Abstract
Claviceps purpurea, a fungal pathogen responsible for ergot diseases in many agriculturally important cereal crops, produces high levels of ricinoleic acid (12-hydroxyoctadec-cis-9-enoic acid) in its sclerotia. It has been believed for many years that the biosynthesis of this fatty acid in C. purpurea involves a hydration process with linoleic acid as the substrate. Using degenerate polymerase chain reaction, we cloned a gene from the sclerotia encoding an enzyme (CpFAH) that has high sequence similarity to the C. purpurea oleate desaturase, but only low similarity to plant oleate hydroxylases. Functional analysis of CpFAH in yeast (Saccharomyces cerevisiae) indicated it acted predominantly as a hydroxylase, introducing hydroxyl groups at the 12-position of oleic acid and palmitoleic acid. As well, it showed Δ12 desaturase activities on 16C and 18C monounsaturated fatty acids and, to a much lesser extent, ω3 desaturase activities on ricinoleic acid. Heterologous expression of CpFAH under the guidance of a seed-specific promoter in Arabidopsis (Arabidopsis thaliana) wild-type and mutant (fad2/fae1) plants resulted in the accumulation of relatively higher levels of hydroxyl fatty acids in seeds. These data indicate that the biosynthesis of ricinoleic acid in C. purpurea is catalyzed by the fungal desaturase-like hydroxylase, and CpFAH, the first Δ12 oleate hydroxylase of nonplant origin, is a good candidate for the transgenic production of hydroxyl fatty acids in oilseed crops.
Hydroxyl fatty acids such as ricinoleic acid (12-hydroxyoctadec-cis-9-enoic acid or 12-OH-18:1-9c) are sporadically found in the storage neutral lipids of certain plants (McKeon et al., 1999) and microorganisms (Morris and Hall, 1966). Recently, there has been a high level of scientific interest in the biosynthesis of these fatty acids due to their specialized industrial uses. In castor (Ricinus communis), where ricinoleic acid can account for up to 90% of the total fatty acids in seeds, biosynthesis of this fatty acid involves a membrane-bound fatty acid hydroxylase-catalyzing hydroxylation at position 12 of oleic acid esterified to the sn-2 position of phosphatidylcholine, using cytochrome b5 and NADH as cofactors (Moreau and Stumpf, 1981; Bafor et al., 1991). The similarities in catalytic mechanisms of membrane-bound oleate desaturases and oleate hydroxylases implies that these enzymes might share sequence similarity. Based on this hypothesis, the first oleate hydroxylase gene was cloned from castor using EST sequencing/similarity searches (van de Loo et al., 1995). Soon afterward, a similar hydroxylase from Lesquerella (Lesquerella fendleri) involved in the first step of lesquerolic acid (14-hydroxyeicos-cis-11-enoic acid or 14-OH-20:1-11c) biosynthesis was isolated by degenerate reverse transcription (RT)-PCR (Broun et al., 1998a). The Lesquerella hydroxylase, like the castor hydroxylase, uses oleic acid as substrate producing ricinoleic acid, which is then elongated to lesquerolic acid by a fatty acid elongase. The proteins encoded by these two hydroxylase genes share high amino acid sequence similarity to an Arabidopsis (Arabidopsis thaliana) oleate desaturase (FAD2; Okuley et al., 1994).
Hydroxyl fatty acids are important feedstocks for industrial products such as lubricants, functional fluids, ink, paints, coatings, nylons, resins, and foams, as well as cosmetics and pharmaceuticals (McKeon et al., 1999; Jaworski and Cahoon, 2003). Currently, castor bean is the only commercially available biological source of hydroxyl fatty acids for the production of these industrial materials. However, due to the poor agronomic performance of the plant and the presence of a highly potent toxin (ricin) in the seed, castor bean is not an ideal source crop. The genetic engineering of high-yielding oilseed crops is viewed as a cost-effective, renewable, and environmentally friendly alternative method to produce various unusual fatty acids. To date, however, transgenic plants carrying genes encoding enzymes catalyzing the synthesis of these fatty acids have produced relatively low levels of targeted fatty acids (Voelker and Kinney, 2001; Jaworski and Cahoon, 2003; Napier, 2007). For example, transgenic plants with a single castor bean or Lesquerella hydroxylase under a strong seed-specific promoter produced hydroxyl fatty acids at levels much lower than those found in native plant species (van de Loo et al., 1995; Broun and Somerville, 1997; Smith et al., 2003; Duak et al., 2007). This implies that additional genes from native species are required for the synthesis and accumulation of high levels of hydroxyl fatty acids in transgenic plants, and extensive efforts have recently been made to identify such genes (Kumar et al., 2006; Lu et al., 2006).
Claviceps purpurea, a fungal pathogen responsible for ergot diseases in plants, parasitizes young flowers of many grasses including agriculturally important cereal crops such as wheat (Triticum aestivum) and barley (Hordeum vulgare; Scheffer and Tudzynski, 2006). During infection, this pathogen forms specialized structures called sclerotia in the ovaries of host plants. Although it has long been known that the sclerotia of this fungus produce high levels of ricinoleic acid (Mantle and Nisbet, 1976), the biosynthetic mechanism for fungal production of this fatty acid has not been established. Early reports proposed that biosynthesis of ricinoleic acid in C. purpurea was catalyzed by a hydratase using linoleic acid as a substrate, and did not require molecular oxygen (Morris et al., 1966; Morris, 1970). This hypothesis has been held for many years. However, a recent study measuring site-specific deuterium abundance suggested that ricinoleic acid is formed in this species via a hydroxylation process, with oleic acid as the substrate (Billault et al., 2004).
