Abstract
Urea is the major nitrogen (N) form supplied as fertilizer in agriculture, but it is also an important N metabolite in plants. Urea transport and assimilation were investigated in Arabidopsis (Arabidopsis thaliana). Uptake studies using 15N-labeled urea demonstrated the capacity of Arabidopsis to absorb urea and that the urea uptake was regulated by the initial N status of the plants. Urea uptake was stimulated by urea but was reduced by the presence of ammonium nitrate in the growth medium. N deficiency in plants did not affect urea uptake. Urea exerted a repressive effect on nitrate influx, whereas urea enhanced ammonium uptake. The use of [15N]urea and [15N]ammonium tracers allowed us to show that urea and ammonium assimilation pathways were similar. Finally, urea uptake was less efficient than nitrate uptake, and urea grown-plants presented signs of N starvation. We also report the first analysis, to our knowledge, of Arabidopsis gene expression profiling in response to urea. Our transcriptomic approach revealed that nitrate and ammonium transporters were transcriptionally regulated by urea as well as key enzymes of the glutamine synthetase-glutamate synthase pathway. AtDUR3, a high-affinity urea transporter in Arabidopsis, was strongly up-regulated by urea. Moreover, our transcriptomic data suggest that other genes are also involved in urea influx.
Urea [CO(NH2)2] is the major nitrogen (N) form supplied as fertilizer in agricultural plant production, ahead of nitrate and ammonium (http://www.fertilizer.org/ifa/statistics/STATSIND/). Urea in soils is hydrolyzed by urease, a nickel-dependent enzyme produced by soil microorganisms (Watson et al., 1994), into ammonium (NH4+), with often a concurrent nitrification to nitrate (NO3−). Thus, fertilization with urea may result in plant roots being simultaneously exposed to urea, ammonium, and nitrate, at least for short periods of time.
The uptake of urea in plant roots and leaves before hydrolysis into carbon dioxide and ammonium has been observed in several independent experiments (Hine and Spent, 1988; Krogmeier et al., 1989; Gerendás et al., 1998). Nevertheless, studies in higher plants have never identified the N form that actually passes the cell membrane. In algae, the use of 14C-labeled urea demonstrated the uptake of urea molecules by two transport systems operating at either high or low urea concentrations (high- or low-affinity urea transport systems; Wilson et al., 1988). These transport systems are clearly different from passive uptake and diffusion-like transport mechanisms and are mediated by probable transmembrane proteins. The molecular basis of urea transport is just beginning to be understood in Arabidopsis (Arabidopsis thaliana). The identification of the high-affinity urea transporter AtDUR3 by Liu et al. (2003a) and of the AtTIP (for tonoplast intrinsic protein) urea permeases (Liu et al., 2003b) led to new insights regarding the molecular basis of urea uptake in plants. Not only members of the TIP subfamily but also PIP-like (for plasma membrane intrinsic protein-like) and NIP-like (for NOD-like intrinsic protein-like) aquaporins may permeate urea (for review, see Kojima et al., 2006). In the same manner, in mammals, four of 11 aquaporins transport urea in addition to water (King et al., 2004). The AtDUR3 gene is predicted to encode an integral membrane protein with 14 transmembrane-spanning domains and to belong to the superfamily of sodium solute symporters (Liu et al., 2003a). Complementation experiments of a urea uptake-defective yeast mutant showed that AtDUR3 mediates high-affinity H+ urea symport. The regulation of AtDUR3 expression is tissue specific and linked to the N status in Arabidopsis (Liu et al., 2003a). This gene is expressed at low levels in shoots and up-regulated during early germination and under N deficiency in roots. It was proposed by Liu et al. (2003a) that AtDUR3 plays a role in urea uptake by plant cells at low external urea concentrations. This hypothesis was recently confirmed, as two T-DNA lines with insertions in the AtDUR3 gene showed impaired growth on urea as the sole N source. Located on the plasma membrane, AtDUR3 has been shown to be the major transporter for high-affinity urea uptake in Arabidopsis roots (Kojima et al., 2007).
Little is known about the interactions between urea and ammonium or nitrate uptake and assimilation. Such interactions have been suggested, since it has been shown that the initial N status of plants and/or the presence of other N metabolites alters the uptake and reduction of nitrate (Crawford and Glass, 1998; Daniel-Vedele et al., 1998; Lejay et al., 1999; Zhuo et al., 1999; Orsel et al., 2002). It is thought that these interactions are mainly operating on transcriptional regulations. Indeed, expression of NRT2.1, the major component of the high-affinity nitrate transport system, is induced by nitrate and repressed by high N status through a negative feedback mechanism, probably mediated by ammonium or amino acids (Filleur and Daniel-Vedele, 1999; Nazoa et al., 2003). It has long been known that the presence of ammonium inhibits nitrate uptake and assimilation (Lee and Drew, 1989; Bloom and Sukrapanna, 1990). Moreover, posttranscriptional control by ammonium supply was also suggested by Fraisier et al. (2000), and the regulation of root nitrate uptake in response to light or N treatment also occurs at the AtNRT2.1 protein level (Wirth et al., 2007). AMT genes, involved in ammonium uptake into plant cells, exhibit a variety of expression patterns in response to N deprivation or after ammonium resupply (Loqué and von Wirén, 2004). In Arabidopsis, four AMT genes expressed specifically in roots are up-regulated to different extents by N starvation (Gazzarrini et al., 1999; Sohlenkamp et al., 2000). Interactions between urea and nitrate or ammonium uptake were also reported by Criddle et al. (1988) and Bradley et al. (1989). In wheat (Triticum aestivum) seedlings, urea was shown to inhibit ammonium and nitrate uptake, while nitrate induced urea absorption. In contrast, addition of ammonium to nitrate not only eliminated the nitrate-mediated enhancement of urea uptake, but ammonium alone was shown to directly inhibit urea influx.
Besides being the predominant N form used for fertilizer application to crops plants, urea is also an important N metabolite in plants. Two major biochemical pathways, Arg degradation and ureide catabolism, together with root and leaf applications of urea, are the three major sources of urea in plants. Since urea accumulates in source leaves of older plants and in germinating seeds (Zonia et al., 1995), the first pathway is supposed to be utilized during N recycling after protein degradation. Urea is synthesized inside mitochondria during Arg degradation via arginase in the Orn or urea cycle (Polacco and Holland, 1993). Arginase-derived urea is then exported to cytoplasm and hydrolyzed by urease. Urea-derived ammonium is assimilated into Gln via Gln synthetase (GS); then, Glu synthase (GOGAT) catalyzes the formation of Glu via the GS-GOGAT cycle. In the second pathway, urea is produced by the catabolism of purines or ureides like allantoin and allantoate, which are used by leguminous species for long-distance translocation of N (Stebbins and Polacco, 1995).
The objectives of the work reported here were (1) to characterize urea uptake and assimilation by Arabidopsis seedlings by following N metabolites of seedlings after exposure to different N fertilization regimes; (2) to examine possible interactions between urea, ammonium, and nitrate on N transport systems by 15N influx measurements; and (3) to better understand the molecular basis for urea uptake and assimilation mechanisms using a transcriptomic approach.
RESULTS
Growth and N Status
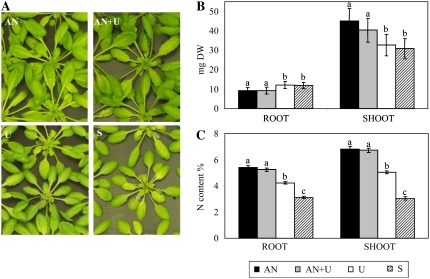
We previously demonstrated that in our hydroponic culture conditions, urea is not hydrolyzed in the nutrient solution (Merigout et al., 2008). In order to characterize the effect of urea on Arabidopsis growth and metabolism, plants were grown in a nutrient solution containing nitrate and ammonium as N sources, and after 5 weeks they were transferred for 1 week in nutrient solutions containing ammonium nitrate (AN), ammonium nitrate + urea (AN+U), or urea alone (U) as N sources or on a solution containing no N (starvation; S). Two independent experiments were performed, and the phenotypes of the plants in one of the experiments are shown in Figure 1A.
