiol 142 1304–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M, Eulenburg K, Krapp A, Daniel-Vedele F (2004) Disruption of the nitrate transporter genes AtNRT2.1 and AtNRT2.2 restricts growth at low external nitrate concentration. Planta 219 714–721 [DOI] [PubMed] [Google Scholar]
- Ortiz-Monasterio JI, Sayre KD, Rajaram S, McMahon M (1997) Genetic progress in wheat yield and nitrogen use efficiency under four nitrogen regimes. Crop Sci 37 898–904 [Google Scholar]
- Paul MJ, Driscoll SP (1997) Sugar repression of photosynthesis: the role of carbohydrates in signalling nitrogen deficiency through source:sink imbalance. Plant Cell Environ 20 110–116 [Google Scholar]
- Peng M, Bi YM, Zhu T, Rothstein SJ (2007. a) Genome-wide analysis of Arabidopsis responsive transcriptome to nitrogen limitation and its regulation by the ubiquitin ligase gene NLA. Plant Mol Biol 65 775–797 [DOI] [PubMed] [Google Scholar]
- Peng M, Hannam C, Gu H, Bi YM, Rothstein SJ (2007. b) A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J 50 320–337 [DOI] [PubMed] [Google Scholar]
- Raun WR, Johnson GV (1999) Improving nitrogen use efficiency for cereal production. Agron J 91 357–363 [Google Scholar]
- Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, Gojon A (2006) A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol 140 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen H (1957) A modified ninhydrin colorimetric analysis for amino acids. Arch Biochem Biophys 67 10–15 [DOI] [PubMed] [Google Scholar]
- Sagi M, Dovrat A, Kipnis T, Lips SH (1997) Ionic balance and the production of biomass and organic nitrogen as affected by salinity and N source in annual ryegrass (Lolium multiflorum Lam). J Plant Nutr 20 1291–1316 [Google Scholar]
- Shapiro BM, Stadtman ER (1970) The regulation of glutamine synthetase in microorganisms. Annu Rev Microbiol 24 501–524 [DOI] [PubMed] [Google Scholar]
- Siedow JN, Day DA (2000) Respiration and photorespiration. In RG Buchanan, W Gruissem, R Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 676–725
- Stitt M (1999) Nitrate regulation of metabolism and growth. Curr Opin Plant Biol 2 178–186 [DOI] [PubMed] [Google Scholar]
- Taira M, Valtersson U, Burkhardt B, Ludwig RA (2004) Arabidopsis thaliana GLN2-encoded glutamine synthetase is dual targeted to leaf mitochondria and chloroplasts. Plant Cell 16 2048–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, Narusaka Y, Narusaka M, Zhu JK, Shinozaki K (2004) Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol 135 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK (2007) Nitrate transporters and peptide transporters. FEBS Lett 581 2290–2300 [DOI] [PubMed] [Google Scholar]
- Volkov V, Amtmann A (2006) Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, has specific root ion channel features supporting K+/Na+ homeostasis under salinity stress. Plant J 48 342–353 [DOI] [PubMed] [Google Scholar]
- Wang R, Liu D, Crawford NM (1998) The Arabidopsis CHL1 protein plays a major role in high affinity nitrate uptake. Proc Natl Acad Sci USA 95 15134–15139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZL, Li PH, Fredricksen M, Gong ZZ, Kim CS, Zhang C, Bohnert HJ, Zhu JK, Bressan RA, Hasegawa PM, et al (2004) Expressed sequence tags from Thellungiella halophila, a new model to study plant salt-tolerance. Plant Sci 166 609–616 [Google Scholar]
- Wilkinson JQ, Crawford NM (1991) Identification of the Arabidopsis CHL3 gene as the nitrate reductase structural gene NIA2. Plant Cell 3 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Purdy S, MacLean JA, Pourtau N (2006) The role of sugars in integrating environmental signals during the regulation of leaf senescence. J Exp Bot 57 391–399 [DOI] [PubMed] [Google Scholar]
- Wirth J, Chopin F, Santoni V, Viennois G, Tillard P, Krapp A, Lejay L, Daniel-Vedele F, Gojon A (2007) Regulation of root nitrate uptake at the NRT2.1 protein level in Arabidopsis thaliana. J Biol Chem 282 23541–23552 [DOI] [PubMed] [Google Scholar]
- Wong CE, Li Y, Labbe A, Guevara D, Paulo N, Whitty B, Dıaz C, Golding GB, Gray GR, Weretilnyk EA, et al (2006) Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol 140 1437–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CE, Li Y, Whitty BR, Dıaz-Camino C, Akhter SR, Brandle JE, Golding GB, Weretilnyk EA, Moffatt BA, Griffith M (2005) Expressed sequence tags from the Yukon ecotype of Thellungiella reveal that gene expression in response to cold, drought and salinity shows little overlap. Plant Mol Biol 58 561–574 [DOI] [PubMed] [Google Scholar]
- Yu X, Sukumaran S, Marton L (1998) Differential expression of the Arabidopsis Nia1 and Nia2 genes. Plant Physiol 116 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Rong H, Pilbeam D (2007) Signalling mechanisms underlying the morphological responses of the root system to nitrogen in Arabidopsis thaliana. J Exp Bot 58 2329–2338 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6 66–72 [DOI] [PubMed] [Google Scholar]
- Zhuo D, Okamoto M, Vidmar JJ, Glass ADM (1999) Regulation of a putative high-affinity nitrate transporter (Nrt2;1At) in roots of Arabidopsis thaliana. Plant J 17 563–568 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.