Based on the recent results (Billault et al., 2004), we hypothesized that the biosynthesis of ricinoleic acid in C. purpurea might occur either by cytochrome P450-like hydroxylation or desaturase-like hydroxylation. In this study, we focused on the later possibility that the biosynthesis of ricinoleic acid in C. purpurea is catalyzed by a desaturase-like hydroxylase. By using degenerate RT-PCR targeted to conserved regions of fungal oleate desaturases, we cloned a cDNA from the sclerotium tissue of C. purpurea encoding an enzyme (CpFAH) with a high level of sequence similarity to fungal desaturases, and only a low level of similarity to plant fatty acid hydroxylases. Functional analysis of CpFAH in yeast (Saccharomyces cerevisiae) indicated its major activity is that of a hydroxylase, introducing a hydroxyl group at position 12 of oleic acid. Heterologous expression of CpFAH in Arabidopsis wild-type and mutant (fad2/fae1) plants revealed a higher level accumulation of hydroxy fatty acids in seeds than was previously observed in Arabidopsis transgenics carrying plant fatty acid hydroxylases.
RESULTS
Cloning of a C. purpurea Gene Encoding a Fatty Acid Hydroxylase
The fungal pathogen C. purpurea produces ricinoleic acid at high levels in its sclerotium tissue (Morris and Hall, 1966). To obtain this tissue, C. purpurea was grown in a sclerotium-inducing medium at room temperature for 4 weeks (Mantle and Nisbet, 1976). Fatty acid analysis of isolated fungal tissues indicated that sclerotium-forming mycelia obtained after 30 d of culture on sclerotium-inducing medium contained approximately 23% of the total fatty acids as ricinoleic acid, whereas this fatty acid was not detectable in non-sclerotium-forming mycelia.
To identify the gene encoding the hydroxylase involved in the synthesis of ricinoleic acid in C. purpurea, we designed several sets of degenerate oligonucleotide primers targeted to conserved domains of known fungal oleate desaturases. RT-PCR amplification with these degenerate primers, using total RNA isolated from sclerotium-forming mycelia as a template, generated three specific products, each showing sequence similarity to fungal Δ12 desaturases.
Subsequently, full-length cDNAs corresponding to each of these products were obtained by 5′ and 3′ RACE. Sequence and functional analyses of full-length clones indicated that one cDNA, CpDes12, encodes a Δ12 desaturase that introduces a Δ12 double bond into oleic and palmitoleic acids. The second clone, CpDesX, encodes a novel desaturase catalyzing Δ12, Δ15, and ω3 desaturation activity, with ω3 desaturation of linoleic acid predominating (Meesapyodsuk et al., 2007). The third gene, CpFAH, which is characterized here, encodes a 477-amino acid polypeptide sharing high amino acid sequence similarity to CpDes12 (86% identity) and CpDesX (80% identity), as well as to fungal Δ12 desaturases from Fusarium (68% identity) and Aspergillus (60% identity). Conversely, CpFAH shares a relatively lower level of sequence similarity with fatty acid hydroxylases from the plant species castor and L. fendleri (39%–40% amino acid identity).
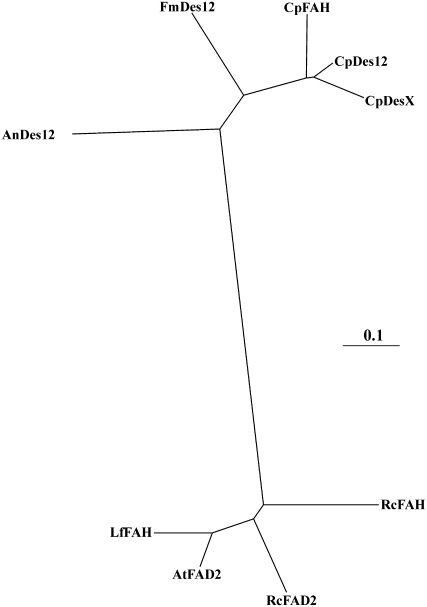
Phylogenetic analysis of CpFAH and related enzymes shows that the three highly homologous enzymes from C. purpurea, CpFAH, CpDes12, and CpDesX, are grouped into a deep subbranch of the phylogenetic tree, belonging to a branch that includes other fungal Δ12 desaturases (Damude et al., 2006; Fig. 1). Within this subbranch, CpFAH and CpDes12 are tightly grouped together. The fatty acid hydroxylases and Δ12 desaturases from plants form a separate distant group in the phylogenetic tree.
Figure 1.
An unrooted phylogenetic analysis based on CpFAH and related enzymes. The GenBank accession numbers of the sequences are indicated in parentheses: CpFAH, C. purpurea 12-hydroxylase (EU661785; this report); CpDes12, C. purpurea Δ12 desaturase (EF536897); CpDesX, C. purpurea novel desaturase (EF536898); AtFAD2, Arabidopsis Δ12 desaturase (P46313); RcFAD2, castor Δ12 desaturase (ABK59093); RcFAH, castor 12-hydroxylase (AAC49010); LfFAH, L. fendleri 12-hydroxylase (AAC32755); FmDes12, Fusarium moniliforme Δ12 desaturase (DQ272515); AnDes12, A. nidulans Δ12 desaturase (XP_658641).
