Abstract
The high light-inducible polypeptides (HLIPs) are critical for survival under high light (HL) conditions in Synechocystis PCC 6803. In this article, we determined the localization of all four HLIPs in thylakoid protein complexes and examined effects of hli gene deletion on the photosynthetic protein complexes. The HliA and HliB proteins were found to be associated with trimeric photosystem I (PSI) complexes and the Slr1128 protein, whereas HliC was associated with PsaL and TMP14. The HliD was associated with partially dissociated PSI complexes. The PSI activities of the hli mutants were 3- to 4-fold lower than that of the wild type. The hli single mutants lost more than 30% of the PSI trimers after they were incubated in intermediate HL for 12 h. The reduction of PSI trimers were further augmented in these cells by the increase of light intensity. The quadruple hli deletion mutant contained less than one-half of PSI trimers following 12-h incubation in intermediate HL. It lost essentially all of the PSI trimers upon exposure to HL for 12 h. Furthermore, a mutant lacking both PSI trimers and Slr1128 showed growth defects similar to that of the quadruple hli deletion mutant under different light conditions. These results suggest that the HLIPs stabilize PSI trimers, interact with Slr1128, and protect cells under HL conditions.
Acclimation to different light environments is crucial for photosynthetic organisms to grow and survive. Low light (LL) limits cell growth, whereas excess light or high light (HL) causes oxidative damage to proteins, lipids, and nucleic acids, ultimately leading to a loss of cell viability (Asada, 1994; Halliwell and Gutteridge, 1999; Niyogi, 1999; Mittler, 2002).
Both microalgae and vascular plants have evolved protective mechanisms against the absorption of excess excitation energy and the unavoidable production of limited amounts of reactive oxygen during photosynthesis (Durnford and Falkowski, 1997; Niyogi, 1999; Foyer and Fletcher, 2001; Mittler, 2002). Well-documented protective mechanisms include (but are by no means limited to) changes in the light-harvesting antenna and/or photosynthetic reaction centers (Demmig-Adams and Adams, 1992, 1996); dissipation of excess excitation energy as harmless heat (Peltier and Schmidt, 1991; Jacob and Lawlor, 1993; Levy et al., 1993); synthesis of antioxidant enzymes such as superoxide dismutase (Perl-Treves and Galun, 1991; Tsang et al., 1991; Bowler et al., 1994), catalase (Redinbaugh et al., 1990; Hertwig et al., 1992; Zhong et al., 1994), and peroxidases (Ort and Baker, 2002; Yabuta et al., 2002; Fryer et al., 2003; Horling et al., 2003); and enhanced accumulation of soluble antioxidants, such as carotenoids, tocopherol, and glutathione that quenches triplet state of chlorophyll and singlet oxygen back to ground state (Di Mascio et al., 1990, 1991).
Photosynthetic organisms also synthesize stress-associated proteins during exposure to HL. These proteins are often important for the acclimation of cells to HL. A family of HL-inducible genes, called hli genes or scp genes (Dolganov et al., 1995; Funk and Vermaas, 1999), encoding the HL-inducible polypeptides (HLIPs) with similarity to the light-harvesting chlorophyll a/b-binding proteins (LHCPs) of plants, were shown to be critical for the survival of cyanobacteria under HL conditions (He et al., 2001). Interestingly, the ferrochelatase in cyanobacteria or the plastidic ferrochelatase of plants contains a C terminus that is similar to HLIP in primary sequences (Funk and Vermaas, 1999; He et al., 2001). In Synechocystis PCC 6803, the levels of the four HLIPs increase in response to HL, low temperature, and nutrient limitation (He et al., 2001), whereas the levels of the ferrochelatase are reduced under these conditions (Q. He and S. Jantaro, unpublished data). HLIPs (HliA–HliD) and ferrochelatase (ScpA) are believed to perform rather distinct functions at least in HL (Funk and Vermaas, 1999; Jantaro et al., 2006). The functions of HLIPs are currently under active investigation, but the results from different groups have been somewhat controversial. The hliA gene was originally found to be a HL-inducible gene (Dolganov et al., 1995). However, Funk and Vermaas (1999) reported that scp (hli) transcripts were not enhanced by HL; rather, they were stimulated by the deletion of PSI. More recent studies demonstrated that HLIPs were indeed induced by HL and by other stress conditions, as well as in either cyanobacterial or plant systems (Heddad and Adamska, 2000; Jansson et al., 2000; He et al., 2001; Suzuki et al., 2001; Mikami et al., 2002; Hsiao et al., 2004; Teramoto et al., 2004; Jantaro et al., 2006). The views about the functions of HLIPs are also divided: HLIPs may serve as chlorophyll carriers (Xu et al., 2004) or promote dissipation of absorbed excess light energy (Havaux et al., 2003). Nevertheless, it is rather clear that HLIPs help cyanobacteria to cope with adverse environmental conditions. HLIPs might perform multiple functions and contribute to specific stress responses in cyanobacteria because groups of hli genes in the organisms were found to be differentially expressed under various stress conditions (Bhaya et al., 2002; Coleman et al., 2006).
