Abstract
A Citrus sinensis spontaneous mutant, navel negra (nan), produces fruit with an abnormal brown-colored flavedo during ripening. Analysis of pigment composition in the wild-type and nan flavedo suggested that typical ripening-related chlorophyll (Chl) degradation, but not carotenoid biosynthesis, was impaired in the mutant, identifying nan as a type C stay-green mutant. nan exhibited normal expression of Chl biosynthetic and catabolic genes and chlorophyllase activity but no accumulation of dephytylated Chl compounds during ripening, suggesting that the mutation is not related to a lesion in any of the principal enzymatic steps in Chl catabolism. Transcript profiling using a citrus microarray indicated that a citrus ortholog of a number of SGR (for STAY-GREEN) genes was expressed at substantially lower levels in nan, both prior to and during ripening. However, the pattern of catabolite accumulation and SGR sequence analysis suggested that the nan mutation is distinct from those in previously described stay-green mutants and is associated with an upstream regulatory step, rather than directly influencing a specific component of Chl catabolism. Transcriptomic and comparative proteomic profiling further indicated that the nan mutation resulted in the suppressed expression of numerous photosynthesis-related genes and in the induction of genes that are associated with oxidative stress. These data, along with metabolite analyses, suggest that nan fruit employ a number of molecular mechanisms to compensate for the elevated Chl levels and associated photooxidative stress.
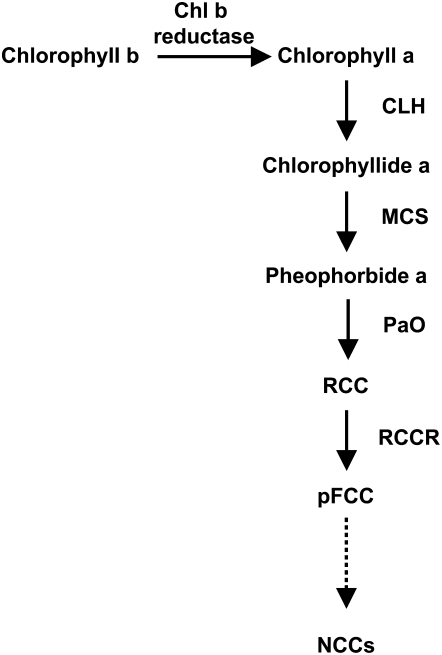
Chlorophyll (Chl) degradation is central to the degreening process that is commonly observed in senescing leaves and the ripening of many fruit, and it has been estimated that approximately 1.2 billion tons of Chl are degraded annually (Hendry et al., 1987). However, despite its importance, the basic steps of the Chl catabolic pathway have only recently been elucidated and a few of the associated genes identified (Hörtensteiner, 2006). The degradation of Chl to a colorless fluorescent intermediate (primary fluorescent Chl catabolite [pFCC]) involves four basic steps, which are apparently common to all plants (Fig. 1): chlorophyllase (CLH) dephytylates Chl a, producing chlorophyllide a; an unknown metal-chelating substance removes magnesium and produces pheophorbide a; and finally, pheophorbide a oxygenase (PaO) and red Chl catabolite reductase (RCCR) convert pheophorbide a to red Chl catabolite (RCC) and then to pFCCs. The conversion of pheophorbide a to RCC has been proposed as the key regulatory step of this pathway, since there is evidence indicating that PaO is the only enzyme of this pathway that is induced during senescence (Thomas et al., 2002), in the form of increased transcript abundance (for review, see Hörtensteiner, 2006). The subsequent reactions are species specific and involve multiple structural modifications of the pFCCs, before they are converted into nonfluorescent Chl catabolites and are finally stored in the vacuole (Hörtensteiner, 2006). Despite excellent progress in characterizing the primary catabolic events, remarkably little is yet known about the factors that regulate the overall rate and extent of Chl breakdown (Hörtensteiner, 2006).
Figure 1.
Scheme of the Chl degradative pathway in higher plants. MCS, Metal-chelating substance; NCCs, nonfluorescent chlorophyll catabolites.
In this regard, important insights are likely to be provided through the identification and characterization of various “stay-green” mutants that exhibit unusual Chl retention during leaf senescence or fruit ripening (Thomas and Howarth, 2000). Those identified to date span a broad taxonomic range, including members of the Gramineae (durum wheat [Triticum durum; Spano et al., 2003]; Festuca pratensis [Thomas, 1987]; rice [Oryza sativa; Cha et al., 2002; Jiang et al., 2007; Kusaba et al., 2007; Park et al., 2007]), Arabidopsis (Arabidopsis thaliana; Woo et al., 2001; Oh et al., 2003, 2004; Ren et al., 2007), and the Leguminosae (soybean [Glycine max; Guiamét and Giannibelli, 1994, 1996; Luquez and Guiamét, 2002]; Phaseolus vulgaris [Ronning et al., 1991; Bachmann et al., 1994]; pea [Pisum sativum; Armstead et al., 2007; Sato et al., 2007]). A stay-green mutant phenotype has also been reported in tomato (Solanum lycopersicum) fruits (the green flesh mutant; Cheung et al., 1993; Akhtar et al., 1999) and pepper (Capsicum annuum; mutant chlorophyll retainer; Efrati et al., 2005; Roca and Mínguez-Mosquera, 2006). The molecular bases of the stay-green phenotype were first characterized in a F. pratensis mutant line showing accumulation of both Chl a and the dephytylated Chl catabolites chlorophyllide a and pheophorbide a (Thomas et al., 2002). After genetic and biochemical analyses, it was concluded that the mutant was affected in either the PaO gene or a specific regulator of this gene. Although biochemical lesions in PaO have been reported for several stay-green mutants (Hörtensteiner, 2006), other types of mutations can also result in stay-green phenotypes, such as Chl b reductase in the rice nyc1 mutant (Kusaba et al., 2007) and genes involved in light-harvesting complex protein II (LHCPII) proteolysis in the ore9 (Woo et al., 2001) and ore10 (Oh et al., 2003, 2004) Arabidopsis stay-green mutants. In addition, it has been shown that RNA interference-mediated knockdown of a soybean senescence-associated receptor-like kinase confers a stay-green phenotype (Li et al., 2006).
A clearer understanding of the genetic basis of the control of Chl degradation has only recently resulted from cloning of STAY-GREEN (SGR) genes from a Lolium/Festuca introgression (Armstead et al., 2006), rice (Jiang et al., 2007; Park et al., 2007), pea (Sato et al., 2007), and tomato and pepper (Barry et al., 2008). Although the biochemical function of SGR proteins is not known, they contain a predicted chloroplast transit peptide, and at least one SGR has been shown to bind LHCPII in vivo (Park et al., 2007). Transcript analyses further indicate that SGR gene expression is closely associated with leaf senescence (Armstead et al., 2006; Hörtensteiner, 2006; Sato et al., 2007), and transgenic Arabidopsis plants, in which both SGR1 and SGR2 genes are suppressed, show a stay-green phenotype, as has been observed in rice (Park et al., 2007). Thus, while SGR genes appear to play an important role in one of the early steps of Chl catabolism, much remains to be learned about their mechanism of action. In addition, several transgenic plant lines have been reported that show stay-green phenotypes, such as those induced by altered hormone status, including increased amounts of cytokinins (Smart et al., 1991; Gan and Amasino, 1995) or decreased ethylene production (John et al., 1995). Clearly, numerous factors remain to be discovered that are directly or indirectly important in regulating Chl breakdown, and it is likely that some of these will be identified through characterizing additional stay-green mutations.
This article describes a spontaneous stay-green mutant, navel negra (nan), from Citrus sinensis ‘Washington Navel’ whose fruit fail to degreen during ripening, although the synthesis of carotenoids is not disrupted. The color change in citrus fruit is particularly evident in the flavedo, the outer colored exocarp of the citrus fruit peel (Davies and Albrigo, 1994), and involves the differentiation of chloroplasts to chromoplasts (Iglesias et al., 2001; Rodrigo et al., 2004) and the biosynthesis of carotenoids. While several studies have described aspects of the latter in citrus fruits (Kato et al., 2004; Rodrigo et al., 2004; Alós et al., 2006), relatively little has been reported about citrus Chl biochemistry, although it has been shown that CHL, which is constitutively expressed during natural fruit development (Jacob-Wilk et al., 1999), is the rate-limiting step of Chl degradation in citrus peel (Harpaz-Saad et al., 2007) and that PaO and geranylgeranyl reductase (CHL P) gene expression correlates with Chl degradation (Alós et al., 2006). We present here an analysis of the Chl metabolite profile and gene expression pattern in nan, which is distinct from previously reported stay-green mutants, that suggests that the mutation is likely associated with an upstream regulatory event rather than with a lesion in a specific step in Chl catabolism. We also describe the use of a citrus microarray and two-dimensional difference gel electrophoresis (2D-DIGE) analysis to contrast the transcriptome and proteome, respectively, of the nan flavedo with that of wild-type orange at preripe and ripe stages in order to gain insights into the consequences of a stay-green mutation on tissue and cellular physiology.
RESULTS
Total Flavedo Pigment Content
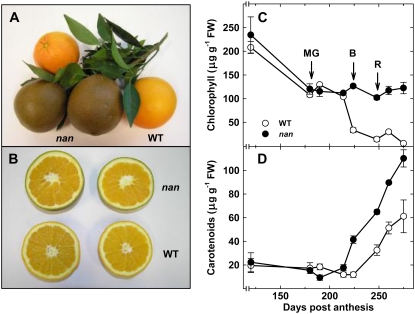
A mutant was identified among a population of C. sinensis trees whose fruit developed a dark brown external color upon ripening, rather than the characteristic orange of the wild type (Fig. 2A). This coloration was confined to the flavedo (Fig. 2B), and no other unusual phenotypes were observed in either the fruit, which were typically green at preripe stages, or vegetative tissues. In order to determine the molecular basis of the abnormal coloration, pigment levels in the flavedo of the nan mutant were compared with those from wild-type fruit at a range of developmental and ripening stages, spanning 120 to 275 DPA. The wild-type and nan fruit showed no differences in the levels of total Chls or carotenoids at the immature and mature green stages (120–180 DPA; Fig. 2, C and D). The apparent Chl depletion in nan and wild-type fruit prior to ripening (120–180 DPA; Fig. 2C) reflects a dilution effect due to cell expansion in the fruit peel (Bain, 1958) rather than Chl degradation, since the ratio of dry weight to fresh weight decreased from 0.30 to 0.14 in the wild type and from 0.32 to 0.16 in nan, resulting in a slight increase in the total amount of Chl (694 ± 42 to 772 ± 32 μg g−1 dry weight in the wild type and 733 ± 122 to 752 ± 68 μg g−1 dry weight in nan). However, while the abundance of Chl decreased during ripening to barely detectable levels in wild-type fruit, this did not occur in nan, and the levels remained unchanged (Fig. 2C) after natural color break (224 DPA). Furthermore, although both varieties showed a characteristic increase in carotenoid levels during ripening, consistent with the development of orange color, they were substantially higher in nan fruit and were approximately double those of the wild type at the fully ripe stage (275 DPA; Fig. 2D).
Figure 2.
External (A) and internal (B) appearance of wild-type and nan fruits at a fully ripe stage (275 DPA), and quantification of total Chls (C) and carotenoids (D) in the flavedo of C. sinensis ‘Washington Navel’ (WT; wild type; white circles) and nan (black circles) during fruit ripening. Data are means ± se (n = 3). Error bars smaller than the symbol size are not visible. MG, Mature green stage (180 DPA); B, breaker stage (224 DPA); R, ripe stage (248 DPA). FW, Fresh weight.
Measurement of Chls, Derivatives, and CLH Activity
The levels of Chls and their derivatives in the flavedo of both varieties were analyzed by HPLC at three developmental stages: mature green (180 DPA), breaker (224 DPA), and ripe (248 DPA; Table I). The concentrations of Chl a (105–120 μg g−1 fresh weight) and Chl b (17–23 μg g−1 fresh weight) showed a typical dramatic decrease during ripening in the wild type (≤98%) but remained high in nan, with almost no Chl degradation. Moreover, Chl dephytylated compounds (chlorophyllides and pheophorbides), which typically accumulate in PaO-affected stay-green mutants during ripening, were not detected in either nan or wild-type flavedo at any stage (Table I; Supplemental Fig. S1). Both varieties had similarly low levels of OH-Chl a, pheophytin a, and OH-Chl b (Table I), although the oxidized Chls were always more abundant in nan fruit. Despite the lack of Chl degradation in nan, no major differences were detected in CLH activity between the wild type and nan in mature green or breaker fruits; indeed, CLH activity was significantly higher in ripe nan fruit (Table I).