Functional Characterization of CpFAH in Yeast
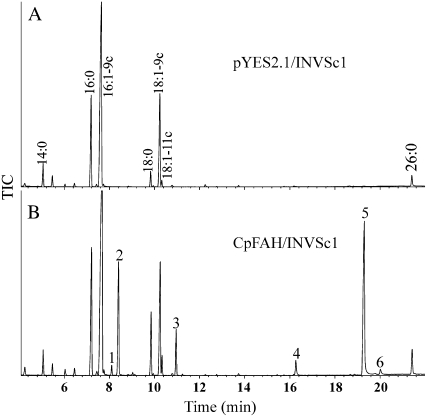
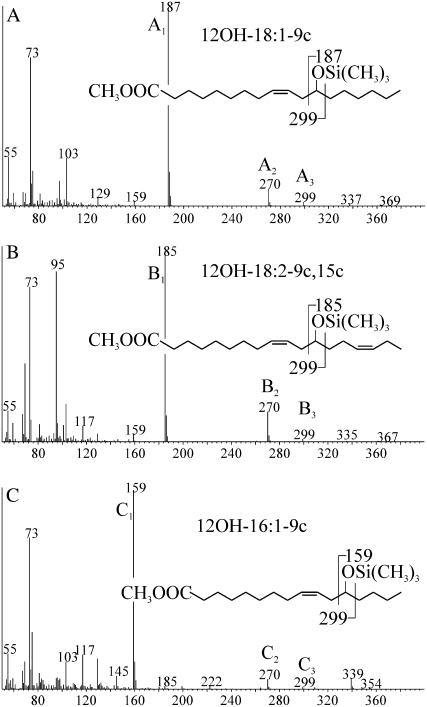
To determine the function of CpFAH, the coding region of the cDNA was cloned into the yeast expression vector pYES2.1 under the control of GAL1 promoter, and the recombinant plasmid was then introduced into the yeast strain INVSc1. Fatty acid analysis of total lipids indicated that yeast transformants expressing CpFAH (CpFAH/INVSc1) produced six new fatty acids compared to the control (pYES2.1/INVSc1; Fig. 2). Three of these fatty acids (peaks 1, 2, and 3) were identified as 16:2-9t,12c, 16:2-9c,12c, and 18:2-9c,12c, respectively, as previously described (Meesapyodsuk et al., 2007). Three new fatty acids having longer retention times (peaks 4, 5, and 6) carried hydroxyl groups able to react with a trimethylsilylating reagent. The chemical structures of these hydroxyl fatty acids were determined by gas chromatography (GC)/mass spectrometry (MS) analysis of fatty acid methyl ester (FAME)-trimethylsilyl (TMS) derivatives. As shown in Figure 3A, the mass spectrum obtained from peak 5, including three diagnostic fragments at mass-to-charge ratio (m/z) 187 (A1), m/z 270 (A2), and m/z 299 (A3), is identical to that previously obtained for a TMS-methylricinoleate standard (van de Loo et al., 1995), indicating that this hydroxyl fatty acid is ricinoleic acid (12-OH-18:1-9c). Figure 3B shows the mass spectrum of FAME-TMS derivatives of peak 6. Two of the three diagnostic fragments, m/z 270 (B2) and m/z 299 (B3), are identical to fragment A2 and A3 of TMS-methylricinoleate, whereas fragment B1 differs from A1 of TMS-methylricinoleate by two mass units, which is due to the loss of two hydrogen atoms. Thus, this hydroxyl fatty acid is densipolic acid (12-OH-18:2-9c,15c), a fatty acid that was also observed as a product of plant hydroxylases (Broun and Somerville, 1997; Broun et al., 1998a). The mass spectrum of the peak 4 derivative is shown in Figure 3C. Two of the diagnostic fragment ions, C2 and C3, are identical to their counterparts A2 and A3 at m/z 270 and m/z 299. The diagnostic fragment ion C1 at m/z 159 differs from its counterpart A1 in the TMS-methylricinoleate spectrum by a reduction of 28 mass units, which corresponds to the mass of two carbon and four hydrogen atoms, indicating this new hydroxyl fatty acid is 12-OH-16:1-9c. This hydroxyl fatty acid was also observed when a Lesquerella oleate hydroxylase was expressed in yeast (Smith et al., 2003).
Figure 2.
GC analysis of FAMEs prepared from yeast transformed with pYES2.1 (A) and CpFAH (B). Peak identification: 1, 16:2-9c,12t; 2, 16:2-9c,12c; 3, 18:2-9c,12c; 4, 12-OH-16:1-9c; 5, 12-OH-18:1-9c; 6, 12-OH-18:2-9c,15c.
Figure 3.
Mass spectra of FAME-TMS derivatives of hydroxyl fatty acid peaks from Figure 2. The identity of the peaks were confirmed as 12-OH-18:1-9c (A), 12-OH-18:2-9c,15c (B), and 12-OH-16:1-9c (C).
CpFAH expression led to the production of densipolic acid, which contains a ω3 double bond. Furthermore, when yeast transformants expressing CpFAH were supplied with linoleic acid (18:2-9c,12c), the increased linoleate levels resulted in the production of the ω3 desaturated product linolenic acid (18:3-9c,12c,15c; data not shown). Thus, the ω3 double bond in densipolic acid is likely derived from a minor ω3 desaturase activity of CpFAH on ricinoleic acid. The ability of ω3 desaturases to introduce a double bond at the ω3 position of ricinoleic acid has also been observed in plants (Meesapyodsuk et al., 2000; Reed et al., 2000).
Among the six new fatty acids produced by CpFAH, ricinoleic acid was the most abundant, accounting for approximately 19% of the total fatty acids in transformed yeast cells. The fatty acids 16:2-9c,12c, 12-OH-16:1-9c, and 18:2-9c,12c were less abundant, accounting for approximately 8%, 4%, and 3% of total fatty acids, respectively. Densipolic acid and 16:2-9c,12t were minor products, in combination representing less than 1.5% of the total fatty acids. The 12-hydroxylation activity of CpFAH was almost 7 times the Δ12 desaturation activity on oleic acid, indicating hydroxylation of oleic acid is the major activity of this enzyme in yeast.