HLIPs contain a single transmembrane helix that is similar to the first and third transmembrane helices of LHCP. They are considered distant relatives of LHCP. Single-helix proteins similar to HLIP, termed OHPs (Jansson et al., 2000), were also found in plants. The expression of OHPs in plants (exclusively expressed under stress conditions) is strikingly similar to the HLIPs in cyanobacteria (He et al., 2001). Interestingly, several other members of the extended LHCP family (Green et al., 1991), including the PsbS protein (Kim et al., 1992; Wedel et al., 1992), the early light-induced proteins (Grimm et al., 1989; Adamska et al., 1992) and the two-helix protein SEPs (Heddad and Adamska, 2000), also show high levels of expression under various stress conditions. These suggest that they not only have a common lineage in evolution, but also might share certain functionality, likely involving aspects of stress defense as was demonstrated in the case of PsbS (Li et al., 2000).
HLIPs are integral thylakoid membrane proteins (He et al., 2001) and attempts were recently made to further locate these proteins within thylakoid protein complexes. In Arabidopsis (Arabidopsis thaliana), the HLIP counterpart (Ohp2) was found to be associated with PSI exclusively (Andersson et al., 2003). Interestingly, studies in Synechocystis PCC 6803 showed a distinctly different result. Reportedly, one of the HLIPs (ScpD [i.e. HliB]) was found in PSII, but not in PSI (Promnares et al., 2006; Yao et al., 2007). However, these localization experiments were mainly carried out either using a Synechocystis mutant overexpressing ScpD (N-terminally His6-tagged) by the constitutive promoter of the psbA2 gene or using the strains of PSI-less background. In this study, we localized all four HLIPs in Synechocystis PCC 6803 in their native or C-terminally His-tagged forms under the control of their native HL-inducible promoters in wild-type background and investigated the impact of HLIP deletion on photosynthetic protein complexes. Our results showed that native HliA and HliB induced by HL treatment were associated with PSI trimers and Slr1128, whereas HliC was associated with PsaL and TMP14. The HliD protein was found to be associated with partially dissociated PSI complexes. In addition, we demonstrated that deletion of HLIPs resulted in diminishing of trimeric PSI complex in HL and reducing PSI activity and chlorophyll fluorescence at 77K. Furthermore, our data showed that PsaL and Slr1128 (with which HLIPs potentially interact) are important for cell survival under HL conditions.