Table I.
Chl, Chl derivative, and CLH activity in the flavedo of wild-type citrus and the nan stay-green mutant at mature green (180 DPA), breaker (224 DPA), and ripe (248 DPA) stages
ND, Not detected. Data are means ± se (n = 3).
| Variable | Mature Green Stage
|
Breaker Stage
|
Ripe Stage
|
|||
|---|---|---|---|---|---|---|
| Wild Type | nan | Wild Type | nan | Wild Type | nan | |
| Pigments (μg g−1 fresh weight) | ||||||
| Chl a | 105.35 ± 8.25 | 120.32 ± 5.21 | 2.64 ± 0.03 | 100.65 ± 2.64 | 1.05 ± 0.01 | 105.34 ± 9.65 |
| OH-Chl a | 0.52 ± 0.02 | 1.42 ± 0.11 | ND | 0.99 ± 0.02 | ND | 5.21 ± 0.02 |
| Pheophytin a | 2.01 ± 0.01 | 1.75 ± 0.05 | 0.09 ± 0.01 | 0.68 ± 0.06 | 0.40 ± 0.04 | 0.65 ± 0.11 |
| Chl b | 23.51 ± 1.57 | 16.57 ± 0.08 | 0.83 ± 0.01 | 21.36 ± 1.12 | ND | 9.65 ± 0.54 |
| OH-Chl b | ND | ND | ND | ND | ND | 1.06 ± 0.09 |
| Total Chls | 131.39 ± 8.64 | 140.24 ± 15.36 | 3.56 ± 0.46 | 123.68 ± 3.21 | 0.07 ± 0.01 | 120.85 ± 8.24 |
| Chlorophyllide a | ND | ND | ND | ND | ND | ND |
| Pheophorbide a | ND | ND | ND | ND | ND | ND |
| CLH activity (μmol h−1 g−1 fresh weight) | 4.5 ± 0.3 | 4.4 ± 0.3 | 2.4 ± 0.1 | 2.1 ± 0.1 | 1.8 ± 0.2 | 2.4 ± 0.2 |
Expression of Genes Involved in Pigment Biosynthesis and Degradation
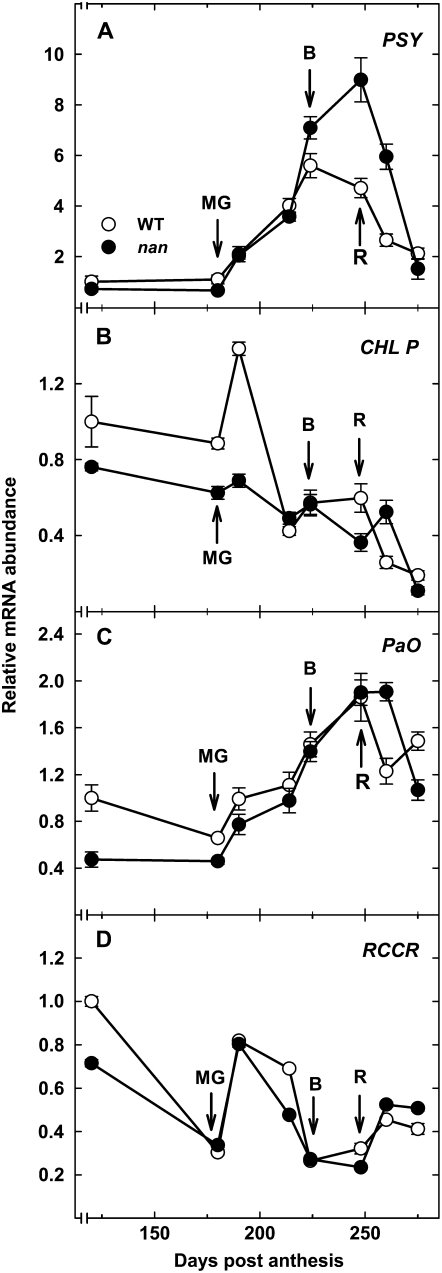
The expression levels of a selection of genes associated with pigment biosynthesis and catabolism were quantified by real-time reverse transcription (RT)-PCR during fruit development and ripening in the flavedo of wild-type and nan fruit (Fig. 3). These included PHYTOENE SYNTHASE (PSY), the first committed step in the carotenoid biosynthesis pathway (Fraser et al., 2002); CHL P, a gene involved in the biosynthesis of the phytyl chain of Chls (Tanaka et al., 1999); and two Chl catabolic genes, PaO and RCCR. Expression of PSY increased rapidly in both wild-type and nan flavedo, although maximal expression in the wild type was seen at the breaker stage, after which PSY transcript abundance decreased, while expression continued to increase during ripening in nan, peaking at the ripe stage, before decreasing during overripening. The expression patterns of the genes involved in Chl biosynthesis and degradation were similar in the wild type and nan in terms of both relative abundance and changes during fruit development (Fig. 3, B–D).
Figure 3.
Expression analysis by real-time RT-PCR of transcripts in the flavedo of the wild type (white circles) and the nan mutant (black circles) of PSY (A), CHL P (B), PaO (C), and RCCR (D) during fruit growth and ripening. An expression value of 1 was arbitrarily assigned to the 120-DPA wild-type sample. MG, Mature green stage (180 DPA); B, breaker stage (224 DPA); R, ripe stage (248 DPA). Data are means ± se (n = 3). Error bars smaller than the symbol size are not visible.
Effect of Ethylene on Pigment Composition and Gene Expression
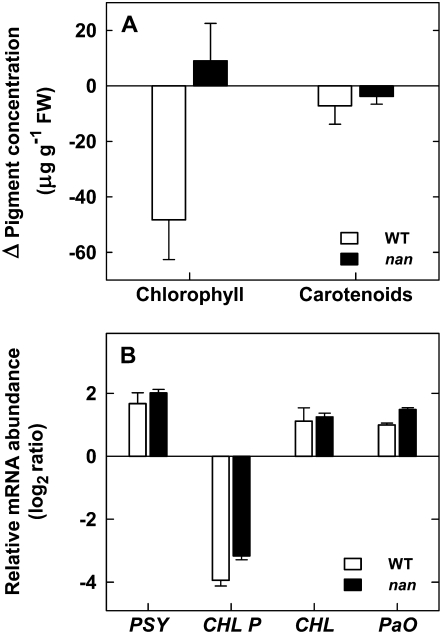
The plant hormone ethylene is known to promote Chl degradation in the citrus flavedo (García-Luis et al., 1986; Jacob-Wilk et al., 1999), so this phenomenon was examined in the nan mutant to determine whether the mutant phenotype might be associated with ethylene insensitivity. To this end, mature green wild-type and nan fruits were treated with 10 μL L−1 ethylene for 72 h, then total Chl and carotenoid levels were assayed and PaO, CHL P, PSY, and CLH expression was analyzed in flavedo tissues (Fig. 4). As expected, the ethylene treatment induced a substantial decrease in total Chl concentration in the wild-type flavedo (approximately 50 μg g−1 fresh weight, which represents almost half of the initial content), while no significant change was detected in the nan flavedo (Fig. 4A). However, the expression patterns of PaO, CHL P, PSY, and CLH showed similar changes in both varieties in response to the ethylene treatment (Fig. 3B), indicating that nan is ethylene responsive but that this response is upstream of, or independent from, the signaling pathway that induces Chl degradation. Furthermore, the treatment did not alter total carotenoid content (Fig. 4A), although PSY expression increased to the same degree in both varieties (Fig. 4B).
Figure 4.
Effects of ethylene treatments on both total pigment composition (A) and changes in mRNA abundance (B) in the flavedo of wild-type (white columns) and nan (black columns) fruits. Fruits were treated with 10 μL L−1 ethylene in controlled-atmosphere postharvest chambers at 20°C for 72 h. Pigment content was expressed as the absolute increment between control and treated samples. Transcript abundance of PSY, CHL P, CHL, and PaO was determined by real-time RT-PCR. The expression ratio between treated and untreated fruits was calculated and log2 transformed. All data are means ± se (n = 3).
Comparative Transcriptome Profiling
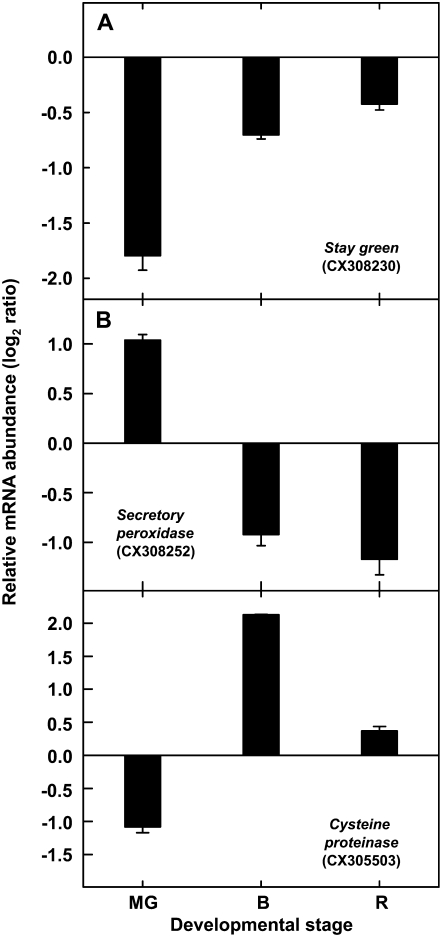
In order to gain insight into the potential basis and consequences of the stay-green phenotype, a survey of transcript expression in wild-type and nan flavedo was made using a 7,000-element citrus cDNA microarray (Forment et al., 2005). Pairwise analyses of RNA extracts from wild-type and nan flavedo tissue at the mature green, breaker, and ripe stages identified 11 distinct genes that were differentially expressed in all three stages, of which five could be annotated based on sequence homology tBLASTx searches against the National Center for Biotechnology Information (NCBI) nonredundant database (Table II). The only EST that showed down-regulation in the nan flavedo at all three developmental stages was a predicted SGR gene homolog, with the highest degree of similarity to an SGR gene from tomato (Table II). In the nan mutant, three other genes, annotated as a miraculin-like protein 2 gene, a secretory peroxidase gene, and a guanyl-nucleotide exchange factor gene, showed up-regulation at the green stage and subsequent down-regulation (Table II). In contrast, the expression level of a Cys protease gene was lower during the green stage but higher at the breaker and ripe stages (Table II). No significant sequence homology was apparent for the other six ESTs (CX299753, CX306150, CX299918, CX299940, CX287589, and CX289503), and, other than CX289503, which was expressed at higher levels in nan at all three stages, they exhibited similar relative expression patterns, with elevated transcript levels in the mutant flavedo at the mature green stage but reduced expression at the breaker and ripe stages (data not shown).
Table II.
Genes showing significant expression changes in the flavedo of the wild type and the nan stay-green mutant at all three stages studied: mature green (180 DPA), breaker (224 DPA), and ripe (248 DPA) stages
Differences in gene expression of at least 2-fold (P ≤ 0.001) in each of three replicates were considered to be statistically significant.
| Citrus EST Accession No. | tBLASTx Best Hit | Identitiesa | Arabidopsis Ortholog | Mature Green Log2 Ratiob | Breaker Log2 Ratiob | Ripe Log2 Ratiob |
|---|---|---|---|---|---|---|
| CX308230 | SGR protein (Solanum lycopersicum) | 86% | At4g22920 | −1.97 ± 0.11 | −1.71 ± 0.12 | −1.67 ± 0.11 |
| CX305230 | Miraculin-like protein 2 (Citrus jambhiri) | 96% | At1g17860 | 1.08 ± 0.17 | −1.72 ± 0.18 | −1.30 ± 0.17 |
| CX308252 | Secretory peroxidase (Nicotiana tabacum) | 89% | At4g21960 | 2.19 ± 0.16 | −1.02 ± 0.18 | −2.76 ± 0.16 |
| CX302192 | Guanyl-nucleotide exchange factor (Arabidopsis thaliana) | 71% | At1g01960 | 1.99 ± 0.14 | −2.14 ± 0.15 | −1.02 ± 0.14 |
| CX305503 | Cys proteinase (Citrus sinensis) | 100% | At4g32940 | −1.05 ± 0.20 | 1.42 ± 0.21 | 1.52 ± 0.20 |
Identities at the amino acid level.