Figure 1.
Distribution of plant biomass and N content. Plants were cultured hydroponically for 5 weeks with a continuous supply of 0.5 mm ammonium nitrate. Arabidopsis plants were then transferred for 1 week to different nutrient conditions in medium containing 1 mm NH4NO3 (AN), 0.5 mm NH4NO3 and 0.5 mm CO(NH2)2 (AN+U), 1 mm CO(NH2)2 (U), or no N (S). A, Plant aerial parts. B, Root and shoot dry weights (DW). The values are means ± sd of 21 replicates. C, Root and shoot N contents (w/w), measured on six replicates. Different letters indicate significant differences between nutrient regimes at P < 0.05.
The analysis of the accumulated dry biomass revealed two groups: the first one corresponded to AN and AN+U treatments, the second to U and S treatments (Fig. 1B). The first group (AN and AN+U) was characterized by a slightly lower root fresh weight and a higher shoot fresh weight (mean 25% increase) than the second group (U and S; Fig. 1B).
This difference in plant growth might reflect differences in N assimilation or uptake related to the N regime. We thus measured the total N content in the two groups of plants. We observed a lower root and shoot N content in the plants grown with urea as the sole N source (Fig. 1C) compared with AN- and AN+U-grown plants, which showed no difference in N content. As expected, this decrease was even more pronounced in N-starved plants (Fig. 1C).
Biomass accumulation of Arabidopsis plants on the different N regimes seemed to be correlated with total N content. In order to better characterize the plant N status, we then measured nitrate, ammonium, urea, and amino acid contents.
N-Related Metabolites
The addition of urea in AN nutrient solution did not significantly affect the NO3− content of plants (Table I). In contrast, in plants fed for 1 week with either urea as the sole N source or without any N source, NO3− almost disappeared from roots and only a small amount of NO3− was still present in shoots. In both treatments, chloride contents increased while sulfate and phosphate contents remain stable (Supplemental Fig. S1). These results show that the duration of the treatment was sufficient to provoke changes in the plant N status. Plants grown on the ureic nutrition contained the highest free NH4+ concentration in both roots and shoots, whereas the N-starved plants had low amounts of NH4+. The two other treatments (AN and AN+U) revealed comparable and intermediate amounts of NH4+ (Table I). With the exception of N-starved plants, in which, as described previously (Kojima et al., 2007), urea concentration was the lowest, analysis of the urea content in roots did not reveal statistically significant differences among the N treatments, even if urea was added to ammonium nitrate. Thus, no dramatic changes in N-related metabolite contents were observed when plants were grown on the different N regimes. We then investigated how N was assimilated into amino acids by plants. The N-starved plants had low concentrations of total soluble amino acid compared with the other nutrient conditions. While addition of urea to ammonium nitrate-containing medium (AN+U) had no effect on the plants' amino acid content compared with AN, plants grown only on urea as the sole N source for 1 week showed a significant increases in amino acid concentration, particularly in roots, where the level was more than twice that of AN and AN+U plants (Table I). The protein content in roots and shoots was lower only in starved plants, but these differences were not statistically significant (Table I).
Table I.
Root and shoot N status of 42-d-old Arabidopsis plants 7 d after their transfer from 0.5 mm NH4NO3 to different N media
Nitrate, ammonium, urea, and total amino acid contents are expressed in μmol g−1 fresh weight. Protein contents are given in percentages of the values obtained in roots of AN-grown plants. Values are means ± sd (n = 3). Different letters indicate significant differences between nutrient regimes at P < 0.05.
| Sample | N Medium | NO3− | NH4+ | CO(NH2)2 | Amino Acids | Proteins |
|---|---|---|---|---|---|---|
| Root | AN | 52.79 ± 0.45a | 1.93 ± 0.27a | 0.85 ± 0.26a | 6.85 ± 1.49b | 100 ± 9a |
| AN+U | 44.30 ± 1.20a | 1.85 ± 0.22a | 1.03 ± 0.16a | 6.48 ± 0.98b | 103 ± 26a | |
| U | 0.46b | 2.31 ± 0.31a | 0.89 ± 0.07a | 15.73 ± 0.86a | 85 ± 6a | |
| S | 0.41b | 0.46 ± 0.14b | 0.31 ± 0.11b | 4.20 ± 0.65c | 64 ± 9a | |
| Shoot | AN | 149.00 ± 8.63a | 1.46 ± 0.29a | 0.19 ± 4.10−3,b | 11.05 ± 0.31b | 80 ± 12a |
| AN+U | 135.34 ± 1.26a | 1.74 ± 0.02a | 0.22 ± 3.10−3,b | 10.98 ± 0.18b | 68 ± 9a | |
| U | 11.41 ± 0.44b | 1.90 ± 0.18a | 0.35 ± 0.03a | 16.09 ± 0.79a | 60 ± 4a | |
| S | 3.72 ± 0.22b | 1.12 ± 0.27b | 0.22 ± 0.02b | 7.36 ± 0.06c | 57 ± 8a |
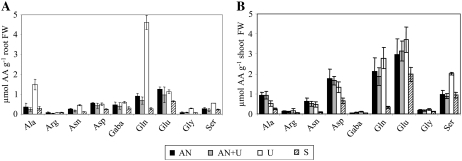
In order to determine which amino acids were responsible for the strong accumulation of total soluble amino acids observed in plants fed with urea as the sole N source, we analyzed the amino acid composition of these plants (Fig. 2). Gln and Ala were 5- and 4-fold more abundant, respectively, in roots of urea-treated plants than in control plants grown on AN solution (Fig. 2A). Similar high Gln and Ala levels were also found in plants fertilized with 1 mm (NH4)2SO4 (data not shown). Gly, Ser, Asn, and γ-aminobutyric acid (GABA) levels were also increased. N-starved plants always had lower concentrations of amino acids, whereas plants grown on AN+U gave similar results to those for AN-grown plants. In the shoots, the concentration of Gln was much higher than in roots, except for N-starved plants, but, unlike in roots, U-grown plants showed only a modest increase in Gln concentration (Fig. 2B). Only Ser levels seemed to augment similarly in roots and shoots when plants were grown on urea as the sole N source and conserved the same ratio between AN and U conditions (Fig. 2B). Mere traces of Orn and citrulline, intermediate products of the urea cycle, could be detected in both plant parts (data not shown).
Figure 2.
Individual amino acids in roots (A) and shoots (B). Amino acid amounts were measured on the same plants as described for Figure 1. The values are means ± sd of three replicates. Different letters indicate significant differences between nutrient regimes at P < 0.05. FW, Fresh weight.
15N Influx and Pattern of 15N Labeling in Amino Acids
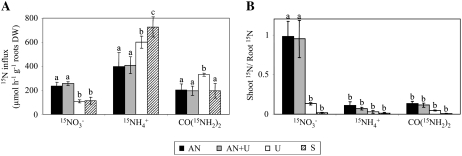
To study urea absorption by root cells, we determined urea influx using 15N-labeled urea. Arabidopsis plants grown on AN, AN+U, U, or S nutrient solutions were incubated with labeled urea for 5 min. N influxes using 15NO3− and 15NH4+ were also measured in order to detect possible effects of urea nutrition on the nitrate and ammonium transport systems. Labeled urea, nitrate, and ammonium were used at high concentrations (10, 6, and 6 mm, respectively) to measure whole uptake processes involving both high- and low-affinity systems.
Urea influx in roots was detected regardless of the N regime of the plants (Fig. 3A). The urea influx reached the highest level when urea was present as the sole N source for 1 week (Fig. 3A). The uptake of nitrate was not affected when urea was used in combination with ammonium nitrate (AN or AN+U conditions) but was reduced in the urea-grown and N-starved plants. Conversely, the influx of ammonium in N-starved plants was 2-fold higher than in AN or AN+U plants. While ammonium and urea seemed to be assimilated directly in roots, nitrate was rapidly (5 min) translocated to the shoots, as the value of the shoot-root 15N ratio was about 1 for the AN and AN+U plants (Fig. 3B). Interestingly, it seems that the presence of nitrate is required during the growth period to induce a high level of nitrate transport to the aerial parts. Indeed, very little labeled nitrate was found in shoots of urea-grown plants and even less in N-starved leaves at the end of the 5-min labeling period (Fig. 3B).
