Abstract
Magnolia grandiflora (Southern Magnolia) is a primitive evergreen tree that has attracted attention because of its horticultural distinctiveness, the wealth of natural products associated with it, and its evolutionary position as a basal angiosperm. Three cDNAs corresponding to terpene synthase (TPS) genes expressed in young leaves were isolated, and the corresponding enzymes were functionally characterized in vitro. Recombinant Mg25 converted farnesyl diphosphate (C15) predominantly to β-cubebene, while Mg17 converted geranyl diphosphate (C5) to α-terpineol. Efforts to functionally characterize Mg11 were unsuccessful. Transcript levels for all three genes were prominent in young leaf tissue and significantly elevated for Mg25 and Mg11 messenger RNAs in stamens. A putative amino-terminal signal peptide of Mg17 targeted the reporter green fluorescent protein to both chloroplasts and mitochondria when transiently expressed in epidermal cells of Nicotiana tabacum leaves. Phylogenetic analyses indicated that Mg25 and Mg11 belonged to the angiosperm sesquiterpene synthase subclass TPS-a, while Mg17 aligned more closely to the angiosperm monoterpene synthase subclass TPS-b. Unexpectedly, the intron-exon organizations for the three Magnolia TPS genes were different from one another and from other well-characterized TPS gene sets. The Mg17 gene consists of six introns arranged in a manner similar to many other angiosperm sesquiterpene synthases, but Mg11 contains only four introns, and Mg25 has only a single intron located near the 5′ terminus of the gene. Our results suggest that the structural diversity observed in the Magnolia TPS genes could have occurred either by a rapid loss of introns from a common ancestor TPS gene or by a gain of introns into an intron-deficient progenote TPS gene.
Many reports on the identification of a new terpene compound or the characterization of a terpene biosynthetic enzyme begin with a comment about the structural complexity and diversity of terpene metabolism in general. That complexity arises, in part, from the genetic and regulatory complexity of the gene families involved (Bohlmann et al., 1998; Trapp and Croteau, 2001; Gang, 2005), but more so from the terpene synthases (TPSs) themselves that initiate the biosynthesis of different classes of terpenes (i.e. monoterpenes, sesquiterpenes, and diterpenes) and impart exquisite regiospecificity and stereospecificity into the biosynthesis of specific members within a terpene family (Davis and Croteau, 2000; Christianson, 2006). Valencene synthase (Sharon-Asa et al., 2003) and 5-epi-aristolochene synthase (Back and Chappell, 1996), for example, are related to one another based on the biosynthesis of sesquiterpenes within the eremophilene class of compounds. Sesquiterpenes in the eremophilane class are fused (bicyclic), decalin ring structures that harbor methyl and isopropenyl substituents in the same regio positions, but with alternative stereo configurations. Valencene also contains a double bond in the A-ring, which is mirrored in the B-ring of aristolochene. One can imagine other possible ring structures (class designations) and alternative arrangements of the various substituents and double bonds, creating a wealth of chemical complexity just within this single class of sesquiterpenes. Equally intriguing to consider are the prospects that the synthase enzymes responsible for the biosynthesis of these similar yet distinct sesquiterpenes are evolutionarily related. As a first approximation, one might assume that these two enzymes arose from a common ancestral gene that underwent a duplication event followed by divergent evolution to give rise to the unique biochemical specificities. If this were so, and we could delineate an evolutionary pathway for how these enzyme specificities arose, then in principle we should be able to use this same pathway/strategy to evolve alternative and new TPS specificities.
One approach to study the evolution of TPSs has been to define the structural features of these enzymes that impart catalytic specificities to the respective enzymes. If regions or domains controlling catalysis can be identified and these in fact serve as “hot spots” for the evolution of catalytic specificities, then these regions of the genes might show different rates of mutation over evolutionary time relative to other regions of the protein not directly related to catalysis. First, such regions or domains must be identified. Two recent studies illustrate efforts in this regard. Yoshikuni et al. (2006) used a random approach to mutate all 19 amino acids within the putative active site of a multifunctional TPS and observed that select mutants in fact reduced the product diversity of these enzymes but did not alter or uncover new product specificities. In contrast, Greenhagen et al. (2006) evaluated the role of second-tier residues on TPS specificities. Second-tier residues were defined as those residues outside the active site, yet oriented in such a manner to influence active site residues, the contour of the active site pocket, and the catalytic cascade yielding reaction product specificities. These investigators were able to demonstrate that reciprocal mutations of eight to nine residues were sufficient to interconvert the catalytic specificity of 5-epi-aristolochene synthase to that of premnaspirodiene synthase and vice versa. Together, these two studies suggest that the evolution of reaction specificity within the TPS gene families could arise as a consequence of mutations within and outside the active site. Moreover, the residues outside the active site that influence catalysis were clearly defined as falling within specific regions and were not the consequence of a random distribution throughout the entire protein.
The notion of divergent evolution rather than convergent evolution for the TPSs is supported by the conservation of sequence similarity and gene structure among the diverse TPSs isolated from gymnosperms and angiosperms to date. Bohlmann et al. (1998) initially categorized TPSs into six subfamilies, TPS-a through TPS-f, based on amino acid sequence similarities and cladograms pointing to a common ancestral TPS gene(s). More recently, Trapp and Croteau (2001) extended this comparison by examining the intron-exon structures for 37 TPSs representing 18 TPS genes in gymnosperm species and 19 genes in angiosperm species. Their analyses included documenting the overall number and positions of introns as well as intron phasing. Class I TPS genes were described as containing 12 to 14 introns and consisting mainly of diterpene synthase genes from gymnosperms and angiosperms. Class II TPS genes contain nine introns inserted in a conserved pattern and are composed predominantly of monoterpene and sesquiterpene synthase genes found in gymnosperms. The class III TPS genes harbor six introns and include monoterpene, sesquiterpene, and diterpene synthase genes all from angiosperm species (Trapp and Croteau, 2001). These authors also suggested that the first progenitor TPS genes contained 14 introns and a highly conserved sequence coding for approximately 200 amino acids within the N-terminal portion of the gene, a sequence initially thought to be a specific sequence for diterpene synthase genes of conifers but subsequently recognized in angiosperm monoterpene and sesquiterpene synthases (Dudareva et al., 1996; Aubourg et al., 2002). Furthermore, they proposed that all TPS genes of extant angiosperms evolved from such an ancient gymnosperm gene through an “intron loss” mechanism. A more recent classification of 40 TPS genes annotated in the Arabidopsis (Arabidopsis thaliana) genome, categorizing 29 TPS genes into class III, three genes into class I, and eight as pseudogenes, has served to extend this model of TPS gene evolution (Aubourg et al., 2002).
To date, many of the TPS genes used in any of the phylogenetic analyses have been isolated from gymnosperms and several relatively advanced families of angiosperms. Moreover, the angiosperm families represent a very limited taxonomic range without much consideration for basal angiosperms (Cronquist, 1988). A more complete phylogeny for the TPS gene families would thus likely benefit from inclusion of TPS genes from putative ancient and diverse lineages. Basal angiosperms include plants within the Magnoliaceae, which are considered ancient because of the primitive organization of their floral parts (Crane et al., 1995), because of the molecular phylogeny of their rbcL, atpB, and 18S rDNA loci (Bremer et al., 2003), and because of their representation within fossil records (Golenberg et al., 1990). For instance, the fossil records suggest that the family Magnoliaceae existed 20 to 25 million years ago, yet the present day rbcL gene found in Magnolia grandiflora exhibits only 17-bp differences relative to the likely progenitor gene found in the fossil species Magnolia latahensis. Such information provides an evolutionary context for comparisons of extant TPS genes in Magnolia species with the TPS genes found in other angiosperms and gymnosperms (Golenberg et al., 1990).
The aim of the work described here was to isolate and functionally characterize TPS genes from M. grandiflora, and then to use this information to assess prevailing models for the evolution of TPS gene structure and function according to the previous studies (Bohlmann et al., 1998; Trapp and Croteau, 2001; Greenhagen et al., 2006). Given these models, we anticipated that the TPS genes of M. grandiflora would more closely resemble the class I genes of gymnosperms in possessing many introns, up to 14, and to encode TPSs catalyzing single to multiple reaction product profiles. What we found is that the three M. grandiflora TPS genes characterized have many fewer introns than expected, yet they encode enzymes catalyzing reactions similar to many other TPS enzymes in yielding multiple, chemically diverse reaction products.
RESULTS
Isolation of TPS cDNAs from the Basal Angiosperm M. grandiflora
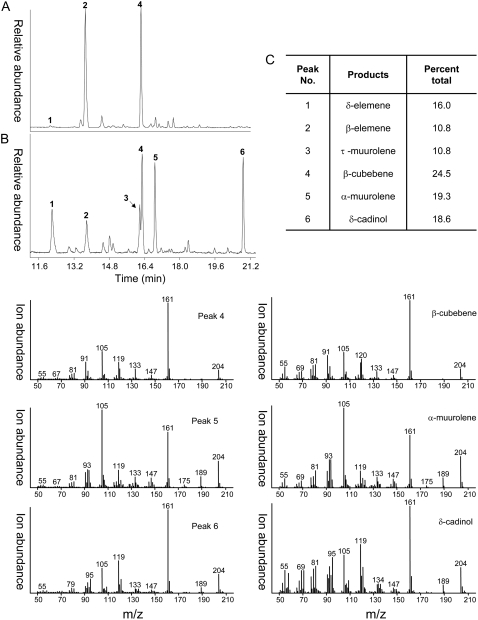
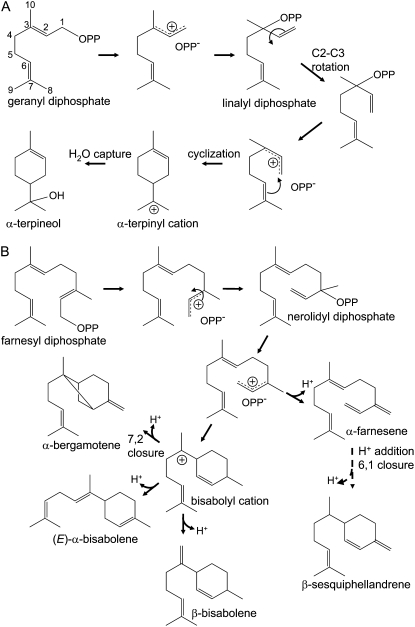
The chemical profile of terpenes found in Magnolia leaves was first assessed to facilitate the design of PCR primers for the isolation of TPSs catalyzing the biosynthesis of particular classes of terpenes. Interestingly, two terpenes dominated the profile, with β-cubebene accounting for 31.3% of the total and β-elemene for 25.3% (Fig. 2A). β-Elemene is not the actual terpene accumulating in the Magnolia leaves. Instead, germacrene A extracted from the leaf tissue undergoes a rapid Cope rearrangement to β-elemene upon injection into the heated sample port of the GC (data not shown; Takeda, 1974; de Kraker et al., 1998). Hence, the dominant terpenes accumulating in Magnolia leaves are β-cubebene and germacrene A, which are biosynthetically related by a predicted germacrenyl cation intermediate (Fig. 3).