Induction fold values are expressed as log-transformed mean ratios of nan to wild-type expression.
Since mutations in the SGR gene are responsible for the phenotypes of several stay-green mutants (Armstead et al., 2007; Jiang et al., 2007; Park et al., 2007; Ren et al., 2007; Sato et al., 2007), a 1.2-kb genomic region, including the promoter and coding regions, of the citrus SGR gene was cloned from wild-type and nan genomic DNA (GenBank accession no. AM922109), but differences were not observed. Furthermore, to determine whether a deletion in one or more copies of the SGR gene could be responsible for the nan phenotype, the relative gene dosage of SGR was assessed in wild-type and nan fruit by real-time PCR using genomic DNA, and again no difference was observed (data not shown).
In addition to the 11 genes that showed differences in transcript abundance at all three stages, a broader group of genes showed significantly different expression levels at one or two stages (Supplemental Table S1). The annotated functions of these genes, which are divided into several classes in Tables III and IV, and their collective expression patterns, suggest two key trends. First, at the mature green stage, a range of genes encoding components of the photosynthetic machinery and ancillary proteins associated with photosynthesis (e.g. NADPH:protochlorophyllide oxidoreductase, Chl a/b-binding protein [Cab], subunits of PSI and PSII, and Rubisco subunit-binding protein) were expressed at substantially lower levels in nan. Second, many genes associated with abiotic stress, and particularly responses to high light and oxidative stress, were upregulated in nan at both the mature green (Table III) and ripe (Table IV) stages. These included genes involved in the synthesis of compounds that ameliorate the effects of oxidative stress and reactive oxygen species (ROS), such as phenylpropanoids, polyamines, and carotenoids, and with the synthesis of jasmonic acid, salicylic acid, and ethylene, which have well-established connections with biotic and abiotic stress-mediated signaling. To further verify that the nan phenotype is associated with increased oxidative stress, the concentration of ascorbic acid, an antioxidant compound that accumulates in response to oxidative stress in many plant species (Mitler et al., 2004), including citrus (Iglesias et al., 2006), was measured in extracts from wild-type and nan ripe flavedo. As predicted, ascorbic acid levels in ripe fruit flavedo were significantly greater in nan than in the wild type (0.43 ± 0.04 and 0.29 ± 0.02 mg g−1 fresh weight, respectively).
Table III.
Examples of genes showing different expression levels in the flavedo of wild-type or nan stay-green mutant fruits at mature green stage (180 DPA), as determined with a citrus microarray, highlighting those associated with oxidative stress, chloroplast function, or senescence
References describing such an association for the Arabidopsis ortholog of the citrus gene are indicated. Differences in gene expression of at least 2-fold (P ≤ 0.001) in each of three replicates were considered to be statistically significant.
| Functional Class | Best BLAST Hit | Higher Expression
|
Citrus EST Accession No. | Arabidopsis Ortholog | Related References | |
|---|---|---|---|---|---|---|
| Wild Type | nan | |||||
| Primary metabolism | Plastidic Glc-6-P dehydrogenase | X | CX293894 | At5g13110 | Debnam et al. (2004) | |
| Chloroplastic Asp aminotransferase | X | CX294238 | At2g22250 | de la Torre et al. (2006) | ||
| Asp synthetase | X | CX306022 | At3g47340 | Lam et al. (1998) | ||
| Photosynthesis, light signaling | RPT2 | X | CX300991 | At2g30520 | Inada et al. (2004) | |
| NADH:protochlorophyllide oxidoreductase | X | CX289541 | At5g54190 | Rossel et al. (2002) | ||
| PSI subunit L | X | CX287010 | At4g12800 | Kimura et al. (2003) | ||
| PSI subunit E | X | CX300489 | At2g20260 | |||
| Putative PSI subunit | X | CX299745 | At4g02770 | Kimura et al. (2003) | ||
| Cab 3 | X | CX287196 | At1g29910 | Kimura et al. (2003) | ||
| PSII type I Cab | X | CX300355 | At2g34430 | Kimura et al. (2003) | ||
| Lhcb2 protein | X | CX300473 | At3g27690 | Kimura et al. (2003) | ||
| Lhcb6 protein | X | CX300344 | At1g15820 | Rossel et al. (2002) | ||
| Lhca2 | X | CX300483 | At3g61470 | Kimura et al. (2003) | ||
| Rubisco subunit-binding protein (chaperonin) | X | CX300392 | At2g28000 | Tosti et al. (2006) | ||
| ROS, redox | Secretory peroxidase | X | CX308252 | At4g21960 | ||
| FAD-linked oxidoreductase | X | CX292465 | At5g44380 | Kim et al. (2005) | ||
| Metallothionein protein | X | CX303735 | At3g15353 | Guo et al. (2004) | ||
| Phenylpropanoids | Phe-ammonia lyase | X | CX308098 | At2g37040 | Rossel et al. (2002) | |
| Chalcone synthase | X | CX308007 | At5g13930 | Rossel et al. (2002) | ||
| Flavonol 3′-O-methyltransferase | X | CX308025 | At5g54160 | Kimura et al. (2003) | ||
| Coumarate 3-hydroxylase | X | CX290216 | At2g40890 | |||
| Polyamines | SAM decarboxylase | X | CX308026 | At3g02470 | Guo et al. (2004) | |
| Protein stability | Chloroplastic heat shock protein | X | CX292078 | At4g27670 | ||
| HSP20 | X | CX309295 | At2g29500 | Kim et al. (2005) | ||
| Heat shock protein | X | CX294165 | At3g52490 | |||
| Defense | Chitinase | X | CX292066 | At3g54420 | ||
| Miscellaneous | β-Amylase | X | CX291892 | At3g23920 | Rossel et al. (2002) | |
| UVI-1 homolog | X | CX301008 | At1g19020 | Gadjev et al. (2006) | ||
| Gly-rich RNA-binding protein | X | CX287915 | At3g26740 | Kimura et al. (2003) | ||
Table IV.
Examples of genes showing different expression levels in the flavedo of wild-type or nan stay-green mutant fruits at the ripe stage (248 DPA), as determined with a citrus microarray, highlighting those associated with oxidative stress, chloroplast function, or senescence
References describing such an association for the Arabidopsis ortholog of the citrus gene are indicated. Differences in gene expression of at least 2-fold (P ≤ 0.001) in each of three replicates were considered to be statistically significant.
| Functional Class | Best BLAST Hit | Higher Expression
|
Citrus EST Accession No. | Arabidopsis Ortholog | Related References | |
|---|---|---|---|---|---|---|
| Wild Type | nan | |||||
| Primary metabolism | Trp synthase | X | CX291092 | At5g54810 | Zhao et al. (1998) | |
| Photosynthesis, light signaling | Lhcb2 protein | X | CX200280 | At2g05100 | Kimura et al. (2003) | |
| ROS, redox | Cytochrome bf6 complex subunit | X | CX37372 | At2g26500 | Kimura et al. (2003) | |
| GST1 | X | CX301042 | At2g30860 | Rossel et al. (2002) | ||
| GST6 | X | CX288839 | At2g47730 | Rossel et al. (2002) | ||
| Peroxidase | X | CX308204 | At4g21960 | Wong et al. (2006) | ||
| Ascorbate oxidase | X | CX289525 | At5g21100 | Yamamoto et al. (2005) | ||
| Phenylpropanoids | Isoflavone reductase-like | X | CX308502 | At4g39230 | ||
| Flavonol 3′-O-methyltransferase | X | CX289068 | At5g54160 | Kimura et al. (2003) | ||
| Polyamines | SAM decarboxylase | X | CX289919 | At3g25570 | Guo et al. (2004) | |
| Isoprenoids, carotenoids | Geranylgeranyl diphosphate synthase | X | CX290062 | At3g14510 | ||
| Salicylic acid | SAM-dependent methyltransferase | X | CX308455 | At3g11480 | Chen et al. (2003) | |
| Jasmonic acid | Lipoxygenase (LOX3) | X | CX288539 | At1g17420 | Mahalingam et al. (2005) | |
| Ethylene | ACC oxidase | X | CX292573 | At1g05010 | Gadjev et al. (2006) | |
| Defense | Chitinase | X | CX291879 | At3g12500 | Bray (2002) | |
| Miscellaneous | Miraculin | X | CX306063 | At1g17860 | Rizhsky et al. (2004) | |
| Cys protease (SAG12) | X | CX287658 | At5g45890 | Guo et al. (2004) | ||
| Glycosyltransferase | X | CX288595 | At3g11340 | Gadjev et al. (2006) | ||
| Glu decarboxylase | X | CX288553 | At5g17330 | |||
| Vacuolar processing enzyme | X | CX287419 | At4g32940 | Gepstein et al. (2003) | ||
In addition to these defined functional categories, a diverse set of genes with previously reported associations with oxidative stress showed differential expression between the varieties (Tables III and IV). For all putative functional categories, where a link between the apparent Arabidopsis ortholog of the citrus gene on the microarray and oxidative stress has been described in the literature, the corresponding reference is cited in Tables III and IV.
To validate the microarray results, quantitative RT-PCR analyses were performed on genes that represented different general patterns of expression (Table II) at the mature green, breaker, and ripe developmental stages. Specifically, these comprised representatives of genes that in the nan mutant either showed reduced expression at all three stages (SGR), higher expression at the mature green stage and then reduced expression during ripening (secretory peroxidase gene), or reduced expression in mature green fruits and increased transcript levels in ripening fruit (Cys protease gene). In all cases, the expression patterns (Fig. 5) were essentially identical to those obtained with the microarray.
Figure 5.
Relative gene expression in the flavedo of wild-type and nan fruits of SGR (A), secretory peroxidase gene (B), and Cys protease gene (C) transcripts determined by real-time RT-PCR. MG, Mature green stage (180 DPA); B, breaker stage (224 DPA); R, ripe stage (248 DPA). Log2-transformed mean ratios of nan to wild-type expression ± se are presented (n = 3). GenBank accession numbers of the ESTs are indicated in parentheses.
2D-DIGE Analysis of Protein Expression
In a parallel analysis, and to complement the microarray study, a comparative proteomic survey was performed on the wild-type and nan flavedo at the same three stages using 2D-DIGE (Rose et al., 2004). The gel comparisons revealed 13 protein spots that were consistently differentially expressed between nan and the wild type in all replicate analyses (arrows in representative gels shown in Supplemental Fig. S2). Differentially expressed spots were defined as those with a volume ratio above or below the 2-fold sd threshold, based on the normalized model curve of the spot volume ratio data set. Subsequent analyses by liquid chromatography-electrospray ionization-tandem mass spectrometry, followed by an interrogation of the NCBI nonredundant Green Plant database with the mass spectra using the Mascot search engine, identified a subset of 11 proteins (corresponding to those labeled 1–11 in Supplemental Fig. S2) with high confidence identification and annotation (Table V). These comprised a manganese superoxide dismutase, a copper zinc superoxide dismutase, the Rubisco large subunit (two spots), two heat shock proteins (HSP19 and HSP21), a lectin-related protein, a Gly-rich RNA-binding protein, a copper chaperone, an early light-induced protein (ELIP), and a protein with unknown function (gi|8778393). At the mature green stage, the only proteins that were detected with reduced expression in nan were HSP21 and the putative Gly-rich protein. HSP21, a protein involved in the chloroplast-to-chromoplast transition of tomato fruits (Neta-Sharir et al., 2005), was consistently less abundant in all three stages (−5.5, −3, and −2.7-fold at the mature green, breaker, and ripe stages, respectively). Conversely, the two superoxide dismutases and the copper chaperone protein were upregulated in nan at the mature green stage. The two spots of the Rubisco large subunit and the lectin-related precursor were less abundant in nan at the breaker and ripe stages, while ELIP and HSP19 were expressed at higher levels than in the wild type during ripening (Table V). This was particularly noticeable with ELIP, which was on average expressed at 14-fold higher levels in nan at the ripe stage.