Figure 3.
N influx in Arabidopsis. Root 15N influx was measured after 5 min of labeling with different nutrient solutions containing 6 mm 15NO3−, 10 mm CO(15NH2)2, or 10 mm 15NH4+. A, 15NO3−, CO(15NH2)2, and 15NH4+ influxes. B, Shoot-root ratio of total 15N amounts. The values are means ± sd of five replicates. Different letters indicate significant differences between nutrient regimes at P < 0.05.
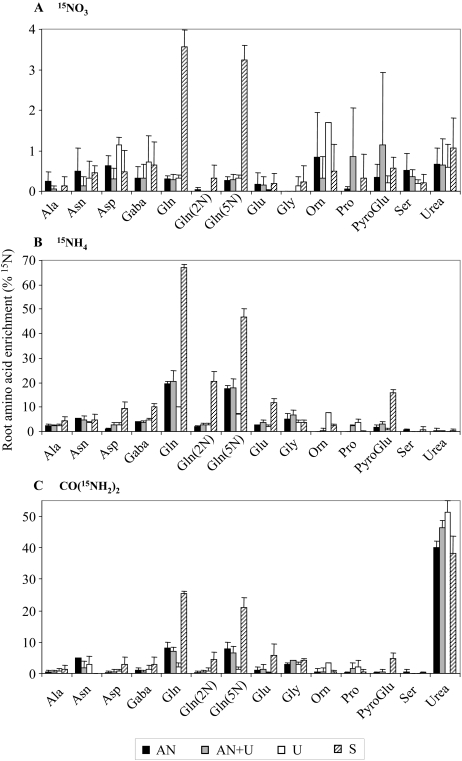
Incorporation of 15N into amino acid in roots was followed by gas chromatography coupled to mass spectrometry (GC-MS) after 5 min of labeling. Feeding the plants with 15NO3 resulted in a very low proportion of labeled amino acids (less than 2%), with a peak for the amide [Gln(5N)] group of Gln after N starvation (Fig. 4A). The low 15N enrichment in 15NO3−-fed plants can be explained by (1) an ineffective reduction of nitrate to ammonium in roots in the 5-min labeling time, (2) a storage of nitrate in root vacuoles, or (3) an export of nitrate in aerial parts via the xylem vessels, as shown in Figure 3B. Conversely, when plants were labeled using 15NH4 or CO(15NH2), the relative level of labeled amino acids was higher, especially for Gln (Fig. 4, B and C). Indeed, around 70% and 25% of the Gln pool in N-starved plants was labeled in 15NH4+- and CO(15NH2)2-fed plants, respectively. This suggests that the Gln pool in roots of N-starved plants is mainly synthesized during the 5-min labeling period and that N is quickly transferred from 15NH4+ or CO(15NH2)2 to Gln. Since urea first needs to be hydrolyzed into 15NH4,+ the rate of Gln synthesis by the GS enzyme may be lower in urea-labeled plants. The ratio of labeled Gln is significantly reduced in roots of plants grown in the presence of nitrate and ammonium and even lower in urea-grown plants when fed with 15NH4+ or CO(15NH2)2. This could be due to a higher Gln concentration at the start of the 15N labeling period, particularly in roots of urea-grown plants (Fig. 2A).
Figure 4.
15N incorporation into amino acids in roots. 15N labeling in amide and amino acid N was determined by GC-MS analyzes on the same plant material as described for Figure 3. A, 15NO3 incorporation. B, 15NH4 incorporation. C, CO(15NH2)2 incorporation. The values are means ± sd of three replicates.
Concerning the CO(15NH2)2 assimilation, the highest 15N labeling was detected as [15N]urea regardless of the N nutrition. This result suggests that most of the urea taken up was not metabolized in roots. The labeled urea pool reached 50% of total 15N in urea-grown plants but was high regardless of the nutrition regime (Fig. 4C). This result was consistent with the measurements of urea influx. Labeled urea was detected in aerial organs (data not shown), suggesting that at least part of the [15N]urea absorbed by roots was transferred to shoots before its hydrolysis and assimilation (see above; Table I). [5-15N]Gln and then [2-15N]Gln were the second and third major sinks, respectively, for 15N from urea. This shows that urea molecules can be taken up and quickly assimilated and/or translocated by Arabidopsis roots. Finally, there was a low incorporation of 15N from urea or ammonium in Gly, Glu, pyro-Glu, Asn, GABA, Asp, and Ala (Fig. 4, B and C). These labeling patterns correspond to those seen in other labeling experiments (Masclaux-Daubresse et al., 2006).
Microarray Analysis
As demonstrated above, the N treatments led to major changes in plant physiology at the levels of both N source uptake and enzymatic activities related to N assimilation. These changes might be linked to differences in gene expression, although global analysis could also reveal other urea-regulated processes. To determine the genomic response of Arabidopsis roots and shoots to urea N nutrition, transcriptome profiling was performed on 42-d-old U- versus AN-grown seedlings with CATMA arrays carrying gene-specific tags (GSTs; Crowe et al., 2003; Hilson et al., 2004). Another comparison consisted of AN+U- versus AN-grown plantlets. Two biological replicates were performed for each comparison with, for each replicate, two reverse-labeling technical replicates. Only genes that showed statistically significant signals and signal ratios in both biological replicates were considered. Genes with differential expression were selected by statistical analysis using the Bonferroni P value threshold of 0.05, as described in “Materials and Methods.” Chloroplastic and mitochondrial probes were omitted.
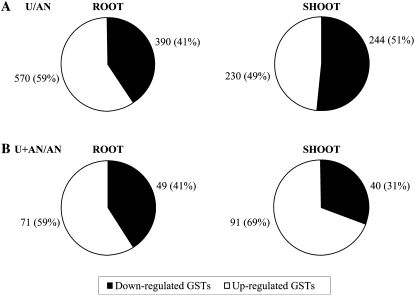
Based on the statistical analysis, 960 (about 4% of total GSTs) and 474 (about 2% of total GSTs) genes were found differentially expressed between U- and AN-grown plants, respectively, in root and shoot parts. Fewer genes were differentially expressed for the comparison between AN+U and AN: 120 (about 0.5% of total GSTs) and 131 in roots and shoots, respectively. These genes are listed in the supplemental data (Supplemental Tables S1, S2, S4, and S5). Figure 5 shows the proportion of up- and down-regulated genes for each comparison. The ratios between up- and down-regulated genes were identical in roots for both comparisons. In contrast, more genes were down-regulated in shoots when ammonium nitrate was mixed with urea (Fig. 5).
Figure 5.
Differential distribution of induced and repressed genes in Arabidopsis according to N nutrition. A statistical cutoff (P < 0.05 after Bonferroni correction) was used to determine which genes were differentially expressed in comparison with the reference NH4NO3 nutrition. Number and percentage (in parentheses) of differentially expressed genes are indicated. A, CO(NH2)2 differentially expressed genes in root and shoot. B, NH4NO3 + CO(NH2)2 differentially expressed genes in root and shoot.
The lists of the urea differentially regulated root and shoot genes are available as Supplemental Tables S1 and S2, respectively. Likewise, the lists of the AN+U differentially regulated root and shoot genes are available as Supplemental Tables S4 and S5, respectively. Genes differentially expressed were classified in functional categories according to the MapMan tool (Thimm et al., 2003; Supplemental Tables S3 and S6).
