Abstract
The circadian coordination of organismal biology with the local temporal environment has consequences for fitness that may become manifest early in development. We directly explored the development of the Arabidopsis (Arabidopsis thaliana) clock in germinating seedlings by monitoring expression of clock genes. Clock function is detected within 2 d of imbibition (hydration of the dried seed). Imbibition is sufficient to synchronize individuals in a population in the absence of entraining cycles of light-dark or temperature, although light-dark and temperature cycles accelerate the appearance of rhythmicity and improve synchrony among individuals. Oscillations seen during the first 2 d following imbibition are dependent on the clock genes LATE ELONGATED HYPOCOTYL, TIMING OF CAB EXPRESSION1, ZEITLUPE, GIGANTEA, PSEUDO-RESPONSE REGULATOR7 (PRR7), and PRR9, although later circadian oscillations develop in mutants defective in each of these genes. In contrast to circadian rhythmicity, which developed under all conditions, amplitude was the only circadian parameter that demonstrated a clear response to the light environment; clock amplitude is low in the dark and high in the light. A circadian clock entrainable by temperature cycles in germinating etiolated seedlings may synchronize the buried seedling with the local daily cycles before emergence from the soil and exposure to light.
The rotation of the earth on its axis subjects most organisms to cycles of light and dark, as well as to cycles of warm and cool temperatures. In many examples, these cycles drive diurnal physiological and molecular rhythmic responses. Other rhythms, however, anticipate the onset of light in the morning or dark in the evening. Many of these rhythms persist in the absence of external time cues with approximately 24-h periods and are thus endogenous and circadian in nature (Dunlap, 1999; Salomé and McClung, 2005b). Recent studies have underscored the extent of circadian control on the Arabidopsis (Arabidopsis thaliana) transcriptome, with estimates of 10% to 15% (Harmer et al., 2000; Edwards et al., 2006; Covington and Harmer, 2007) and as high as 80% to 90% (Michael et al., 2008) of genes, effectively representing thousands of genes affecting all aspects of seedling and plant development, exhibiting clock control. By convention, genes whose expression is under the control of the circadian clock, yet do not by themselves participate in clock function, are called output genes. A small group of genes, termed “clock genes,” orchestrates the daily circadian oscillations witnessed in output genes. Intense forward and reverse genetic screens have helped uncover many, but unlikely all, of these so-called clock genes.
Interlocked transcription-translation feedback loops have been shown to be at the heart of the circadian mechanism in eukaryotic model systems (Bell-Pedersen et al., 2005). Current models of the Arabidopsis circadian clock postulate three to four interlocked feedback loops (Locke et al., 2006; McClung, 2006; Zeilinger et al., 2006). A pair of single Myb-domain transcription factors, CIRCADIAN CLOCK ASSOCIATED1 (CCA1; Wang and Tobin, 1998) and LATE ELONGATED HYPOCOTYL (LHY; Schaffer et al., 1998), play central roles in two loops. In one loop, CCA1 and LHY repress the expression of the pseudo-response regulator (PRR) gene TIMING OF CAB EXPRESSION1 (TOC1; also known as PRR1; Strayer et al., 2000). TOC1 closes the first loop by inducing CCA1 and LHY transcription for the next cycle (Alabadí et al., 2001). Proper regulation of CCA1 and LHY requires other evening-expressed clock genes, including EARLY FLOWERING4 (ELF4; Doyle et al., 2002; Kikis et al., 2005; McWatters et al., 2007), which encodes a protein of unknown function, and LUX ARRHYTHMO/PHYTOCLOCK1 (LUX/PCL1), which encodes a Myb domain transcription factor (Hazen et al., 2005; Onai and Ishiura, 2005). In a second loop, two TOC1-related genes, PRR7 and PRR9, are induced by and subsequently repress CCA1 and LHY (Farré et al., 2005; Nakamichi et al., 2005; Salomé and McClung, 2005a). In a third loop, GIGANTEA (GI) is a positive regulator of TOC1 (Makino et al., 2002). GI itself is negatively regulated by both CCA1/LHY and TOC1 (Locke et al., 2005), although the mechanism by which CCA1 and LHY influence GI expression remains unclear. Indeed, overexpression of CCA1 increases GI mRNA levels, whereas LHY overexpression represses GI (Fowler et al., 1999). This third loop is important in temperature compensation (Gould et al., 2006). Although current mathematical models of the Arabidopsis clock posit that CCA1 and LHY are equivalent and interchangeable for the sake of simplification (Locke et al., 2006; Zeilinger et al., 2006), their relative importance in maintaining clock function varies according to ambient temperature (Gould et al., 2006).
While our view of the oscillator is becoming increasingly sophisticated, little is known of the requirements for onset of rhythmicity in Arabidopsis. How early in development can circadian rhythms be detected? Is entrainment required to show rhythms? Is exposure to light necessary? Is circadian amplitude in gene expression affected by growth conditions? This topic of early circadian rhythmicity has been recently reviewed (Vallone et al., 2007). In mammals, the fetal clock is synchronized with the outside environment by maternal signals (Reppert and Schwartz, 1983; Reppert, 1995). However, even in the absence of a functioning maternal clock, individual pups still develop circadian rhythms, although littermates are not synchronized with one another (Jud and Albrecht, 2006). In zebrafish, entrainment is critical for onset of circadian rhythmicity (Kaneko and Cahill, 2005). A single light exposure 12 h after fertilization is sufficient to allow detection of an oscillation as early as 2 (Vuilleumier et al., 2006) or 3 (Ziv and Gothilf, 2006) d postfertilization, and temperature cycles may substitute for light-dark (LD) cycles in constant darkness (Lahiri et al., 2005). In Drosophila, as in mice, entrainment is dispensable for rhythmicity but not for synchrony among individuals (Sehgal et al., 1992). A light treatment given at the first larval stage is sufficient to synchronize individuals; this stage is when strong cycling of the period protein, a critical clock component, is first detected in lateral neurons (Kaneko et al., 1997).
The onset of circadian rhythmicity in plants has been the subject of some controversy. We previously inferred that a circadian system is running in etiolated seedlings maintained in the dark for 10 d (Zhong et al., 1998). Although we were unable to observe oscillations in the clock-controlled CAT2 or CAT3 steady-state mRNA levels in etiolated wild-type seedlings (Zhong et al., 1994, 1997, 1998), we observed a circadian rhythm in the amplitude of the acute induction of CAT2 mRNA in response to light (Zhong et al., 1998). From this, we deduced that a circadian clock must be running in etiolated seedlings and gating the acute induction of CAT2 mRNA by light. Because groups of seedlings were assayed for changes in CAT2 mRNA levels, we concluded that the clocks of individual seedlings were synchronized within a population, probably by imbibition, the hydration of the seeds. However, we were unable to address whether clock genes such as CCA1, LHY, and TOC1 were themselves cycling in etiolated seedlings. In addition, the timing of the first circadian oscillation could not be deduced from our results; at best, we could conclude that the onset of circadian rhythmicity in etiolated seedlings occurs within 10 d after hydration of the seed (Zhong et al., 1998).
Kikis and colleagues (Kikis et al., 2005) recently reported that mRNA abundances for the core clock components CCA1, LHY, and TOC1 fail to oscillate in etiolated seedlings, apparently contradicting our detection of circadian function in etiolated seedlings. Kikis et al. (2005) found that rhythms are established and maintained for several days following a single transition from darkness into constant red light, which would provide a strong entraining cue. For the circadian clock to gate the acute response of CAT2 expression to light in the absence of apparent oscillations of genes comprising the core oscillator poses a conundrum. Kikis et al. (2005) speculated that the acute response of CAT2 to light might be under the control of a clock that is not dependent on the core CCA1/LHY/TOC1 oscillator. Multiple oscillators have been proposed to explain the different free-running periods of the CHALCONE SYNTHASE and LHCB1*1 genes (Thain et al., 2002) and the differential response of the LHCB1*1 and CAT3 genes to photocycles versus temperature cycles (Michael et al., 2003a). Alternatively, a failure to detect circadian rhythms in steady-state mRNA levels does not preclude the existence of a rhythm in the transcription of the same gene. For instance, CAT3 mRNA levels lose their rhythmic pattern upon transfer into constant darkness, yet rhythmic transcription of the CAT3 gene persists, as evidenced by rhythmic CAT3∷LUC activity (Zhong et al., 1997; Michael and McClung, 2002). Similarly, circadian rhythms in the clock output genes LHCB1*1 and CAT2 are lost upon transfer into constant darkness, although the clock genes themselves remain rhythmic in the dark (Michael and McClung, 2002; Salomé and McClung, 2005a). Therefore, there may be rhythmic expression of the clock genes in etiolated seedlings, possibly with low overall amplitude, that might not have been detected by northern-blot analysis of total mRNA. In addition, as summarized above, posttranscriptional regulation is commonly encountered in many clock systems, and it is possible that clock function is occurring in the absence of evident oscillations in clock gene steady-state mRNA abundance.