Table V.
Relative changes in the abundance of 11 protein spots in the flavedo of wild-type and nan mutant fruits at mature green (180 DPA), breaker (224 DPA), and ripe (248 DPA) stages, as determined using 2D-DIGE
Differentially expressed spots were defined as those with a volume ratio above or below the 2 sd threshold based on the normalized model curve of the spot volume ratio data set. Protein annotation was performed after a search in the NCBI nonredundant Green Plant database. The accession numbers and corresponding protein scores are listed. Data are means ± se (n = 4) of the normalized spot volume nan to wild-type ratio. Arrows (→) indicate no significant changes in relative protein abundance.
| Spot | Accession No. | Best Hit | Protein Score | Fold Change
|
||
|---|---|---|---|---|---|---|
| Mature Green | Breaker | Ripe | ||||
| 1 | gi|54292100 | Manganese superoxide dismutase (Camellia sinensis) | 103 | 2 ± 0.05 | 2 ± 0.2 | → |
| 2 | gi|14158 | Heat shock protein 21 (Petunia × hybrida) | 101 | −5.5 ± 2.1 | −3.0 ± 0.6 | −2.7 ± 0.6 |
| 3 | gi|30039180 | Copper chaperone (Solanum lycopersicum) | 53 | 4.8 ± 0.5 | → | → |
| 4 | gi|2274917 | Copper/zinc superoxide dismutase (Citrus sinensis) | 61 | 5 ± 0.3 | → | 7.1 ± 0.7 |
| 5 | gi|6911142 | Gly-rich RNA-binding protein 1 (Catharanthus roseus) | 42 | −7.4 ± 0.1 | → | → |
| 6 | gi|4206520 | Ribulose-1,5-bisphosphate carboxylase (Severinia buxifolia) | 851 | → | −2.2 ± 0.1 | −2.7 ± 0.1 |
| 7 | gi|4206520 | Ribulose-1,5-bisphosphate carboxylase (Severinia buxifolia) | 711 | → | −2.3 ± 0.1 | −3.4 ± 0.1 |
| 8 | gi|11596188 | Lectin-related protein precursor (Citrus × paradisi) | 329 | → | −3.8 ± 0.6 | −3.2 ± 1.0 |
| 9 | gi|7447856 | Early light-inducible protein precursor (Glycine max) | 56 | → | 6.6 ± 0.7 | 14 ± 3.7 |
| 10 | gi|8778393 | F16A14.17 (Arabidopsis thaliana) | 47 | → | 2.8 ± 0.6 | → |
| 11 | gi|30575572 | HSP19 class I (Citrus × paradisi) | 168 | → | → | 2.8 ± 0.02 |
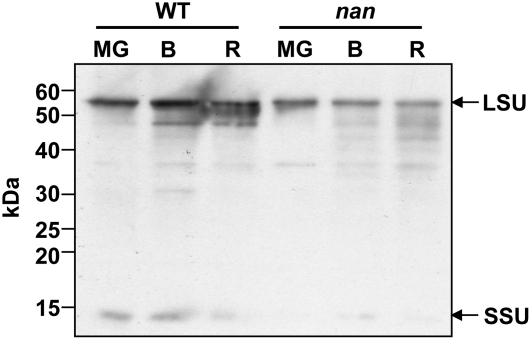
The 2D-DIGE screens were further validated by western-blot analysis of protein extracts from the flavedo of wild-type and nan fruits at all three stages with a Rubisco antiserum. The immunoblot analysis indicated that the Rubisco large subunit and small subunit were expressed at lower levels in nan, in agreement with the 2D-DIGE analysis, and that protein abundance declined during ripening (Fig. 6; Table V). It should be noted that cDNAs for the Rubisco large subunit were not represented on the citrus microarray, but those for the small subunit were present and the array analysis indicated that expression was statistically lower in breaker fruit. However, data generated by similar analyses with RNA from the mature green and ripe stages did not reach the P value threshold for a statistically significant comparison (Supplemental Table S1).
Figure 6.
Immunoblot analysis of Rubisco expression in flavedo of wild-type and nan fruits. Bands corresponding to the large subunit (LSU) and the small subunit (SSU) are highlighted. MG, Mature green stage (180 DPA); B, breaker stage (224 DPA); R, ripe stage (248 DPA). Molecular mass markers are indicated.
The 2D-DIGE data were compared with the microarray study in an attempt to identify targets that were common to both analyses. Most of the microarray elements corresponding to the 10 proteins (other than Rubisco) found in the 2D-DIGE study either did not generate expression data with significant P value thresholds in the microarray analyses or were not represented on the array. Thus, only two differentially expressed putative unigenes in the citrus chip matched a high-confidence best hit from the proteomic analyses (Table V; Supplemental Fig. S2): a lectin-related protein and HSP19. For the lectin-related protein, statistically significant data in the two surveys indicated repression in the nan mutant of both mRNA (Supplemental Table S1) and protein (Table V) at the ripe stage. However, for HSP19, transcript abundance was lower in nan at the mature green stage (Supplemental Table S1), while protein levels appeared to be higher (Table V) at the ripe stage.
Gene Expression of ELIP
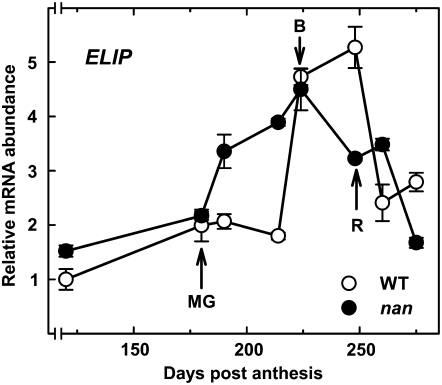
The transcript levels corresponding to ELIP, the protein with the greatest difference in expression between wild-type and nan flavedo (Table V), were quantified in the two varieties by real-time RT-PCR (Fig. 7). Expression of ELIP in the wild type was relatively low until the breaker stage (214 DPA), when levels increased dramatically, before peaking at the ripe stage and declining thereafter. In contrast, ELIP mRNA levels increased earlier in nan, prior to the breaker stage, and similarly declined somewhat earlier than the wild type during ripening.
Figure 7.
Transcript levels of the ELIP gene during fruit growth and ripening in the flavedo of wild-type (white circles) and nan (black circles) fruits. Analyses were performed by real-time RT-PCR, and an expression value of 1 was arbitrarily assigned to the 120-DPA wild-type sample. MG, mature green stage (180 DPA); B, breaker stage (224 DPA); R, ripe stage (248 DPA). Data are means ± se (n = 3). Error bars smaller than the symbol size are not visible.
DISCUSSION
The most striking phenotype of the nan mutant, which resulted in its initial identification, is the unusual brown color of the flavedo in the ripe fruit (Fig. 2A). An evaluation of the flavedo pigment composition indicated that nan can be defined as a type C stay-green variant (Thomas and Howarth, 2000), since Chl levels remained elevated during ripening rather than showing a typical decrease, while other ripening-related processes, such as carotenoid synthesis, proceeded normally (Fig. 2A; Table I). The brown coloration of nan fruit thus reflects the cumulative accumulation of Chl and carotenoids. This is further evidenced by the observation that PaO, RCCR, and CHL P transcript levels remained relatively constant, or showed only small decreases, during this period (Fig. 3, B–D).
Descriptions of stay-green mutants with major reductions in PaO activity from a range of plant species consistently report the accumulation of dephytylated intermediates in the Chl catabolic pathway during senescence (Hörtensteiner, 2006), and specifically elevated levels of pheophorbide a (Vicentini et al., 1995; Thomas et al., 1996; Roca et al., 2004), while no such accumulation has been detected in corresponding wild-type plants. These stay-green mutants are therefore impaired in particular steps in Chl degradation, rather than showing down-regulation of the entire pathway. To compare nan with the previously characterized stay-green mutants, and to determine whether there are similarly distinct defects in specific steps in Chl catabolism, we measured the levels of Chl, Chl dephytylated compounds, and CLH activity in the flavedo of nan and wild-type fruits (Table I). Neither contained detectable levels of chlorophyllides or pheophorbides, although oxidized Chl derivatives were present in both varieties. Relatively low levels of OH-Chl a were measured at the mature green stage in wild-type and nan fruits, but they were only detected in the mutant at the ripe stage and OH-Chl b was also only observed in ripe nan flavedo. The accumulation of such hydroxylated Chl derivatives has been noted previously in several stay-green mutants and has been attributed to nonspecific oxidation resulting from the elevated Chl levels, although the exact mechanism leading to their production is unknown (Roca and Mínguez-Mosquera, 2006). The patterns of CLH activity (Table I) and PaO and RCCR gene expression (Fig. 2) were similar in both varieties, and the patterns of expression of PSY, CHL P, and PaO were in close agreement with similar studies of other citrus species (Alós et al., 2006).
The response of the nan mutant to ethylene was also evaluated, since this hormone regulates leaf senescence (Kao and Yang, 1983; Choe and Whang, 1986) and color change in many fruit species, including citrus, through the activation of Chl degradation (García-Luis et al., 1986; Trebitsh et al., 1993; Jacob-Wilk et al., 1999) and carotenoid biosynthesis (Young and Jahn, 1972; Eilati et al., 1975). The results indicated that rapid ethylene-induced Chl loss was impaired in the nan mutant (Fig. 4), as has been reported previously in tomato and soybean stay-green mutants (Guiamét and Giannibelli, 1994; Akhtar et al., 1999). However, nan is ethylene responsive, because ethylene-induced gene expression, including transcripts involved in Chl synthesis and degradation or carotenoid biosynthesis, was similar in both varieties. While PSY expression was upregulated in nan and the wild type (Fig. 4B), the time course of the analysis was not sufficiently long to generate significant changes in carotenoid levels (Fig. 4A).
These data suggest that the nan mutation is not directly related to a single disruption in any of the principal established enzymatic steps (CLH, PaO, and RCCR) of Chl catabolism and is thus distinct from previously reported stay-green mutants. This hypothesis was explored by profiling transcript expression in the flavedo of wild-type and nan fruit at the mature green, breaker, and ripe stages. It was reasoned that genes whose expression was disrupted throughout fruit ontogeny might be more directly related to the function of the nan mutation, while those that showed differential expression only at later ripening stages might reflect secondary effects. Therefore, particular attention was paid to identifying transcripts with significantly different expression levels at all three stages. Interestingly, of the 11 genes that matched this criterion, the only one that was consistently downregulated was an SGR homolog that appears to be orthologous to Arabidopsis SGR1 (Table II). Cloning and sequencing of the citrus SGR gene revealed no differences between the wild type and nan, and no difference in gene dosage was detected in the two varieties. Taken together, the lack of Chl catabolite accumulation and the suppressed expression of the citrus SGR gene suggest that the nan mutation is associated with an early regulatory step that modulates SGR expression, rather than directly exerting a downstream effect on a specific aspect of catabolism.
In addition to characterizing the potential basis of the nan mutation, comparative transcriptomic and proteomic profiling were performed to assess the consequences of a stay-green mutation on flavedo tissue physiology and to provide insights into the metabolic pathways that are affected. To date, the only reported attempt at a larger scale study of differential gene expression in a stay-green mutant focused on three genes (Rubisco activase, soluble starch synthase, and Gly decarboxylase genes), which were identified through a differential display screen of a durum wheat stay-green mutant (Rampino et al., 2006). These genes, together with those for the Rubisco small subunit and Cab, were expressed at higher levels or underwent a slower decline in abundance during senescence in the mutant. In addition, a study of the stay-green green flesh tomato mutant suggested that the expression of several photosynthesis-related transcripts, or their cognate proteins, underwent a slower decrease in abundance in senescing leaves of the mutant than the wild type (Akhtar et al., 1999). Global transcript expression was monitored in the flavedo of nan and wild-type fruit at the mature green and ripe stages to identify genes that were up- or down-regulated in the mutant. Published Arabidopsis microarray data were then analyzed manually, or using tools such as Genevestigator (www.genevestigator.ethz.ch), to identify developmental processes or stimuli that similarly influence transcript accumulation of the Arabidopsis orthologs of the differentially expressed citrus clones. A similar procedure was followed using differentially expressed proteins (Table V).