Urea versus Ammonium Nitrate
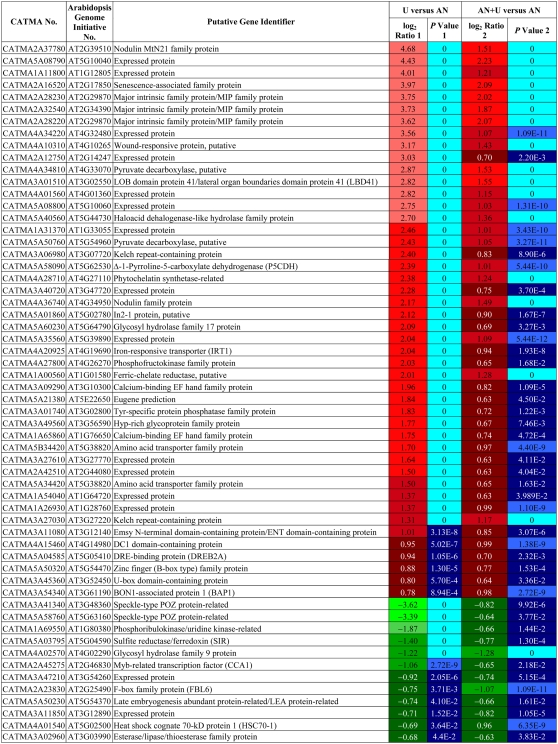
In roots, the functional group with the highest number of genes differentially expressed (122) concerned transcription. Genes involved in RNA machinery were more induced (62%) than repressed (38%). Among these induced genes, those encoding zinc finger family proteins were well represented. The second most represented category was transport, and a similar number of genes involved in transport were induced and repressed. This category encompassed the most induced genes in response to urea as the unique source of N, like At5g45380 (AtDUR3; a high-affinity urea/H+ symporter; Liu et al., 2003a), At3g24290 (a putative ammonium transporter), At1g12940 (AtNrt2.5; a high-affinity nitrate transporter), and aquaporins (MIP family). The high-affinity nitrate transporter NRT2.4 (At5g60770) and the high-affinity ammonium transporters AMT2 (At2g38290), AMT1.3 (At3g24300), and AMT1.1 (At4g13510) were also induced, as well as several amino acids and sugar transporters. Concerning the repressed genes involved in transport, we noted sulfate transport genes, aquaporins (PIP, MIP, and TIP), the chloride channel protein CLC-a (At5g40890; recently demonstrated to act as a vacuolar nitrate transporter; De Angeli et al., 2006), the low-affinity nitrate/chlorate transporter NRT1.1 (CHL1; At1g12110), and six GSTs of the proton-dependent oligopeptide transport protein family. The next gene categories that were differentially expressed are, in decreasing order, protein machinery, stress, hormone metabolism, amino acid metabolism, development, and signaling. In the stress and signaling classes, most of the genes were up-regulated by urea.
In shoots, the most represented category, the RNA machinery (58 genes), was the same as that for roots. However, a similar number of genes were up- and down-regulated. With a difference of only 10 genes, protein (synthesis and degradation) was the second functional group, and more genes of this class were induced than repressed. Other genes found to respond to urea belonged to hormone metabolism, amino acid metabolism, transport, and stress. Ninety-one percent of genes of the amino acid metabolism class were up-regulated. For hormone metabolism, stress, and transport (aquaporins), the majority of genes were repressed.
Ammonium Nitrate + Urea versus Ammonium Nitrate
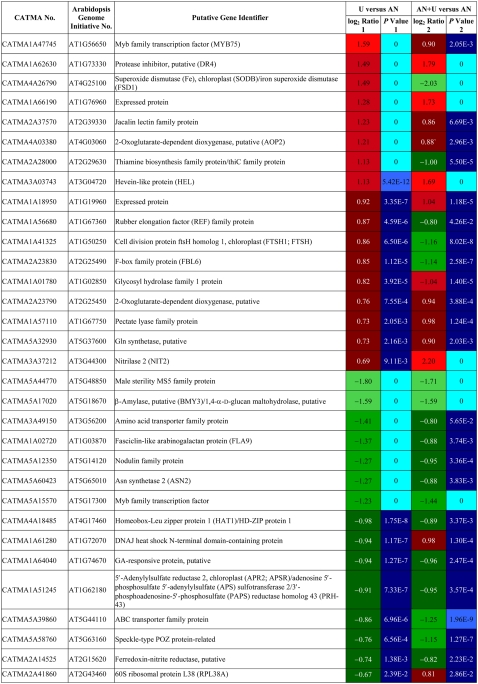
Transcription, protein machinery, and transport were the most representative categories of genes differentially expressed, irrespective of the organ. Among transport genes, we recognized some amino acid transporters induced in roots and shoots as well as the same two MIP family genes induced in roots of urea-fed plants. Surprisingly, the high-affinity AtNRT2.1 nitrate transporter (At3g45060) was up-regulated in shoots, while it is mainly expressed in roots (Orsel et al., 2002). A permease of the purine and allantoin pathway, a source of urea in plants, was repressed in shoots.
Specific Genes Transcriptionally Regulated by Urea
The comparison of results for the AN+U and U conditions with the AN reference allowed us to distinguish genes for which transcription was specifically regulated by urea. We found 58 and 32 common genes differentially regulated in roots and shoots, respectively, between the AN+U and U groups compared with the reference AN group. These genes are listed in Tables II and III. The patterns of regulation of gene expression in root for U and AN+U-grown plants were perfectly identical (Table II). This was less true for shoots (Table III), but globally, the regulation profiles were similar for the majority of the genes. In roots, 46 of the common genes were induced, and only 12 were repressed. These 58 genes were mainly distributed between transcription and transport categories. Two MIPs, At2g29870 and At2g34390, were included in the 46 induced genes (Table II). In shoots, 17 and 15 genes were up- and down-regulated, respectively. We found genes implicated in N and amino acid metabolism. While the transcription of a putative GS (At5g37600) was induced, genes coding for nitrite reductase (At2g15620) and for the Asn synthetase ASN2 (At5g65010) were negatively regulated (Table III).
Table II.
List of the genes differentially regulated in roots that are common between the urea and ammonium nitrate + urea nutrient regimes
CATMA numbers are indicated, together with Arabidopsis Genome Initiative numbers. The putative function of the gene is indicated according to The Institute for Genomic Research Arabidopsis gene index (release 5, January 2004). Positive ratio indicates that the gene is specifically overexpressed in exposure to urea (red boxes); negative ratio indicates that the gene is specifically repressed in exposure to urea (green boxes). P values are Bonferroni corrected. Genes were considered differentially regulated at P < 0.05.
Table III.
List of the genes differentially regulated in shoots that are common between urea and ammonium nitrate + urea nutrient regimes
CATMA numbers are indicated, together with Arabidopsis Genome Initiative numbers. The putative function of the gene is indicated according to The Institute for Genomic Research Arabidopsis gene index (release 5, January 2004). Positive ratio indicates that the gene is specifically overexpressed in exposure to urea (red boxes); negative ratio indicates that the gene is specifically repressed in exposure to urea (green boxes). P values are Bonferroni corrected. Genes were considered differentially regulated at P < 0.05.
DISCUSSION
Arabidopsis Plants Absorb Urea as an External N Source
To our knowledge, this study is the first to associate physiological and molecular analyses of Arabidopsis plants treated with different N supplies including urea. Hydroponic culture conditions were chosen in order to circumvent any conversion of the N forms supplied during the course of the experiments. Indeed, our results demonstrate that, under our conditions, urea is directly taken up by root cells prior to hydrolysis.