We sought to resolve the apparent contradiction between the observations of Kikis et al. (2005) and our earlier work (Zhong et al., 1998) by directly monitoring gene expression in etiolated seedlings. By far the most studied circadian clock-controlled gene in Arabidopsis is LHCB1*1 (e.g. Anderson and Kay, 1995; Anderson et al., 1997; Somers et al., 1998; Devlin and Kay, 2000), and many studies have used an LHCB1*1∷LUC reporter in which the firefly luciferase gene (LUC) is placed under the control of the LHCB1*1 promoter (Millar et al., 1992). However, LHCB1*1 transcription is also strongly induced by light (Chory et al., 1989) and chloroplast development (Strand et al., 2003; Koussevitzky et al., 2007) that has largely limited the use of LHCB1*1∷LUC to light-grown Arabidopsis seedlings. In particular, LHCB1*1∷LUC expression is low in etiolated (dark-grown) seedlings and damps to low levels in light-grown seedlings transferred to darkness. LHCB1*1∷LUC expression is arrhythmic in etiolated Arabidopsis seedlings, although in etiolated tobacco (Nicotiana tabacum) seedlings, LHCB oscillations are detected (Millar et al., 1992; Anderson et al., 1994). Accordingly, we felt that other genes might be more suited to our study.
The recent availability of LUC reporters for multiple clock genes as well as for other clock-controlled genes now offers the advantage of assaying rhythmicity in single seedlings with very high sensitivity and has allowed us to address the initiation of clock function in individual etiolated and light-grown seedlings. We germinated individual seeds in 96-well plates and detected LUC activity of individual seedlings within hours of seed hydration (imbibition). This experimental system allowed the systematic investigation of signals required for onset of rhythmicity in Arabidopsis seedlings and conditions under which oscillations in clock genes can be detected. We establish that rhythms in germinating seedlings can be readily detected with a number of LUC reporters, including reporters for the clock genes CCA1, LHY, and TOC1, even in the absence of prior entraining cycles. Remarkably, etiolated seedlings exhibit rhythmic expression of the clock genes as early as 2 d following seed hydration, although at a lower amplitude than in light-grown plants. We also provide genetic evidence that several components, including LHY and TOC1, of the oscillator characterized in light-grown seedlings are required for proper onset of rhythmicity in etiolated seedlings. We propose that seed hydration is sufficient for both onset of rhythmicity and synchronization among individuals. The existence of a running oscillator in etiolated seedlings also suggests that germinating seedlings may use entraining cues first from temperature cycles and then from LD cycles to synchronize with the outside environment as they emerge from the soil.
RESULTS
Plants Grown in Constant Light Are Rhythmic
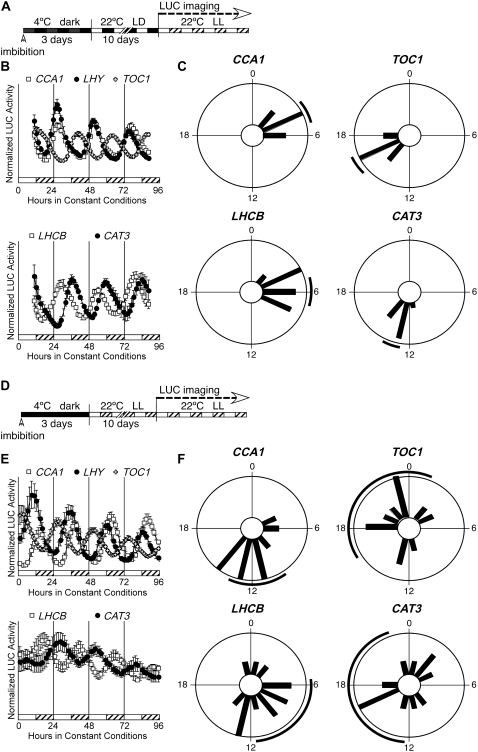
A typical circadian experiment in Arabidopsis entails sowing dry seeds onto plates of growth medium. Plated seeds are then stratified at 4°C in total darkness for 3 to 4 d to enhance germination before release into an entraining regimen of LD or temperature (HC) cycles. Seedlings entrained to LD cycles (Fig. 1A) express the clock genes CCA1, LHY, and TOC1 at their expected phases (Fig. 1B; Table I), and the phases among individual seedlings are synchronized (Fig. 1C; see Supplemental Fig. S1 for individual traces). Similarly, the clock-regulated output genes LHCB1*1 and CAT3 show strong and synchronized rhythms in expression (Fig. 1, B and C; Table I). An identical orchestrated rhythmic expression is observed following HC cycles (not shown; see also Salomé and McClung, 2005a).
Figure 1.
LD cycles are dispensable for rhythmicity but enhance synchronization among individuals. A, Diagram representing the growth conditions used in B and C. Seeds were hydrated at noon on the first day, then stratified for 3 d. At noon on the fourth day, plates were released into LD cycles (lights on at noon; lights off at midnight). Individual seedlings (12 per LUC reporter) were transferred to 96-well plates at noon (subjective dawn) after 10 d in LD cycles and moved to constant conditions of LL and constant temperature (22°C) for LUC activity measurement. Phase estimations by fast Fourier transform-nonlinear least squares were obtained for a 72-h window, starting 12 h after release into constant conditions. B, Expression of clock (CCA1, LHY, and TOC1) and clock-regulated (LHCB1*1 and CAT3) genes in LL after LD cycles. To facilitate phase comparison between genes of different absolute expression levels, LUC activities for each seedling at each time point are expressed relative to the mean LUC activity for that seedling and are presented as “normalized LUC activity.” White bars represent subjective day and hatched bars represent subjective night. Mean expression of each gene is given ± se, n = 12; expression from individual seedlings for all reporters tested is shown in Supplemental Figure S1. Top, White squares, CCA1∷LUC; black circles, LHY∷LUC; white crosses, TOC1∷LUC. Bottom, White squares, LHCB1*1∷LUC; black circles, CAT3∷LUC. C, Phase distributions of individual seedlings following entrainment by LD cycles. The phase of individual seedlings for each reporter is plotted in 100-min time bins; the black line outside of the circle corresponds to the 95% confidence interval (±2 se) associated with the distribution. D, Diagram representing the growth conditions used in E and F. Seeds were hydrated at noon on the first day then stratified for 3 d. At noon on the fourth day, plates were released into LL. Individual seedlings (12 per LUC reporter) were transferred to 96-well plates after 10 d in constant conditions of LL and constant temperature (22°C) before LUC activity measurement on days 11 through 14. Phase estimations by fast Fourier transform-nonlinear least squares were obtained for a 72-h window, starting 12 h after release into constant conditions. E, Expression of clock (CCA1, LHY, and TOC1) and clock-regulated (LHCB1*1 and CAT3) genes in LL in the absence of entrainment by LD cycles. White bars represent subjective day and hatched bars represent subjective night, where subjective dawn is defined by release from stratification. Top, Mean expression of each gene (±se, n = 12); expression from individual seedlings for CCA1 and TOC1 is shown in the bottom segments. Top, White squares, CCA1∷LUC; black circles, LHY∷LUC; white crosses, TOC1∷LUC. Bottom, White squares, LHCB1*1∷LUC; black circles, CAT3∷LUC. F, Phase distributions of individual seedlings in the absence of prior entrainment. The phase of individual seedlings for each reporter is plotted in 100-min time bins; the black line outside of the circle corresponds to the 95% confidence interval (±2 se) associated with the distribution.
Table I.
Relative synchronization of phase among a population of seedlings is enhanced by, but does not require, prior entrainment
| Gene | Condition | n | Period ± sda | RAE ± sdb | Phase ± sd | F-Test P Valuec |
|---|---|---|---|---|---|---|
| h | h | |||||
| LHCB1*1 | LD | 11 | 24.4 ± 0.9 | 0.23 ± 0.07 | 5.7 ± 1.5 | |
| LL | 12 | 25.4 ± 0.3* | 0.21 ± 0.07 | 8.5 ± 4.0 | <0.001 | |
| LHCB1*1 | DD HC | 11 | 26.2 ± 0.4 | 0.23 ± 0.13 | 4.7 ± 0.7 | |
| DD | 12 | 27.2 ± 2.9 | 0.32 ± 0.18 | 9.3 ± 3.1 | <0.001 | |
| CAT3 | LD | 12 | 23.8 ± 0.8 | 0.22 ± 0.09 | 13.2 ± 0.9 | |
| LL | 12 | 25.7 ± 1.0* | 0.29 ± 0.13 | 18.5 ± 4.7 | <0.001 | |
| CCA1 | LD | 12 | 24.2 ± 0.7 | 0.19 ± 0.07 | 4.3 ± 1.2 | |
| LL | 12 | 24.3 ± 0.6 | 0.17 ± 0.03 | 11.0 ± 3.6 | 0.001 | |
| CCA1 | DD HC | 12 | 26.7 ± 1.7 | 0.29 ± 0.20 | 4.5 ± 1.6 | |
| DD | 12 | 27.3 ± 2.0 | 0.18 ± 0.07 | 8.3 ± 1.5 | 0.9 | |
| TOC1 | LD | 11 | 24.3 ± 0.4 | 0.20 ± 0.06 | 15.8 ± 1.0 | |
| LL | 12 | 24.7 ± 0.8 | 0.15 ± 0.09 | 13.7 ± 6.1 | <0.001 | |
| TOC1 | DD HC | 11 | 26.5 ± 2.0 | 0.22 ± 0.06 | 21.9 ± 3.1 | |
| DD | 11 | 27.0 ± 2.7 | 0.23 ± 0.14 | 1.6 ± 6.7 | 0.15 |
Period for rhythms measured in LL and LD were compared by a Student's homoscedastic t test. Asterisk indicates statistically significant difference (P < 0.002).