Expression of CpFAH in Arabidopsis
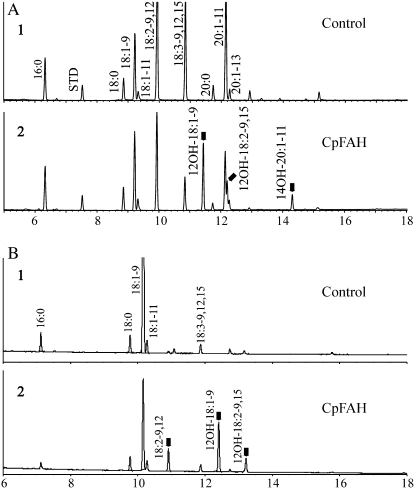
To assess the capability of the fungal hydroxylase to synthesize hydroxyl fatty acids in planta, CpFAH under the control of the seed-specific napin promoter was transformed into wild-type and fad2/fae1 mutant Arabidopsis plants, along with the DsRed2 reporter gene. The Arabidopsis mutant line (fad2/fae1), which carries mutations in both the FAD2 and FAE1 genes, produces a high level of substrate fatty acid (oleate) that can be used for the synthesis of ricinoleic acid (Smith et al., 2003). Use of the DsRed2 reporter gene permits the easy identification of transgenic seeds by examination under green illumination using a red filter (Lu et al., 2006). A floral-dip transformation approach (Clough and Bent, 1998) was used to obtain 11 transgenic plants from each of the two genotypes used in this study. Typical chromatograms from GC analysis of FAME derivatives of transgenic seeds are shown in Figure 4. The most abundant fatty acid in untransformed wild-type Arabidopsis seeds was 18:2-9c,12c, followed by 20:1-11c, 18:3-9c,12c,15c, and 18:1-9c. Minor components included 16:0 and 18:0. Expression of CpFAH in wild-type Arabidopsis resulted in the production of three hydroxyl fatty acids; among these, ricinoleic acid, which is produced via the direct hydroxylation of oleic acid by CpFAH, was the most abundant, followed by densipolic acid, derived from the ω3 desaturation of ricinoleic acid, and lesquerolic acid, a 20C hydroxyl fatty acid derived from the elongation of ricinoleic acid (Fig. 4A; Table I). However, unlike yeast, transformed wild-type Arabidopsis did not produce a detectable level of 12-OH-16:1-9c, which may have been due to the low availability of the 16:1-9c substrate.
Figure 4.
GC analysis of FAMEs prepared from mature seeds of Arabidopsis. A, Wild-type (1) untransformed, and (2) transformed with CpFAH. B, fad2/fae1 double mutant untransformed (1) and transformed with CpFAH (2).
Table I.
Primers used in this study
The restriction sites used for cloning are underlined.
| Primers Used |
|---|
| Degenerate primers for cloning of CpFAH |
| DM34: 5′-GCICAYGARTGYGGICAYSRIGCITT-3′ |
| DM36: 5′-TAIGTDATIGCIACIARCCARTGRTKIACCCA-3′ |
| For 5′ and 3′ RACE experiments |
| DM58: 5′-GGAGCGCGACATGGTCTTTCTGCCTCAG-3′ |
| DM59: 5′-CAGACAGCCATGTTGGCCCAGCCGAAT-3′ |
| For full-length cDNA amplification |
| DM61: 5′-CACTAGGGCAACGAATTACTCTGC-3′ |
| DM62: 5′-GGACGCCATCGTTGACTTCC-3′ |
| For yeast and plant expression |
| DM63: 5′-GCGAATTCGAAATGGCTTCCGCTACTCC-3′ |
| DM64: 5′-GCGAATTCCTACTGAGTCTTCATTGAAATGG-3′ |
| DM68: 5′-TCTAGAGAAATGGCTTCCGCTACTCC-3′ |
| DM69: 5′-TCTAGACTACTGAGTCTTCATTG-3′ |
| DM81: 5′-CGCGGATCCATGGCCTCCTCCGAGAAC-3′ |
| DM82: 5′-CGCGGATCCTCACAGGAACAGGTGGTGGC-3′ |
The untransformed mutant Arabidopsis, because it lacks activities of both the Δ12 desaturase (FAD2) and a condensing enzyme or elongase (FAE1), accumulated substantial amounts of oleic acid. Expression of CpFAH in this line resulted in the production of two hydroxyl fatty acids (ricinoleic acid and densipolic acid) and one Δ12 desaturated fatty acid (linoleic acid) with ricinoleic acid predominating (Fig. 4B). Although oleic acid was still the most abundant fatty acid, its level was dramatically reduced from more than 80% in the untransformed control to approximately 45% in the transgenic plants (Table I). The production of linoleic acid in transformed mutant Arabidopsis seeds confirmed that CpFAH exhibited Δ12 desaturase activity in plants, as was also observed in yeast.
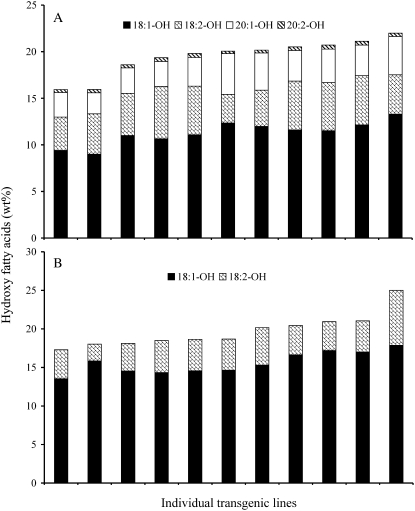
The amount of hydroxyl fatty acids produced in transgenic lines is depicted in Figure 5. The total hydroxyl fatty acids in transformed wild-type Arabidopsis ranged from 15.9% to 22.0%. Heterologous expression of CpFAH in the fad2/fae1 genotype led to the accumulation of 17.3% to 25% of hydroxyl fatty acids (Table I), with the highest level in a single seed reaching 29.1%. These are considerably higher levels than were seen with the expression of plant hydroxylases in either wild-type or mutant Arabidopsis plants (Broun and Somerville, 1997; Smith et al., 2003). A large number of Arabidopsis plants carrying either castor or Lesquerella hydroxylases in different genetic backgrounds (wild type, fad3, fad2/fae1, fad3/fae1) have been produced; however, hydroxyl fatty acids have rarely exceeded 20% of the total fatty acids in seeds in these lines (Smith et al., 2003).
Figure 5.
Hydroxyl fatty acid content of mature seeds from individual transgenic Arabidopsis plants expressing CpFAH. A, Wild type. B, fad2/fae1 double mutant. Transgenic seeds were screened using the DsRed2 fluorescent marker. Values are measurements of five T2 seeds.