One particularly clear trend that emerged from the analysis was that a broad set of photosynthesis-related genes and proteins were downregulated in nan at the mature green stage (Tables III and IV), a phenomenon that is closely associated with high light-induced stress (Rossel et al., 2002; Kimura et al., 2003). Plants can adjust the size of the light-harvesting antenna complex under such conditions, and it is thought that this occurs in order to reduce the production of ROS, which are generated under excess light conditions (Escoubas et al., 1995; Heddad and Adamska, 2000). Oxidative stress-inducing treatments have also been reported to trigger the down-regulation of photosynthesis-associated genes (Tosti et al., 2006); thus, there appears to be feedback between cellular redox status and oxidative stress that represses photosynthetic function. In addition, the proteomic analysis identified rbcL as one of the major proteins with lower abundance in nan (Figs. 5 and 6; Table V), which differs from the results seen with a rice sgr mutant (Sato et al., 2007) and the retention of Rubisco in leaves of the tomato green flesh mutant (Akhtar et al., 1999) but agrees with the previously observed reduced Rubisco protein levels in a P. vulgaris stay-green mutant (Bachmann et al., 1994).
If photosynthetic capacity is insufficient under conditions of excess absorbed light, free Chl can generate ROS, which in turn can cause extensive oxidative damage to the thylakoid membrane (Barber and Andersson, 1992; Niyogi, 1999). Accordingly, a second pattern that emerged from the proteomic and transcriptomic profiling was the association of genes that were among the most substantially up- or down-regulated in nan with abiotic stress, and specifically with high light or oxidative stress (Tables III and IV; Supplemental Table S1). While it is not practical to describe all the relevant genes and biochemical or physiological processes here, examples are shown in Tables III and IV, together with associated reports in the literature linking the specific Arabidopsis orthologs with stress responses or senescence. The nan flavedo had elevated levels of transcripts or proteins associated with enzymatic mechanisms for scavenging ROS, such as superoxide dismutase, and the biosynthesis of phenylpropanoids, polyamines, and isoprenoids, which provide nonenzymatic antioxidative protection (Jordan, 2002; Apel and Hirt, 2004; Kuehn and Phillips, 2005). Furthermore, the elevated carotenoid levels, higher ascorbic acid content, and higher levels of OH-Chls in nan after the breaker stage of ripening (Fig. 2) were likely a response to ROS, as reported previously (Bouvier et al., 1998; Mitler et al., 2004). Another set of pathways that showed apparent upregulation in nan were those leading to the synthesis of ethylene, jasmonic acid, and salicylic acid (Table III). While these hormones have a well-established role in regulating defense responses following microbial challenge, there is increasing evidence that they are involved in substantial cross talk between biotic and abiotic stress pathways and ROS-triggered molecular events (Fujita et al., 2006).
The comparative proteomic analyses also provided insights into the diversity of processes that were influenced by the nan mutation and, importantly, revealed different gene targets from the microarray analyses; therefore, the two approaches were complementary. The only identified protein that was expressed at lower levels in all three stages (Table V) was HSP21, a heat shock protein with chaperone-like activity (Lee et al., 1997). Although its biochemical function in fruit has yet to be fully elucidated, this chloroplastic protein has proposed roles in the protection of PSII from photooxidative stress and in the conversion of chloroplasts to chromoplasts during fruit ripening (Neta-Sharir et al., 2005). The reduced expressed of HSP21 in nan, therefore, may be associated with the abnormal levels of Chl or altered plastid ontogeny in the mutant, for example, if it participates in the disassembly of thylakoid membrane proteins and binding to partially folded or denatured proteins (Sun et al., 2002).
In contrast, the protein that showed the most relatively elevated expression in nan (Table V) was highly homologous to ELIPs, which are thought to play a photoprotective role. ELIPs are known to bind to Chl (Adamska et al., 1999) and are expressed during processes involving both thylakoid assembly (Adamska et al., 1993) and disassembly (Adamska et al., 1992; Bartels et al., 1992; Adamska and Kloppstech, 1994; Bhalerao et al., 2003), including the chloroplast-to-chromoplast transition (Bruno and Wetzel, 2004). Their accumulation in the thylakoid also correlates closely with the production of ROS and light stress (Adamska et al., 1992, 1993; Adamska and Kloppstech, 1994; Hutin et al., 2003), in parallel with the reduced expression of Cab genes/proteins (Montané and Kloppstech, 2000; Kimura et al., 2003), as was observed in nan (Table III). In the nan mutant, a distinct time lag occurred between elevated levels of ELIP transcripts, which were seen at 190 DPA, long before color break or an equivalent increase in the wild type (Fig. 7), and increased levels of ELIP protein, which were not seen until color break and ripening (Fig. 5; Table V). However, a lack of correlation between changes in steady-state levels of ELIP transcripts and proteins was also noted in studies in Arabidopsis (Heddad et al., 2006). A recent report describing an ELIP gene knockout in Arabidopsis suggested that it did not affect tolerance to photoinhibition and photooxidative stress (Rossini et al., 2006), although the authors acknowledged that the many potential compensatory responses complicates the interpretation of their data. Overall, a large body of evidence, including the coordinated up-regulation of ELIP mRNA and protein levels with those of a range of adaptations to oxidative stress, supports a role for ELIPs in protection against photooxidative damage.
In conclusion, the gene and protein expression profiling analyses and metabolite analyses reveal that the nan mutant shows numerous hallmarks of oxidative stress. Some of these are readily apparent, in the form of genes or proteins with a defined role in senescence or in providing protection against ROS (Table III). In other examples, no clear mechanistic relationship with a response to oxidative stress is apparent, even though there is precedence for an association based on previous microarray analyses, as is the case with β-amylase (Rossel et al., 2002; Table III). It is notable that nan shows substantial changes in gene and protein expression at the mature green stage, prior to the normal onset of Chl degradation or any detectable difference in Chl levels, compared with the wild type. This observation, together with the metabolite data, suggests that nan represents a new class of stay-green mutant with a lesion in a regulatory pathway that is upstream of SGR and that the mutant exhibits symptoms of oxidative stress prior to the onset of the normal degreening process.
MATERIALS AND METHODS
Plant Material and Ethylene Treatments
Fruits of Citrus sinensis ‘Washington Navel’ (wild type) and the nan stay-green mutant (‘Navel Negra’) were harvested from trees at the Instituto Valenciano de Investigaciones Agrarias and a commercial orchard, respectively. Sampling dates were July 26, 2004 (120 DPA), and then 180 (mature green stage), 190, 214, 224 (breaker stage), 248 (ripe stage), 260, and 275 DPA. For ethylene treatments, fruits were harvested at 180 DPA, when high levels of Chl were present in the flavedo, and treated with 10 μL L−1 ethylene at 20°C in a sealed chamber, and samples were taken at 0 and 72 h. Flavedo tissue from all samples was frozen in liquid nitrogen, powdered, and stored at −80°C until pigment analysis or RNA extractions. Aliquots of the frozen ground tissue were lyophilized prior to protein extraction.
Total Chl and Carotenoid Extraction and Quantification
Chls and carotenoids were extracted with methanol and chloroform as described by Rodrigo et al. (2003). Chl a, Chl b, and total Chl contents were calculated as described by Smith and Benítez (1955) after measuring the A644 and A662. The pigment ethereal solution was then dried and saponified using 6% (w/v) KOH:methanol. Carotenoids were subsequently reextracted with diethyl ether until the hypophase was colorless. An aliquot of this extract was used for total carotenoid content quantification by measuring the A450 and using the extinction coefficient of β-carotene (ɛ1% = 2,500; Davies, 1976).
All steps were carried out on ice and under dim light to prevent photodegradation, isomerization, and structural changes in the carotenoids. At least three independent extractions were performed for each sample.
Chl and Derivative Extraction and HPLC Analysis
Chls and derivatives were extracted as described by Mínguez-Mosquera and Garrido-Fernández (1989), and Chls, Chl derivatives, and xanthophylls were retained in the dimethyl formamide phase and analyzed by HPLC. Chl a and b standards were purchased from Sigma. Chlorophyllide a was formed by enzymatic deesterification of Chl a (Mínguez-Mosquera et al., 1994). The C-13 epimer of Chl a was prepared by treatment with chloroform (Watanabe et al., 1984), and 132-OH-Chl a and b were obtained as described by Laitalainen et al. (1990). All standards were purified by normal-phase and reverse-phase TLC (Mínguez-Mosquera et al., 1991, 1993). The separation and quantification of Chl degradation products were carried out by HPLC (HP1100 Hewlett-Packard liquid chromatograph fitted with a HP1100 automatic injector) using a stainless-steel column (25 × 0.46 cm i.d.) packed with a 5-μm C18 Superior ODS-2 (Teknokroma). Separation was performed using an elution gradient (2 mL min−1) with the mobile phases water:ion pair reagent:methanol (1:1:8, v/v/v) and methanol:acetone (1:1, v/v), as described by Mínguez-Mosquera et al. (1991). Sequential detection was performed with a photodiode array detector at 650 nm for series b and 666 nm for series a compounds. Data were collected and processed with an LC HP ChemStation (Rev.A.05.04). Pigments were identified by cochromatography with authentic samples and from their spectral characteristics. The on-line UV-visible spectra were recorded from 350 to 800 nm with the photodiode array detector. At least three independent extractions were performed for each sample.
CLH Activity
The method was an adaptation of that used by Terpstra and Lambers (1983). Flavedo tissue (5–10 g) was homogenized with 20 volumes of acetone at −20°C. The supernatant was removed by filtration, and the residue was treated again with 8 volumes of acetone. This operation was repeated until the supernatant was colorless. Finally, the precipitate was collected by vacuum filtration and left to dry at ambient temperature (20°C–25°C). From each 1 g of fruit, approximately 0.15 g of acetone powder was obtained. Extraction of the enzyme was carried out according to Johnson-Flanagan and Thiagarajah (1990). The acetone powder (0.5 g) was extracted with 15 mL of 5 mm sodium phosphate buffer (pH 7) containing 50 mm KCl and 0.24% (w/v) Triton X-100.
After centrifugation, the supernatant was used as a crude enzyme extract. The substrate, Chl a, was isolated from fresh spinach leaves by pigment extraction using acetone (Holden, 1976), followed by TLC separation as described by Mínguez-Mosquera and Garrido-Fernández (1989). The standard reaction mixture (1.1 mL) contained approximately 0.1 μmol of Chl a in acetone, 100 mm Tris buffer (pH 8.5) containing 0.24% (w/v) Triton-X-100, and enzyme extract in a 1:5:5 ratio. Chlorophyllide a levels were quantified as described by Mínguez-Mosquera et al. (1994), and the results are expressed as μmol h−1 g−1 fresh weight. At least three independent extractions were performed for each sample.
Ascorbic Acid Quantification
Discs of flavedo tissue were excised, frozen in liquid nitrogen, and ground to a fine powder, and 500 mg of each sample was homogenized in 5 mL of 2% metaphosphoric acid. After centrifugation (5,000g, 4°C, 10 min) and filtration, the ascorbic acid content of the supernatant was spectrophotometrically measured as described by Takahama and Oniki (1992) with a Cary 50 Bio spectrophotometer (Varian). Concentration was determined by monitoring the absorbance decrease at 265 nm due to the oxidation of ascorbate to deydroascorbate catalyzed by ascorbate oxidase (EC 1.10.3.3; from Cucurbita spp.). At least three independent determinations per sample were performed.