RAE for rhythms measured in LL and LD are not statistically different, as determined by a Student's homoscedastic t test (for LHCB1*1, CAT3, and CCA1) or Student's heteroscedastic t test (for TOC1).
Differences in variance for phase of each gene assayed with (LD or DD HC) or without (LL or DD) prior entrainment was tested using the F test. P values are indicated.
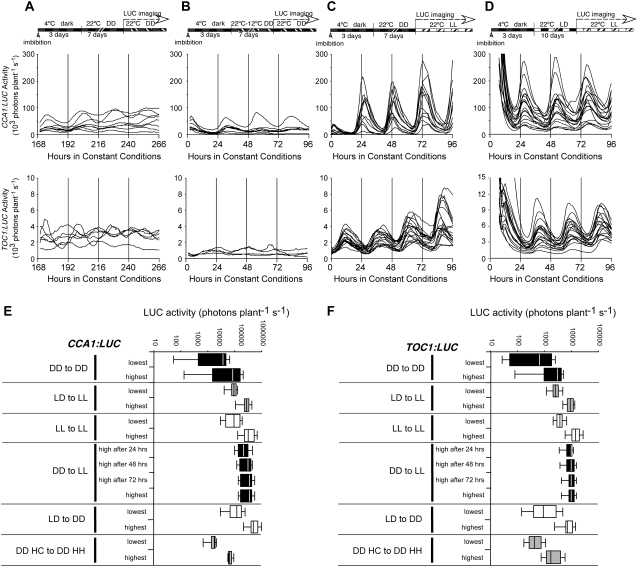
We tested whether seedlings grown in constant light (in the absence of either LD or HC cycles) were individually rhythmic and, if so, whether the phases of clock-regulated genes would be timed properly. To this end, we stratified seeds for 3 d in the dark before releasing them in constant light and a constant temperature of 22°C (Fig. 1D). After 10 d, seedlings were transferred to 96-well plates and LUC activity was recorded. The clock genes CCA1, LHY, and TOC1 were strongly rhythmic in individual seedlings under these conditions (Fig. 1E; Table I). Relative synchronization among seedlings was observed as well (Fig. 1F; Supplemental Fig. S1), although the circadian phase in continuous light (LL) was almost 180° out of phase with seedlings grown under LD cycles before release into LL. We observed the same change in phase when cotyledon movement was recorded (data not shown). In the absence of entrainment, the observed phases of the clock output genes LHCB1*1 and CAT3 were more poorly synchronized (Fig. 1F; Table I; Supplemental Fig. S1) than those of the clock genes. From these results, we conclude that entraining cycles following release from stratification are dispensable for the generation of rhythmic circadian gene expression in light-grown seedlings. This contrasts with zebrafish, in which exposure to LD cycles is required for the later observation of rhythmic expression (Kaneko and Cahill, 2005). In Arabidopsis, entraining cycles enhance synchronization among individuals, as indicated by significantly decreased variance in entrained populations (Table I). It is clear, however, that imbibition followed by stratification is sufficient to set the phase of clock gene expression. Both the temperature step from 4°C to 22°C and the dark to light transition would be expected to provide strong entraining cues sufficient to synchronize individuals in a population.
Stratified, Etiolated Seedlings Are Rhythmic
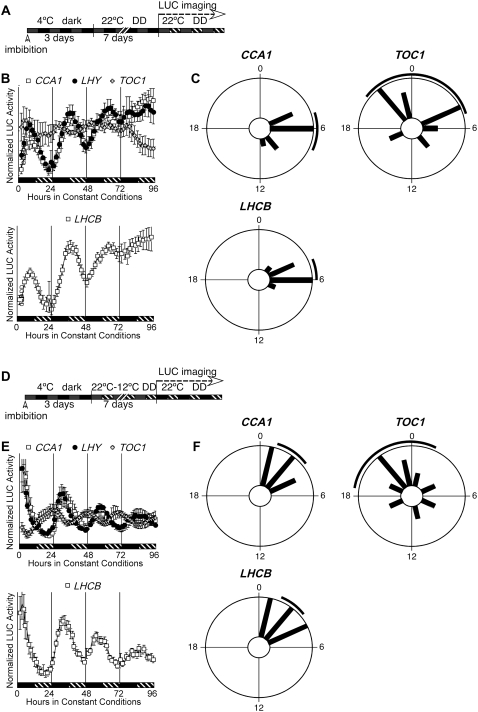
We asked whether the high sensitivity of LUC reporter activity would allow a clear determination of rhythmicity in etiolated seedlings. We examined the expression of the CCA1, LHY, and TOC1 clock genes, as well as the output gene LHCB1*1, in etiolated seedlings following the protocol depicted in Figure 2A. Following release from stratification, seedlings were maintained in darkness (DD) for 7 d before LUC recording in DD. We detected rhythmic expression of each of the clock genes (Fig. 2B), demonstrating that 7-d-old etiolated seedlings possess a functional circadian clock including the clock components CCA1, LHY, and TOC1. Entraining cycles are therefore dispensable for rhythmicity. Remarkably, we were also able to detect oscillations in LHCB1*1 with a period of approximately 27 h (Fig. 2B), characteristic of rhythms seen in LD-grown seedlings transferred into DD (Salomé and McClung, 2005a). The rhythmic expression of LHCB1*1 in etiolated seedlings indicates that clock function in light-grown seedlings transferred into DD may differ from that in etiolated seedlings. Light exposure might be required to couple LHCB1*1 expression to both clock- and light-regulated signaling pathways (Kolar et al., 1995). Relative synchronization within a population of etiolated seedlings was observed for CCA1, LHY, and LHCB1*1, although not for TOC1 (Fig. 2, B and C; Supplemental Fig. S2).
Figure 2.
Etiolated seedlings are rhythmic. A, Diagram representing the growth conditions used in B and C. Seeds were hydrated at noon on the first day and sown directly into 96-well plates, then stratified in the dark at 4°C for 3 d. At noon on the fourth day, plates were released into DD. LUC activity was recorded starting at the beginning of the 11th day. Phase values were determined for data collected between ZT0 and ZT72. B, Expression of clock (CCA1, LHY, and TOC1) and clock-regulated (LHCB1*1) genes in DD in the absence of entrainment. Mean expression of each gene is given ± se, n = 12. Black bars represent subjective day and hatched bars represent subjective night, where subjective dawn is defined by release from stratification. Top, White squares, CCA1∷LUC; black circles, LHY∷LUC; white crosses, TOC1∷LUC. Bottom, White squares, LHCB1*1∷LUC. C, Phase distributions of individual etiolated seedlings in the absence of prior entrainment or light exposure. The phase of individual seedlings for each reporter is plotted in 100-min time bins; the black line outside of the circle corresponds to the 95% confidence interval associated with the distribution. D, Diagram representing the growth conditions used in E and F. Seeds were hydrated at noon on the first day and sowed directly into 96-well plates, then stratified for 3 d. At noon on the fourth day, plates were released into HC (22°/12°C) cycles in DD. After 7 d, plates were released into constant conditions of continuous dark and constant temperature (22°C), and LUC activity was recorded. Phase values were determined for data collected between ZT0 and ZT72. E, Expression of the clock genes CCA1, LHY, and TOC1 and of the clock-regulated gene LHCB1*1 in DD after entrainment by HC cycles. Mean expression of each gene is given ± se, n = 12. Black bars represent subjective day and hatched bars represent subjective night, where subjective dawn is defined by onset of 22°C during entrainment. Top, White squares, CCA1∷LUC; black circles, LHY∷LUC; white crosses, TOC1∷LUC. Bottom, White squares, LHCB1*1∷LUC. F, Phase distributions of individual etiolated seedlings following entrainment by HC cycles. The phase of individual seedlings for each reporter is plotted in 100-min time bins; the black line outside of the circle corresponds to the 95% confidence interval associated with the distribution.