DISCUSSION
Although it has long been known that the fungus C. purpurea produces substantial amounts of ricinoleic acid (Morris and Hall, 1966), the mechanism for the biosynthesis of this fatty acid has yet to be elucidated. It has been believed for almost 40 years that its biosynthesis in C. purpurea involves a hydration process that introduced a water molecule into linoleic acid, giving rise to ricinoleic acid (Morris et al., 1966; Morris, 1970). However, a recent study using deuterium NMR rebutted this hypothesis and showed that the synthesis of ricinoleic acid in C. purpurea follows an oxygen-dependent hydroxylation process using oleic acid as the substrate (Billault et al., 2004), although the exact nature of the hydroxylase enzyme was not determined. Two possibilities existed; the hydroxylation of oleic acid could be catalyzed by a desaturase-like hydroxylase as occurs in plants or by a cytochrome P450-like hydroxylase as is seen in some fungal species. For example, the fungus Aspergillus nidulans uses a family of oxygenated long-chain fatty acids called psi factors (precocious sexual inducers), such as psiBα (8-hydroxy-18:2-9,12) and psiCα (5,8-dihydroxy-18:2-9,12), to modulate sexual and asexual spore development. Both these hydroxyl fatty acids are synthesized by cytochrome P450-like fatty acid oxygenases (Tsitsigiannis et al., 2004, 2005). In this study, we focused on the possibility that the biosynthesis of ricinoleic acid in C. purpurea resembles that in plants, where the oleate hydroxylase has high sequence similarity to oleate desaturases (van de Loo et al., 1995). We designed two degenerate primers targeting fungal oleate desaturases and used RNA isolated from C. purpurea sclerotia as the template for RT-PCR. By this method we were able to identify a cDNA (CpFAH) from C. purpurea that encodes an enzyme that preferentially introduces a hydroxyl group into position 12 of oleic acid. CpFAH represents the first 12-oleate hydroxylase of nonplant origin. Compared to its plant counterparts, CpFAH appears to have less strict substrate specificity. As well as showing strong hydroxylase activity on oleate and palmitoleate, CpFAH also catalyzes Δ12 desaturation of oleate and palmitoleate and, to a limited extent, is able to desaturate linoleate and ricinoleate at the ω3 position.
Sequence analysis of CpFAH indicates it shares approximately 80% amino acid identity with both CpDes12 (a Δ12 desaturase) and CpDesX (a novel desaturase) from C. purpurea (Meesapyodsuk et al., 2007), but only approximately 40% amino acid identity with its plant counterparts—castor bean and Lesquerella hydroxylases. Phylogenetic analysis suggests that the CpFAH gene of C. purpurea might have evolved independently from its own prototype Δ12 desaturase, rather than from an ancient common ancestral 12-hydroxylase or from a Δ12 desaturase shared by plants and fungi. Many common features of fatty acid desaturases and fatty acid hydroxylases, such as their strikingly similar hydropathy profiles that predict similar membrane topology, and conserved His-rich motifs thought to be responsible for diiron binding at the catalytic center, point to a common evolutionary relationship for these two types of enzymes (Shanklin and Cahoon, 1998; Sperling et al., 2003). Indeed, site-directed mutagenesis experiments have shown that as few as four amino acid substitutions can convert an Arabidopsis oleate desaturase to an oleate hydroxylase (Broun et al., 1998b; Broadwater et al., 2002). In C. purpurea, CpDes12, CpDesX, and CpFAH share striking similarity to each other in primary sequence but possess distinct functions. CpDes12 is a typical Δ12 desaturase active on monounsaturated fatty acids such as oleic and palmitoleic acids. CpDesX is a desaturase with Δ12, Δ15, and ω3 types of desaturation activities, although ω3 activity on linoleic acid predominates. The primary activity of CpFAH is that of an oleic acid hydroxylase, although it is also capable of introducing Δ12 double bonds to oleic and palmitoleic acids. The high sequence similarity of these enzymes coupled with their distinct functions provides a unique opportunity to investigate the influence of individual amino acids on substrate specificity, regioselectivity, and function.
Metabolic engineering of plants to produce unusual fatty acids for industrial applications is the ultimate goal of studies on the biosynthesis and bioassembly of unusual fatty acids. Many genes encoding enzymes involved in the synthesis of these fatty acids have recently been identified from native plant species. However, to date the production of unusual fatty acids through the introduction of these genes into oilseed crops has met with only partial success. Although proof of concept has been achieved by the production of many unusual fatty acids in transgenic crops, commercially viable levels have generally yet to be attained (Cahoon et al., 2007; Napier, 2007). The use of genetically engineered plants to produce ricinoleic acid has drawn a particularly high level of interest due to its specialized industrial use. Three oleate hydroxylase genes from castor and Lesquerella have been transformed into Arabidopsis, tobacco (Nicotiana tabacum), and Brassica napus; however, the hydroxyl fatty acid content in transgenic seeds has rarely exceeded 20% of the total fatty acids (van de Loo et al., 1995; Broun and Somerville, 1997; Broun et al., 1998a; Smith et al., 2003). Although it is not yet known why the level of hydroxyl fatty acid in transgenic plants is so much lower than that in native species, it has been hypothesized that contributing factors may include the inefficient removal of newly synthesized unusual fatty acids from membrane phospholipids (Thomæus et al., 2001; Cahoon et al., 2006) and the inefficient transfer of newly synthesized unusual fatty acids to storage lipids (Cahoon et al., 2007; Napier, 2007).
Expression of the C. purpurea CpFAH in transgenic plants resulted in the accumulation of substantial amounts of hydroxyl fatty acids and a small amount of linoleic acid in the double mutant Arabidopsis line. The latter is derived from the minor Δ12 desaturase activity of the transgene on oleic acid. The hydroxyl fatty acids in the transgenic mutant Arabidopsis reached 25% of the total fatty acids in bulked seeds, with a maximum level of 29.1% in a single seed of a T2 transgenic plant. Thus, it appears that selection of genes from diverse sources, especially from nonplant sources, can make a positive contribution to transgenic production of unusual fatty acids. The reason for the better performance of the fungal hydroxylase is currently unknown, but might be simply that CpFAH itself has higher hydroxylase activity. Furthermore, since the fungal oleate hydroxylase has less similarity than plant counterparts to endogenous homologous desaturase genes, CpFAH might be subject to less stringent regulatory control by the host plant. Regardless of the cause, the higher hydroxylase activity of CpFAH in both yeast and plants makes this gene a good candidate for use in metabolic engineering of hydroxyl fatty acids in oilseed crops.