Figure 2.
Total ion chromatograms of the sesquiterpene products generated by Mg25. The Mg25 cDNA was expressed in bacteria and bacterial lysates used as the source of enzyme for in vitro assays, which consisted of incubations with FPP and profiling the reaction products by GC-MS. A, GC trace of terpenes extracted from M. grandiflora leaves. B, GC trace of in vitro reaction products generated by the Mg25 enzyme. C, Table of relative amounts of products generated by the Mg25 enzyme, calculated from the chromatograph in B. The peaks labeled in the GC traces (A and B) were identified by comparison of their mass spectra with those in the NIST mass spectra library and authentic standards, as illustrated in the panels at bottom for β-cubebene, α-muurolene, and δ-cadinol and in Supplemental Figure S1 for δ-elemene, β-elemene, and τ-muurolene.
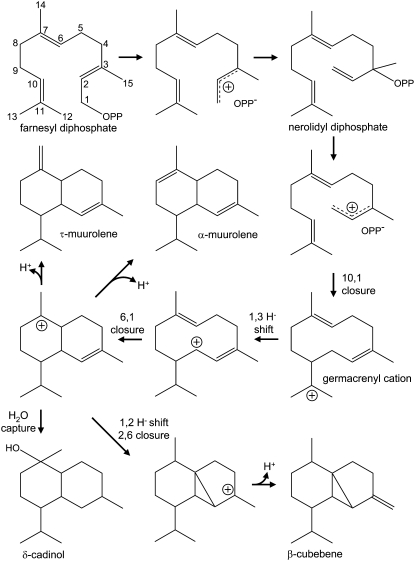
Figure 3.
Proposed reaction mechanisms rationalizing the reaction products generated by Mg25 upon incubation with the substrate FPP.
The initial PCR primers were designed to conserved regions near the C-terminal Asp-rich motif (DDXXD) found in association with germacrene A to D synthases functionally characterized from plants in the Solanaceae and Asteraceae (Colby et al., 1998; van der Hoeven et al., 2000; Bennett et al., 2002; Bouwmeester et al., 2002; Prosser et al., 2002, 2004; Arimura et al., 2004; Iijima et al., 2004). Seven fragments between 100 and 250 bp in length were amplified using these degenerate primers in reverse transcription (RT)-PCR assays with mRNA extracted from young leaf tissue. All seven fragments exhibited sequence similarities to TPS genes (E ≤ e-19) and were considered further. 5′ and 3′ RACE approaches were then used to isolate three full-length cDNAs, and the other four cDNAs were subsequently disregarded because they possessed several internal stop codons, were unusually short (1–1.2 kb), or were missing obvious 5′ start sequences.
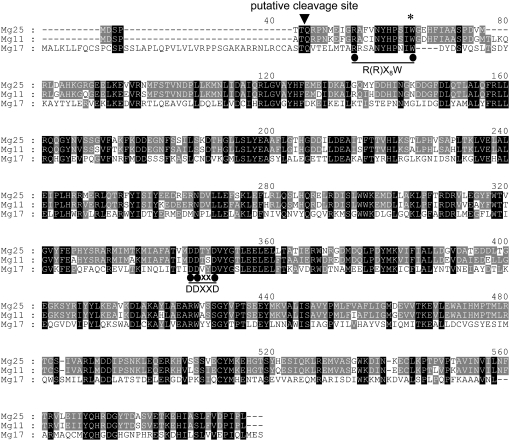
Mg25 (551 residues, 1,653 bp) and Mg11 (555 residues, 1,779 bp) showed considerable sequence similarities to one another, with 91.7% identity at the nucleotide level and 91.5% and 87.2% similarity and identity, respectively, at the predicted amino acid level (Fig. 1). No additional evidence for N-terminal sequences upstream of the R(R)X8W motif was obtained for either of these cDNAs even after multiple 5′ RACE attempts. Highly conserved regions among sesquiterpene synthases were also found in these putative sesquiterpene synthases from the basal angiosperm Magnolia. For example, the Asp-rich substrate binding motif known as DDXXD (Fig. 1) and the R(R)X8W motif (Fig. 1) were found in spatially conserved positions. Mg25 and Mg11 were most similar to the germacrene D synthase gene from grape vine (Vitis vinifera), with amino acid identity in excess of 48% and similarity greater than 66% (Lucker et al., 2004). Mg11 contained an internal stop codon corresponding to residue 25, which was mutagenized to a Trp codon, a conserved amino acid at this position among sesquiterpene synthase, prior to functional analysis. In contrast, Mg17 (592 residues, 1,779 bp) harbored not only the DDXXD motif and the RRX8W (residues 54–64) but an additional N-terminal sequence (43 amino acids) characteristic of known monoterpene synthases (Bohlmann et al., 1998; Williams et al., 1998; Lucker et al., 2002). Moreover, the Mg17 N-terminal sequence upstream of the RRX8W motif was predicted as a putative transit peptide for chloroplast targeting by several computational algorithms (ChloroP, Predotar, and PSORT). Mg17 most closely resembled the grape vine α-terpineol synthase, with an amino acid sequence identity of 60% and similarity score of 75% (Martin and Bohlmann, 2004).
Figure 1.
A comparison of the deduced amino acid sequences for TPSs cloned from M. grandiflora. All three genes have a conserved Asp-rich domain, DDXXD, that coordinates substrate binding via the formation of divalent cation salt bridges (Starks et al., 1997). The monoterpene synthase, Mg17, contains a R(R)X8W motif, which is a characteristic domain for all monoterpene synthase, as well as a putative N-terminal signal sequence (approximately 43 amino acids at the N-terminal region), which putatively targets the mature protein to intracellular organelles. The putative cleavage site (arrowhead) for Mg17 was predicted by the ChloroP server (http://www.cbs.dtu.dk/services/ChloroP/). The asterisk denotes the location of an internal stop codon in Mg11 that was removed by site-directed mutagenesis, creating a Trp (W) residue instead.
Functional Characterization of Mg25 as a β-Cubebene Synthase
To determine the functional activities encoded by the putative TPS cDNAs, each was expressed in bacteria and bacterial lysates were tested for their ability to convert geranyl diphosphate (GPP), farnesyl diphosphate (FPP), and geranylgeranyl diphosphate (GGPP) into terpene reaction products. Hydrocarbon and oxygenated reaction products generated by incubations with the recombinant enzymes were analyzed by gas chromatography-mass spectrometry (GC-MS) and identified by a combination of retention time and mass spectra in comparison with authentic standards and National Institute of Standards and Technology (NIST) library matches (Rising et al., 2000; Martin et al., 2004). Mg25 was capable of utilizing FPP as a substrate, but not GPP (negligible activity) or GGPP (no activity). The dominant reaction products generated by Mg25 were β-cubebene (24.5%), α-muurolene (19.3%), δ-cadinol (18.6%), δ-elemene (16.0%), τ-muurolene (10.8%), and β-elemene (10.8%; Fig. 2B). The multifunctional nature of Mg25 is not unique (Steele et al., 1998), and the multiple reaction products can readily be rationalized as arising from a common reaction intermediate, nerolidyl diphosphate, through a series of hydride shifts, ring closures, deprotonations, and a hydroxide capture (Fig. 3). Not surprisingly, the mixture of reaction products was pH dependent (Supplemental Fig. S1A). Maximal activity was observed at pH 7.5 and dominated by four products: β-cubebene, α-muurolene, δ-cadinol, and τ-muurolene. However, at pH 5.5, no β-cubebene was evident; it was only detected from pH 6.5 to pH 8.5. In comparison, δ-cadinol was readily detected from pH 5.5 to 7.5 but not at pH values above this range. Maximal activity was observed at 24°C to 26°C, with an apparent Km value for FPP of 1.07 ± 0.22 μm and a Kcat value of 0.35 × 10−3 ± 0.02 s−1 (Supplemental Fig. S1B). Mg25 also exhibited a divalent cation requirement for activity, with maximal activities observed at Mg2+ concentrations of 1 to 10 mm and Mn2+ concentrations of 1 mm or less (Supplemental Fig. S1C).
Functional Characterization of Mg11 Failed
Although full-length cDNAs for Mg11 were isolated repeatedly in separate experimental attempts, a stop codon was invariably observed at the position corresponding to codon 25. In an attempt to express and functionally characterize Mg11, the stop codon was substituted with that for Trp (W), which is a highly conserved residue at this position among TPSs. Unfortunately, no enzyme activity was ever associated with lysates derived from bacteria overexpressing the modified Mg11 cDNA, despite attempts with a variety of expression vectors (pET100/D-TOPO, pET28, pET32, and pET42) and expression conditions, including the induction treatment, the induction time period, and the temperature at which the bacteria were grown. In all cases, the Mg11 protein was detected in the insoluble fraction (inclusion bodies), and even attempts to solubilize the Mg11 protein using chaotropic reagents like urea were unsuccessful in uncovering any TPS activity with GPP, FPP, and GGPP as substrates.