RESULTS
Effect of hli Deletion on the Sensitivity of PSII and PSI to HL
The hli deletion mutants do not exhibit any detectable defects in growth or photosynthetic activities under standard LL conditions (40 μmol of photon m−2 s−1 at 30°C), but they are abnormal in photosynthetic activities under intermediate HL conditions and are severely impaired in growth or fitness in HL (He et al., 2001; Havaux et al., 2003). To investigate whether the enhanced light sensitivity of hli deletion mutants is due to the sensitivity of PSII to excess light, we grew cultures under LL conditions and transferred cells to intermediate HL, under which conditions all strains grew reasonably well despite the moderate excess excitation. Samples were taken at different time intervals of intermediate HL treatment, and the change of the maximum quantum yield of PSII photochemistry (Fv/Fm) of the wild-type and mutant strains was monitored. We found that both the wild-type and mutant strains showed signs of photoinhibition during the first 6 h of the intermediate HL treatment as indicated by approximately a 40% decrease in Fv/Fm, but they were able to acclimate to the growth conditions within 24 h as indicated by the recovery of Fv/Fm. No statistically significant difference in Fv/Fm was observed between the wild-type and any of the mutant strains at any given time points (data not shown). We also measured the steady-state level of the PSII electron transport rates of thylakoid membranes isolated from cells treated by intermediate HL for 24 h (see “Materials and Methods” for experiment details). As shown in Figure 1, no statistically significant difference in O2 evolution level was observed between the wild-type and any of the mutant strains examined. These results suggest that the sensitivity of PSII to moderate excess excitation was not affected by the deletion of the hli genes.
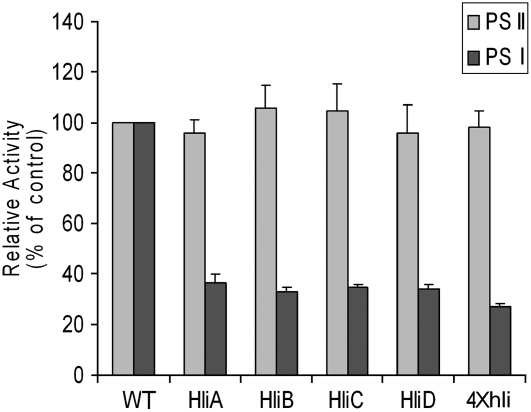
Figure 1.
Relative PSI and PSII activities of thylakoid membranes. Thylakoid membranes were isolated from cells incubated in intermediate HL for 12 h. The activities of PSI and PSII were evaluated by measuring oxygen evolution resulting from electron transport from DCIP/ascorbic acid via PSI to MV and from H2O via PSII to phenyl-p-benzoquinone, respectively. Thylakoid membranes were added to the reaction mixture to the final concentration of 15 μg/mL of chlorophyll. The electron transport rates in the wild-type cells were normalized to 100%. WT, Wild type; HliA, hliA deletion mutant; HliB, hliB deletion mutant; HliC, hliC deletion mutant; HliD, hliD deletion mutant; 4×hli, the quadruple hli deletion mutant. The 100% activities of PSI and PSII are 735.7 ± 27.6 and 176.6 ± 10.4 μmol O2 h−1 mg−1 chlorophyll.
To evaluate the impact of hli deletion on PSI activity under intermediate HL conditions, we estimated the steady-state level of the PSI electron transport rates by measuring methyl viologen (MV)-dependent O2 consumption of thylakoid membranes isolated from cells treated by intermediate HL for 24 h. As shown in Figure 1, the PSI electron transport rates of thylakoid membranes of the hli mutants were significantly lower than that of the wild type, whereas the PSII rates were similar. The PSI activities of the single mutants were about one-third of the wild-type level, and the quadruple mutant was about one-fourth of the wild-type level. In contrast, the maximum PSII activity measured in the presence of exogenous electron acceptors was at a similar level for all strains. This suggests that each Hli protein is important for optimal PSI activity in intermediate HL.