Our study on CO(15NH2)2 influx, measured after 5 min, provides direct evidence that (1) urea is able to pass through the root cell membrane as an intact molecule regardless of the initial N status of the plants, (2) this uptake is stimulated when plants are grown on urea as the sole N source compared with AN, and (3) this uptake is inhibited by ammonium or nitrate (Fig. 3A). The concentration of urea in influx experiments (10 mm) was five times higher than that in the hydroponic urea treatment (2 mm). At this high concentration, a combination of high- and low-affinity urea transport systems could operate. At least four Arabidopsis genes coding for TIPs are able to complement a Saccharomyces cerevisiae mutant unable to use urea as the sole N source. Among them, AtTIP2.1 expression in Xenopus laevis oocytes also promotes urea accumulation even when urea concentrations are raised to 30 mm, suggesting a putative role as a low-affinity transporter (Liu et al., 2003b). Furthermore, recent work using T-DNA insertion mutants demonstrated that AtDUR3 does indeed represent the major transporter for high-affinity urea uptake in Arabidopsis (Kojima et al., 2007). These genes were significantly expressed in roots of urea-treated plants (Supplemental Table S1) and could have been involved in the urea uptake process. Nutrient uptake processes and their molecular components are often regulated in response to two major parameters, nutrient availability and plant nutritional status (Buchner et al., 2001, 2004; Glass et al., 2002). Accordingly, urea uptake indeed seems to be induced by its own substrate, but only in the absence of other N sources (Fig. 3A). The role of AtDUR3 in this process is further reinforced by the correlation between the increase of urea uptake and AtDUR3 relative expression levels in plants treated with urea versus ammonium nitrate (Fig. 3A; Supplemental Table S1). Such a correlation between the expression of genes coding for transporters and real uptake is not always the case. For example, this correlation did not occur when plants were starved for N, as no increase in urea influx was observed in starvation conditions, whereas AtDUR3 was already shown to be up-regulated by N starvation (Liu et al., 2003a). The facilitated urea TIP transporters AtTIP1.1, AtTIP1.2, AtTIP2.1, and AtTIP4.1 (Liu et al., 2003b) were also not differentially expressed under urea nutrition in our transcriptome study. Surprisingly, when ammonium nitrate was present together with urea in the culture medium, urea influx was no longer induced and AtDUR3 expression was not induced (Supplemental Tables S4 and S5). These results suggest that ammonium nitrate could inhibit the transcription of AtDUR3, leading to a reduced urea influx.
The detection of labeled urea in roots after urea influx using mass spectrometry (Fig. 4C) provides the definitive proof of intact urea uptake by root cells. Urea uptake via the AtDUR3 high-affinity transporter was already described in Arabidopsis (Liu et al., 2003a; Kojima et al., 2007), but to our knowledge, no 15N-enriched urea tracing experiments in intact hydroponically grown plants have yet been reported.
Urea Assimilation in Arabidopsis Plants
The stable protein content and the much higher levels of ammonium and total amino acids in roots after exposure to the urea solution compared with other N treatments (Table I) indicate efficient assimilation of urea N in roots. We assume, therefore, that urea, like ammonium but in contrast to nitrate (Loqué and von Wirén, 2004), was principally metabolized in the roots. However, the increased urea content in the shoots of these plants (Table I), the 15N shoot-root ratio (Fig. 3B), as well as the presence of 15N-labeled urea in shoots (data not shown) suggest that a substantial part (20%) of urea absorbed by the roots was translocated to the aerial parts before its hydrolysis by root cytoplasmic ureases. Large amounts of urea were also found in rice (Oryza sativa) shoots grown with urea (Gerendás et al., 1998), suggesting a transport of urea molecules from the roots to the aerial parts of the plant.
In order to dissect the assimilation pathways of urea, we examined the individual amino acids in the different N-treated plants (Fig. 2), and we followed the route of 15N-radiolabeled urea by GC-MS in comparison with 15N from enriched nitrate and ammonium (Fig. 4). After 1 week of exposure to the urea solution, plant roots contained high levels of Gln and Ala. This suggests that in the root tissue, urea is first degraded by cytosolic ureases and then ammonium is incorporated via the GS-GOGAT cycle (Miflin and Lea, 1980). Thus, it is conceivable that urea and its ammonium product were utilized in the same manner as the ammonium derived from other processes, photorespiration, or nitrate assimilation (Fig. 6). This hypothesis is strengthened further by the fact that ammonium treatment during 7 d led to the same growth and metabolic characteristics, apart from a more pronounced accumulation of amino acids, than urea treatment (data not shown). This contrasts with previous reports from maize (Zea mays) and wheat, in which urea was assimilated directly via the reversal of the Orn cycle or another mechanism (van Beusichem et al., 1982; Bradley et al., 1989). This assimilation pathway did not occur in our study, as only traces of Orn or citrulline were detected in our chromatography analysis. Moreover, our 15N-labeling study confirms that urea and ammonium shared the same assimilatory pathway in Arabidopsis roots (Fig. 4, B and C). Indeed, 15N from ammonium or urea presented similar 15N-labeling patterns. 15N labeling was detected as [5-15N]Gln, then as [2-15N]Gln, [2-15N]Glu, and other amino acids such as GABA, Asp, Asn, and Ala, and this labeling seemed to take place in only 5 min (time course of the experiment). These labeling rates correlate with the high efficiency of GS activity. [15N]Ammonium and [15N]ammonium derived from 15N-enriched urea were rapidly incorporated into [5-15N]Gln by GS. Then, ferredoxin- and/or NADH-GOGAT transferred 5-15N of Gln to [2-15N]Glu. As soon as it is synthesized by GOGAT, [2-15N]Glu can be used by GS to form [2,5-15N]Gln. The Asp aminotransferase could then catalyze the transfer of the 15N amino group of Glu to oxaloacetate, yielding [2-15N]Asp. [2,4-15N]Asn was synthesized from [15N]ammonium or [2,5-15N]Gln and [2-15N]Asp by Asn synthetase (Masclaux-Daubresse et al., 2006).
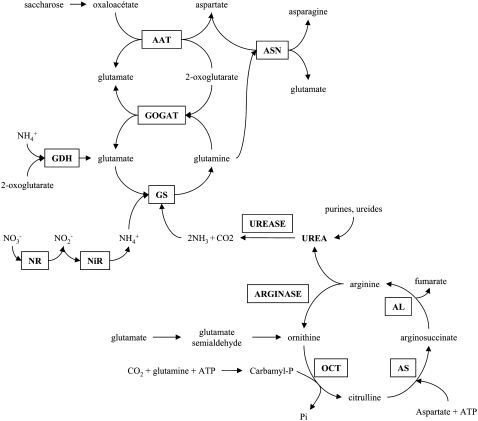
Figure 6.
Proposed urea assimilatory pathway in Arabidopsis. Urea is synthesized inside mitochondria during Arg degradation via arginase in the Orn or urea cycle. Arginase-derived urea is then exported to cytoplasm and hydrolyzed by urease. Urea-derived ammonium is assimilated into Gln via GS; then, GOGAT catalyzes the formation of Glu via the GS-GOGAT cycle. AAT, Asp aminotransferase; AL, arginosuccinate lyase; AS, arginosuccinate synthetase; ASN, Asn synthetase; GDH, Glu dehydrogenase; NiR, nitrite reductase; NR, nitrate reductase; OCT, Orn carbamyl transferase.
Some genes of the GS-GOGAT cycle were differentially expressed in plants grown with urea (Supplemental Tables S1 and S2), and their regulation correlates with the proposed pathway for urea N assimilation (see above). At1g66200, At5g37600 (GS1 putative), and At2g41220 (GLU2) were up-regulated by urea nutrition. GDH1 (At5g18170) was negatively regulated by urea treatment. Some genes involved in primary ammonium assimilation into amino acids were also regulated by the N status of plants. The expression levels of Asp aminotransferase 2 (At5g19550), Ala aminotransferase (At1g17290), as well as Asn synthetase 1 (At3g47340; ASN1) were increased with urea. These increases were clearly consistent with the increase of Asp, Ala, and Asn in roots supplied with urea. In contrast to ASN1, ASN2 (At5g65010) was down-regulated by urea, but it is already know that ASN1 and ASN2 are regulated in a reciprocal fashion by carbon or metabolites like amino acids (Lam et al., 1998), which is in agreement with our transcriptome results. As far as the enzymes of the urea cycle were concerned, no transcriptional regulation by urea was observed for urease, arginase, Orn carbamyl transferase, arginosuccinate synthetase, or arginosuccinate lyase. Only a urease accessory protein, UREG (At2g34470), was up-regulated by urea in roots of urea-grown plants. AtUREG, along with AtURED and AtUREF, has been demonstrated to be required for urease activation in Arabidopsis (Witte et al., 2005).