MATERIALS AND METHODS
Organisms and Culture Conditions
To produce sclerotium-forming mycelia, Claviceps purpurea was routinely maintained at 25°C for 14 d in medium C or for 20 to 30 d in amino acid medium (Mantle and Nisbet, 1976; Meesapyodsuk et al., 2007).
Cloning of CpFAH cDNA from C. purpurea
Total RNA was extracted from sclerotium-forming mycelia of C. purpurea using TRIzol reagent (Invitrogen), and 5 μg was used to synthesize first-strand cDNA using the SuperScript III first-strand synthesis system (Invitrogen). Two microliters of the first-strand cDNA was used as a template for PCR amplification with two degenerate oligonucleotide primers (DM34 and DM36) that were designed based on conserved amino acid sequences of Δ12 and bifunctional Δ12/Δ15 desaturases from fungi (Damude et al., 2006; Meesapyodsuk et al., 2007). The detailed sequences of all primers used in this study are shown in Table II. The forward primer (DM34) corresponds to the first conserved His box and has the amino acid sequence AHECGH(G/Q)AF, while the reverse primer (DM36) lies between His boxes 2 and 3 and corresponds to the amino acid sequence WV(N/H)HWLVAITY. Amplified products having the expected size of approximately 600 bp were gel purified, then cloned into the pCR4-TOPO vector (Invitrogen) and sequenced.
Table II.
Fatty acid composition of single seeds of Arabidopsis wild-type (WT) and mutant (fad2/fae1) transgenics
The values are derived from GC analysis of FAME-TMS derivatives (mean ± se).
| Fatty Acids | WT
|
fad2/fae1
|
||
|---|---|---|---|---|
| Nontransformed | CpFAH | Nontransformed | CpFAH | |
| % | % | % | % | |
| 16:0 | 8.35 ± 0.56 | 10.38 ± 0.75 | 5.6 ± 0.23 | 4.18 ± 0.48 |
| 16:1 | 1.04 ± 0.77 | 0.07 ± 0.05 | – | – |
| 18:0 | 5.12 ± 0.55 | 5.82 ± 0.54 | 3.71 ± 0.21 | 6.56 ± 0.30 |
| 18:1-9 | 10.79 ± 0.32 | 13.76 ± 1.46 | 81.62 ± 0.56 | 45.04 ± 1.22 |
| 18:1-11 | 5.14 ± 0.86 | 5.48 ± 0.90 | 3.24 ± 0.05 | 5.51 ± 0.27 |
| 18:2-9,12 | 25.50 ± 0.54 | 24.69 ± 0.72 | 1.58 ± 0.34 | 8.96 ± 0.23 |
| 18:3-9,12,15 | 15.97 ± 0.74 | 5.22 ± 0.24 | 2.48 ± 0.06 | 3.85 ± 0.06 |
| 20:0 | 2.55 ± 0.06 | 1.43 ± 0.08 | 1.08 ± 0.02 | 0.3 ± 0.02 |
| 20:1-11 | 19.97 ± 0.80 | 10.16 ± 0.36 | 0.70 ± 0.02 | 0.2 ± 0.01 |
| 20:1-13 | 2.17 ± 0.05 | 2.62 ± 0.24 | – | – |
| 20:2 | 1.90 ± 0.08 | 0.51 ± 0.10 | – | – |
| 22:1 | 1.68 ± 0.13 | 0.23 ± 0.08 | – | – |
| 12-OH-18:1-9 | 0 | 12.65 ± 0.41 | – | 17.86 ± 1.09 |
| 12-OH-18:2-9,15 | 0 | 3.43 ± 0.32 | – | 7.54 ± 0.39 |
| 14-OH-20:1-11 | 0 | 3.42 ± 0.24 | – | – |
| 14-OH-20:2-11,17 | 0 | 0.15 ± 0.05 | – | – |
| Total hydroxyl fatty acids | 0 | 19.65 ± 0.73 | – | 25.00 ± 1.43 |
The 5′ and 3′ ends of the CpFAH cDNA were obtained using the Marathon cDNA amplification kit (BD Biosciences, CLONTECH) following the manufacturer's recommendations. Primers DM58 and DM59 were used to obtain the 3′ and 5′ ends, respectively. The full-length sequence, including both untranslated and coding regions, was then amplified using Pfx50 DNA polymerase (Invitrogen) with specific primers DM61 and DM62. The resulting PCR products were cloned into the pCR4-TOPO vector, producing construct pDM15.
Phylogenetic Analysis
Homologous sequences of CpFAH from fungi and plants were identified by BLASTP searches using CpFAH as a query against public databases at the National Center for Biotechnology Information. Six highly homologous sequences of fungal Δ12 and bifunctional Δ12/Δ15 desaturases from Aspergillus nidulans and Fusarium moniliforme, along with four well-known Δ12 desaturases and hydroxylases from the plants Arabidopsis (Arabidopsis thaliana), castor (Ricinus communis), and Lesquerella (Lesquerella fendleri) were chosen for construction of the phylogenetic tree. Multiple alignment of amino acid sequences was performed using ClustalW multiple alignment software (DNAstar) with default parameters. An unrooted phylogenetic tree was generated from the alignment and displayed using TreeView (Page, 1996).
Functional Analysis of CpFAH in Yeast
The coding region of the CpFAH cDNA was amplified from pDM15 using Pfx50 DNA polymerase with primers DM63 and DM64. After a brief Taq DNA polymerase (Invitrogen) treatment, the amplified fragment was cloned directly into the pYES2.1-TOPO expression vector (Invitrogen). The resulting plasmid construct, pDM16, was introduced into yeast (Saccharomyces cerevisiae) strain INVSc1 (MATa his3Δ1 leu2 trp1-289 ura3-52 MATα his3Δ1 leu2 trp1-289 ura3-52; Invitrogen) using the S.C. EasyComp transformation kit (Invitrogen). For expression studies, yeast transformed with either pDM16 or the empty vector pYES2.1 were grown at 28°C for 2 d in synthetic dropout medium containing 0.17% (w/v) yeast nitrogen base, 0.5% ammonium sulfate, 2% (w/v) dextrose, and 0.06% (w/v) dropout supplement lacking uracil. Following two washes with sterile water, the expression of the transgene was induced using synthetic dropout medium containing 2% Gal with or without 0.25 mm substrate fatty acid supplementation in the presence of 0.1% Tergitol (Nonidet P-40). Cells were incubated for another 2 d at 20°C before being harvested for fatty acid analysis.