77K Fluorescence Emission of Isolated Thylakoid Membranes
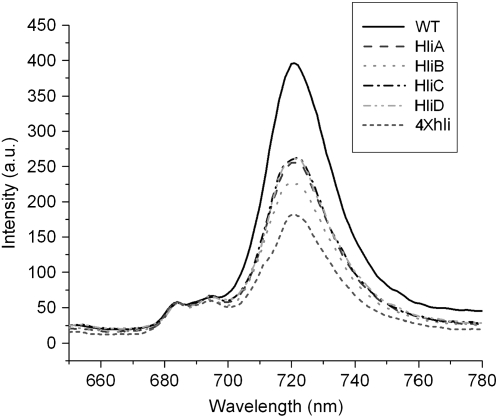
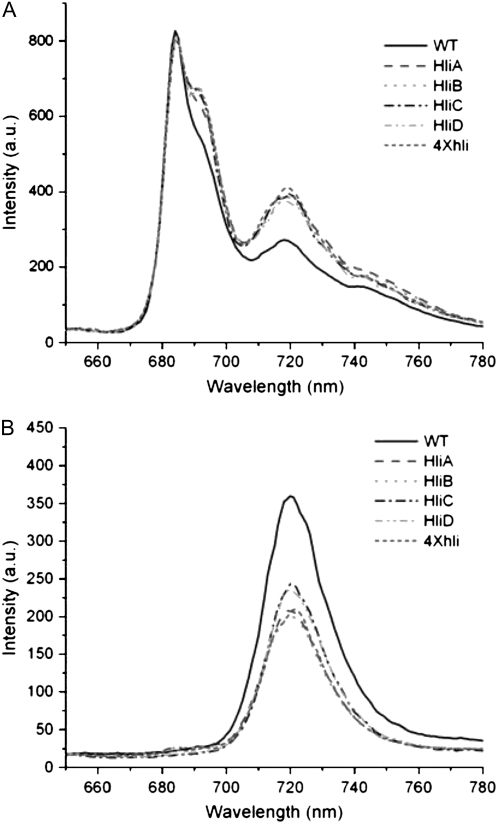
The impact of hli deletion on the stoichiometry of photosystems was diagnostically investigated by 77K fluorescence spectroscopy. Thylakoid membranes were isolated from cells grown under intermediate HL conditions, and their chlorophyll fluorescence emission spectra were recorded in liquid nitrogen. Figure 2 shows typical 77K fluorescence emission spectra of the wild-type and mutant thylakoid membranes excited at 430 nm. The thylakoid membranes of all strains showed a major peak at 720 nm, which corresponds to PSI-associated chlorophyll, and two smaller peaks at 685 and 695 nm, which are originated mainly from PSII-associated chlorophyll. The relative fluorescence of PSI chlorophyll (FPSI) of the mutant strains was significantly lower than that of the wild type when normalized at 685 nm. The F720:F685 ratio of the hli single mutants ranges from 4 to 5, whereas the ratio for the wild type is about 7 (Table I). The quadruple hli mutant (4×hli) had a very low F720:F685 ratio of about 3.19. In cyanobacteria, the F720:F685 ratio generally correlates well with the relative content of PSI and PSII (Murakami et al., 1997). Therefore, the lowered F720:F685 seen in the hli mutant strains suggests that hli deletions affected the stoichiometry of photosystems.
Figure 2.
77K fluorescence of thylakoid membranes of the Synechocystis hli deletion strains. Thylakoid membranes were isolated from cells treated by intermediate HL for 12 h. The fluorescence emission spectra of the thylakoid membranes were recorded with an excitation wavelength of 430 nm at 77K. All samples correspond to 15 μg of chlorophyll/mL. The spectra were normalized at 685 nm. WT, Wild type; HliA, hliA deletion mutant; HliB, hliB deletion mutant; HliC, hliC deletion mutant; HliD, hliD deletion mutant; 4×hli, the quadruple hli deletion mutant.
Table I.