Etiolated Seedlings Can Be Synchronized by Temperature Cycles
Using etiolated seedlings, we tested whether HC cycles consisting of 12 h at 22°C followed by 12 h at 12°C could synchronize the phases of the clock genes CCA1 and TOC1 and the output gene LHCB1*1. After seven HC cycles, seedlings were released into constant temperature (22°C) and DD (Fig. 2, D–F). As with light-grown seedlings (Salomé and McClung, 2005a), all three genes were well synchronized by the temperature cycles, demonstrating that light is not required for proper entrainment by temperature and that, by extension, HC cycles are sufficient to synchronize the clock in etiolated seedlings following release from stratification. Identical results were obtained with weaker HC cycles (18°C–22°C; not shown).
Rhythms Can Be Detected during the Second Day after Imbibition
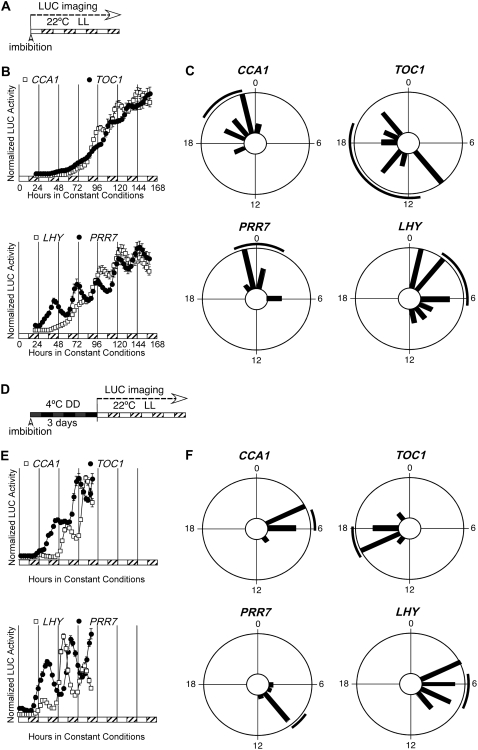
Our measurement of the Arabidopsis circadian clock has thus far begun approximately 10 d after germination. As Figures 1 and 2 indicate, circadian rhythms develop some time between seed hydration and the start of LUC imaging, even in the absence of entraining cycles. Estimating the precise timing of the establishment of a circadian rhythm has not been attempted in Arabidopsis. Early detection of rhythms may be in part limited by the choice of the proper clock-controlled reporter. We tested several LUC reporter constructs (including a new reporter with the CAT2 promoter, which behaves in a manner very similar to LHCB1*1; compare Supplemental Fig. S3 with Fig. 1B of this article and with figure 5 of Millar et al. [1992]) in the assay protocol shown in Figure 3A. Seeds were directly sown onto Murashige and Skoog agar medium in 96-well plates and LUC activity was recorded in LL for 7 d. Under these conditions, LUC activity is not detected for most reporter constructs (e.g. LHCB1*1, CAT2, and CAT3) for 2 d after imbibition (Supplemental Fig. S4). The first clear circadian cycle in CCA1 expression is seen 4 d after imbibition and is followed 12 h later by the first clear circadian cycle in TOC1 expression (Fig. 3B). Remarkably, the clock gene PRR7 shows a clear rhythm as early as the second day following imbibition, 2 d earlier than the first strong oscillation in CCA1 expression (Fig. 3B). We conclude that a functional circadian oscillator is present in germinating seeds as early as 2 d following imbibition. In addition, individual germinating seeds appear well synchronized immediately following imbibition (Fig. 3C), indicating that hydration of seeds in LL is sufficient to set the phase of the clock and to synchronize individual seedlings in a population. However, 5 d after imbibition, a loss of synchrony among individuals is evident in the expression profiles of CCA1, TOC1, and PRR7 (Supplemental Fig. S4). Seedlings that have been directly germinated in LL, without stratification, are therefore rhythmic and initially set to the same phase but quickly lose synchrony in the absence of entraining cycles.
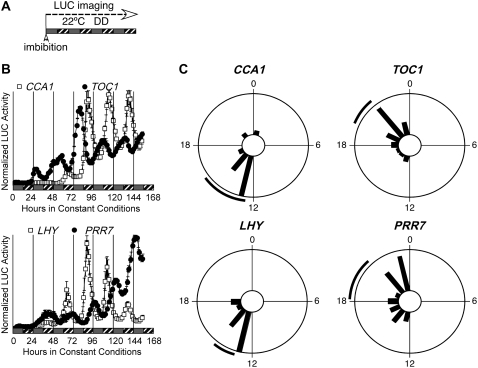
Figure 5.
Stratification is dispensable for rhythmicity of etiolated seedlings. A, Diagram representing the growth conditions used in B and C. Seeds were hydrated at noon on the first day and sown directly into 96-well plates, then released into darkness. LUC activity was recorded immediately for 7 d. Gray bars represent subjective day and hatched bars represent subjective night, where subjective dawn is defined by seed imbibition. B, Expression of the clock genes CCA1, LHY, PRR7, and TOC1 in DD in the absence of entrainment and stratification. Segments show mean expression of each gene (±se, n = 24). Top, White squares, CCA1∷LUC; black circles, TOC1∷LUC. Bottom, White squares, LHY∷LUC; black circles, PRR7∷LUC. C, Phase distributions of individual germinating seeds for CCA1, LHY, PRR7, and TOC1 in the absence of entrainment. The phase of individual seedlings for each reporter is plotted in 100-min time bins; the black line outside of the circle corresponds to the 95% confidence interval (±2 se) associated with the distribution.
Figure 3.
Germinating seeds are rhythmic. A, Diagram representing the growth conditions used in B and C. Seeds were hydrated at noon on the first day and sown directly into 96-well plates. Plates were then released into LL within 4 h of hydration and LUC activity was recorded. B, Expression of the clock genes CCA1, LHY, TOC1, and PRR7 in germinating seeds. Mean expression is shown ± se, n = 12. Top, White squares, CCA1∷LUC; black circles, TOC1∷LUC. Bottom, White squares, LHY∷LUC; black circles, PRR7∷LUC. White bars represent subjective day and hatched bars represent subjective night, where subjective dawn is defined by hydration of the seed. C, Phase distributions of individual germinating seeds for CCA1, LHY, TOC1, and PRR7 in the absence of entrainment. D, Diagram representing the growth conditions used in E and F. Seeds were hydrated at noon on the first day and sown directly into 96-well plates. Plates were stratified for 3 d, released into LL at 22°C, and LUC activity was recorded. E, Expression of the clock genes CCA1, LHY, TOC1, and PRR7 in germinating seeds. Mean expression is shown ± se, n = 12. Top, White squares, CCA1∷LUC; black circles, TOC1∷LUC. Bottom, White squares, LHY∷LUC; black circles, PRR7∷LUC. White bars represent subjective day and hatched bars represent subjective night, where subjective dawn is defined by hydration of the seed. F, Phase distributions of individual seedlings for CCA1, LHY, TOC1, and PRR7 in the absence of entrainment, following release from stratification.
LHY Expression in Etiolated Seedlings Is Primarily in the Cotyledons
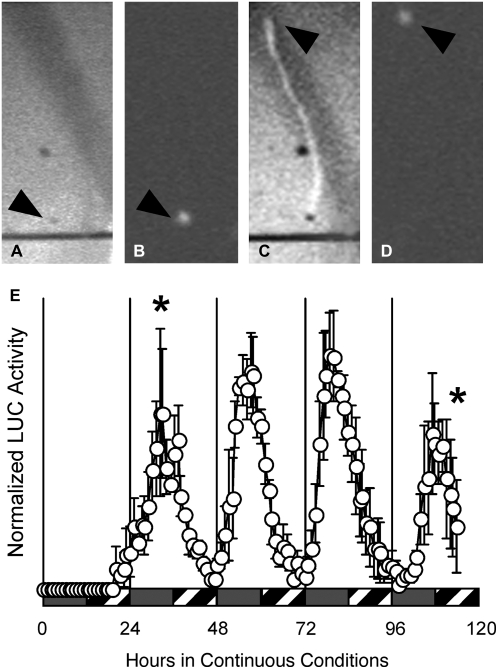
The experiments described in Figures 1 to 3 used a Packard TopCount luminometer, which counts photons but does not capture images. These experiments therefore address clock function at the level of the whole seedling without providing any spatial resolution. To determine which organs are rhythmic, we used a CCD camera to image etiolated seedlings carrying the LHY∷LUC transgene grown in the conditions described in Figure 3A. No light production above background was detected for 20 h but, thereafter, light production was detected in the cotyledons (Fig. 4; see Supplemental Video S1). We could detect no LUC activity in either the hypocotyl or the roots, although it is possible that LUC activity is present in those organs but falls below the threshold of detection. We conclude that the primary site of LHY expression in etiolated seedlings is the cotyledon. In parallel experiments with seedlings carrying the TOC1∷LUC transgene, we failed to detect LUC activity, as it fell below the detection level of our CCD camera.