Functional Characterization of Mg17 as an α-Terpineol Synthase
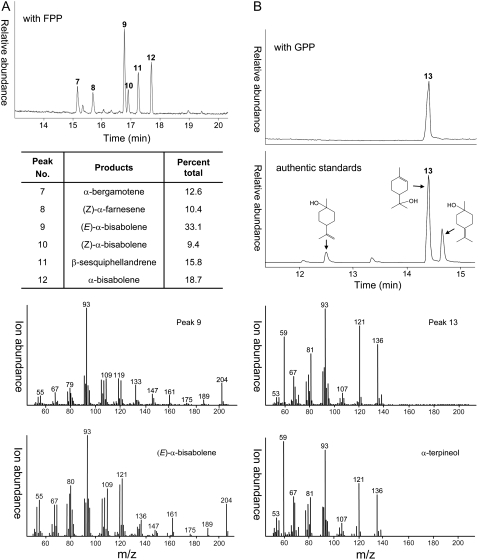
When overexpressed in Escherichia coli, N-terminal truncated Mg17 encoded a bifunctional TPS that readily utilized GPP and FPP as substrates, producing α-terpineol and (E)-α-bisabolene, respectively, as the major reaction products (Fig. 4). Deletion of the DNA sequence coding for the putative signal peptide (43 amino acids) at the N terminus was necessary in order to obtain soluble enzyme activity, similar to what has been observed for many monoterpene synthases (Bohlmann et al., 1998); otherwise, an insoluble form of the Mg17 protein was observed. With FPP as the substrate, bisabolenoid sesquiterpenes were produced by the Mg17 enzyme. These included (E)-α-bisabolene (33.1%) as the principal product, α-bisabolene (18.7%), β-sesquiphellandrene (15.8%), β-bergamotene (12.6%), (Z)-α-farnesene (10.4%), and (Z)-α-bisabolene (9.4%; Fig. 4). With GPP as the substrate, only a single monocyclic alcohol, α-terpineol, was biosynthesized. This contrasts with the results obtained with α-terpineol synthases isolated from grape vine (Martin and Bohlmann, 2004) and pine (Pinus spp.; Phillips et al., 2003). Grape vine α-terpineol synthase catalyzes the conversion of GPP into 14 monoterpene products, including both enantiomers of α-terpineol, together constituting greater than 50% of the total reaction products (Martin and Bohlmann, 2004). Phillips et al. (2003) likewise demonstrated that the loblolly pine monoterpene synthase Pt10 converted GPP into at least six reaction products, with multiple enantiomeric forms of α-terpineol accounting for 57% of the total reaction products.
Figure 4.
Total ion chromatograms of the monoterpene (A) and sesquiterpene (B) products generated by Mg17. An N-terminal signal sequence corresponding to 43 amino acids was deleted from the Mg17 cDNA before expressing this gene in bacteria. Bacterial lysates were subsequently used as the source of enzyme for in vitro assays, which consisted of incubations with GPP (A) or FPP (B) and profiling the reaction products by GC-MS. Labeled peaks were either identified by MS comparisons with authentic standards or matched to the mass spectra of compounds reported in the NIST library: 7, β-bergamotene; 8, (Z)-α-farnesene; 9, (E)-α-bisabolene; 10, (Z)-α-bisabolene; 11, β-sesquiphellandrene; 12, α-bisabolene; 13, α-terpineol. Mass spectra for the dominant products formed with FPP or GPP are shown at bottom, while the spectra for the other peaks are provided in Supplemental Figure S2.
Because of Mg17's higher sequence similarity to monoterpene synthases than sesquiterpene synthases and its putative N-terminal chloroplast targeting sequence, Mg17 was classified as a monoterpene synthase. This was in spite of reports that monoterpene synthases typically have a more restricted substrate specificity (Steele et al., 1998). Nonetheless, plausible mechanisms for how Mg17 might utilize either GPP or FPP as substrate can be reconciled with chemical rationalization for partial reactions, including initial ionization followed by isomerization in common with both substrates (Fig. 5).
Figure 5.
Proposed reaction mechanisms to rationalize the monoterpene (A) and sesquiterpene (B) reaction products generated by Mg17 upon incubation with the substrates GPP and FPP.
Steady-State mRNA Accumulation in Various Tissues
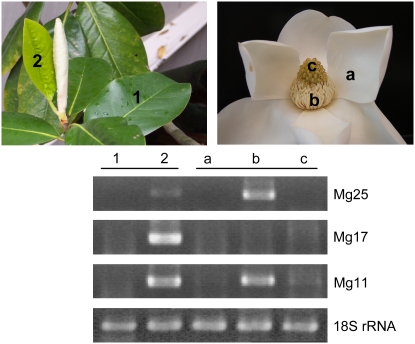
A quantitative RT-PCR method was employed to measure the steady-state transcript levels for the three TPS genes in leaves at different developmental stages and in flower organs, including tepals (sepals + petals), stamens, and carpels. PCR primer pairs were designed to selectively amplify each gene (Supplemental Fig. S3) and normalized relative to an amplicon for 18S rRNA in each sample (Fig. 6). Transcripts for all three genes, Mg25, Mg17, and Mg11, were much more evident in young developing leaf tissues than in older, more mature leaves (Fig. 6). And while the transcript levels for all three TPS genes were low or below detection limits in tepal and carpel tissues, the levels of Mg25 and Mg11 mRNA were greatly elevated in stamens.
Figure 6.
Measurements of the steady-state mRNA levels for the M. grandiflora TPS genes Mg25, Mg17, and Mg11 in different tissues. Total RNAs were isolated from the indicated tissues and used as the templates for gene-specific RT-PCR assays. Gene-specific primers were designed and evaluated by sequencing of the respective amplicons (data not shown). Amplification of an 18S ribosomal RNA fragment was used as the PCR control. A, The different developmental stages of leaves: 1, mature leaf; 2, developing leaf. B, The primitive floral organs of M. grandiflora: a, tepals (sepals + petals); b, stamens; c, carpels.
The N-Terminal Sequence of Mg17 Functions as a Dual Targeting Signal Sequence
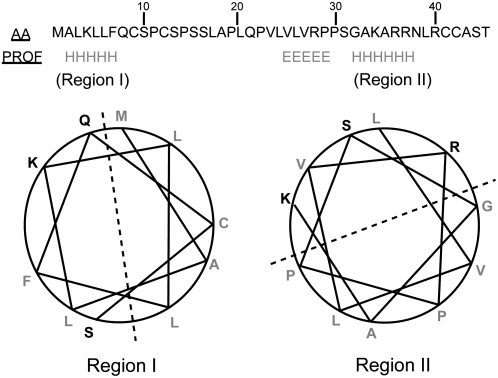
Several signal sequence analysis programs, including Predotar, PSORT, and ChloroP, predicted that the N-terminal extension of Mg17 could be targeting this enzyme to both the chloroplast and the mitochondrial compartments. The Mg17 transit peptide (Mg17tp) sequence also satisfied the criteria that were derived from the previously characterized transit peptides (Roise et al., 1988; Akashi et al., 1998). Mg17tp lacks acidic residues (0%) but has a significant proportion of basic (13.3%), hydrophilic (31.1%), and hydrophobic (37.7%) residues, making this peptide sequence potentially amphiphilic. The amphiphilic nature of signal sequences was noted earlier by Cunillera et al. (1997) to give targeting sequences sidedness when projected in helical formats. Secondary structure predictions for the 43 residues making up the putative signal sequence of Mg17 likewise suggested two possible amphiphilic structures, from residues 1 to 9 and residues 24 to 34, with each displaying hydrophobic residues on one face of the helix and polar/basic residues on the other (Fig. 7).
Figure 7.
Secondary structures and helical wheel predictions of the Mg17 transit peptide (Mg17tp). Secondary structures (PROF) of the Mg17tp were predicted by the PredictProtein program (Rost et al., 2004) and are noted below the amino acid sequence (H, α-helix; E, β-sheet). Helical wheel predictions of the Mg17tp were drawn according to PEPWHEEL (http://bioweb.pasteur.fr/seqanal/interfaces/pepwheel.html). Letters in gray represent hydrophobic residues.
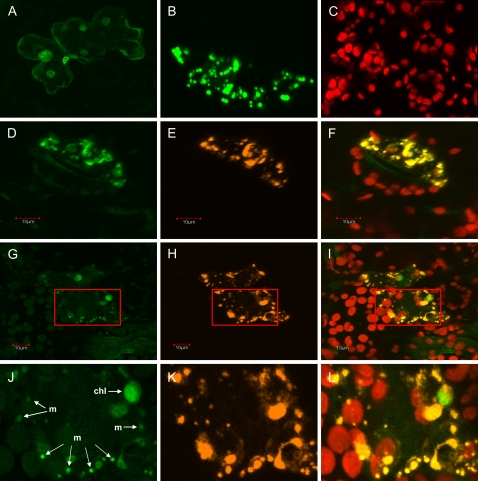
To functionally test the targeting nature of the Mg17 leader sequence, the coding sequence for the 43 N-terminal residues was fused to the GFP reporter gene, and transient expression of the construct was monitored after particle bombardment into epidermal cells of tobacco (Nicotiana tabacum) leaves by laser confocal microscopy (Fig. 8). Expression controls included the Arabidopsis RbcS transit peptide fused to GFP as a plastidic marker (Fig. 8A) and the Arabidopsis CoxIV signal sequence fused with DsRFP (for Discosoma red fluorescent protein) as a mitochondrial signal sequence control (Fig. 8B; Dinkins et al., 2003). When the recombinant Mg17tp:GFP and CoxIV:DsRFP were coexpressed, Mg17tp-GFP appeared localized to two populations of organelles resembling mitochondria and chloroplasts (Fig. 8, D, G, and J). Evidence for mitochondrial targeting was provided by colocalization of CoxIV-DsRFP to the smaller organelles (Fig. 8, E, H, and K), which overlapped with the Mg17tp-GFP images (Fig. 8, F, I, and L). Evidence that the fluorescence localized to the larger organelles in cells expressing Mg17tp-GFP was associated with chloroplasts was evident from the autofluorescence (red organelles in Fig. 8, F, I, and L) coincident with the GFP fluorescence and independent localization of RbcS-GFP and Mg17tp-GFP to these same organelles in separate determinations (data not shown).
Figure 8.
Confocal images of tobacco leaf epidermal cells showing the localization of reporter proteins to the chloroplasts and mitochondria when fused with the N-terminal signal peptide (43 amino acids) from the M. grandiflora monoterpene synthase Mg17. At least eight different cells were examined for each construct, but only representative images for targeting controls and two independent experimental images for Mg17tp-GFP are presented here. A, The Arabidopsis RbcS chloroplast signal sequence fused with GFP. B, The Arabidopsis CoxIV mitochondrial signal sequence fused with DsRFP. C, Background autofluorescence resulting from untransformed chloroplasts. D and G, Mg17 transit peptide fused with GFP. E and H, The CoxIV transit peptide fused to DsRFP. F, I, and L, Overlaid images (D + E, G + H, and J + K, respectively). J to L, Magnified images of corresponding zones indicated in G to I. Arrows labeled “chl” and “m” point to chloroplasts and mitochondria, respectively. Boxes in G through I were positioned on the identical area in each image.