PSI:PSII ratio of thylakoid membrane and the fraction 2 of Suc gradient
Thylakoid membranes from cells treated by intermediate HL for 12 h were isolated, solubilized, and fractionated by Suc gradient as described in “Materials and Methods.” The thylakoid membranes and the fraction 2 (mixture of monomeric PSI and PSII) of Suc gradient were then analyzed by fluorescence emission scan at 77K with an excitation wavelength of 440 nm. The ratio of the fluorescence intensity originated from PSI (at 720 nm) and PSII (685 nm) was used as an estimate of PSI:PSII ratio. TK, Thylakoid membrane; F2, fraction 2.
| Strain |
F720/F685
|
|
|---|---|---|
| TK | F2 | |
| Wild type | 7.09 ± 0.12 | 0.33 ± 0.01 |
| hliA− | 4.51 ± 0.11 | 0.49 ± 0.02 |
| hliB− | 4.18 ± 0.23 | 0.46 ± 0.02 |
| hliC− | 4.79 ± 0.20 | 0.50 ± 0.01 |
| hliD− | 4.85 ± 0.70 | 0.47 ± 0.02 |
| 4×hli | 3.19 ± 0.15 | 0.49 ± 0.01 |
Association of HLIPs with Pigment Complexes
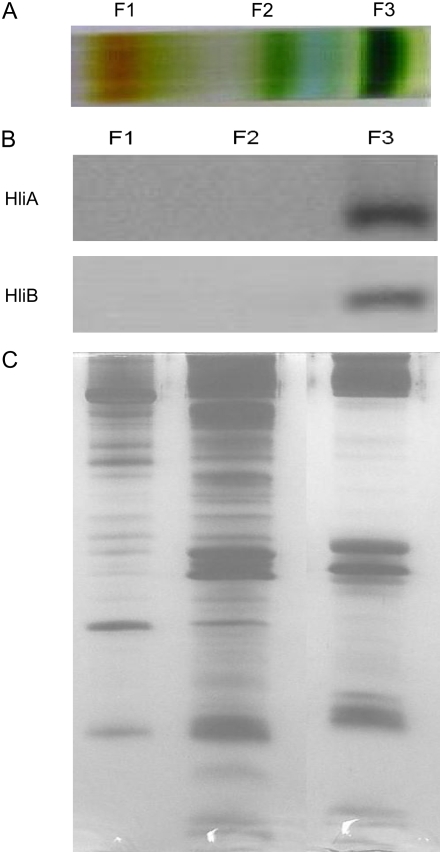
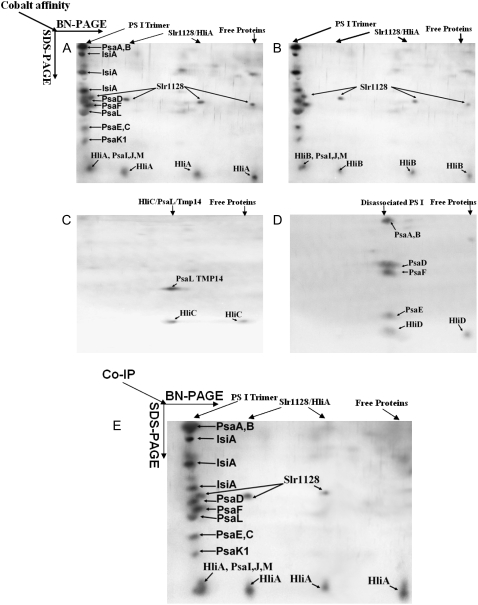
The observed effects of hli deletion on photosynthetic characteristics prompted us to investigate whether HLIPs are present in photosynthetic pigment complexes. To this aim, thylakoid membranes from the wild-type and mutant cells were solubilized by dodecyl maltoside, and the pigment complexes were separated by Suc gradient ultracentrifugation. This resulted in the separation of PSI trimers (fraction 3) from another green band, which consists of the PSII and the monomeric PSI complexes (fraction 2; Fig. 3A). The pigment complexes were then denatured and fractionated by a 12% to 20% SDS-PAGE with 4 m urea. The HLIPs were tracked using HLIP-specific antibodies. As shown in Figure 3B, HliA and HliB were detected mainly in the trimeric PSI fraction. Low amounts of these two HLIPs were also detected in fraction 2, when large amounts of proteins (50 μg/lane) were loaded and excess antibodies were used (data not shown). However, no HLIPs were detected in the monomeric PSI or PSII complexes prepared by blue native (BN) gel (data not shown). HliC and HliD were not found in either fraction 2 or fraction 3 of Suc gradient or in the PSII or the monomeric PSI complexes prepared by BN gel (data not shown). This suggests that HliC and HliD are not associated with intact photosystem protein complexes or their level of presence in these complexes is lower than the detection limit.