Figure 4.
LHY∷LUC is expressed in the cotyledons of etiolated seedlings. Seeds bearing the LHY∷LUC transgene were stratified on plates for 3 d in darkness. LUC activity was then recorded in constant darkness at 22°C by a Hamamatsu digital CCD camera. A, Bright-field image of an individual hydrated seed taken immediately after release from stratification. Exposure, 10 ms. B, LUC activity is restricted to the hydrated seed 32 h after release from stratification. Exposure, 30 min. C, Bright-field image of the individual etiolated seedling, taken 114 h after release from stratification. Exposure, 10 ms. D, LUC activity is restricted to the cotyledons of the etiolated seedling, 114 h after release from stratification. Exposure, 30 min. E, Mean LHY∷LUC activity from three etiolated germinating seedlings. Data shown as mean ± se.
Stratification and the Onset of Rhythmicity
A stratification period is typically imposed to enhance germination before release into the regimen of interest, and, accordingly, we tested the effect of stratification on the onset of rhythmic expression. We sowed seeds directly in 96-well plates and stratified at 4°C in the dark for 3 d (Fig. 3D). At the end of stratification, plates were released into constant light at 22°C and LUC activity was recorded. Again, rhythmic PRR7 expression is clear during the second day after release (Fig. 3E). Clear and well-synchronized oscillations are also detected in TOC1 and LHY expression 2 d (Fig. 3E) and in LHCB1*1, CAT3, and CCA1 expression (Supplemental Fig. S4) 3 (and possibly 2) d after release from stratification. We conclude that stratification allows the earlier detection of oscillations in the clock and clock-output genes described in this study when compared to nonstratified seedlings. Stratification also helps to synchronize individuals within a population (compare Fig. 3, C and F). This improved synchronization is likely associated with the onset of light and/or the step up in temperature, both strong entraining cues, associated with the release from stratification.
Early Circadian Expression in Nonstratified Etiolated Seedlings
Results shown in Figure 3D cannot exclude a role for release from stratification as a major synchronizing signal for individual clocks of germinating seeds. We therefore tested whether circadian rhythms could develop in germinating seeds maintained in the dark in the absence of stratification. Seeds were sown directly onto medium in 96-well plates, and LUC activity was measured during subsequent incubation in the dark. Seeds spent at most 1 h in the light after imbibition before being transferred into the dark. Under these conditions, strong circadian rhythms develop for CCA1, LHY, TOC1, and PRR7 within 2 d of imbibition (Fig. 5B), with circadian periods of 25.0 ± 0.8 h for CCA1 (n = 15; relative amplitude error [RAE] = 0.25 ± 0.09; RAE is a measure of the strength of the rhythm, where RAE = 0 defines a perfect sine wave and RAE = 1 defines the limit of a statistically significant rhythm) and 24.7 ± 0.9 h for LHY (n = 18; RAE = 0.36 ± 0.08). Such periods are consistent with those reported for LHCB expression in etiolated seedlings not exposed to entraining cycles (Kolar et al., 1998). Moreover, the deviation of these periods from 24 h is important, as it excludes inadvertent entrainment due to small daily temperature fluctuations in the growth chambers, which would be expected to have a 24-h (diurnal) period. Based on clock gene expression, synchronization is also very clear among individual seedlings within a population (Fig. 5C; Supplemental Fig. S5). These results therefore demonstrate that imbibition by itself is sufficient to synchronize the initial clock phase of seeds germinating in darkness. The expression patterns of the clock output genes CAT2, CAT3, and LHCB1*1 appear weakly rhythmic under the same conditions (Supplemental Fig. S5).
Effects of Light on Amplitude
Circadian rhythmicity can be readily detected within 48 h of seed imbibition, regardless of light status or cold pretreatment. Therefore, light exposure or entraining cycles are dispensable for onset of rhythmicity or synchronization among individuals. However, we noticed a strong increase in the expression strength and in the amplitude of the circadian rhythm in expression of all genes tested in response to light. The amplitude of clock genes in etiolated seedlings is much lower than that seen in the light (compare Fig. 6, A and B with C and D). Temperature cycles synchronize etiolated seedlings with one another but do not increase circadian amplitude (Fig. 6B). To distinguish between direct effects of light on amplitude and effects of light on overall seedling morphology, we tested the effect of a single dark-to-light transition on circadian amplitude. Seeds were stratified for 3 d, then released in constant darkness at 22°C, grown for 7 d, released into constant white light (fluence rate approximately 20–25 μmol m−2 s−1) for LUC activity measurement (Fig. 6C). A rapid acute response to light onset is detected in CCA1 but not TOC1 expression (Fig. 6C), consistent with previous results (Kikis et al., 2005). In addition, seedlings are synchronized by the dark to light transition. Notably, the amplitudes of CCA1 and TOC1 increase within 24 h of illumination to values similar to the amplitudes seen in light-grown seedlings (Fig. 6, C and D). Although TOC1 expression is not directly induced by light, it has been postulated that GI, itself light induced, is a positive regulator of TOC1 (Makino et al., 2002). Seedlings illuminated after 7 d of growth in the dark retained the elongated hypocotyl characteristic of etiolation, but the cotyledons greened (not shown). Therefore, the higher amplitude in CCA1 expression likely reflects a direct effect of light.
Figure 6.
Comparison of absolute expression levels of CCA1 and TOC1 among all conditions tested in this study. Summary diagrams of all conditions depicted in A to D are shown above the corresponding expression profiles of CCA1 and TOC1. A, Absolute expression levels from individual CCA1∷LUC (top) and TOC1∷LUC (bottom) seedlings grown in DD without HC entrainment and released into DD. B, Absolute expression levels from individual CCA1∷LUC (top) and TOC1∷LUC (bottom) seedlings grown in DD with entrainment to HC cycles and released into DD. C, Absolute expression levels from individual CCA1∷LUC (top) and TOC1∷LUC (bottom) seedlings grown in DD without entrainment and released into LL. D, Absolute expression levels from individual CCA1∷LUC (top) and TOC1∷LUC (bottom) seedlings entrained in LD cycles in constant temperature and released into LL. E and F, LUC activity from CCA1∷LUC (E) and TOC1∷LUC (F) in the conditions described in Figures 1 to 5. Lowest and highest LUC activity is given in box plot format (box indicates 25%–75% with the vertical line at the mean; error bars indicate ±2 sd from the mean), from 12 to 36 seedlings in each condition, on a log scale.
When surveyed under all the conditions described in Figures 1 to 6, amplitude and overall LUC activity levels are higher in light-grown seedlings relative to etiolated seedlings (Fig. 6, E and F). Taken together, our results suggest that light exposure is dispensable for rhythmicity and synchronization among seedlings but leads to a 10-fold increase in expression of the clock genes CCA1 and TOC1 (Fig. 6) and of the output genes CAT2 and CAT3 (not shown).
Early Circadian Rhythmicity in Circadian Mutants
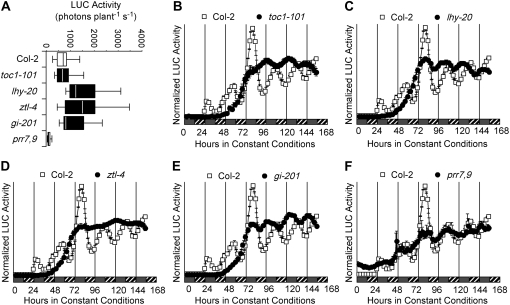
Rhythmic expression in the clock genes is observed after 48 h in etiolated seedlings not subjected to stratification (Fig. 5). Proper control of TOC1 expression requires the complex interaction of clock genes, among them CCA1, LHY, TOC1, ZEITLUPE (ZTL), GI, PRR7, and PRR9 (Park et al., 1999; Más et al., 2003; Farré et al., 2005; Nakamichi et al., 2005). To determine whether early circadian oscillations in etiolated seedlings depend on the same molecular mechanisms as in light-grown seedlings, we characterized TOC1∷LUC expression pattern in a number of clock mutants (toc1-101, lhy-20, ztl-4, gi-201, and prr7,9). All mutants, except prr7,9, displayed LUC activity levels close to those seen in wild type (Fig. 7A). TOC1∷LUC expression was reduced about 10-fold in prr7,9 (Fig. 7, A and F), demonstrating that PRR7 and PRR9 act as critical positive regulators of TOC1 expression in the dark at this stage of seedling development. Although the prr7,9 double mutant was previously shown to be arrhythmic in light-grown seedlings transferred into the dark (Salomé and McClung, 2005a), we observed sustained albeit low amplitude rhythmicity in etiolated prr7,9 seedlings (Fig. 7F), suggesting that clock function differs between light-grown and etiolated seedlings.