RNA Extraction and Real-Time RT-PCR
Total RNA was isolated from frozen flavedo using the RNeasy Plant Mini Kit (Qiagen) and treated with ribonuclease-free DNase (Qiagen) according to the manufacturer's instructions. UV light absorption spectrophotometry and agarose gel electrophoresis were performed to test RNA quality as described by Sambrook et al. (1989). Quantitative real-time RT-PCR was performed with a LightCycler 2.0 Instrument (Roche) equipped with LightCycler Software version 4.0 (Roche) as described by Alós et al. (2006). Specificity of the amplification reactions was assessed by postamplification dissociation curves and by sequencing the reaction products. To transform fluorescence intensity measurements into relative mRNA levels, a 10-fold dilution series of a RNA sample was used as a standard curve. Reproducible data were obtained after normalization to total RNA amounts accurately quantified with the RNA-specific fluorescent dye Ribogreen (Molecular Probes; Bustin, 2002; Hashimoto et al., 2004). Each sample was analyzed in triplicate and means ± se were calculated. Induction values of 1-fold were arbitrarily assigned to the 120-DPA sample for the natural ripening-associated gene expression. In the ethylene treatment experiment, the expression ratio between the treated and untreated fruits was calculated and log transformed. To validate the microarray data, real-time PCR of three selected genes was performed in triplicate and expression levels were log transformed. The sequences of the primers used for the real-time RT-PCR and associated references that were used for primer design are shown in Supplemental Table S2.
Genomic DNA Extraction and Real-Time PCR
Genomic DNA was isolated from frozen leaves using the DNeasy Plant Mini Kit (Qiagen) according to the manufacturer's instructions. UV light absorption spectrophotometry and agarose gel electrophoresis were performed to test DNA quality as described by Sambrook et al. (1989). DNA concentration was accurately quantified with the DNA-specific fluorescent dye Picogreen (Molecular Probes).
To determine the relative gene dosage of SGR in wild-type and nan fruits by quantitative real-time PCR, two specific oligonucleotide primers, sgrZCF (5′-AGTTTGGTTGCTGCTCTTGG-3′) and sgrZCR (5′-AGTGCGTTTTGCTGCTCATA-3′), were designed corresponding to positions 93 to 112 and 160 to 141, respectively, in the 566-bp SGR cDNA insert (accession no. CX308230). PCR was carried out with 1 ng of genomic DNA by adding 2 μL of LC FastStart DNA MasterPLUS SYBR Green I (Roche) and 2.5 pmol of each primer in a total volume of 10 μL. Incubations were carried out at 95°C for 10 min followed by 40 cycles at 95°C for 2 s, 55°C for 10 s, and 72°C for 15 s. Fluorescence intensity data were acquired during the 72°C extension step. Specificity of the amplification reactions was assessed by postamplification dissociation curves and by sequencing the reaction products. To transform fluorescence intensity measurements into relative DNA levels, a 10-fold dilution series of a DNA sample was used as a standard curve. Each sample was analyzed in triplicate and means ± se were calculated.
Cloning and Sequencing the SGR Genomic Region
The promoter region of the citrus SGR gene was cloned from wild-type genomic DNA using the GenomeWalker Universal Kit (Clontech) following the manufacturer's instructions, except that six GenomeWalker libraries were constructed using the restriction enzymes DraI, EcoRV, HincII, PvuII, ScaI, and SmaI. Two gene-specific oligonucleotide primers, sgrA (5′-CTCTGACTGAGTGGGAGAG-3′) and sgrB (5′-GTTGAAACGACCTGAC-3′), were designed corresponding to positions 66 to 48 and 31 to 16, respectively, in the 5′ end of the 566-bp SGR cDNA insert (accession no. CX308230). After two nested PCRs, a single 800-bp product was amplified from the DraI library, cloned into the pCR2.1 vector (Invitrogen), and fully sequenced from both ends.
The genomic region containing the promoter and coding region of the SGR gene was cloned by PCR using genomic DNA from both nan and wild-type fruits using a forward primer specific for the 5′ end of the promoter region (sgrF, 5′-CTGACTCCCAGCGCAATTAC-3′) and a reverse primer specific for the 3′ end of the cDNA, adjacent to the poly(A) tail (sgrR, 5′-TCAAGATTCCATCTCAAAAGCTC-3′). The PCR mix consisted of 5 ng of genomic DNA, 1 μL of 10 mm dNTP mix, 5 pmol of each oligonucleotide, 2.5 μL of 10× reaction buffer, and 0.5 μL of Advantage 2 polymerase mix (Clontech). Touch-down PCR was carried out under the following conditions: 5 min at 95°C; 10 cycles of 30 s at 95°C, 1 min at the annealing temperature, decreasing by 1°C per cycle from 65°C to 55°C, and 3 min at 72°C; then 35 cycles of 30 s at 95°C, 1 min at 55°C, and 3 min at 72°C; and a final extension step at 72°C for 5 min. A single band of 1.2 kb was amplified from each of the genomic DNA samples. The products were cloned into the pCR2.1 vector, and plasmid DNA from 12 independent clones of each product was fully sequenced from both ends.
Microarray Hybridization and Analysis
Sample labeling, microarray hybridization and washing, and data acquisition and analysis were performed as described by Cercós et al. (2006). RNA was extracted from each sample with at least three biological replicates and independently processed, labeled, and hybridized to different microarrays. Differences in gene expression were considered to be significant when the P value was <0.001 and the induction or repression ratio was equal to or higher than 2-fold. Only high-quality PCR spots (Forment et al., 2005) were used for analyses.
2D-DIGE Analysis
Proteins were extracted from wild-type and nan flavedo tissue of mature green (180 DPA), breaker (224 DPA), and ripe (248 DPA) fruits as described by Saravanan and Rose (2004). Protein concentrations were quantified with the Bio-Rad protein assay using BSA as a standard. Protein labeling (50 μg per extract) was performed using Cy Dye DIGE fluors (Amersham Biosciences) according to the manufacturer's instructions. Immobilized pH gradient strips (24 cm, linear pH 4–7; Bio-Rad ReadyStrip; Bio-Rad) were rehydrated overnight with 450 μL of isoelectric focusing buffer containing the Cy Dye-labeled protein mixture described above and focused using a Protean IEF Cell (Bio-Rad) at 20°C. The following program was applied: a linear increase from 0 to 500 V over 1 h, 500 V to 104 V over 5 h, and then held at 104 V for a total of 100 kVh. After focusing, the proteins were reduced by incubating the immobilized pH gradient strips with 2% (w/v) dithiothreitol for 10 min and alkylated with 2% (w/v) iodoacetamide in 10 mL of equilibration buffer (6 m urea, 20% [v/v] glycerol, 3% [w/v] SDS, and 375 mm Tris-HCl, pH 8.8) for 10 min. The strips were then transferred to 12.5% SDS-PAGE gels for second-dimension electrophoresis with the Ettan Dalt six gel system (Amersham Biosciences) using SDS electrophoresis buffer (25 mm Tris, 192 mm Gly, and 0.1% [w/v] SDS) at 2 W per gel for 16 h. Four independent extracts were made from each sample, resulting in four replicate gels for each developmental stage. In each comparison, wild-type samples were labeled with the Cy3 dye in three gels and a dye swap was performed in the fourth.
Cy3 and Cy5 images were collected using a Typhoon scanner (Amersham Biosciences) in fluorescence mode at 100-μm resolution. Images were analyzed using ImageQuant version 5.2 (Amersham Biosciences) and Decyder version 4.0 (Amersham Biosciences). Spot volumes were determined after background subtraction and volume ratio values were normalized, so that the modal peak of volume ratios was zero. Differentially expressed spots were defined as those with a volume ratio above or below the 2 sd threshold. Final spot ratio values are means ± se of four independent biological replicates for each developmental stage.
Protein Identification
Gels were fixed in water:methanol:acetic acid (83:10:7, v/v/v) for 2 h and subsequently stained with colloidal Coomassie Brilliant Blue G-250. Gel plugs containing protein spots of interest were manually excised, washed with 50 μL of water for 5 min and with 50 μL of acetonitrile and 50 mm ammonium bicarbonate (1:1, v/v) for 10 min, rehydrated in 15 μL of trypsin solution (10 ng μL−1 in 25 mm ammonium bicarbonate), and covered with 10 μL of 50 mm ammonium bicarbonate. After overnight incubation at 37°C, the supernatant was collected and peptides were reextracted sequentially with 60 μL of acetonitrile:formic acid (20:1, v/v) and 30 μL of acetonitrile:formic acid (180:1, v/v). The supernatants were combined and dried in a Speed-Vac (ThermoSavant).
The samples were reconstituted in 10 μL of 0.1% formic acid and 2% acetonitrile (v/v) for liquid chromatography-electrospray ionization-tandem mass spectrometry analysis. The CapLC was carried out with an LC Packings Ultimate integrated capillary HPLC system equipped with a Switchos valve switching unit (Dionex). The gel-extracted peptides (6.4 μL) were injected using a Famous autosampler (Dionex) onto a C18 μ-precolumn cartridge (5 μm, 300 μm × 5 mm; Dionex) for on-line desalting and then separated on a PepMap C-18 RP capillary column (3 μm, 300 μm i.d. × 150 mm; Dionex). Peptides were eluted in a 30-min gradient of 5% to 45% acetonitrile in 0.1% formic acid at 4 μL min−1. The CapLC was connected in-line to a hybrid triple-quadrupole linear ion trap mass spectrometer (4000 Q Trap; ABI/MDS Sciex) equipped with a Turbo V source. Data acquisition was performed using Analyst 1.4 software (Applied Biosystems) in the positive ion mode for information-dependent acquisition analysis. In information-dependent acquisition analysis, after each survey scan from m/z 400 to 1,600, an enhanced resolution scan was performed followed by tandem mass spectrometry (MS/MS) of the three highest intensity ions with multiple charge states. The MS/MS data were submitted to Mascot 1.9 for a database search against the NCBI nonredundant Green Plant database. The search was performed allowing one trypsin miscleavage site, and the peptide tolerance and MS/MS tolerance values were set to 0.8 and 2 D, respectively. Only significant scores defined by a Mascot probability analysis (www.matrixscience.com/help/scoring_help.html#PBM) greater than “identity” were considered for assigning protein identity.
Immunoblot Analysis
Immunoblot analysis was carried with the same flavedo tissue samples used for the 2D-DIGE experiments. Ground flavedo tissue (500 mg) was resuspended in 2 mL of 50 mm Tris-HCl, pH 7.5, and 20 μL of 100 mm phenylmethylsulfonyl fluoride, incubated for 30 min at 2°C, and centrifuged at 10,000g at 4°C. The supernatant was collected and quantified by the Bradford assay (see above). Protein extracts (5 μg per lane) were separated by SDS-PAGE on 12.5% polyacrylamide gels (Bio-Rad). Gels were stained with SyproRuby (Bio-Rad) to confirm equivalent sample loading or transferred to Hybond ECL membranes, according to the manufacturer's instructions (Amersham Life Science). Immunoblot analysis was performed after blocking the membranes in 3% (w/v) bovine serum albumin and 0.02% (w/v) sodium azide in sterile phosphate-buffered saline (PBS)-Tween (1× PBS and 0.1% [v/v] Tween 20), using the ECL western-blotting kit, according to the manufacturer's instructions (Amersham Life Science). The blot was incubated with Rubisco antiserum (diluted 1:10,000 in PBS-Tween) followed by a 1:2,000 dilution of the horseradish peroxidase-conjugated secondary antibody. After each incubation with antiserum, the membrane was washed three times in PBS-Tween.
All of the microarray data described in this study were deposited in the ArrayExpress database (accession no. E-MEXP-967). The sequence data described in this study were deposited in GenBank (accession no. AM922109).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. HPLC results of Chls and derivatives in wild-type and nan ripe fruit.
Supplemental Figure S2. Overlay images from 2D-DIGE analysis.
Supplemental Table S1. Microarray results for genes that showed statistically significant differences in expression levels between nan flavedo and Washington Navel wild-type flavedo at the mature green and ripe stages.
Supplemental Table S2. Oligonucleotides used as primers for real-time RT-PCR.
Supplementary Material
Acknowledgments
We thank Isabel Sanchis, Israel Morte, and Angel Boix for technical support, and Tal Isaacson and Eric Schaffler for their help with two-dimensional gel analysis.
This work was supported by the Spanish Ministerio de Ciencia y Tecnología (grant nos. AGL2003–08502–C04–01 and GEN2001–4885–C05–03) and the Instituto Nacional de Investigaciones Agrarias (grant nos. RTA03–106, RTA04–013, and RTA05–247). E.A. was the recipient of an Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA) predoctoral fellowship, and D.J.I. and M.C. were the recipients of INIA-Comunidades Autónomas contracts. J.K.C.R. was supported by the National Science Foundation's Plant Genome Program (award no. DBI 0606595).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Manuel Talón (talon_man@gva.es).