Genomic Organization of TPS Genes in Magnolia
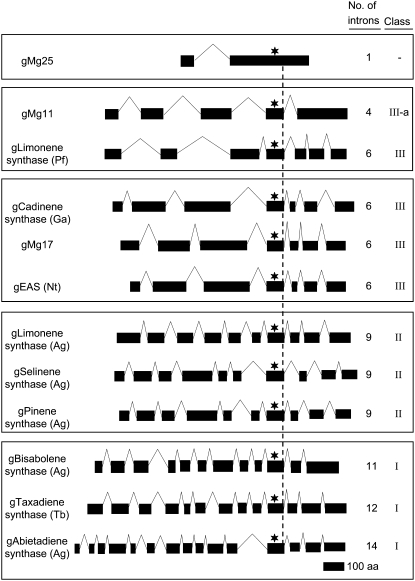
To examine the intron-exon organization of the Magnolia TPS genes, overlapping amplicons for genomic clones were assembled and aligned with each of the corresponding cDNAs. The genomic clones were then oriented relative to the highly conserved DDXXD motif to illustrate the relative lengths and positions of exons and introns, and were arrayed along with archetypical representatives of class I to III TPS genes defined according to Trapp and Croteau (2001; Fig. 9; Table I). Interestingly, all three Magnolia genes exhibited different intron-exon organizations. Mg17 resembled a typical class III TPS gene, with seven exons and six introns, much like 5-epi-aristolochene synthase from tobacco (Facchini and Chappell, 1992) or (+)-δ-cadinene synthase from cotton (Gossypium hirsutum; Chen et al., 1995). Also important, the exon and intron sizes are relatively well conserved between these class III genes and Mg17, except for exon 1 of Mg17, which contains an N-terminal signal sequence of an extra 129 bp. In contrast, Mg11 consists of five exons and four introns, with the first four exons organized similar to monoterpene synthase genes found within class III but differing by having a single C-terminal exon encompassing the equivalent of the last three exons of the class III genes. Because the Mg11 gene structure may represent a variant of the class III organization, it was designated as a class III-a member. The Mg25 genomic gene represents a more unique exon-intron organization, with only two exons interrupted by a single large intron. While the size of the 5′ exon does resemble those for other sesquiterpene synthase genes in the class III family, no similar single-intron-containing TPS gene from higher plants has yet to be reported. A fungal TPS gene harboring a single intron, however, has been observed (Trapp et al., 1998).
Figure 9.
Comparison of the genomic organization of Magnolia TPS genes with well-known and characterized TPS genes. Mg17 was assigned to TPS class III based on sequence alignments. However, Mg25 and Mg11 showed unique genomic structures in numbers and positions of exons and introns. Black blocks and open arrowheads represent exons and introns, respectively. Asterisks represent the position of the DDXXD motif. Gene classification is based on the intron-exon organization according to Trapp and Croteau (2001). aa, Amino acids; Ag, Abies grandis; Ga, Gossypium arboreum; Nt, N. tabacum; Pf, Perilla frutescens; Tb, Taxus brevifolia.
Table I.
A summary of TPS genes used to infer structural and functional features of the M. grandiflora TPS genes
Genomic sequences for Mg25, Mg11, and Mg17 were verified by comparing these to their corresponding cDNA clones, deriving the number, position, and size of introns.
| Product | Species | Class | GenBank Accession No. | Genomic DNAa | Amino Acid | No. of Introns |
|---|---|---|---|---|---|---|
| bp | ||||||
| β-Cubebene | M. grandiflora | Sesquiterpene | EU366429 | 2,339 | 551 | 1 |
| – | M. grandiflora | Sesquiterpeneb | EU366431 | 4,357 | 555 | 4 |
| α-Terpineol | M. grandiflora | Monoterpene | EU366430 | 2,783 | 593 | 6 |
| Trichodiene | M. roridum | Sesquiterpene | AF009416 | 1,215 | 385 | 1 |
| (−)-Limonene | Perilla frutescens | Monoterpene | AB005744 | 3,996 | 603 | 6 |
| 5-epi-Aristolochene | N. tabacum | Sesquiterpene | L04680 | 2,341 | 550 | 6 |
| (−)-Limonene | Abies grandis | Monoterpene | AF326518 | 3,059 | 637 | 9 |
| δ-Selinene | A. grandis | Sesquiterpene | AF326513 | 3,351 | 582 | 9 |
| (E)-α-Bisabolene | A. grandis | Sesquiterpene | AF326515 | 4,647 | 814 | 11 |
| Taxadiene | Taxus brevifolia | Diterpene | AF326519 | 3,999 | 862 | 12 |
| Abietadiene | A. grandis | Diterpene | AF326516 | 4,623 | 868 | 14 |
The size of each gene (genomic DNA) was calculated from the start codon to the termination codon.
Although an internal stop codon was substituted with a conserved amino acid (Trp) at this position, no enzyme activity was detected with a variety of substrates. Based on sequence homology and characteristic domains within the ORF, this gene was considered a putative sesquiterpene synthase pseudogene.
All three Magnolia genes contained intron-exon junction sites that followed standard GT/AG boundary rules (Brown and Simpson, 1998). Moreover, the phasing of the intron insertion sites appeared equally well conserved. Intron phasing is analogous to open reading frames (ORFs), refers to how codons are split across intron boundaries, and is defined as phase 0, 1, or 2 depending on the number of nucleotides split between two exons (Long et al., 1995). Based on the spatial positioning and phasing of each intron, the introns in all three Magnolia genes appear very well conserved relative to one another as well as with all other angiosperm and gymnosperm TPS genes examined (Table II).
Table II.
Phases of introns within Magnolia TPS genes examined, and comparison with those of known angiosperm and gymnosperm TPS genes
Roman numerals represent intron numbers. Intron phase numbers 0, 1, and 2 refer to the intron insertion before the first nucleotide or after the first or second nucleotide of a codon, respectively. Dashes indicate the absence of an intron. LS (Pf) refers to limonene synthase from Perilla frutescens; CS (Ga) refers to δ-cadinene synthase from G. arboreum; ES (Nt) refers to 5-epi-aristolochene synthase from N. tabacum; LS (Ag) refers to limonene synthase from Abies grandis; SS (Ag) refers to selinene synthase from A. grandis; PS (Ag) refers to (−)-pinene synthase from A. grandis; BS (Ag) refers to α-bisabolene synthase from A. grandis; TS (Tb) refers to taxadiene synthase from Taxus brevifolia; and AS (Ag) refers to abietadiene synthase from A. grandis. All data except for that from Magnolia genes were adapted from the literature (Trapp and Croteau 2001). CON represents the conserved phasing of introns between Magnolia and the other TPS genes listed.
| Type | Intron | I | II | III | IV | V | VI | VII | VIII | IX | X | XI | XII | XIII | XIV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Basal angiosperm | Mg11 | – | – | 0 | – | – | – | – | 1 | – | – | 2 | 2 | – | – |
| Mg17 | – | – | 0 | – | – | – | – | 1 | – | – | 2 | 2 | 0 | 0 | |
| Angiosperms | LS (Pf) | – | – | 0 | – | – | – | – | 1 | – | – | 2 | 2 | 0 | 0 |
| CS (Ga) | – | – | 0 | – | – | – | – | 1 | – | – | 2 | 2 | 0 | 0 | |
| ES (Nt) | – | – | 0 | – | – | – | – | 1 | – | – | 2 | 2 | 0 | 0 | |
| Gymnosperms | LS (Ag) | – | – | 0 | – | – | – | 2 | 1 | 0 | 2 | 2 | 2 | 0 | 0 |
| SS (Ag) | – | – | 0 | – | – | – | 2 | 1 | 0 | 2 | 2 | 2 | 0 | 0 | |
| PS (Ag) | – | – | 0 | – | – | – | 2 | 1 | 0 | 2 | 2 | 2 | 0 | 0 | |
| BS (Ag) | – | – | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 2 | 2 | 2 | 0 | – | |
| TS (Tb) | – | – | 0 | 1 | 2 | 1 | 2 | 1 | 1 | 2 | 2 | 2 | 0 | 0 | |
| AS (Ag) | 1 | 1 | 0 | 1 | 2 | 1 | 2 | 1 | 0 | 2 | 2 | 2 | 0 | 1 | |
| CON | – | – | 0 | – | – | – | – | 1 | – | – | 2 | 2 | 0 | 0 |
DISCUSSION
Functional Characterization of Magnolia TPSs
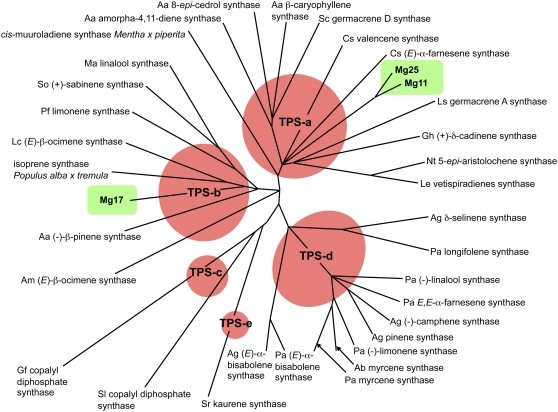
In this study, three new TPS genes were isolated from M. grandiflora, an ancestral or basal angiosperm, and further characterized biochemically and genetically. All three Magnolia genes were able to form clades with known TPSs, consistent with other studies for this class of genes (Bohlmann et al., 1998). In an unrooted phylogenetic tree of TPSs assembled by the nearest-neighbor-joining method, Mg25, a β-cubebene synthase, and Mg11, a highly expressed likely pseudogene, were most closely related to a group of well-characterized angiosperm sesquiterpene synthases making up the TPS-a clade (Fig. 10; Bohlmann et al., 1998). In contrast, the Magnolia α-terpineol synthase (Mg17) fell into the TPS-b clade of synthases, a clade dominated by monoterpene synthases from angiosperms. This is also consistent with the designation of Mg17 as a monoterpene synthase, rather than a sesquiterpene synthase, even though it could utilize both monoterpene (GPP) and sesquiterpene (FPP) substrates. As noted above, designating Mg17 as a monoterpene synthase is also more consistent with the signal sequence it harbors at its N terminus. No such signal sequences have been reported for any sesquiterpene synthases, and there are no reports of FPP biosynthesis occurring naturally in chloroplasts. Hence, Mg17 targeted to the chloroplast would not have access to FPP and likely would be associated only with monoterpene biosynthesis. However, Mg17 targeted to the mitochondria could be associated with sesquiterpene biosynthesis there, because FPP synthesis is known to occur in this organelle to some extent (Cunillera et al., 1997; Martin et al., 2007).