Plant Vector Construction
The plant expression vector pDM22 with CpFAH from C. purpurea and DsRed2 from Discosoma sp. encoding a red fluorescent protein, each under control of a seed-specific napin promoter, was constructed as described previously (Wu et al., 2005). DsRed2 was PCR amplified from plasmid pSAT6-DsRed2-C1 (Tzfira et al., 2005) with primers DM81 and DM82, and CpFAH was PCR amplified from pDM16 with primers DM68 and DM69. Primers DM81 and DM82 introduce BamHI restriction sites, while primers DM68 and DM69 introduce XhoI restriction sites. Amplified fragments were first cloned into the pCR4-TOPO vector and sequenced. The fragments were then released by digestion with the appropriate restriction enzymes and sequentially subcloned into pUC19 between napin promoters and octopine synthase terminators. The two-gene cassette was removed from pUC19 by EcoRI digestion and inserted into the EcoRI site of a plant-expression vector, producing construct pDM22.
Production of Transgenic Arabidopsis
The construct pDM22 was introduced into Agrobacterium tumefaciens strain GV3101 (pMP90) by electroporation. Arabidopsis wild-type (Columbia ecotype) and fad2/fae1 mutant lines (Smith et al., 2003) were transformed using the floral-dip method (Clough and Bent, 1998). Selection of transformants was based on either kanamycin resistance (T1 seeds) or DsRed2 expression (T2 seeds). Plants were grown in a growth chamber at 22°C under a 16-h-light (120 μE m−2 s−1)/8-h-dark photoperiod.
Fatty Acid Analysis
Yeast fatty acids were extracted and methylated as previously described (Meesapyodsuk et al., 2007). Arabidopsis transgenic seeds identified by DsRed2 expression were analyzed by GC or GC-MS of FAMEs and/or TMS derivatives. Seeds were crushed with glass rod and FAMEs were derived using 1% H2SO4 in methanol and extracted with water and hexane. The total FAMEs in 50 μL (five seeds) or 15 μL (single seed) of hexane were trimethylsilylated by adding 30 μL (five seeds) or 10 μL (single seed) of N,O-bis(TMS)-acetamide:pyridine (1:1) and heating at 90°C for 30 min. Two-microliter samples of total FAMEs and/or TMS derivatives were analyzed on an Agilent 6890N gas chromatograph equipped with a DB-23 column (30 m × 0.25 mm) with 0.25-μm film thickness (J&W Scientific). The column temperature was maintained at 160°C for 1 min, then increased to 240°C at a rate of 4°C/min. For MS, the mass selective detector was run under standard electron impact conditions (70 eV), scanning an effective m/z range of 40 to 700 at 2.26 scans/s. Fatty acids were identified by comparison of retention times with authentic fatty acid standards, whereas hydroxylated derivatives of fatty acids were identified by MS on the basis of their fragmentation patterns as previously reported (van de Loo et al., 1995; Broun and Somerville, 1997). Fatty acid compositions were calculated as a weight percentage of the total fatty acids.
The nucleotide sequence reported in this article has been submitted to GenBank with accession number EU661785.
Acknowledgments
We are grateful to Patricia Vrinten for critical reading of the manuscript and editorial assistance. We thank Yu Chen for the C. purpurea strain, Mark Smith for seeds of the Arabidopsis fad2/fae1 mutant, Guohai Wu for pUC19 with two napin expression cassettes, Darwin Reed and Mike Giblin for technical discussion, Li Tan for the assistance in fatty acid analysis, and Patrick Covello and Mark Smith for reviewing the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Xiao Qiu (xiao.qiu@usask.ca).
Open Access articles can be viewed online without a subscription.
References
- Bafor M, Smith MA, Jonsson L, Stobart K, Stymne S (1991) Ricinoleic acid biosynthesis and triacylglycerol assembly in microsomal preparations from developing castor-bean (Ricinus communis) endosperm. Biochem J 280 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billault I, Mantle PG, Robins RJ (2004) Deuterium NMR used to indicate a common mechanism for the biosynthesis of ricinoleic acid by Ricinus communis and Claviceps purpurea. J Am Chem Soc 126 3250–3256 [DOI] [PubMed] [Google Scholar]
- Broadwater JA, Whittle E, Shanklin J (2002) Desaturation and hydroxylation: Residues 148 and 324 of Arabidopsis FAD2, in addition to substrate chain length, exert a major influence in partitioning of catalytic specificity. J Biol Chem 277 15613–15620 [DOI] [PubMed] [Google Scholar]
- Broun P, Boddupalli S, Somerville C (1998. a) A bifunctional oleate 12-hydroxylase: desaturase from Lesquerella fendleri. Plant J 13 201–210 [DOI] [PubMed] [Google Scholar]
- Broun P, Shanklin J, Whittle E, Somerville C (1998. b) Catalytic plasticity of fatty acid modification enzymes underlying chemical diversity of plant lipids. Science 282 1315–1317 [DOI] [PubMed] [Google Scholar]
- Broun P, Somerville C (1997) Accumulation of ricinoleic, lesquerolic, and densipolic acids in seeds of transgenic Arabidopsis plants that express a fatty acyl hydroxylase cDNA from castor bean. Plant Physiol 113 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon EB, Dietrich CR, Meyer K, Damude HG, Dyer JM, Kinney AJ (2006) Conjugated fatty acids accumulate to high levels in phospholipids of metabolically engineered soybean and Arabidopsis seeds. Phytochemistry 67 1166–1176 [DOI] [PubMed] [Google Scholar]