Figure 7.
An intact oscillator is required for initial circadian rhythmicity. TOC1∷LUC was introduced into the toc1, lhy, ztl, and gi mutants by crossing with one line in which the transgene had integrated into Col-2 through transformation. TOC1∷LUC was independently transformed into prr7,9. Seeds for each line were imbibed at noon on the first day and sown directly into 96-well plates. Plates were then released in DD at 22°C and LUC activity was recorded for 7 d. A, Mean ± se TOC1∷LUC LUC activity (n = 24–26) for Col-2, toc1-101, lhy-20, ztl-4, gi-201, and prr7,9. Time courses of mean TOC1∷LUC expression in Col-2 (white squares) and toc1-101 (black circles in B), lhy-20 (black circles in C), ztl-4 (black circles in D), gi-201 (black circles in E), and prr7,9 (black circles in F) in DD. For B to F, time of imbibition is taken as ZT0. Gray bars mark subjective day and hatched bars represent subjective night, where dawn is defined by imbibition.
Absence of functional LHY, TOC1, ZTL, or GI results in a loss of the first two oscillations in TOC1∷LUC expression (Fig. 7, B–E), although rhythmic expression becomes apparent on the fourth day following imbibition. The timing of the first peak in all mutants coincides with the third peak in wild type, although the mutations are known to affect the clock differently in the light; mutations in TOC1 and LHY shorten the period, while mutations in ZTL lengthen period (Somers et al., 2000; Strayer et al., 2000; Mizoguchi et al., 2002). gi-201, a T-DNA insertion allele of GI, causes a great decrease in circadian amplitude, but does not affect period length in the light (Martin-Tryon et al., 2007). New gi alleles were isolated based on a short period phenotype conferred on the CCR2∷LUC reporter in constant darkness, indicating that a short period can be expected for gi mutants in DD (Martin-Tryon et al., 2007). Following the emergence of a rhythm, all mutants subsequently develop circadian oscillations with their characteristic short (toc1-101, 22.7 ± 2.7 h; lhy-20, 23.4 ± 3.2 h; gi-201, 23.1 ± 2.2 h) or long (ztl-4, 36.2 ± 4.5 h) period phenotype, relative to wild type (Columbia [Col-2], 26.2 ± 1.8 h). These results are in agreement with previous observations that mutations in ZTL lengthen (Somers et al., 2004) and mutations in TOC1 shorten the period in the dark (Strayer et al., 2000).
In conclusion, imbibition is necessary and sufficient to initiate circadian oscillations in germinating etiolated seedlings, and the clock proteins LHY, TOC1, ZTL, and GI are required for the proper onset of rhythmicity following imbibition.
DISCUSSION
How early in the development of an organism can rhythmic gene expression be detected? In most circadian systems, this question is difficult to answer because of experimental limitations to the ability to measure gene expression or any rhythmic behavior during early development. In contrast with other organisms, plants suspend the development of their progeny in the form of a seed until more auspicious growth conditions are met. The seed allows the dissemination of the population and protects the embryo against environmental insults like cold, fire, and drought. Typically, a plant seed remains dormant until imbibition. Therefore, freshly hydrated seeds can be viewed as a developmentally synchronized population and provide an excellent system to study onset of circadian rhythmicity in plants. We show here that Arabidopsis seedlings possess the ability to manifest circadian gene expression as early as 2 d following seed imbibition without any entraining cycles or prolonged light exposure. At this stage of development, the radicle (root) is just emerging from the seed coat, and cotyledons have yet to emerge. Therefore, circadian rhythmicity is evident at very early stages in germinating seeds. Kikis et al. (2005) failed to detect oscillations in steady-state mRNA abundance for the clock genes CCA1, LHY, and TOC1 in etiolated seedlings and concluded that the central oscillator, as defined by these genes, is not oscillating in etiolated Arabidopsis seedlings. However, we show that transcription of each of these genes is rhythmic in etiolated seedlings and, moreover, that this rhythmic transcription is perturbed by mutations eliminating function of LHY, TOC1, and other clock genes. One possible explanation of the inability of Kikis et al. (2005) to detect rhythms is that northern-blot analysis requires considerable tissue and measures mRNA derived from populations of seedlings harvested sequentially. This could incorrectly resemble arrhythmicity if individual seedlings were not synchronized with one another, as our data predict would be the case (for example, note the loss of synchrony among individuals in the later points in the time course in Supplemental Fig. S2). Similar results were recently published for zebrafish cell lines (Carr and Whitmore, 2005) and mouse fibroblasts (Welsh et al., 2004); although mean LUC activity appears arrhythmic, recording of single cells demonstrated robust oscillations, albeit with varying circadian periods and phases. On the basis of this molecular and genetic evidence, we conclude that there is a circadian oscillator functioning in etiolated seedlings as early as 2 d following imbibition and that this oscillator is composed of many of the same gene products as that in light-grown plants.
The overall synchrony of the CCA1, LHY, and TOC1∷LUC reporters is greater among etiolated seedlings than among seedlings grown in continuous light without stratification (compare Fig. 5C with Fig. 3C). This implies that light either interferes with synchronization among a population of seedlings or may override a prior synchronizing signal. The phase of the Arabidopsis LHCB1*1 promoter cannot be reset by light pulses provided up to 12 h after imbibition, demonstrating that freshly imbibed seeds are initially unable to process light information for the control of circadian gene expression (Kolar et al., 1998). Only 60 h following imbibition did LHCB phase respond to light pulses (Kolar et al., 1998). We speculate that the lower degree of synchronization among seedlings germinated directly in continuous light stems from the successive phase setting information provided first by the act of seed imbibition and second by the onset of phase setting in response to light. If the onset of light resetting varied slightly from seedling to seedling, the synchrony of phases of a group of light-grown seedlings would be reduced relative to etiolated seedlings, as we have observed in our experiments.
In all experiments, we have limited the amount of light, both in fluence rate and duration, received by the imbibed seeds during manipulation and transfer into 96-well plates. It is known that germination can be induced by phytochrome B (phyB) as early as 1 h following the onset of seed imbibition, indicating that light-responsive phy proteins are present in the seed at the time of imbibition (Shinomura et al., 1994). This seed pool of phy proteins may not be coupled to the control of clock and output gene expression, as light pulses given during the first 12 h following onset of imbibition failed to induce the expression of LHCB1*3 (Brusslan and Tobin, 1992) or reset the phase of LHCB (Kolar et al., 1998). Light responsiveness in gene expression only becomes apparent at least 24 h after onset of imbibition (Brusslan and Tobin, 1992; Kolar et al., 1998). We therefore suspect that onset of circadian rhythmicity and initial synchronization among etiolated seedlings stem from seed imbibition and not phy photoconversion while handling imbibed seeds into 96-well plates.
In the fungus Neurospora crassa, entraining cycles are dispensable for rhythmicity. Synchronization of cells within the suspension (in liquid culture) or growth front (on race tubes) is accomplished by maintaining cells in LL for a few hours before transfer into DD. At the molecular level, LL drives the continuous induction of the clock gene frq by the White Collar Complex (WCC; Crosthwaite et al., 1995). Upon transfer into DD, FRQ binds to the WCC and represses the activity of the complex, resulting in a decrease in frq message and protein. In Arabidopsis, initiation of rhythmicity by imbibition may follow a similar mechanism, whereby imbibition induces the expression of one of the clock genes. Clock gene mRNAs accumulate to high levels in seeds (Supplemental Fig. S6), and mutants carrying loss-of-function alleles of these genes fail to initiate rhythms during the first 2 d following imbibition (Fig. 7), demonstrating their essential involvement in the early onset of circadian rhythmicity. However, our results cannot distinguish between a possible maternal contribution and imbibition-induced transcription. The germinating seed might not require any transcription of clock genes early but instead would make use of the clock mRNAs deposited in the seed on the mother plant. Alternatively, clock proteins could represent the maternal contribution, bypassing the need for de novo transcription or translation. Because our CLOCK GENE∷LUC reporters include promoter and 5′ untranslated region sequences, we cannot discriminate between early rhythms in transcription, mRNA stability, or translation and each may contribute to the oscillations in LUC activity. In zebrafish, zper3 mRNA levels appeared rhythmic as early as 1 d postfertilization, but zper3∷luc (which contains both promoter and 5′ untranslated region sequences) is not rhythmic before 4 to 5 d postfertilization (Delaunay et al., 2000; Kaneko and Cahill, 2005). Early rhythmic mRNA levels in zebrafish embryos may in fact reflect circadian regulation of mRNA stability, rather than rhythmic de novo transcription during early larval development.