Effects of Urea Nutrition on Different N Fluxes
Both N-minus treatment and urea as the sole N source stimulated ammonium uptake. The gene AMT1.1 makes a major contribution to this process in roots (Loqué and von Wirén, 2004). Up-regulation by N starvation of ammonium HATS activity and AtAMT1 transcript levels, correlated with the increase of influx in roots, was reported previously (Gazzarrini et al., 1999; Rawat et al., 1999). Nevertheless, these treatments inhibited nitrate transport, both the uptake of nitrate by root cells and the nitrate translocation within the whole plant between roots and shoots. Nitrate HATS and LATS are mediated primarily by the genes AtNRT2.1 and AtNRT1.1, respectively (Okamoto et al., 2003). Lejay et al. (1999) investigated the response of both NRT2.1 and NRT1 to changes in the N status of Arabidopsis. After a transient increase of the NRT2.1 transcript levels, NRT2.1, NRT1.1 transcripts, and the HATS and LATS activities decreased very rapidly after N starvation. Our results concerning the effect of N starvation on nitrate uptake are consistent with this latter observation. The repressive effect of urea as the sole N source on nitrate influx could be explained by a direct inhibition, but also by the high Gln content in root cells of urea-grown plants, as it is already known that NRT2.1 transcription and nitrate influx at low external nitrate concentrations are under feedback repression by N metabolites resulting from nitrate repression (Lejay et al., 1999; Zhuo et al., 1999). We have shown here that nitrate translocation from roots to shoots is repressed by N starvation, and we can speculate that the gene(s) involved in this process, such as AtNRT1.4 (Chiu et al., 2004), are down-regulated both by N starvation and by the use of urea as the sole N source. However, urea is not a repressive factor for ammonium or nitrate uptake when it is used in combination with ammonium and nitrate. This result is important, as urea, when used as a fertilizer, is often mixed with other N sources.
Urea Use Efficiency
If the supply of urea in conjunction with ammonium nitrate did not affect the growth and the N status of plants, plants treated with urea as the sole N source showed symptoms of N starvation, such as a severe drop of nitrate. After 7 d, nitrate was nearly undetectable in the roots and in very low abundance in the shoots. The root development of the urea-grown plants was also favored at the expense of the shoots, as in plants starved for N (Fig. 1B). Nevertheless, total N contents were statistically different in roots and shoots between urea-grown and N-starved plants (Fig. 1B). Thus, symptoms of N starvation after an exposure to the urea solution seem to reflect a less efficient but still active uptake and/or assimilation of the N in the ureic form compared with ammonium or nitrate by Arabidopsis. Comparable results were previously reported for a number of other species (Kirkby and Mengel, 1967; van Beusichem et al., 1982; Bradley et al., 1989). The limited capacity for urea transport into roots in our culture conditions could result from a too low concentration of urea (2 mm). It would be interesting to monitor the same experiment with higher urea concentrations in the nutrient solutions.
In conclusion, the data presented here demonstrate the capacity of Arabidopsis to absorb and assimilate urea as the sole N source. Urea uptake was regulated by urea itself and ammonium nitrate, but not by N starvation, as is the case for nitrate or ammonium uptake. Urea uptake was less efficient than nitrate uptake, and urea-grown plants presented signs of N starvation. Urea assimilation, however, was similar to the ammonium assimilation pathway. When mixed in equal proportion with ammonium nitrate, urea did not affect plant growth or N uptake, and nitrate content in plants tended to be lower. Thus, fertilization with urea in combination with ammonium nitrate fertilizers represents an alternative to reduce nitrate in crops. A comprehensive physiological and molecular investigation of urea transport and assimilation in planta is required not only to better understand the importance of urea for plants but also to improve its utilization as a root- and leaf-applied N fertilizer in agricultural crop production.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seed stocks of Arabidopsis (Arabidopsis thaliana) from the Columbia ecotype were used for all experiments. Plants were grown under hydroponic culture conditions in a growth chamber with an 8-h-light/16-h-dark cycle at 21°C/17°C, respectively, 80% relative humidity, and 150 μmol m−2 s−1 irradiation. Seeds were sterilized and stratified at 4°C for 5 d. Plant growth in hydroponic conditions was performed as described earlier (Orsel et al., 2004). Plants were supplied with basic nutrient medium supplemented with 0.5 mm NH4NO3. Basic medium contained 1 mm MgSO4, 1 mm KH2PO4, 2.5 mm K2SO4, 2.2 mm CaCl2, 10 μm MnSO4, 24 μm H3BO3, 3 μm ZnSO4, 0.9 μm CuSO4, 0.04 μm (NH4)6Mo7O24, and 10 mg L−1 iron-EDTA (Sequestrene; Ciba-Geigy). Nutrient solution was renewed every 2 d and, during the 2 first weeks, used at half-strength. At the age of 5 weeks, and 7 d before the experiment, the plants were transferred to basic medium in which N was supplied as 1 mm NH4NO3, or 0.5 mm NH4NO3 supplemented with 0.5 mm CO(NH2)2, or only 1 mm CO(NH2)2, each nutrient medium offering 2 mm N. A fourth treatment consisted of complete N starvation. Nutrient solutions were renewed daily during these 7 d to ensure constant N availability and concentration and pH stability. Plants were harvested at 42 d after sowing and analyzed further. Shoots and roots were weighed separately and frozen in liquid N. For each nutritional condition, three plants were pooled and homogenized to a powder for metabolite analysis. Two independent experiments were performed, and results from a representative one are shown. In the second experiment, a fifth source of N in the form of 1 mm (NH4)2SO4 was added.
Analysis of N-Related Metabolites
An aliquot of the powder was weighed (50 mg fresh weight) and extracted with a two-step ethanol-water procedure, and supernatants were analyzed for anion concentration by HPLC on a DX-120 apparatus (Dionex) as described by Loudet et al. (2003). Ethanol-water extracts were also subjected to the determination of free NH4+ by the phenol hypochlorite colorimetric method (Berthelot reaction) using (NH4)2SO4 as a reference.
Free amino acids were determined after extraction of an aliquot of powder (100 mg fresh weight) in 1 mL of a 2% (w/v) solution of sulfosalicylic acid. Supernatants were assayed for total amino acid determination by ninhydrin (Rosen, 1957). Individual amino acids were separated by ion-exchange chromatography using the AminoTac JLC-500/V amino acid analyzer according to the instructions of the manufacturer (JEOL).
For urea extraction (method according to Killingsbaeck, 1975; modified by N. von Wirén, personal communication), 1 mL of 10 mm ice-cold formic acid was added to about 100 mg of ground fresh sample. Each extract was vortexed twice and centrifuged (15 min, 13,200 rpm, 4°C), and the supernatant was transferred in a fresh tube.
For urea measurements, 1 mL of color development reagent was added to 30 μL of the formic acid extract in a microcentrifuge tube. The color development reagent consisted of 1:1:1 mixed acid reagent (20% [v/v] H2SO4 and 0.06% [v/v] 74 mm ferric chloride hexahydrate in 9% [v/v] ortho-phosphoric acid), mixed color reagent (7% [v/v] 0.2 m diacetylmonoxime and 7% [v/v] 0.05 m thiosemicarbazide), and water. Tubes were incubated for 15 min at 99°C. The samples were cooled for 5 min on ice, and the A540 was determined with a spectrophotometer.
15N Labeling Experiment, 15N Influx Studies, and GC-MS Measurements
N influxes were performed by 15N labeling as described by Orsel et al. (2004). Liquid medium for influx studies contained basic N-free medium supplemented with 6 mm 15NO3−, 10 mm 15NH4+, or 10 mm CO(15NH2)2 (atom % 15N, 98%). Influx of 15NO3−, 15NH4+, and CO(15NH2)2 was calculated from the 15N content of the roots and shoots (about 2 mg dry weight). The 15N analyses were performed using an integrated system for continuous flow isotope ratio mass spectrometry (Euro-EA elemental analyzer [EuroVector] and Isoprime mass spectrometer [GV Instruments]). The values are means of five replicates.