- Cahoon EB, Shockey JM, Dietrich CR, Gidda SK, Mullen RT, Dyer JM (2007) Engineering oilseeds for sustainable production of industrial and nutritional feedstocks: solving bottlenecks in fatty acid flux. Curr Opin Plant Biol 10 236–244 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Damude HG, Zhang H, Farrall L, Ripp KG, Tomb JF, Hollerbach D, Yadav NS (2006) Identification of bifunctional delta12/omega3 fatty acid desaturases for improving the ratio of omega3 to omega6 fatty acids in microbes and plants. Proc Natl Acad Sci USA 103 9446–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duak M, Lam P, Kunst L, Smith MA (2007) A FAD2 homologue from Lesquerella lindheimeri has predominantly fatty acid hydroxylase activity. Plant Sci 173 43–49 [Google Scholar]
- Jaworski J, Cahoon EB (2003) Industrial oils from transgenic plants. Curr Opin Plant Biol 6 178–184 [DOI] [PubMed] [Google Scholar]
- Kumar R, Wallis JG, Skidmore C, Browse J (2006) A mutation in Arabidopsis cytochrome b5 reductase identified by high-throughput screening differentially affects hydroxylation and desaturation. Plant J 48 920–932 [DOI] [PubMed] [Google Scholar]
- Lu C, Fulda M, Wallis JG, Browse J (2006) A high-throughput screen for genes from castor that boost hydroxy fatty acid accumulation in seed oils of transgenic Arabidopsis. Plant J 45 847–856 [DOI] [PubMed] [Google Scholar]
- Mantle PG, Nisbet LJ (1976) Differentiation of Claviceps purpurea in axenic culture. J Gen Microbiol 93 321–334 [DOI] [PubMed] [Google Scholar]
- McKeon TA, Lin JT, Stafford AE (1999) Biosynthesis of ricinoleate in castor oil. Adv Exp Med Biol 464 37–47 [DOI] [PubMed] [Google Scholar]
- Meesapyodsuk D, Reed DW, Covello PS, Qiu X (2007) Primary structure, regioselectivity, and evolution of the membrane-bound fatty acid desaturases of Claviceps purpurea. J Biol Chem 282 20191–20199 [DOI] [PubMed] [Google Scholar]
- Meesapyodsuk D, Reed DW, Savile CK, Buist PH, Ambrose SJ, Covello PS (2000) Characterization of the regiochemistry and cryptoregiochemistry of a Caenorhabditis elegans fatty acid desaturase (FAT-1) expressed in Saccharomyces cerevisiae. Biochemistry 39 11948–11954 [DOI] [PubMed] [Google Scholar]
- Moreau RA, Stumpf PK (1981) Recent studies of the enzymic synthesis of ricinoleic acid by developing castor beans. Plant Physiol 67 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris LJ (1970) Mechanisms and stereochemistry in fatty acid metabolism. Biochem J 118 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris LJ, Hall SW (1966) The structure of the glycerides of ergot oils. Lipids 1 188–196 [DOI] [PubMed] [Google Scholar]
- Morris LJ, Hall SW, James AT (1966) The biosynthesis of ricinoleic acid by Claviceps purpurea. Biochem J 100 29C–30C [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier JA (2007) The production of unusual fatty acids in transgenic plants. Annu Rev Plant Biol 58 295–319 [DOI] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J (1994) Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12 357–358 [DOI] [PubMed] [Google Scholar]
- Reed DW, Schafer UA, Covello PS (2000) Characterization of the Brassica napus extraplastidial linoleate desaturase by expression in Saccharomyces cerevisiae. Plant Physiol 122 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer J, Tudzynski P (2006) In vitro pathogenicity assay for the ergot fungus Claviceps purpurea. Mycol Res 110 465–470 [DOI] [PubMed] [Google Scholar]
- Shanklin J, Cahoon EB (1998) Desaturation and related modifications of fatty acids. Annu Rev Plant Physiol Plant Mol Biol 49 611–641 [DOI] [PubMed] [Google Scholar]
- Smith MA, Moon H, Chowrira G, Kunst L (2003) Heterologous expression of a fatty acid hydroxylase gene in developing seeds of Arabidopsis. Planta 217 507–516 [DOI] [PubMed] [Google Scholar]
- Sperling P, Ternes P, Zank TK, Heinz E (2003) The evolution of desaturases. Prostaglandins Leukot Essent Fatty Acids 68 73–95 [DOI] [PubMed] [Google Scholar]
- Thomæus S, Carlsson AS, Stymne S (2001) Distribution of fatty acids in polar and neutral lipids during seed development in Arabidopsis genetically engineered to produce acetylenic, epoxy and hydroxy fatty acids. Plant Sci 161 997–1003 [Google Scholar]
- Tsitsigiannis DI, Kowieski TM, Zarnowski R, Keller NP (2004) Endogenous lipogenic regulators of spore balance in Aspergillus nidulans. Eukaryot Cell 3 1398–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis DI, Kowieski TM, Zarnowski R, Keller NP (2005) Three putative oxylipin biosynthetic genes integrate sexual and asexual development in Aspergillus nidulans. Microbiology 151 1809–1821 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57 503–516 [DOI] [PubMed] [Google Scholar]
- van de Loo FJ, Broun P, Turner S, Somerville C (1995) An oleate 12-hydroxylase from Ricinus communis L. is a fatty acyl desaturase homolog. Proc Natl Acad Sci USA 92 6743–6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker T, Kinney AJ (2001) Variations in the biosynthesis of seed-storage lipids. Annu Rev Plant Physiol Plant Mol Biol 52 335–361 [DOI] [PubMed] [Google Scholar]
- Wu G, Truksa M, Datla N, Vrinten P, Bauer J, Zank T, Cirpus P, Heinz E, Qiu X (2005) Stepwise engineering to produce high yields of very long-chain polyunsaturated fatty acids in plants. Nat Biotechnol 23 1013–1017 [DOI] [PubMed] [Google Scholar]