Early recording of LUC activity during seed germination supports results from mammalian cell culture studies in which circadian rhythms were maintained across cell divisions (Nagoshi et al., 2004). Although cell division is ongoing during plant growth, it has remained unclear whether rhythms recorded from 10- to 17-d-old seedlings emanated from dividing cells, nondividing differentiated cells, or both. We detected rhythmic expression of PRR7 as early as 2 d after imbibition, followed by LHY in the next cycle. During this developmental time, imbibed seeds undergo numerous cell divisions; between 36 and 63 h after release from stratification, about 30% to 40% of all cells exhibit a 4C ploidy level indicative of cells in the G2 stage of mitosis (Masubelele et al., 2005). Although we cannot absolutely rule out circadian expression exclusively from nondividing cells during this window of time, it seems likely that circadian phase is maintained across cell divisions.
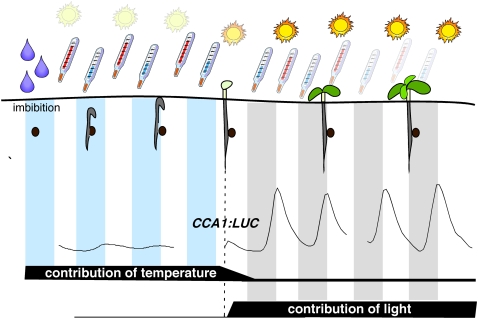
We have shown here that etiolated seedlings possess a functional circadian clock, albeit one with low amplitude oscillations (Fig. 6). What advantage would young etiolated seedlings gain from initiating oscillations in their clock components when a single dark-to-light transition is sufficient in starting high amplitude oscillations? A germinating seed will likely be buried in the soil and will need to break the surface before starting photomorphogenesis, development in the light. While in the soil, the germinating seed will be subjected to daily variations in temperature, and these will help to synchronize the endogenous clock with the external thermo- and photoperiod (Fig. 8). Light is a much stronger zeitgeber than temperature in most systems, but the clock in germinating seedlings cannot be reset by light immediately following imbibition (Kolar et al., 1998), which may ensure that circadian phase will not be reset by light before cotyledons have truly emerged from the soil surface. In a shallowly buried seed that is directly exposed to light, this may ensure that light resetting of phase does not occur immediately upon imbibition but rather is delayed until a dark-to-light transition at the subsequent true dawn. At this stage, light perception will raise amplitude of clock-regulated genes whose phases peak in the morning, like CCA1 (Fig. 6C). Because seeds may germinate months or even years after leaving the mother plant, circadian synchronization with maternal rhythms is unlikely to be effective. Instead, plants have been obliged to use imbibition, the first step in germination, for synchronization during the onset of circadian rhythms.
Figure 8.
Integration of signals from imbibition and thermo- and photocycles in synchronization of germinating seeds. Imbibition initiates the circadian clock in the germinating seed. While the newly emerging seedling grows in the soil and, therefore, in the dark, thermocycles from the outside environment are sufficient to entrain their circadian oscillator as circadian amplitude is low. Upon release into the light, amplitude greatly increases, thereby diminishing the relative contribution to entrainment from thermocycles, instead switching the main resetting signals to photocycles.
MATERIALS AND METHODS
LUC Activity Recordings
Seeds were surface sterilized overnight by vapor-phase sterilization (Clough and Bent, 1998). This dry sterilization technique, in contrast to other sterilization methods like immersion in bleach or ethanol and subsequent rinsing, does not hydrate the seeds and allows for precise experimental control of the timing of imbibition. Seeds were hydrated at noon the following day, either by sowing seeds on petri plates containing Murashige and Skoog salts supplemented with 2% Suc and 0.8% agar, or by sowing seeds into the wells of 96-well plates containing 280 μL of Murashige and Skoog salts, 2% Suc, 0.8% agar, and 30 μL of 2.5 mm d-luciferin, potassium salt. For stratification treatments, 96-well plates were covered in two layers of aluminum foil and kept in a cold room for 3 d at 4°C before being released at noon on the fourth day into LL, DD, 12-h-light/12-h-dark (LD), or 12-h-warm/12-h-cold (HC) cycles. For experiments without stratification, seeds were sown as described above and released directly into LL or DD, at which time LUC activity measurements started. In all cases, with or without stratification, hydrating seeds spent at most 1 h under the light of the sterile hood (fluence = 10–15 μmol m−2 s−1) before advancing to the next step of the protocol (Figs. 1, A and D; 2, A and D; 3, A and D; 5A; and 6, A–D). When seeds were released into constant conditions, we arbitrarily designated ZT0 as time of release from stratification (Figs. 1D and 2A) or of seed hydration (Fig. 3).
LUC measurements were carried out as described (Salomé et al., 2002). Briefly, LUC activity was detected with a TopCount luminometer (Perkin Elmer Life Sciences). Controlled temperature environment was provided by a walk-in AC-40 “Bioroom” growth chamber (Biochambers) or by a Percival I-66 incubator (Percival Scientific). Raw data were imported to Excel with the help of import and analysis, and circadian parameters extracted by fast-Fourier transform nonlinear least square analysis (Plautz et al., 1997). Phase plots were drawn with the Oriana 2.0 software package (Kovach Computing Services). Whole seedling LUC imaging was performed using a Hamamatsu ORCA II ER CCD camera (C4742-98 ERG; Hamamatsu Photonics, http://www.hamamatsu.com), with data collected every 1 h with 30-min exposure times. Images were analyzed with Metamorph software (Molecular Devices, http://www.moleculardevices.com/pages/software/metamorph.html).
To facilitate comparison among genes of different absolute expression levels, LUC activities for each seedling at each time point are expressed relative to the mean LUC activity for that seedling and are presented as “normalized LUC activity,” except where otherwise indicated. Thus, absolute expression levels cannot be inferred from the figures except where directly presented (Figs. 5 and 6A).
Plant Genotypes
Transgenic lines in the Col-2 background were generated previously (Salomé and McClung, 2005a). Details about the promoters are given in Supplemental Table S1. Multiple (4–10) T2 lines were investigated for consistent circadian expression (based on overall expression level, period, and phase) and were all found to behave similarly. Therefore, the first T2 line for each reporter construct was carried on to the T3 generation and used throughout. Plants harboring the LHCB1*1 and CAT3 promoters fused to LUC were described previously (Anderson et al., 1994; Michael and McClung, 2002). For the generation of the CAT2∷LUC fusion, the CAT2 promoter was PCR amplified from bacterial artificial chromosome clone M4E13 and cloned into pPZP LUCΩ+ (Schultz et al., 2001).
Mutants in circadian clock components described in this study are toc1-101 (Kikis et al., 2005), lhy-20 (Michael et al., 2003b), ztl-4 (Michael et al., 2003b), prr7,9 (Michael et al., 2003b; Salomé and McClung, 2005a), and gi-201 (Martin-Tryon et al., 2007). With the exception of prr7,9, the TOC1∷LUC transgene was introduced into the mutants via genetic crossing, and selection of F2 plants with the appropriate circadian phenotype in LL after LD entrainment. All SALK lines were generated in the Col-0 background; we have not detected any effect due to potential natural variation between the two accessions in F1, F2, and F3 progeny of a cross between Col-2 TOC1∷LUC and Col-0 (P.A. Salomé, unpublished data).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this study are as follows: CAT2 (At4g35090), CAT3 (At1g20620), CCA1 (At2g46830), CCR2 (At2g21660), ELF4 (At2g40080), GI (At1g22770), LHCB1*1 (At1g29920), LHCB1*3 (At1g29930), LHY (At1g01060), LUX/PCL (At3g46640), PRR7 (At5g02810), PRR9 (At2g46790), TOC1 (At5g61380), and ZTL (At5g57360).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. LD cycles are dispensable for rhythmicity but not for synchronization between individuals.
Supplemental Figure S2. Etiolated seedlings are rhythmic.
Supplemental Figure S3. Description of the CAT2∷LUC reporter.
Supplemental Figure S4. Germinating seeds are rhythmic.
Supplemental Figure S5. Stratification is dispensable for rhythmicity of etiolated seedlings.
Supplemental Figure S6. Expression of the clock genes CCA1, LHY, TOC1, ZTL, PRR7, and PRR9 in the developmental series of the AtGenExpress tool.
Supplemental Movie S1. LHY∷LUC is expressed in the cotyledons of etiolated seedlings.
Supplemental Table S1. Promoter∷LUC fusions used in this study.
Supplementary Material
Acknowledgments
We thank Jerry Hayes, Perkin Elmer support engineer, for technical support over the years. We are also indebted to Detlef Weigel for allowing some of the experiments to be conducted in his laboratory. We acknowledge the Arabidopsis Biological Resource Center (The Ohio State University, Columbus, OH) for DNA clones, and SIGnAL (Salk Institute, La Jolla, CA) for providing the sequence-indexed Arabidopsis T-DNA insertion mutants.