Figure 10.
An unrooted phylogenetic tree of TPS genes analyzed by the neighbor-joining method and depicting the estimation of pair-wise distances at the amino acid level. The red circles represent groups, families, or clades of related genes. The M. grandiflora genes characterized in this study are highlighted by green boxes. For the monoterpenes, putative signal sequences were deleted for the analysis. Classification of TPS genes a to e followed the previous analyses of Bohlmann et al. (1998). The analysis was generated with ClustalW (http://www.ch.embnet.org/software/ClustalW.html), and the tree was drawn with the TREEVIEW program (Page, 1996). Aa, Artemisia annua; Ag, Abies grandis; Am, Antirrhinum majus; Cs, Cucumis sativus; Gf, Gibberella fujikuroi; Gh, G. hirsutum; Lc, Lotus corniculatus; Le, Lycopersicon esculentum; Ls, Lactuca sativa; Ma, Mentha aquatica; Nt, N. tabacum; Pa, Picea abies; Pf, Perilla frutescens; Sc, Solidago canadensis; Sl, Solanum lycopersicum; So, Salvia officinalis; Sr, Stevia rebaudiana.
Intriguing observations of dual targeting of a single gene product to both mitochondria and chloroplasts have been reported previously, and two mechanisms have been proposed to account for this (Karniely and Pines, 2005). In one case, alternative translation start sites can provide different targeting signal sequences, yielding different peptides that are targeted to different intracellular locales or organelles. The Arabidopsis THI1 gene associated with thiamine biosynthesis utilizes such an alternative targeting signal sequence system to achieve differential targeting (Chabregas et al., 2001). The other case is for a single signal sequence to serve as a signal for two different targets, a so-called “ambiguous targeting signal” that receptors of two organelles can recognize. Both histidyl-tRNA synthetase and methioninyl-tRNA synthetase from Arabidopsis, which play critical roles in protein translation, rely on an ambiguous targeting signal to direct these proteins to the mitochondria and chloroplasts (Akashi et al., 1998; Menand et al., 1998). With the available sequence information, Mg17 does not appear to contain alternative translation start sites, so if this enzyme does colocalize to the mitochondria and chloroplast, then it might occur via the action of an ambiguous signal sequence mechanism.
The catalytic abilities of Mg17 to generate both the monoterpene and the sesquiterpene synthases may not be related to any physiological targeting of the enzyme to multiple intracellular compartments but may simply reflect the evolutionary recruitment of a sesquiterpene biosynthetic gene to monoterpene metabolism. This could have entailed a molecular exchange of the original N-terminal exon of Mg17 with that for another monoterpene synthase, creating a “chimeric” gene: exon 1 from a monoterpene synthase and exons 2 to 7 from the original Mg17 sesquiterpene synthase. The ability of sesquiterpene synthases to utilize GPP as a substrate has been documented previously and makes sense from the perspective that GPP simply represents a smaller substrate than the normal FPP substrate. The converse does not appear to have precedence. Monoterpene synthases have not been reported to utilize FPP as a substrate, possibly because the active site cannot accommodate the larger substrate. If the suggestion of exon exchange is correct in accounting for the origin of Mg17, then one might suspect that additional examples of exon 1 swapping will be found in large phylogenetic types of analyses in which putative “progenitor” TPS genes found in ancient, basal plant species are compared with “similar” genes found in evolutionarily advanced genomes.
Evolutionary Inferences from the Magnolia TPS Genes
The trichodiene synthase gene found in the fungus Myrothecium roridum contains a single intron (GenBank accession no. AF009416), but until now, no counterpart TPS gene in the plant kingdom had been described (Trapp et al., 1998). Trapp and Croteau (2001) used this type of information, the intron-exon organization, to suggest that plant and fungal TPS genes might have had different evolutionary origins. The β-cubebene synthase gene, Mg25, described here from Magnolia possesses a single intron positioned near the 5′ region of the gene, similar to the first intron in all other class III TPSs from plants. The intron found in the fungal trichodiene synthase gene is inserted into the middle of the trichodiene synthase gene and is not spatially oriented similar to the insertion site of the first intron in any of the plant genes, including Mg25. Hence, our data are consistent with those of Trapp and Croteau (2001) in arguing for separate evolutionary pathways for the fungal and plant TPS gene families.
Bohlmann et al. (1998) and Trapp and Croteau (2001) also hypothesized earlier that angiosperm TPSs might have evolved from an ancestral TPS gene in common with gymnosperms, especially one containing 14 introns. This is an appealing hypothesis because it is easier to envision the loss of introns from a common ancestral gene giving rise to the apparently conserved intron-exon organization and intron phasing observed in the extant TPS genes of angiosperms and gymnosperms than the conservation of a mechanism able to insert introns into spatially conserved positions over evolutionary time and between plant families. However, the unique exon-intron organization of Mg11 and Mg25 observed here suggests that such intron loss proceeded relatively more rapidly in Magnolia than in other branches of the angiosperms, which is not wholly consistent with the relatively modest changes observed at other genetic loci within the Magnolia species (Golenberg et al., 1990).
The findings reported here suggest that alternative mechanisms for TPS gene evolution might also be considered. For example, if the angiosperm TPS genes evolved from a gene in common with gymnosperms, sequence similarities and overall conservation of gene structure might be anticipated, especially with genes from a basal angiosperm like Magnolia in comparison with those in gymnosperms. None of the Magnolia TPS genes described here fall into associations with the class I or II TPS genes, classes in which all of the gymnosperm TPS genes fall. Instead, the Magnolia genes map to class III, which includes most angiosperm TPS genes, or appear to represent distinct or unique classes.
The greatest obstacle in elucidating the evolutionary pathway of present-day genes is that what we can document today is only the final format to have arisen by the gain or loss of introns. Moreover, with the limited data sets that we now possess, one can envision angiosperm TPS genes evolving independently from gymnosperm genes, including “gain of intron” mechanisms (Lecharny et al., 2003; Coughlan and Wolfe, 2004) as well as “loss of intron” mechanisms as suggested by Trapp and Croteau (2001). Perhaps a progenote TPS gene had a single long intron (600–900 bp) in a specific position, for example, the position occupied by present-day intron 3. Through the course of evolution, this intron might have undergone modifications allowing it to be partitioned into smaller pieces and inserted into neighboring regions and TPS genes, creating multiple introns. The basic six-intron organization of TPS genes observed today in angiosperms thus formed could then have multiplied and diverged by gene duplication and exon shuffling to create diverse TPS functionalities. This alternative hypothesis to the intron-loss model of Trapp and Croteau (2001) might be referred to as the “intron partitioning and distribution theory.” Without a doubt, such a discussion based on so few genes from so few taxa makes any such suggestion highly speculative. Hence, to further sort through these possibilities, TPS genes from other extant basal angiosperms (Laurales and Piperales, for examples) and intermediate taxa (between Magnolia and advanced species in the molecular phylogeny tree) must be examined, as well as even earlier land plants, such as mosses, liverworts, and hornworts.
MATERIALS AND METHODS
Plant Material, Reagents, and Standards
Plant leaves and flowers were collected from June through August, 2005 to 2007, from Magnolia grandiflora (Southern Magnolia trees) grown on the University of Kentucky campus at variable times throughout the day. Light-green leaves with white pubescence on the abaxial side were classified as developing leaf tissue. Larger leaves with dark-green color were considered the more mature leaves, which had a brown pubescence on the abaxial side. All solvents were from Fisher Scientific unless stated otherwise.
Terpene Analyses
To extract terpene compounds, 2 g of young Magnolia leaf tissue was first pulverized in liquid nitrogen with a mortar and pestle, then extracted sequentially with 5 mL of hexane:ethyl acetate (85:15) twice. The extracts were combined and purified by two rounds of silica column chromatography. The extract was applied to a silica column (7 mm × 146 mm) followed by washing with 6 mL of fresh hexane:ethyl acetate (85:15). The collected eluent was then applied to a second silica column (7 mm × 146 mm), followed by 3 mL of the hexane:ethyl acetate (85:15) to elute hydrocarbons and oxygenated compounds. The final eluant (approximately 20 mL) was concentrated to 2 mL under nitrogen before analysis by GC-MS.
Aliquots of the various samples were analyzed for terpene constituents with a Thermo Finnigan DSQ GC/MS system (Thermo Fisher Scientific) equipped with a Restec Rtx-5 capillary column (30 m × 0.32 mm, 0.25-mm phase thickness). For sesquiterpene analyses, the injector port was maintained at 220°C in the splitless mode and the initial oven temperature of 70°C (0.5 min) was increased in a 4°C min−1 gradient to 180°C followed by a 20°C min−1 gradient to 300°C. Mass spectra were recorded at 70 eV, scanning from 35 to 300 atomic mass units. For monoterpene analysis, the oven temperature was programmed to 40°C for 1 min followed by a ramp of 4°C min−1 to 150°C and then a ramp of 20°C min−1 to 300°C.
All compounds detected from the leaf extracts or the enzyme assays were confirmed by comparing retention time and mass spectra with authentic standards, or by comparison with mass spectra reported in the NIST mass spectra library version 2.0 and MassFinder 2.3 software (edited by Dr. D.H. Hochmuth). Authentic standards were obtained from Sigma Chemical Company (for α-terpineol), Bedoukian Research (for bisabolenes), and Firmenich (for β-cubebene).