The online version of this article contains Web-only data.
References
- Adamska I, Kloppstech K (1994) Low temperature increases the abundance of early light-inducible transcript under light stress conditions. J Biol Chem 269 30221–30226 [PubMed] [Google Scholar]
- Adamska I, Kloppstech K, Ohad I (1993) Early light-inducible protein in pea is stable during light stress but is degraded during recovery at low light intensity. J Biol Chem 268 5438–5444 [PubMed] [Google Scholar]
- Adamska I, Ohad I, Kloppstech K (1992) Synthesis of the early light-inducible protein is controlled by blue light and related to light stress. Proc Natl Acad Sci USA 89 2610–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamska I, Roobol-Bóza M, Lindahl M, Andersson B (1999) Isolation of pigment-binding early light-inducible proteins from pea. Eur J Biochem 260 453–460 [DOI] [PubMed] [Google Scholar]
- Akhtar MS, Goldschmidt EE, John I, Rodoni S, Matile P, Grierson D (1999) Altered patterns of senescence and ripening in gf, a stay-green mutant of tomato (Lycopersicon esculentum Mill.). J Exp Bot 336 1115–1122 [Google Scholar]
- Alós E, Cercós M, Rodrigo MJ, Zacarías L, Talón M (2006) Regulation of color break in Citrus fruits: changes in pigment profiling and gene expression induced by gibberellins and nitrate, two ripening retardants. J Agric Food Chem 54 4888–4895 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress and signal transduction. Annu Rev Plant Biol 55 373–399 [DOI] [PubMed] [Google Scholar]
- Armstead I, Donnison I, Aubry S, Harper J, Hörtensteiner S, James C, Mani J, Moffet M, Ougham H, Roberts L, et al (2006) From crop to model to crop: identifying the genetic basis of the stay-green mutation in the Lolium/Festuca forage and amenity grasses. New Phytol 172 592–597 [DOI] [PubMed] [Google Scholar]
- Armstead I, Donnison I, Aubry S, Harper J, Hörtensteiner S, James C, Mani J, Moffet M, Ougham H, Roberts L, et al (2007) Cross-species identification of Mendel's I locus. Science 315 73. [DOI] [PubMed] [Google Scholar]
- Bachmann A, Fernández-López J, Ginsburg S, Thomas H, Bouwkamp JC, Solomos T, Matile P (1994) Stay-green genotypes of Phaseolus vulgaris L.: chloroplast proteins and chlorophyll catabolites during foliar senescence. New Phytol 126 593–600 [Google Scholar]
- Bain JM (1958) Morphological, anatomical and physiological changes in the developing fruit of the Valencia orange Citrus sinensis (L) Osbeck. Aust J Bot 6 1–24 [Google Scholar]
- Barber J, Andersson B (1992) Too much of a good thing: light can be bad for photosynthesis. Trends Biochem Sci 17 61–66 [DOI] [PubMed] [Google Scholar]
- Barry CS, McQuinn RP, Chung MY, Besuden A, Giovannoni JJ (2008) Amino acid substitutions in homologs of the STAY-GREEN (SGR) protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol 147 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D, Hanke C, Schneider K, Michel D, Salamini F (1992) A desiccation-related ELIP-like gene from the resurrection plant Crateostigma plantagineum is regulated by light and ABA. EMBO J 11 2771–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao R, Keskitalo J, Sterky F, Erlandsson R, Björkbacka H, Birve SJ, Karlsson J, Gardeström P, Gustafsson P, Lundeberg J, et al (2003) Gene expression in autumn leaves. Plant Physiol 131 430–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier F, Backaus RA, Camara B (1998) Induction and control of chromoplast-specific carotenoid genes by oxidative stress. J Biol Chem 273 30651–30659 [DOI] [PubMed] [Google Scholar]
- Bray EA (2002) Classification of genes differentially expressed during water-deficit stress in Arabidopsis: an analysis using microarray and differential expression data. Ann Bot (Lond) 89 803–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno AK, Wetzel CM (2004) The early light-inducible protein (ELIP) gene is expressed during chloroplast-to-chromoplast transition in ripening tomato fruit. J Exp Bot 408 2541–2548 [DOI] [PubMed] [Google Scholar]
- Bustin SA (2002) Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29 23–39 [DOI] [PubMed] [Google Scholar]
- Cercós M, Soler G, Iglesias DJ, Gadea J, Forment J, Talón M (2006) Global analysis of gene expression during development and ripening of Citrus fruit flesh: a proposed mechanism for citric acid utilization. Plant Mol Biol 62 513–527 [DOI] [PubMed] [Google Scholar]
- Cha KW, Lee YJ, Koh HJ, Lee BM, Nam YW, Paek NC (2002) Isolation, characterization, and mapping of the stay green mutant in rice. Theor Appl Genet 104 526–532 [DOI] [PubMed] [Google Scholar]
- Chen F, D'Auria JC, Tholl D, Ross JR, Gershenzon J, Noel JP, Pichersky E (2003) An Arabidopsis gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J 36 577–588 [DOI] [PubMed] [Google Scholar]
- Cheung AY, McNellis T, Piekos B (1993) Maintenance of chloroplast components during chromoplast differentiation in the tomato mutant green flesh. Plant Physiol 101 1223–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe HC, Whang M (1986) Effects of ethephon on aging and photosynthetic activity in isolated chloroplasts. Plant Physiol 80 305–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BH (1976) Carotenoids. In TW Goodwin, ed, Chemistry and Biochemistry of Plant Pigments. Academic Press, New York, pp 38–165
- Davies FS, Albrigo LG (1994) Citrus. CAB International, Oxon, UK, pp 202–215
- Debnam PM, Fernie AR, Leisse A, Golding A, Bowsher CG, Grimshaw C, Knight JS, Emes MJ (2004) Altered activity of the P2 isoform of plastidic glucose 6-phosphate dehydrogenase in tobacco (Nicotiana tabacum cv. Samsun) causes changes in carbohydrate metabolism and response to oxidative stress in leaves. Plant J 38 49–59 [DOI] [PubMed] [Google Scholar]
- de la Torre F, De Santis L, Suárez MF, Crespillo R, Cánovas FM (2006) Identification and functional analysis of a prokaryotic-type aspartate aminotransferase: implications for plant amino acid metabolism. Plant J 46 414–425 [DOI] [PubMed] [Google Scholar]
- Efrati A, Eyal Y, Paran I (2005) Molecular mapping of the chlorophyll retainer (cl) mutation in pepper (Capsicum spp.) and screening for candidate genes using tomato ESTs homologous to structural genes of the chlorophyll catabolism pathway. Genome 48 347–351 [DOI] [PubMed] [Google Scholar]
- Eilati SK, Budowski P, Monselise SP (1975) Carotenoid changes in the Shamouti orange peel during chloroplast-chromoplast transformation on and off the tree. J Exp Bot 26 624–632 [Google Scholar]
- Escoubas JM, Lomas M, Laroche J, Falkowski PG (1995) Light-intensity regulation of Cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA 92 10237–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forment J, Gadea J, Huerta L, Abizanda L, Agustí J, Alamar S, Alós E, Andrés F, Arribas R, Beltrán JP, et al (2005) Development of a citrus genome-wide EST collection and cDNA microarray as resources for genomic studies. Plant Mol Biol 57 375–391 [DOI] [PubMed] [Google Scholar]
- Fraser PD, Romer S, Shipton CA, Mills PB, Kiano JW, Misawa N, Drake RG, Schuch W, Bramley PM (2002) Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc Natl Acad Sci USA 99 1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9 436–442 [DOI] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Van Breusegem F (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S, Amasino RM (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270 1986–1988 [DOI] [PubMed] [Google Scholar]
- García-Luis A, Fornes F, Guardiola JL (1986) Effects of gibberellin A3 and cytokinins on natural post-harvest, ethylene-induced pigmentation of Satsuma mandarin peel. Physiol Plant 68 271–274 [Google Scholar]
- Gepstein S, Sabehi G, Carp MJ, Hajouj T, Nesher MFO, Yariv I, Dor C, Bassani M (2003) Large-scale identification of leaf senescence-associated genes. Plant J 36 629–642 [DOI] [PubMed] [Google Scholar]
- Guiamét JJ, Giannibelli MC (1994) Inhibition of the degradation of chloroplast membranes during senescence in nuclear ‘stay green’ mutants of soybean. Physiol Plant 91 395–402 [Google Scholar]
- Guiamét JJ, Giannibelli MC (1996) Nuclear and cytoplasmic ‘stay-green’ mutations of soybean alter the loss of leaf soluble proteins during senescence. Physiol Plant 96 655–661 [Google Scholar]
- Guo Y, Cai Z, Gan S (2004) Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ 27 521–549 [Google Scholar]
- Harpaz-Saad S, Azoulay T, Arazi T, Ben-Yaakov E, Mett A, Shiboleth YM, Hörtensteiner S, Gidoni D, Gal-On A, Goldschmidt EE, et al (2007) Chlorophyllase is a rate-limiting enzyme in chlorophyll catabolism and is posttranslationally regulated. Plant Cell 19 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto JG, Beadles-Bohling AS, Wiren KM (2004) Comparison of RiboGreen and 18S rRNA quantitation for normalizing real-time RT-PCR expression analysis. Biotechniques 36 54–60 [DOI] [PubMed] [Google Scholar]
- Heddad M, Adamska I (2000) Light stress-regulated two-helix proteins in Arabidopsis related to the chlorophyll a/b binding gene family. Proc Natl Acad Sci USA 97 3741–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddad M, Norén H, Reisewr V, Dunaeva M, Andersson B, Adamska I (2006) Differential expression and localization of early light-induced proteins in Arabidopsis. Plant Physiol 142 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry GAF, Houghton JD, Brown SB (1987) The degradation of chlorophyll: a biological enigma. New Phytol 107 255–302 [DOI] [PubMed] [Google Scholar]
- Holden M (1976) Analytical methods chlorophylls. In TW Goodwin, ed, Chemistry and Biochemistry of Plant Pigments, Vol II. Academic Press, London, pp 1–37
- Hörtensteiner S (2006) Chlorophyll degradation during senescence. Annu Rev Plant Biol 57 55–77 [DOI] [PubMed] [Google Scholar]
- Hutin C, Nussaume L, Moise N, Moya I, Kloppstech K, Havaux M (2003) Early light-induced proteins protect Arabidopsis from photooxidative stress. Proc Natl Acad Sci USA 100 4921–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias DJ, Calatayud A, Barreno E, Primo-Millo E, Talon M (2006) Responses of citrus plants to ozone: leaf biochemistry, antioxidant mechanisms and lipid peroxidation. Plant Physiol Biochem 44 125–131 [DOI] [PubMed] [Google Scholar]
- Iglesias DJ, Tadeo FR, Legaz F, Primo-Millo E, Talon M (2001) In vivo sucrose stimulation of colour change in citrus fruits epicarps: interactions between nutritional and hormonal signals. Physiol Plant 112 244–250 [DOI] [PubMed] [Google Scholar]
- Inada S, Ohgishi M, Mayama T, Okada K, Sakai T (2004) RPT2 is a signal transducer involved in phototropic response and stomatal opening by association with phototropin 1 in Arabidopsis. Plant Cell 16 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob-Wilk D, Holland D, Goldschmidt EE, Riov J, Eyal Y (1999) Chlorophyll breakdown by chlorophyllase: isolation and functional expression of the Chlase1 gene from ethylene-treated Citrus fruit and its regulation during development. Plant J 20 653–661 [DOI] [PubMed] [Google Scholar]
- Jiang H, Li M, Ling N, Yan H, Xu X, Liu J, Xu Z, Chen F, Wu G (2007) Molecular cloning and function analysis of the stay green gene in rice. Plant J 52 197–209 [DOI] [PubMed] [Google Scholar]
- John I, Drake R, Farrell A, Cooper W, Lee P, Horton P, Grierson D (1995) Delayed leaf senescence in ethylene deficient ACC-oxidase antisense tomato plants: molecular and physiological analysis. Plant J 7 483–490 [Google Scholar]