This work was supported by the National Science Foundation (grant no. MCB–0343887 to C.R.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: C. Robertson McClung (mcclung@dartmouth.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293 880–883 [DOI] [PubMed] [Google Scholar]
- Anderson SL, Kay SA (1995) Functional dissection of circadian clock- and phytochrome-regulated transcription of the Arabidopsis CAB2 gene. Proc Natl Acad Sci USA 92 1500–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SL, Somers DE, Millar AJ, Hanson K, Chory J, Kay SA (1997) Attenuation of phytochrome A and B signaling pathways by the Arabidopsis circadian clock. Plant Cell 9 1727–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SL, Teakle GR, Martino-Catt SJ, Kay SA (1994) Circadian clock- and phytochrome-regulated transcription is conferred by a 78 bp cis-acting domain of the Arabidopsis CAB2 promoter. Plant J 6 457–470 [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ (2005) Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6 544–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusslan JA, Tobin EM (1992) Light-independent developmental regulation of cab gene expression in Arabidopsis thaliana seedlings. Proc Natl Acad Sci USA 89 7791–7795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr AJF, Whitmore D (2005) Imaging of single light-responsive clock cells reveals fluctuating free-running periods. Nat Cell Biol 7 319–321 [DOI] [PubMed] [Google Scholar]
- Chory J, Peto CA, Ashbaugh M, Saganich R, Pratt L, Ausubel F (1989) Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. Plant Cell 1 867–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Covington MF, Harmer SL (2007) The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5 e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosthwaite SK, Loros JJ, Dunlap JC (1995) Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell 81 1003–1012 [DOI] [PubMed] [Google Scholar]
- Delaunay F, Thisse C, Marchand O, Laudet V, Thisse B (2000) An inherited functional circadian clock in zebrafish embryos. Science 289 297–300 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Kay SA (2000) Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12 2499–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419 74–77 [DOI] [PubMed] [Google Scholar]
- Dunlap JC (1999) Molecular bases for circadian clocks. Cell 96 271–290 [DOI] [PubMed] [Google Scholar]
- Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JCW, Lynn JR, Straume M, Smith JQ, Millar AJ (2006) FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15 47–54 [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several membrane-spanning domains. EMBO J 18 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould PD, Locke JCW, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, et al (2006) The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jud C, Albrecht U (2006) Circadian rhythms in murine pups develop in absence of a functional maternal circadian clock. J Biol Rhythms 21 149–154 [DOI] [PubMed] [Google Scholar]
- Kaneko M, Cahill GM (2005) Light-dependent development of circadian gene expression in transgenic zebrafish. PLoS Biol 3 e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Helfrich-Forster C, Hall JC (1997) Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: newly identified pacemaker candidates and novel features of clock gene product cycling. J Neurosci 17 6745–6760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis EA, Khanna R, Quail PH (2005) ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J 44 300–313 [DOI] [PubMed] [Google Scholar]
- Kolar C, Ádám É, Schäfer E, Nagy F (1995) Expression of tobacco genes for light-harvesting chlorophyll a/b binding proteins of photosystem II is controlled by two circadian oscillators in a developmentally regulated fashion. Proc Natl Acad Sci USA 92 2174–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar C, Fejes E, Ádám É, Schäfer E, Kay S, Nagy F (1998) Transcription of Arabidopsis and wheat Cab genes in single tobacco transgenic seedlings exhibits independent rhythms in a developmentally regulated fashion. Plant J 13 563–569 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316 715–719 [PubMed] [Google Scholar]
- Lahiri K, Vallone D, Gondi SB, Antoriello C, Dickmeis T, Foulkes NS (2005) Temperature regulates transcription in the zebrafish circadian clock. PLoS Biol 3 e351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JCW, Kozma-Bognár L, Gould PD, Fehér B, Kevei É, Nagy F, Turner MS, Hall A, Millar AJ (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2: 59 [DOI] [PMC free article] [PubMed]
- Locke JCW, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS, Millar AJ (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Sys Biol 1 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T (2002) The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol 43 58–69 [DOI] [PubMed] [Google Scholar]
- Martin-Tryon EL, Kreps JA, Harmer SL (2007) GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol 143 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P, Kim W-Y, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426 567–570 [DOI] [PubMed] [Google Scholar]
- Masubelele NH, Dewitte W, Menges M, Maughan S, Collins C, Huntley R, Nieuwland J, Scofield S, Murray JA (2005) D-type cyclins activate division in the root apex to promote seed germination in Arabidopsis. Proc Natl Acad Sci USA 102 15694–15699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR (2006) Plant circadian rhythms. Plant Cell 18 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWatters HG, Kolmos E, Hall A, Doyle MR, Amasino RM, Gyula P, Nagy F, Millar AJ, Davis SJ (2007) ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol 144 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, McClung CR (2002) Phase-specific circadian clock regulatory elements in Arabidopsis thaliana. Plant Physiol 130 627–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, et al (2008) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules PLoS Genet 4 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Salomé PA, McClung CR (2003. a) Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc Natl Acad Sci USA 100 6878–6883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Salomé PA, Yu HJ, Spencer TR, Sharp EL, Alonso JM, Ecker JR, McClung CR (2003. b) Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302 1049–1053 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Short SR, Chua NH, Kay SA (1992) A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell 4 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song H-R, Carré IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2 629–641 [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U (2004) Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119 693–705 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Ito S, Sato E, Yamashino T, Mizuno T (2005) PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol 46 686–698 [DOI] [PubMed] [Google Scholar]
- Onai K, Ishiura M (2005) PHYTOCLOCK1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10 963–972 [DOI] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285 1579–1582 [DOI] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA (1997) Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12 204–217 [DOI] [PubMed] [Google Scholar]
- Reppert SM (1995) Interaction between the circadian clocks of mother and fetus. In DJ Chadwick, K Ackrill, eds, Circadian Clocks and Their Adjustment, CIBA Foundation Symposium Vol 183. Wiley, Chichester, UK, pp 198–211 [DOI] [PubMed]
- Reppert SM, Schwartz WJ (1983) Maternal coordination of the fetal biological clock in utero. Science 220 969–971 [DOI] [PubMed] [Google Scholar]
- Salomé PA, McClung CR (2005. a) PRR7 and PRR9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, McClung CR (2005. b) What makes Arabidopsis tick: light and temperature entrainment of the circadian clock. Plant Cell Environ 28 21–38 [Google Scholar]
- Salomé PA, Michael TP, Kearns EV, Fett-Neto AG, Sharrock RA, McClung CR (2002) The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol 129 1674–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93 1219–1229 [DOI] [PubMed] [Google Scholar]
- Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA (2001) A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13 2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Price J, Young MW (1992) Ontogeny of a biological clock in Drosophila melanogaster. Proc Natl Acad Sci USA 89 1423–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Chory J, Furuya M (1994) The induction of seed germination in Arabidopsis thaliana is regulated principally by photochrome B and secondarily by phytochrome A. Plant Physiol 104 363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Devlin P, Kay SA (1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282 1488–1490 [DOI] [PubMed] [Google Scholar]
- Somers DE, Kim WY, Geng R (2004) The F-Box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101 319–329 [DOI] [PubMed] [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrin IX. Nature 421 79–83 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289 768–771 [DOI] [PubMed] [Google Scholar]
- Thain SC, Murtas G, Lynn JR, McGrath RB, Millar AJ (2002) The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol 130 102–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallone D, Lahiri K, Dickmeis T, Foulkes NS (2007) Start the clock! Circadian rhythms and development. Dev Dyn 236 142–155 [DOI] [PubMed] [Google Scholar]
- Vuilleumier R, Besseau L, Boeuf G, Piparelli A, Gothilf Y, Gehring WG, Klein DC, Falcon J (2006) Starting the zebrafish pineal circadian clock with a single photic transition. Endocrinology 147 2273–2279 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217 [DOI] [PubMed] [Google Scholar]
- Welsh DK, Yoo S-H, Liu AC, Takahashi JS, Kay SA (2004) Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol 14 2289–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilinger MN, Farré EM, Taylor SR, Kay SA, Doyle FJ III (2006) A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol Syst Biol 2 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong HH, Painter JE, Salomé PA, Straume M, McClung CR (1998) Imbibition, but not release from stratification, sets the circadian clock in Arabidopsis seedlings. Plant Cell 10 2005–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong HH, Resnick AS, Straume M, McClung CR (1997) Effects of synergistic signaling by phytochrome A and cryptochrome 1 on circadian clock-regulated catalase expression. Plant Cell 9 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong HH, Young JC, Pease EA, Hangarter RP, McClung CR (1994) Interactions between light and the circadian clock in the regulation of CAT2 expression in Arabidopsis. Plant Physiol 104 889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv L, Gothilf Y (2006) Circadian time-keeping during early stages of development. Proc Natl Acad Sci USA 103 4146–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.