Both the evolution of amino acid contents and respective 15N enrichments were examined by GC-MS. Amino acids and ammonium from an aliquot of the root and shoot powder (about 150 mg fresh weight) were extracted with 1 mL of 2% (w/v) sulfosalicylic acid. Extracts were centrifuged at 17,500g for 20 min to eliminate cellular debris. To purify amino acids, extracts were applied to a column (AG 50W-X8 resin, 100–200 mesh, H+ form, 5 × 0.5 cm; Bio-Rad Laboratories), washed with 2 mL of water, and eluted with 2.5 mL of 6 m NH4OH. The amino acid fraction was frozen, evaporated, and rinsed twice with 500 μL of water, then evaporated. Amino acid samples were resuspended in 0.1 n HCl, dried under N2, and derivatized with N-methyl-N-(tert-butyldimethylsilyl)-trifluoroacetamide in acetonitrile at 75°C for 30 min (Chaves Das Neves and Vasconcelos, 1987) or N,N-dimethylformamide at 125°C for 1 h (Williams and Wolf, 1994). The atom percent of 15N of each amino acid was then determined by GC-MS analysis (model MD800; Fisons). For 15N ammonium measurement, 150 μL of sulfosalicylic acid extracts was mixed with 1 mL of 5% NaHCO3 to adjust the pH to 8 before adding 3 μL of pentafluorobenzoyl chloride (PFB-Cl; Sigma). After shaking twice for 1 min, the mixture was incubated at room temperature for 15 min. Pentafluorobenzamide was formed by reacting ammonium with PFB-Cl and extracted with 1 mL of ethyl acetate. The organic and aqueous phases were separated by centrifugation, and the organic phase was mixed with 200 μL of 6% H3PO4 to eliminate NaHCO3 and PFB-Cl. The two phases were separated by centrifugation, and the organic phase was dehydrated by adding CuSO4. The organic phase containing the pentafluorobenzamide was derivatized with N-methyl-N-(tert-butyldimethylsilyl)-trifluoroacetamide in acetonitrile at 75°C for 30 min and analyzed by GC-MS as described for amino acids.
Statistical Analysis
Statistical analyses were performed with the ANOVA function in XLStat-Pro 7.5 (Addinsoft).
Transcriptome Studies
Microarray analysis was carried out at the Research Unit in Plant Genomics using the CATMA array (Crowe et al., 2003; Hilson et al., 2004) containing 24,576 GSTs from Arabidopsis. RNA samples from two independent biological replicates were used. For each biological repetition, RNA samples for a condition were obtained by pooling RNAs from six plants. For each comparison, one technical replication with fluorochrome reversal was performed for each biological replicate (i.e. four hybridizations per comparison). The reverse transcription of RNA in the presence of Cy3-dUTP or Cy5-dUTP (Perkin-Elmer-NEN Life Science Products), the hybridization of labeled samples to the slides, and the scanning of the slides were performed as described by Lurin et al. (2004).
Statistical Analysis of Microarray Data
Statistical analysis was based on two dye swaps (i.e. four arrays, each containing 24,576 GSTs and 384 controls) as described by Lurin et al. (2004). Controls were used for assessing the quality of the hybridizations but were not included in the statistical tests or the graphic representation of the results. For each array, the raw data comprised the logarithm of median feature pixel intensity at wavelengths 635 nm (red) and 532 nm (green). No background was subtracted. In the following description, log ratio refers to the differential expression between two conditions. It is either log2 (red/green) or log2 (green/red) according to the experimental design. Array-by-array normalization was performed to remove systematic biases. First, we excluded spots that were considered badly formed features. Then, we performed global intensity-dependent normalization using the Loess procedure to correct the dye bias. Finally, for each block, the log ratio median calculated over the values for the entire block was subtracted from each individual log ratio value to correct print tip effects on each metablock. To determine differentially expressed genes, we performed a paired t test on the log ratios, assuming that the variance of the log ratios was the same for all genes. Spots displaying extreme variance (too small or too large) were excluded. The raw P values were adjusted by the Bonferroni method, which controls the familywise error rate. We considered as being differentially expressed the genes with a familywise error rate value of 5%. We use the Bonferroni method (with a type I error equal to 5%) in order to keep a strong control of the false positives in a multiple-comparison context (Ge et al., 2003).
Microarray data from this article were deposited at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession no.GSE9258) and at CATdb (http://urgv.evry.inra.fr/CATdb/; project RA05-09_UREA) according to the Minimum Information about a Microarray Experiment standards.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Root and shoot anion contents.
Supplemental Table S1. List of the genes that are differentially regulated in roots between CO(NH2)2- and NH4NO3-grown Arabidopsis plants.
Supplemental Table S2. List of the genes that are differentially regulated in shoots between CO(NH2)2- and NH4NO3-grown Arabidopsis plants.
Supplemental Table S3. Functional category distributions of the CO(NH2)2-induced and -repressed genes.
Supplemental Table S4. List of the genes that are differentially regulated in roots between NH4NO3 + CO(NH2)2- and NH4NO3-grown Arabidopsis plants.
Supplemental Table S5. List of the genes that are differentially regulated in shoots between NH4NO3 + CO(NH2)2- and NH4NO3-grown Arabidopsis plants.
Supplemental Table S6. Functional category distributions of the NH4NO3 + CO(NH2)2-induced and -repressed genes.
Supplementary Material
Acknowledgments
The help of S. Boutet and M.-T. Leydecker with amino acid and nitrate analyses, respectively, is gratefully acknowledged. We thank Hoai-Nam Truong for assistance with MapMan and Helen North for carefully rereading the manuscript. We are grateful to Joël Talbotec and François Gosse for taking care of plant materials.
This work was supported by BiotechMarine.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Françoise Daniel-Vedele (vedele@versailles.inra.fr).
The online version of this article contains Web-only data.
References
- Bloom AJ, Sukrapanna SS (1990) Effects of exposure to ammonium and transplant shock upon the induction of nitrate absorption. Plant Physiol 94 85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley DP, Morgan MA, O'Toole P (1989) Uptake and apparent utilization of urea and ammonium nitrate in wheat seedlings. Fert Res 20 41–49 [Google Scholar]
- Buchner M, Rausch C, Daram P (2001) Molecular and biochemical mechanisms of phosphorus uptake into plants. J Plant Nutr Soil Sci 164 209–217 [Google Scholar]
- Buchner P, Takahashi H, Hawkesford MJ (2004) Plant sulphate transporters: co-ordination of uptake, intracellular and long-distance transport. J Exp Bot 55 1765–1774 [DOI] [PubMed] [Google Scholar]
- Chaves Das Neves HJ, Vasconcelos AM (1987) Capillary gas chromatography of amino acids, including asparagine and glutamine: sensitive gas chromatographic-mass spectrometric and selected ion monitoring gas chromatographic-mass spectrometric detection of the N,O(S)-tert-butyldimethylsilyl derivatives. J Chromatogr 392 249–258 [DOI] [PubMed] [Google Scholar]
- Chiu CC, Lin CS, Hsia AP, Su RC, Lin HL, Tsay YF (2004) Mutation of a nitrate transporter, AtNRT1:4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol 45 1139–1148 [DOI] [PubMed] [Google Scholar]
- Crawford NM, Glass ADM (1998) Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci 3 389–395 [Google Scholar]
- Criddle RS, Ward MR, Huffaker RC (1988) Nitrogen uptake by wheat seedlings, interactive effects of four nitrogen sources: NO3−, NO2−, NH4+, and urea. Plant Physiol 86 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe MLL, Serizet C, Thareau V, Aubourg S, Rouze P, Hilson P, Beynon J, Weisbeek P, van Hummelen P, Reymond P, et al (2003) CATMA: a complete Arabidopsis GST database. Nucleic Acids Res 31 56–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel-Vedele F, Filleur S, Caboche M (1998) Nitrate transport: a key step in nitrate assimilation. Curr Opin Plant Biol 1 235–239 [DOI] [PubMed] [Google Scholar]