RNA Isolation
Leaves were collected, frozen immediately in liquid nitrogen, and then pulverized to make a fine powder using a chilled mortar and pestle. Total RNA was isolated using a sodium isoascorbate acid-guanidium-phenol-chloroform method with minor modifications (Suzuki et al., 2003), and 200 to 250 μg of total RNA was usually obtained from 1 g of leaf tissues. One gram of a leaf was pulverized in a chilled mortar and pestle. Five volumes of extraction buffer (500 mm sodium isoascorbate, 100 mm Tris-HCl, pH 8.0, 10 mm EDTA, 5% [v/v] β-mercaptoethanol, and 2% [w/v] SDS) was added directly into the powder, making a buffer-powder slush. About 500 μL of the slush was transferred to a fresh 1.5-mL Eppendorf tube, and an equal volume of chloroform:isoamyl alcohol (24:1, v/v) was added. After phase separation by centrifugation for 20 min at 4°C, the supernatant was extracted a second time using an equal volume of chloroform:isoamyl alcohol (24:1, v/v). The supernatant was then mixed with 0.5 volume of 66.7% (w/v) guanidine isothiocyanate, an equal volume of water-saturated phenol, and 0.15 volume of 3 m sodium acetate, pH 5.2. After a 3-min incubation at room temperature, 0.2 volume (of the added phenol) of chloroform:isoamyl alcohol (24:1, v/v) was added and shaken vigorously by hand. After incubation on ice for 15 min, the mixture was centrifuged for 20 min at 4°C. To remove polysaccharides from the supernatant, one-third volume of 1.2 m NaCl and 0.8 m sodium citrate was added, incubated for a short time, and then centrifuged for 10 min at full speed. Total RNA was precipitated from the resulting supernatant with two-thirds volume of isopropanol. After 10 min of incubation, total RNA was pelleted by centrifugation for 20 min at 4°C. The pellet was washed with 75% ethanol twice and dissolved in ribonuclease-free water. Total RNA was quantified by spectrophotometry. When necessary, mRNA was purified from 100 μg of total RNA isolated using the Oligotex mRNA mini kit (Qiagen) according to the manufacturer's recommendations.
Isolating Initial TPS cDNA Fragments
First-strand cDNAs were synthesized from poly(A+) RNA using SuperScript II reverse transcriptase (Invitrogen) and oligo(dT) primer (Invitrogen) in a total volume of 20 μL at 42°C for 50 min according to the manufacturer's recommendations. PCRs were performed in a total volume of 50 μL containing 2 μL of first-strand cDNA, 0.25 mm primers, 0.2 mm dNTPs, 1× Pfu PCR buffer, and 2.5 units of Pfu Ultra High-Fidelity DNA Polymerase (Stratagene). Several combinations of degenerate oligonucleotides were used to amplify TPS cDNA fragments. Three forward oligonucleotides were designed: PrimerA-F, 5′-GAGCTTAGC(C/G)A(A/T)(C/G)TTTCAAAGTGGTGG-3′; PrimerB-F, 5′-TAGC(C/G)A(A/T)(C/G)TTTCAAAGTGGTGGAA-3′; and PrimerC-F, 5′-TGGATATTAGGAGT(C/G)TACTT(C/T)GAGCC-3′; three reverse primers were also designed: PrimerA-R, 5′-GTAGCATA(A/G)GCATCATA(A/C/T)GTGTCGTC-3′; PrimerB-R, 5′-ATC(A/C)AT(A/G)GCA(C/G)TAATTTCCCACCT-3′; and PrimerC-R, 5′-GCAATCTTCATATACTC(A/C/T)TCAATGTTGG-3′. Amplicons were (dA)-tailed, ligated in pGEM-T Easy vector (Promega), and transformed into DH5-α–competent cells. Cloned inserts were sequenced in both directions using the BigDye Terminator Cycle Sequencing Kit (Perkin-Elmer) with an automated sequencing analyzer (ABI PRISM 310 Genetic Analyzer; Applied Biosystems). DNA sequence information was analyzed by BLASTX searches of the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/BLAST). Fragments exhibiting significant matches (E ≤ e-4) to TPSs in the GenBank database were considered significant. All custom oligonucleotides were synthesized by Integrated DNA Technologies.
5′ and 3′ RACE for Mg25, Mg17, and Mg11 Genes
A BD SMART RACE cDNA Amplification Kit (Clontech) was used to obtain the sequence information necessary to obtain full-length cDNA clones for the prioritized Magnolia TPS cDNA fragments. Adaptor-ligated double-stranded cDNAs were generated, and an adaptor-specific element provided with the kit was used for amplifications combined with gene-specific oligonucleotides. In some cases, a gene-specific first-strand cDNA was synthesized to increase the corresponding cDNA pools using gene-specific oligonucleotides instead of 5′ or 3′ anchor primers provided by the manufacturer. The 3′ and 5′ ends were amplified with the Advantage 2 PCR Kit (Clontech). The reaction mixtures in 50 μL included 5 μL of 10× PCR buffer, 200 nm dNTP mix, 1 μL of Advantage 2 Polymerase Mix, 200 μm adaptor-specific primer, 200 μm gene-specific primer, and 2.5 μL of 100- to 300-fold diluted RACE-ready cDNA. The first amplification was completed in conditions as follows: 95°C for 3 min, then 30 cycles of 30 s at 95°C, 40 s at 60 to 65°C, and 2 to 3 min at 72°C. Sequentially, nested PCR was performed with 5 μL of 200-fold diluted cDNA from the first amplification combined with nested oligonucleotide sets at 2°C to 3°C lower annealing temperature. The amplified products were cloned and sequenced as described above.
In order to obtain 3′ end amplicons, gene-specific forward primers were designed: for Mg25, 5′-ACCTATGACGTATACGGCACATTGGAGG-3′ and 5′-GAAATATATACTACTGCTATTGAGAGATGGG-3′; for Mg17, 5′-CTTGCATTGGAGGGTACTAAGATTAGAGGC-3′; and for Mg11, 5′-CAATTTCAGTGCAATATTAAGCAGCGACACACACG-3′. To obtain specific fragments, nested gene-specific primers were considered: 5′-GGGCTTCTGAATGCAGTCGATCAAATC-3′ for Mg25; for Mg17, 5′-AACATCGTTCAAAATGTATACCAGGGACAAGTCAG-3′; and for Mg11, 5′-TCTTTCACCACAGTGGCACCTCAAATCTGC-3′. Based on the sequence information from 3′ end amplicons, three gene-specific reverse oligonucleotides were designed for 5′ RACE: 5′-CATCGTCCTTCCCATTGATGTGGTCATCATAC-3′ for Mg25, 5′-GTAGCCTTTGGCCAAATTCGTAATAACTTTCTTTATGTAGGG-3′ for Mg17, and 5′-GTCAAAACCATCATCATTCCCATTGATGTGG-3′ for Mg11. Nested gene-specific primers were 5′-TAGGCTACACCAAGGCGTTGAATTGC-3′ for Mg25, 5′-GTATTCGGTAGGATTTTCCTTCCCTGC-3′ for Mg17, and 5′-CGCATAAAGGACAGGGTGAAGAACTGAAAGG-3′ for Mg11. Met codons served to locate the most upstream start site, and the various ORFs were assessed according to those putative start sites. The first stop codon in any ORF was regarded as the translation stop codon.
Isolating Full-Length Mg25, Mg17, and Mg11 cDNAs
Mg25 full-length cDNA was amplified in the reaction containing Pfu Ultra High-Fidelity DNA Polymerase with forward (5′-CACCATGGATAGTCCCACTACTCAAAGGCCAAACATGGAG-3′) and reverse (5′-TTAAAGTGGAATAGGGTCCACAAACAGTGATG-3′) primers using 3′ RACE Ready cDNA as a template. PCR conditions were as follows: 3 min at 95°C; 30 cycles of 30 s at 95°C, 30 s at 52°C, and 100 s at 72°C; and a final extension for 5 min at 72°C. The amplified cDNA (1.6 kb) was ligated into pET100/D-TOPO vector (Invitrogen) and transformed into DH5-α-competent cells for plasmid amplification. Insert DNAs were sequenced in both directions using automated DNA sequencing. For Mg17 harboring a putative signal sequence, two different cDNA fragments were amplified and cloned. Full-length Mg17 (1.8 kb) was amplified with forward primer 5′-ATGGCACTTAAGCTCCTCTTCCAATGC-3′ and reverse primer 5′-TTAGCTCTCCATTAATTGAATAGGTTCC-3′; for signal sequence truncated cDNA (1.6 kb), forward primer 5′-ATGCGACGCTCGGCTAATTACC-3′ was used with the same reverse primer used for full-length amplification. The signal sequence domain was hence removed and a Met was added to the N terminus of the truncated ORF fragment, creating an artificial translation start site. Both cDNAs were ligated into pET100/D-TOPO or pET43(a) for functional characterization. For Mg11 (1.6 kb), forward primer 5′-ATGGATAGTCCTGCTACTCAAAGG-3′ and reverse primer 5′-CTAAAGTGGAATAGGGTCCAC-3′ were used to amplify full-length cDNA, and site-directed mutagenesis was performed using the QuikChange methodology (Stratagene) to modify an internal stop codon near the N terminus to Trp (W), a conserved amino acid at this position among TPSs. The mutagenized cDNA was cloned and expressed in three different expression vectors: pET100/D-TOPO, pET28(a), and pET43(a). Sequence alignments were performed with ClustalW (http://www.ch.embnet.org/software/ClustalW.html) and visualized with GENEDOC (http://www.psc.edu/biomed/genedoc).
Heterologous Expression of TPS cDNAs in Escherichia coli and Enzyme Assays
Full-length cDNAs cloned into various expression vectors were transformed into BL21 Star(DE3) (Invitrogen). Cell cultures initiated from single transformation colonies were grown to an optical density at 600 nm of 0.6 at 37°C before the addition of 0.4 mm isopropylthio-β-galactoside for pET43(a), or 1 mm isopropylthio-β-galactoside for pET100/D-TOPO and pET28(a), then incubated overnight at 22°C. The cells were separated from the medium by centrifugation and the pellets were stored at −80°C if not used immediately. The pellets were resuspended and sonicated six times for 10 s in lysis buffer (80 mm potassium phosphate, pH 7.0, 10 mm meta-bisulfate, 10 mm sodium ascorbic acid, and 1 mm phenylmethylsulfonyl fluoride), and the clarified lysate was collected by centrifugation at 12,000g for 15 min. The lysate was then loaded onto a Ni2+ affinity resin (Novagen) to purify His-tagged recombinant protein. Purified proteins were dialyzed against a buffer containing 20 mm HEPES, pH 7.5, 5 mm MgCl2, 1 mm dithiothreitol, used immediately for enzyme assays, or otherwise stored as a 50% glycerol stock at −80°C. Protein concentrations were estimated by the Bradford assay (Bio-Rad) using bovine serum albumin as the standard. Bacterial lysates and purified enzyme samples were also examined on SDS-polyacrylamide gels (10%) stained with Coomassie Blue.