- Johnson-Flanagan A, Thiagarajah M (1990) Degreening in canola (Brassica napus, cv Westar) embryos under optimum conditions. J Plant Physiol 136 180–186 [Google Scholar]
- Jordan BR (2002) Molecular responses of plant cells to UV-B stress. Funct Plant Biol 29 909–916 [DOI] [PubMed] [Google Scholar]
- Kao CH, Yang SF (1983) Role of ethylene in the senescence of detached rice leaves. Plant Physiol 73 881–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Ikoma Y, Matsumoto H, Sugiura M, Hyodo H, Yano M (2004) Accumulation of carotenoids and expression of the carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol 134 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Yu Y, Snesrud EC, Moy LP, Linford LD, Haas BJ, Nierman WC, Quackenbush J (2005) Transcriptional divergence of the duplicated oxidative stress-responsive genes in the Arabidopsis genome. Plant J 41 212–220 [DOI] [PubMed] [Google Scholar]
- Kimura M, Yamamoto YY, Seki M, Sakurai T, Sato M, Abe T, Yoshida S, Manabe K, Shinozaki K, Matsui M (2003) Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem Photobiol 77 226–233 [DOI] [PubMed] [Google Scholar]
- Kuehn GD, Phillips GC (2005) Role of polyamines in apoptosis and other recent advances in plant polyamines. CRC Crit Rev Plant Sci 24 123–130 [Google Scholar]
- Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, et al (2007) Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19 1362–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitalainen T, Pitkänen J, Hynninen P (1990) Diastereoselective 132-hydroxylation of chlorophyll a with SeO2. In Abstracts of the 8th International IUPAC Conference on Organic Synthesis. IUPAC, Helsinki, p 246
- Lam H-M, Hsieh M-H, Coruzzi G (1998) Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis. Plant J 16 345–353 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Roseman AM, Saibil HR, Vierling E (1997) A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J 16 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XP, Gan R, Li PL, Ma YY, Zhang LW, Zhang R, Wang Y, Wang NN (2006) Identification and functional characterization of a leucine-rich repeat receptor-like kinase gene that is involved in regulation of soybean leaf senescence. Plant Mol Biol 61 829–844 [DOI] [PubMed] [Google Scholar]
- Luquez VM, Guiamét JJ (2002) The stay green mutations d1 and d2 increase water stress susceptibility in soybeans. J Exp Bot 376 1421–1428 [DOI] [PubMed] [Google Scholar]
- Mahalingam R, Shah N, Scrymgeour A, Fedoroff N (2005) Temporal evolution of the Arabidopsis oxidative stress response. Plant Mol Biol 57 709–730 [DOI] [PubMed] [Google Scholar]
- Mínguez-Mosquera MI, Gallardo-Guerrero L, Gandul-Rojas B (1993) Characterization and separation of oxidized derivatives of pheophorbide a and b by thin-layer and high-performance liquid chromatography. J Chromatogr 633 295–299 [Google Scholar]
- Mínguez-Mosquera MI, Gandul-Rojas B, Gallardo-Guerrero L (1994) Measurement of chlorophyllase activity in olive fruit (Olea europaea). J Biochem 116 263–268 [DOI] [PubMed] [Google Scholar]
- Mínguez-Mosquera MI, Gandul-Rojas B, Montaño-Asquerino A, Garrido-Fernández J (1991) Determination of chlorophylls and carotenoids by HPLC during olive lactic fermentation. J Chromatogr 585 259–266 [Google Scholar]
- Mínguez-Mosquera MI, Garrido-Fernández J (1989) Chlorophyll and carotenoid presence in olive fruit (Olea europaea L.). J Agric Food Chem 37 1–7 [DOI] [PubMed] [Google Scholar]
- Mitler R, Vanderauwera S, Gollery M, Breusegem FV (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9 490–498 [DOI] [PubMed] [Google Scholar]
- Montané MH, Kloppstech K (2000) The family of light-harvesting-related proteins (LHCs, ELIPs, HLIPs): was the harvesting of life their primary function? Gene 258 1–8 [DOI] [PubMed] [Google Scholar]
- Neta-Sharir I, Isaacson T, Lurie S, Weiss D (2005) Dual role for tomato heat shock protein 21: protecting photosystem II from oxidative stress and promoting color changes during fruit maturation. Plant Cell 17 1829–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK (1999) Photoprotection revisited. Annu Rev Plant Physiol Plant Mol Biol 50 333–359 [DOI] [PubMed] [Google Scholar]
- Oh MH, Kim JH, Moon YH, Lee CH (2004) Defects in a proteolytic step of light-harvesting complex II in an Arabidopsis stay-green mutant, ore10, during dark-induced leaf senescence. J Plant Biol 47 330–337 [Google Scholar]
- Oh MH, Moon YH, Lee CH (2003) Increased stability of LHCII by aggregate formation during dark-induced leaf senescence in the Arabidopsis mutant, ore 10. Plant Cell Physiol 44 1368–1377 [DOI] [PubMed] [Google Scholar]
- Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, Lee SK, Jeong SW, Seo HS, Koh HJ, et al (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19 1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampino P, Spano G, Pataleo S, Mita G, Napier JA, Di Fonzo N, Shewry PR, Perrotta C (2006) Molecular analysis of a durum wheat ‘stay green’ mutant: expression pattern of photosynthesis-related genes. J Cereal Sci 43 160–168 [Google Scholar]
- Ren G, An K, Liao Y, Zhou X, Cao Y, Zhao H, Ge X, Kuai B (2007) Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol 144 1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R (2004) The zinc-finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem 279 11736–11743 [DOI] [PubMed] [Google Scholar]
- Roca M, James J, Pruzinska A, Hortensteiner S, Thomas H, Ougham H (2004) Analysis of the chlorophyll catabolism pathway in leaves of an introgression senescence mutant of Lolium temulentum. Phytochemistry 65 1231–1238 [DOI] [PubMed] [Google Scholar]
- Roca M, Mínguez-Mosquera MI (2006) Chlorophyll catabolism pathway in fruits of Capsicum annuum (L.): stay-green versus red fruits. J Agric Food Chem 54 4035–40 [DOI] [PubMed] [Google Scholar]
- Rodrigo MJ, Marcos J, Alférez F, Mallent MD, Zacarías L (2003) Characterization of Pinalate, a novel Citrus sinensis mutant with a fruit-specific alteration that results in yellow pigmentation and decreased ABA content. J Exp Bot 54 727–738 [DOI] [PubMed] [Google Scholar]
- Rodrigo MJ, Marcos JF, Zacarías L (2004) Biochemical and molecular analysis of carotenoid biosynthesis in flavedo of orange (Citrus sinensis L.) during fruit development and maturation. J Agric Food Chem 52 6724–6731 [DOI] [PubMed] [Google Scholar]
- Ronning CM, Bouwkamp JC, Solomos T (1991) Observations on the senescence of a mutant non-yellowing genotype of Phaseolus vulgaris L. J Exp Bot 235 235–241 [Google Scholar]
- Rose JKC, Bashir S, Giovannoni JJ, Jahn MM, Saravanan RS (2004) Tackling the plant proteome: practical approaches, hurdles and experimental tools. Plant J 39 715–733 [DOI] [PubMed] [Google Scholar]
- Rossel JB, Wilson IW, Pogson BJ (2002) Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol 130 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossini S, Casazza AP, Engelmann EC, Havaux M, Jennings RC, Soave C (2006) Suppression of both ELIP1 and ELIP2 in Arabidopsis does not affect tolerance to photoinhibition and photooxidative stress. Plant Physiol 141 1264–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fitsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Saravanan RS, Rose JKC (2004) A critical evaluation of sample extraction techniques for enhanced proteomic analysis of recalcitrant plant tissues. Proteomics 4 2522–2532 [DOI] [PubMed] [Google Scholar]
- Sato Y, Morita R, Nishimura M, Yamaguchi H, Kusaba M (2007) Mendel's green cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway. Proc Natl Acad Sci USA 104 14169–14174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart CM, Scofield SR, Bevan MW, Dyer TA (1991) Delayed leaf senescence in tobacco plants transformed with tmr, a gene for cytokinin production in Agrobacterium. Plant Cell 3 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JHC, Benítez A (1955) Chlorophylls. In K Paech, MM Tracey, eds, Modern Methods of Plant Analysis. Springer, Berlin, pp 142–196
- Spano G, Di Fonzo N, Perrotta C, Platani C, Ronga G, Lawlor DW, Napier JA, Shewry PR (2003) Physiological characterisation of ‘stay green’ mutants in durum wheat. J Exp Bot 386 1415–1420 [DOI] [PubMed] [Google Scholar]
- Sun W, van Montagu M, Verbruggen N (2002) Small heat shock proteins and stress tolerance in plants. Biochim Biophys Acta 1577 1–9 [DOI] [PubMed] [Google Scholar]
- Takahama U, Oniki T (1992) Regulation of peroxidase-dependent oxidation of phenolics in the apoplast of spinach leaves by ascorbate. Plant Cell Physiol 33 379–387 [Google Scholar]
- Tanaka R, Oster U, Kruse E, Rudiger W, Grimm B (1999) Reduced activity of geranylgeranyl reductase leads to loss of chlorophyll and tocopherol and to partially geranylgeranylated chlorophyll in transgenic tobacco plants expressing antisense RNA for geranylgeranyl reductase. Plant Physiol 120 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpstra W, Lambers J (1983) Interactions between chlorophyllase, chlorophyll a, plants lipids and Mg2+. Biochim Biophys Acta 746 23–31 [Google Scholar]
- Thomas H (1987) Sid: a Mendelian locus controlling thylakoid membrane disassembly in senescing leaves of Festuca pratensis. Theor Appl Genet 73 551–555 [DOI] [PubMed] [Google Scholar]
- Thomas H, Howarth CJ (2000) Five ways to stay green. J Exp Bot 51 329–337 [DOI] [PubMed] [Google Scholar]
- Thomas H, Ougham H, Canter P, Donnison I (2002) What stay-green mutants tell us about nitrogen remobilization in leaf senescence. J Exp Bot 53 801–808 [DOI] [PubMed] [Google Scholar]
- Thomas H, Schellenberg M, Vicentini F, Matile P (1996) Gregor Mendel's green and yellow pea seeds. Bot Acta 109 3–4 [Google Scholar]
- Tosti N, Pasqualini S, Borgogni A, Ederli L, Falistocco E, Crispi S, Paolocci F (2006) Gene expression profiles of O3-treated Arabidopsis plants. Plant Cell Environ 29 1686–1702 [DOI] [PubMed] [Google Scholar]
- Trebitsh T, Goldschmidt EE, Riov J (1993) Ethylene induces de novo synthesis of chlorophyllase, a chlorophyll degrading enzyme, in Citrus fruit peel. Proc Natl Acad Sci USA 90 9441–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicentini F, Hörtensteiner S, Schellenberg M, Thomas H, Matile P (1995) Chlorophyll breakdown in senescent leaves: identification of the biochemical lesion in a stay-green genotype of Festuca pratensis Huds. New Phytol 129 247–252 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Hongu A, Honda K, Nakazato M, Konno M, Saithoh S (1984) Preparation of chlorophylls and pheophytins by isocratic liquid chromatography. Anal Chem 56 251–256 [Google Scholar]
- Wong CE, Li Y, Labbe A, Guevara D, Nuin P, Whitty B, Diaz C, Golding GB, Gray GR, Weretilnyk EA, et al (2006) Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol 140 1437–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Bhuiyan NMH, Watidee R, Tanaka Y, Esaka M, Oba K, Jagendorf AT, Takabe T (2005) Suppressed expression of the apoplastic ascorbate oxidase gene increases salt tolerance in tobacco and Arabidopsis plants. J Exp Bot 56 1785–1796 [DOI] [PubMed] [Google Scholar]
- Young R, Jahn O (1972) Ethylene-induced carotenoid accumulation in citrus fruit rinds. J Am Soc Hortic Sci 97 258–261 [Google Scholar]
- Zhao J, Williams CC, Last RL (1998) Induction of Arabidopsis tryptophan pathway enzymes and camalexin by amino acid starvation, oxidative stress, and an abiotic elicitor. Plant Cell 10 359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.