- De Angeli A, Monachello D, Ephritikine G, Frachisse JM, Thomine S, Gambale F, Barbier-Brygoo H (2006) The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 442 939–942 [DOI] [PubMed] [Google Scholar]
- Filleur S, Daniel-Vedele F (1999) Expression analysis of a high-affinity nitrate transporter isolated from Arabidopsis thaliana by differential display. Planta 207 461–469 [DOI] [PubMed] [Google Scholar]
- Fraisier V, Gojon A, Tillard P, Daniel-Vedele F (2000) Constitutive expression of a putative high-affinity nitrate transporter in Nicotiana plumbaginifolia: evidence of a post-transcriptional regulation by a reduced nitrogen source. Plant J 23 489–496 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Lejay L, Gojon A, Ninnemann O, Frommer WB, von Wirén N (1999) Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium in Arabidopsis roots. Plant Cell 11 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Dudoit S, Speed TP (2003) Resampling-based multiple testing for microarray data analysis. Test 12 1–77 [Google Scholar]
- Gerendás J, Zhu Z, Sttelmacher B (1998) Influence of nitrogen and Ni supply on nitrogen metabolism and urease activity in rice (Oryza sativa L.). J Exp Bot 49 1545–1554 [Google Scholar]
- Glass ADM, Britto DT, Kaiser BN, Kinghorn JR, Kronzucker J, Kumar A, Okamoto M, Rawat S, Siddiqi MY, Unckles SE, et al (2002) The regulation of nitrate and ammonium transport systems in plants. J Exp Bot 53 855–864 [DOI] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hine JC, Spent JI (1988) Growth of Phaseolus vulgaris on various nitrogen sources: the importance of urease. J Exp Bot 39 1505–1512 [Google Scholar]
- Killingsbaeck A (1975) Extraction and colorimetric determination of urea in plants. Acta Agric Scand 25 109–112 [Google Scholar]
- King SL, Kozono D, Agre P (2004) From structure to disease: the evolving tale of aquaporin biology. Nat Rev Mol Cell Biol 5 687–698 [DOI] [PubMed] [Google Scholar]
- Kirkby EA, Mengel K (1967) Ionic balance in different tissues of the tomato plant in relation to nitrate, urea, or ammonium nutrition. Plant Physiol 42 6–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Bohner A, Gassert B, Yuan L, von Wirén N (2007) AtDUR3 represents the major transporter for high-affinity urea transport across the plasma membrane of nitrogen-deficient Arabidopsis roots. Plant J 52 30–40 [DOI] [PubMed] [Google Scholar]
- Kojima S, Bohner A, von Wirèn N (2006) Molecular mechanisms of urea transport in plants. J Membr Biol 212 83–91 [DOI] [PubMed] [Google Scholar]
- Krogmeier MJ, McCarty GW, Bremner JM (1989) Phytotoxicity of foliar-applied urea. Proc Natl Acad Sci USA 86 8189–8191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Hsieh MH, Corruzi G (1998) Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana. Plant J 16 345–353 [DOI] [PubMed] [Google Scholar]
- Lee RB, Drew MC (1989) Rapid, reversible inhibition of nitrate influx in barley by ammonium. J Exp Bot 40 741–752 [Google Scholar]
- Lejay L, Tillard P, Lepetit M, Olive F, Filleur S, Daniel-Vedele F, Gojon A (1999) Molecular and functional regulation of two NO3− uptake systems by N- and C-status of Arabidopsis plants. Plant J 18 509–519 [DOI] [PubMed] [Google Scholar]
- Liu LH, Ludewig U, Frommer WB, von Wirén N (2003. a) AtDUR3 encodes a new type of high-affinity urea/H+ symporter in Arabidopsis. Plant Cell 15 790–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LH, Ludewig U, Gassert B, Frommer WB, von Wirén N (2003. b) Urea transport by nitrogen-regulated tonoplast intrinsic proteins in Arabidopsis. Plant Physiol 133 1220–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqué D, von Wirén N (2004) Regulatory levels for the transport of ammonium in plant roots. J Exp Bot 55 1293–1305 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Merigout P, Talbotec J, Daniel-Vedele F (2003) Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis. Plant Physiol 131 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Reisdorf-Cren M, Pageau K, Lelandais M, Grandjean O, Kronenberger J, Valadier MH, Feraud M, Jouglet T, Suzuki A (2006) Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol 140 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merigout P, Gaudon V, Quilleré I, Briand X, Daniel-Vedele F (2008) Urea use efficiency of hydroponically grown maize and wheat plants. J Plant Nutr 31 427–443 [Google Scholar]
- Miflin BJ, Lea PJ (1980) Ammonia assimilation. In BJ Miflin, ed, The Biochemistry of Plants, Vol 5. Academic Press, New York, pp 169–202
- Nazoa P, Vidmar J, Tranbarger T, Mouline K, Damiani I, Tillard P, Zhuo D, Glass ADM, Touraine B (2003) Regulation of the nitrate transporter gene AtNRT2.1 in Arabidopsis thaliana: responses to nitrate, amino acids and developmental stage. Plant Mol Biol 52 689–703 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Vidmar JJ, Glass AD (2003) Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: responses to nitrate provision. Plant Cell Physiol 44 304–317 [DOI] [PubMed] [Google Scholar]
- Orsel M, Eulenburg K, Krapp A, Daniel-Vedele F (2004) Disruption of the nitrate transporter genes AtNRT2.1 and AtNRT2.2 restricts growth at low external nitrate concentration. Planta 219 714–721 [DOI] [PubMed] [Google Scholar]
- Orsel M, Krapp A, Daniel-Vedele F (2002) Analysis of the NRT2 nitrate transport family in Arabidopsis: structure and gene expression. Plant Physiol 129 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacco JC, Holland MA (1993) Roles of urease in plant cells. Int Rev Cytol 145 65–103 [Google Scholar]
- Rawat SR, Silim SN, Kronzucker HJ, Siddiqi MY, Glass AD (1999) AtAMT1 gene expression and NH4+ uptake in roots of Arabidopsis thaliana: evidence for regulation by root glutamine levels. Plant J 19 143–152 [DOI] [PubMed] [Google Scholar]
- Rosen H (1957) A modified ninhydrin colorimetric analysis for amino acids. Arch Biochem Biophys 67 10–15 [DOI] [PubMed] [Google Scholar]
- Sohlenkamp C, Shelden M, Howitt S, Udvardi M (2000) Characterization of Arabidopsis AtAMT2, a novel ammonium transporter in plants. FEBS Lett 467 273–278 [DOI] [PubMed] [Google Scholar]
- Stebbins NE, Polacco JC (1995) Urease is not essential for ureide degradation in soybean. Plant Physiol 109 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2003) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37 914–39 [DOI] [PubMed] [Google Scholar]
- van Beusichem ML, Neeteson JJ (1982) Urea nutrition of young maize and sugar-beet plants with emphasis on ionic balance and vascular transport of nitrogenous compounds. Neth J Agric Sci 30 317–330 [Google Scholar]
- Watson CJ, Miller H, Poland P, Kilpatrick DJ, Allen MDB, Garret MK, Christianson CB (1994) Soil properties and the ability of the urease inhibitor N-(N-butyl)thiophosphoric triamide (NBTPT) to reduce ammonia volatilization from surface-applied urea. Soil Biol Biochem 26 1165–1171 [Google Scholar]
- Williams BD, Wolf RR (1994) Determination of amino- and amide-15N glutamine enrichment with tertiary butyldimethylsilyl derivatives. Biol Mass Spectrom 23 682–688 [DOI] [PubMed] [Google Scholar]
- Wilson MR, O'Donoghue SI, Walker NA (1988) The transport and metabolism of urea in Chara australis: III. Two specific transport systems. J Exp Bot 39 763–774 [Google Scholar]
- Wirth J, Chopin F, Santoni V, Viennois G, Tillard P, Krapp A, Lejay L, Daniel-Vedele F, Gojon A (2007) Regulation of root nitrate uptake at the NRT2.1 protein level in Arabidopsis thaliana. J Biol Chem 282 23541–23552 [DOI] [PubMed] [Google Scholar]
- Witte CP, Rosso MG, Romeis T (2005) Identification of three urease accessory proteins that are required for urease activation in Arabidopsis. Plant Physiol 139 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo D, Okamoto M, Vidmar JJ, Glass AD (1999) Regulation of a putative high-affinity nitrate transporter (Nrt2;1At) in roots of Arabidopsis thaliana. Plant J 17 563–568 [DOI] [PubMed] [Google Scholar]
- Zonia LE, Stebbins NE, Polacco JC (1995) Essential role of urease in germination of nitrogen-limited Arabidopsis thaliana seeds. Plant Physiol 107 1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.