Enzymatic properties evaluated included incubation time, temperature, pH, and cofactor requirement in an assay using radiolabeled substrates. Typically, cell lysates or purified enzymes were incubated with labeled substrates, 0.5 μCi of [1-3H]GPP (20 Ci mmol−1; American Radiolabeled Chemicals), [1-3H]FPP (26 Ci mmol−1; Perkin-Elmer), or [1-3H]GGPP (23 Ci mmol−1; Perkin-Elmer) with reaction conditions specific for monoterpene (25 mm HEPES, pH 7.2, 100 mm KCl, and 10 mm MnCl2), sesquiterpene (25 mm Tris-HCl, pH 7.5, and 20 mm MgCl2), and diterpene (25 mm HEPES, pH 7.2, 100 mm KCl, 10 mm MgCl2, and 10 μm MnCl2; Rising et al., 2000; Martin et al., 2004) biosynthesis. For kinetic analyses, purified enzyme was incubated with unlabeled FPP (2 mg mL−1; Sigma Chemical Company) ranging from 0.115 to 23 μm in 1-mL reaction volumes. Reactions were incubated for 10 min at 25°C before adding 100 ng of α-cedrene as an internal standard and then extracting twice with 2 mL of pentane. Extracts were combined, concentrated to 50 μL under nitrogen, and then analyzed by GC-MS as described previously (Takahashi et al., 2005). For Mg17 enzyme assays, both FPP (2 mg/mL; Sigma Chemical Company) and GPP (1 mg/mL; Sigma Chemical Company) were evaluated.
Steady-State mRNA Detection by RT-PCR
Steady-state transcript levels for the three genes were examined in various tissues: young and mature leaves as well as specified floral organs such as tepals (sepals + petals), stamens, and carpels. Total RNA was isolated from each tissue using the same method described above, and first-strand cDNAs were synthesized using SuperScript II reverse transcriptase (Invitrogen) and oligo(dT) primer (Invitrogen) following the manufacturer's protocol. Oligonucleotides for PCR were designed to unique, gene-specific regions for each of the three genes. Gene-specific primers were verified by showing that primers designed for one Magnolia gene could not be used to amplify DNA fragments for either of the other two cDNA clones. The amount of 18S ribosomal RNA was used as the qualitative and quantitative control.
Phylogenetic Analysis
To examine the relatedness of genes within phylogenetic trees, the deduced amino acid sequences of full-length Magnolia TPS genes were aligned with those of known TPS genes (Trapp and Croteau, 2001; Martin et al., 2004) in ClustalW (http://www.ch.embnet.org/software/ClustalW.html), and the phylogenetic tree was redrawn by the neighbor-joining method in the TREEVIEW program (Page, 1996). For monoterpene synthases, signal sequences were removed prior to analysis because these sequences show low similarities among all of the genes.
Reporter Protein Localization Studies
The N-terminal sequence of Mg17 was analyzed with computer-based prediction servers such as ChloroP (http://www.cbs.dtu.dk/services/ChloroP/; Emanuelsson et al., 1999), PSORT (old version, http://psort.nibb.ac.jp; Nakai and Horton, 1999), and Predotar (http://www.inra.fr/predotar/; Small et al., 2004). The secondary structure of the Mg17 transit peptide was predicted with the PredictProtein program of Rost et al. (2004), and the helical wheel structure was drawn with PEPWHEEL (http://bioweb.pasteur.fr/seqanal/interfaces/pepwheel.html).
The putative signal sequence was amplified with Pfu Ultra High-Fidelity DNA Polymerase with 10 pmol of forward primer (HindIII; 5′-ATGGCACTTAAGCTCCTCTTC-3′) and reverse primer (SstI; 5′-AGTACTCGCACAGCACCG-3′). The amplicon was digested with HindIII/SstI restriction enzymes. The fragments were gel purified and cloned into pGEM-T Easy vector, followed by DNA sequencing analysis. All inserts were then digested and integrated in pKYLX80-GFP, a pKYLX71 derivative. This vector was generated from pBluescript KS(+) vector armed with a 35S2 promoter, a multiple cloning site, a GFP reporter gene, and an rbcS 3′ terminator. In order to examine localization sites, constructs of Mg17 and targeting control constructs were used alone or premixed together and cobombarded onto the abaxial side of tobacco leaves (Nicotiana tabacum ‘KY160’) according to Dinkins et al. (2003). For a chloroplast- and a mitochondria-targeting control, the Arabidopsis (Arabidopsis thaliana) signal sequences of rbcS and CoxIV were fused with GFP and DsRFP, respectively (Dinkins et al., 2003).
The fluorescence of transiently expressed reporter proteins was monitored within 24 to 48 h after bombardment with an Olympus FV1000 laser-scanning confocal microscope (Olympus America). An intervein section (0.5 cm × 1.5 cm) of leaf tissue was placed on glass slides in water and covered with a glass coverslip, and six different cells in regions of interest were examined. Initial focus was performed with an Olympus water-immersion PLAP40X WLSM-NA1.0 microscope with 488-nm (GFP) or 543-nm (RFP) laser lines from a multiline argon laser set at 10% or 22%, respectively. Images were acquired with a resolution of 512 × 512 pixels and a scan rate of 4 μs pixel−1. Acquired files were exported as TIFF files from Olympus FluoView software version 1.5, and final images were rendered with Adobe Photoshop 7.0.
DNA Extraction and Structural Gene Analysis
Prior to extraction, the pubescence material of young leaf tissue was removed by scrubbing because it was inhibitory to subsequent DNA manipulations. Genomic DNA was extracted using a method adapted from Li et al. (2002). Briefly, 100 mg (1 volume) of frozen leaf was pulverized in a prechilled mortar to fine powder, and immediately, 5 volumes of extraction buffer (25 mm Tris-HCl, pH 8.0, 50 mm EDTA, 0.5% [w/v] SDS, 2% [v/v] β-mercaptoethanol, and insoluble polyvinylpolypyrrolidone) was added to the powder and mixed. After 15 min of incubation at 56°C with gentle mixing by inversion, 5 volumes of phenol:chloroform (1:1, v/v) was added and mixed gently again. Debris was removed by centrifugation, and the aqueous phase was extracted a second time. An equal volume of chloloform:isoamyl alcohol (24:1, v/v) was mixed with the aqueous phase and incubated on ice for 15 min. The aqueous phase was then separated by centrifugation, mixed with 0.1 volume of 3 m sodium acetate (pH 5.2) and 2.5 volumes of ice-cold isopropanol, and incubated at −20°C for 15 min, and the precipitated nucleic acids were collected by centrifugation for 15 min. The precipitated nucleic acids were washed with 75% ethanol twice and dissolved in deionized water or 0 mm Tris, pH 7.5, 1 mm EDTA buffer followed by a ribonuclease A treatment. The quality and yield of genomic DNA were estimated by spectrophotometry.
Initial PCRs were performed using primers that annealed to the start and stop sites of the respective cDNAs. If these amplification reactions failed, the ORFs of the respective cDNAs were divided into two to three segments (>500 bp) that overlapped each other minimally by 100 nucleotides. The PCRs were performed with 50 to 500 ng of genomic DNA, 10 pmol of primers, and Pfu Ultra High-Fidelity DNA Polymerase in a total volume of 50 μL with typical reaction conditions of 94°C for 3 min; seven cycles of 94°C for 30 s, 58 to 61°C for 30 s, and 72°C for 1 to 2 min; 30 cycles of 94°C for 30 s, 56 to 59°C for 30 s, and 72°C for 2 to 3 min; with a final extension for 5 min at 72°C. The amplicons were purified with the QIAquick Gel Extraction Kit (Qiagen) and cloned into pGEM-T Easy vector (Promega) for DNA sequencing. DNA sequences were assembled and aligned with corresponding cDNA sequences using Vector NTI 10.3.0 (Invitrogen). The three Magnolia genes were classified by following the method of Trapp and Croteau (2001).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EU366429 to EU366431.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Enzymatic properties of Mg25.
Supplemental Figure S2. Additional mass spectra from GC peaks shown in Figures 2 and 4.
Supplemental Figure S3. Nucleotide sequence alignments of Mg25, Mg11, and Mg17 used to design gene-specific PCR primers.
Supplementary Material
Acknowledgments
We thank all the members of the Chappell laboratory for useful discussions, especially Dr. Shuiqin Wu for help with the GC-MS analyses and Drs. Randy Dinkins and Nihar R. Nayak for assistance with the confocal microscopy.
This work was supported by grants from the Kentucky Tobacco Research and Development Center and the National Science Foundation (grant no. IBN–0136034). S.L. was supported by the University of Kentucky Philip Morris Scholarship Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Joseph Chappell (chappell@uky.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Akashi K, Grandjean O, Small I (1998) Potential dual targeting of an Arabidopsis archaebacterial-like histidyl-tRNA synthetase to mitochondria and chloroplasts. FEBS Lett 431 39–44 [DOI] [PubMed] [Google Scholar]
- Arimura G-i, Huber DPW, Bohlmann J (2004) Forest tent caterpillars (Malacosoma disstria) induce local and systemic diurnal emissions of terpenoid volatiles in hybrid poplar (Populus trichocarpa × deltoides): cDNA cloning, functional characterization, and patterns of gene expression of (−)-germacrene D synthase, PtdTPS1. Plant J 37 603–616 [DOI] [PubMed] [Google Scholar]
- Aubourg S, Lecharny A, Bohlmann J (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genomics 267 730–745 [DOI] [PubMed] [Google Scholar]
- Back K, Chappell J (1996) Identifying functional domains within terpene cyclases using a domain-swapping strategy. Proc Natl Acad Sci USA 93 6841–6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MH, Mansfield JW, Lewis MJ, Beale MH (2002) Cloning and expression of sesquiterpene synthase genes from lettuce (Lactuca sativa L.). Phytochemistry 60 255–261 [DOI] [PubMed] [Google Scholar]
- Bohlmann J, Meyer-Gauen G, Croteau R (1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA 95 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester HJ, Kodde J, Verstappen FWA, Altug IG, de Kraker JW, Wallaart TE (2002) Isolation and characterization of two germacrene A synthase cDNA clones from chicory. Plant Physiol 129 134–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer B, Bremer K, Chase MW, Reveal JL, Soltis DE, Soltis PS, Stevens PF, Anderberg AA, Fay MF, Goldblatt P, et al (2003) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc 141 399–436 [Google Scholar]
- Brown JWS, Simpson CG (1998) Splice site selection in plant pre-mRNA splicing. Annu Rev Plant Physiol Plant Mol Biol 49 77–95 [DOI] [PubMed] [Google Scholar]
- Chabregas SM, Luche DD, Farias LP, Ribeiro AF, van Sluys MA, Menck CFM, Silva MC (2001) Dual targeting properties of the N-terminal signal sequence of Arabidopsis thaliana THI1 protein to mitochondria and chloroplasts. Plant Mol Biol 46 639–650 [DOI] [PubMed] [Google Scholar]
- Chen XY, Chen Y, Heinstein P, Davisson VJ (1995) Cloning, expression, and characterization of (+)-delta-cadinene synthase: a catalyst for cotton phytoalexin biosynthesis. Arch Biochem Biophys 324 255–266 [DOI] [PubMed] [Google Scholar]
- Christianson DW (2006) Structural biology and chemistry of the terpenoid cyclases. Chem Rev 106 3412–3442 [DOI] [PubMed] [Google Scholar]
- Colby SM, Crock J, Dowdle-Rizzo B, Lemaux PG, Croteau R (1998) Germacrene C synthase from Lycopersicon esculentum cv. VFNT Cherry tomato: cDNA isolation, characterization, and bacterial expression of the multiple product sesquiterpene cyclase. Proc Natl Acad Sci USA 95 2216–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan A, Wolfe KH (2004) Origins of recently gained introns in Caenorhabditis. Proc Natl Acad Sci USA 101 11362–11367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane PR, Friis EM, Pedersen KR (1995) The origin and early diversification of angiosperms. Nature 374 27–33 [Google Scholar]
- Cronquist A (1988) The Evolution and Classification of Flowering Plants. New York Botanical Garden, New York
- Cunillera N, Boronat A, Ferrer A (1997) The Arabidopsis thaliana FPS1 gene generates a novel mRNA that encodes a mitochondrial farnesyl-diphosphate synthase isoform. J Biol Chem 272 15381–15388 [DOI] [PubMed] [Google Scholar]
- Davis ED, Croteau R (2000) Cyclization enzymes in the biosynthesis of monoterpenes, sesquiterpenes, and diterpenes. Top Curr Chem 209 53–95 [Google Scholar]
- de Kraker JW, Franssen MCR, de Groot A, Konig WA, Bouwmeester HJ (1998) (+)-Germacrene A biosynthesis: the committed step in the biosynthesis of bitter sesquiterpene lactones in chicory. Plant Physiol 117 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkins RD, Conn HM, Dirk LMA, Williams MA, Houtz RL (2003) The Arabidopsis thaliana peptide deformylase 1 protein is localized to both mitochondria and chloroplasts. Plant Sci 165 751–758 [Google Scholar]
- Dudareva N, Cseke L, Blanc VM, Pichersky E (1996) Evolution of floral scent in Clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell 8 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini PJ, Chappell J (1992) Gene family for an elicitor-induced sesquiterpene cyclase in tobacco. Proc Natl Acad Sci USA 89 11088–11092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang DR (2005) Evolution of flavors and scents. Annu Rev Plant Biol 56 301–325 [DOI] [PubMed] [Google Scholar]
- Golenberg EM, Giannasi DE, Clegg MT, Smiley CJ, Durbin M, Henderson D, Zurawski G (1990) Chloroplast DNA-sequence from a Miocene Magnolia species. Nature 344 656–658 [DOI] [PubMed] [Google Scholar]
- Greenhagen BT, O'Maille PE, Noel JP, Chappell J (2006) Identifying and manipulating structural determinates linking catalytic specificities in terpene synthases. Proc Natl Acad Sci USA 103 9826–9831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima Y, Davidovich-Rikanati R, Fridman E, Gang DR, Bar E, Lewinsohn E, Pichersky E (2004) The biochemical and molecular basis for the divergent patterns in the biosynthesis of terpenes and phenylpropenes in the peltate glands of three cultivars of basil. Plant Physiol 136 3724–3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniely S, Pines O (2005) Single translation-dual destination: mechanisms of dual protein targeting in eukaryotes. EMBO Rep 6 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecharny A, Boudet N, Gy I, Aubourg S, Kreis M (2003) Introns in, introns out in plant gene families: a genomic approach of the dynamics of gene structure. J Struct Funct Genomics 3 111–116 [PubMed] [Google Scholar]
- Li Y, Su Z, Chen F (2002) Rapid extraction of genomic DNA from leaves and bracts of dove tree (Davidia involucrate). Plant Mol Biol Rep 20 185a–185e [Google Scholar]
- Long MY, Rosenberg C, Gilbert W (1995) Intron phase correlations and the evolution of the intron exon structure of genes. Proc Natl Acad Sci USA 92 12495–12499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucker J, Bowen P, Bohlmann J (2004) Vitis vinifera terpenoid cyclases: functional identification of two sesquiterpene synthase cDNAs encoding (+)-valencene synthase and (−)-germacrene D synthase and expression of mono- and sesquiterpene synthases in grapevine flowers and berries. Phytochemistry 65 2649–2659 [DOI] [PubMed] [Google Scholar]
- Lucker J, El Tamer MK, Schwab W, Verstappen FWA, van der Plas LHW, Bouwmeester HJ, Verhoeven HA (2002) Monoterpene biosynthesis in lemon (Citrus limon): cDNA isolation and functional analysis of four monoterpene synthases. Eur J Biochem 269 3160–3171 [DOI] [PubMed] [Google Scholar]
- Martin D, Piulachs MD, Cunillera N, Ferrer A, Belles X (2007) Mitochondrial targeting of farnesyl diphosphate synthase is a widespread phenomenon in eukaryotes. Biochim Biophys Acta 1773 419–426 [DOI] [PubMed] [Google Scholar]
- Martin DM, Bohlmann J (2004) Identification of Vitis vinifera (−)-alpha-terpineol synthase by in silico screening of full-length cDNA ESTs and functional characterization of recombinant terpene synthase. Phytochemistry 65 1223–1229 [DOI] [PubMed] [Google Scholar]
- Martin DM, Faldt J, Bohlmann J (2004) Functional characterization of nine Norway spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol 135 1908–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Marechal-Drouard L, Sakamoto W, Dietrich A, Wintz H (1998) A single gene of chloroplast origin codes for mitochondrial and chloroplastic methionyl-tRNA synthetase in Arabidopsis thaliana. Proc Natl Acad Sci USA 95 11014–11019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24 34–35 [DOI] [PubMed] [Google Scholar]
- Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12 357–358 [DOI] [PubMed] [Google Scholar]
- Phillips MA, Wildung MR, Williams DC, Hyatt DC, Croteau R (2003) cDNA isolation, functional expression, and characterization of (+)-alpha-pinene synthase and (−)-alpha-pinene synthase from loblolly pine (Pinus taeda): stereocontrol in pinene biosynthesis. Arch Biochem Biophys 411 267–276 [DOI] [PubMed] [Google Scholar]
- Prosser I, Altug IG, Phillips AL, Konig WA, Bouwmeester HJ, Beale MH (2004) Enantiospecific (+)- and (−)-germacrene D synthases, cloned from goldenrod, reveal a functionally active variant of the universal isoprenoid-biosynthesis aspartate-rich motif. Arch Biochem Biophys 432 136–144 [DOI] [PubMed] [Google Scholar]
- Prosser I, Phillips AL, Gittings S, Lewis MJ, Hooper AM, Pickett JA, Beale MH (2002) (+)-(10R)-germacrene A synthase from goldenrod, Solidago canadensis: cDNA isolation, bacterial expression and functional analysis. Phytochemistry 60 691–702 [DOI] [PubMed] [Google Scholar]
- Rising KA, Starks CM, Noel JP, Chappell J (2000) Demonstration of germacrene A as an intermediate in 5-epi-aristolochene synthase catalysis. J Am Chem Soc 122 1861–1866 [Google Scholar]
- Roise D, Theiler F, Horvath SJ, Tomich JM, Richards JH, Allison DS, Schatz G (1988) Amphiphilicity is essential for mitochondrial presequence function. EMBO J 7 649–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu JF (2004) The PredictProtein server. Nucleic Acids Res 32 W321–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon-Asa L, Shalit M, Frydman A, Bar E, Holland D, Or E, Lavi U, Lewinsohn E, Eyal Y (2003) Citrus fruit flavor and aroma biosynthesis: isolation, functional characterization, and developmental regulation of Cstps1, a key gene in the production of the sesquiterpene aroma compound valencene. Plant J 36 664–674 [DOI] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4 1581–1590 [DOI] [PubMed] [Google Scholar]
- Starks CM, Back K, Chappell J, Noel JP (1997) Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science 277 1815–1820 [DOI] [PubMed] [Google Scholar]
- Steele CL, Crock J, Bohlmann J, Croteau R (1998) Sesquiterpene synthases from grand fir (Abies grandis): comparison of constitutive and wound-induced activities, and cDNA isolation, characterization and bacterial expression of delta-selinene synthase and gamma-humulene synthase. J Biol Chem 273 2078–2089 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Hibino T, Kawazu T, Wada T, Kihara T, Koyama H (2003) Extraction of total RNA from leaves of eucalyptus and other woody and herbaceous plants using sodium isoascorbate. Biotechniques 34 988–993 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Zhao YX, O'Maille PE, Greenhagen BT, Noel JP, Coates RM, Chappell J (2005) Kinetic and molecular analysis of 5-epiaristolochene 1,3-dihydroxylase, a cytochrome P450 enzyme catalyzing successive hydroxylations of sesquiterpenes. J Biol Chem 280 3686–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K (1974) Stereospecific cope rearrangement of the germacrene-type sesquiterpenes. Tetrahedron 30 1525–1534 [Google Scholar]
- Trapp SC, Croteau RB (2001) Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 158 811–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp SC, Hohn TM, McCormick S, Jarvis BB (1998) Characterization of the gene cluster for biosynthesis of macrocyclic trichothecenes in Myrothecium roridum. Mol Gen Genet 257 421–432 [DOI] [PubMed] [Google Scholar]
- van der Hoeven RS, Monforte AJ, Breeden D, Tanksley SD, Steffens JC (2000) Genetic control and evolution of sesquiterpene biosynthesis in Lycopersicon esculentum and L. hirsutum. Plant Cell 12 2283–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DC, McGarvey DJ, Katahira EJ, Croteau R (1998) Truncation of limonene synthase preprotein provides a fully active ‘pseudomature’ form of this monoterpene cyclase and reveals the function of the amino-terminal arginine pair. Biochemistry 37 12213–12220 [DOI] [PubMed] [Google Scholar]
- Yoshikuni Y, Ferrin TE, Keasling JD (2006) Designed divergent evolution of enzyme function. Nature 440 1078–1082 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.