Figure 3.
Presence of HLIPs in PSI trimer fractions. A, Protein complexes of thylakoid membranes isolated from wild-type cells treated by intermediate HL were fractionated by Suc gradient ultracentrifugation. B, Fraction 1 (mostly carotenoids and free proteins), fraction 2 (PSII and monomeric PSI complexes), and fraction 3 (PSI trimers) were collected and separated by SDS-PAGE (12%–20%) with 6 m urea. HLIPs were tracked using specific antibodies against each individual Hli polypeptide. C, Loading control. For fraction 1, 1.5 μg of total carotenoids was loaded onto each lane; for fractions 2 and 3, 3 μg of chlorophyll was loaded onto each lane. [See online article for color version of this figure.]
Impact of hli Deletion on Trimeric PSI
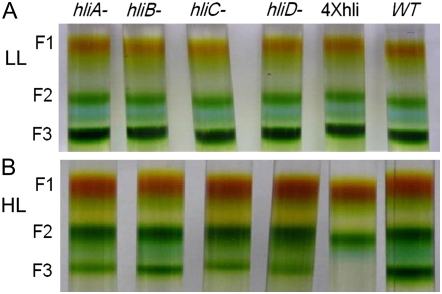
Thylakoid membranes isolated from cells grown in LL or treated by intermediate HL or HL for 12 h were solubilized and fractionated by Suc gradient ultracentrifugation. Figure 4 shows the typical fraction 2 and fraction 3 of LL (Fig. 4A) and HL (Fig. 4B) samples of various strains. We estimated the amounts of the trimeric PSI in the wild type and the quadruple hli mutant by measuring the chlorophyll content in fraction 2 and fraction 3. The results are shown in Table II. In cells grown in LL, the majority of chlorophyll (approximately two-thirds of the total chlorophyll) was associated with trimeric PSI fraction for the wild type and any of the hli mutants examined. After cells were incubated in intermediate HL or HL for 12 h, the wild-type cells showed very little change in the content of PSI trimers. In contrast, the hli single mutants lost more than 30% of the PSI trimers after they had been incubated in intermediate HL for 12 h. The reduction of PSI trimers were further augmented in these cells by the increase of light intensity; hli single mutants retained only one-third of PSI trimers after they were treated by HL for 12 h. The quadruple mutant contained less than one-half of PSI trimers following a 12-h incubation in intermediate HL; it lost essentially all of the PSI trimers after being incubated in HL for 12 h.
Figure 4.
Suc gradient fractions of thylakoid protein complexes from the wild-type and hli strains. Thylakoid membranes were isolated from the wild-type and hli deletion strains grown in LL (A) or treated by HL (B) for 12 h. Thylakoid protein complexes were separated by step Suc gradient ultracentrifugation. The lack of trimeric PSI in HL-treated 4×hli cells was visually demonstrated (B).
Table II.
Relative content of trimeric PSI complex in the hli deletion strains of Synechocystis PCC 6803
Thylakoid membranes from cells grown in low light (LL) or treated by intermediate high light (IHL) or high light (HL) for 12 h were isolated, solubilized, and fractionated by Suc gradient. The chlorophyll content of fraction 3 (trimeric PSI) and fraction 2 (mixture of monomeric PSI and PSII) was determined. The trimeric PSI content was estimated as percentage of chlorophyll associated with fraction 3 (chlorophyll content in fraction 3 divided by the total chlorophyll content in fraction 2 and fraction 3).
| Strain | Trimeric PSI Content
|
||
|---|---|---|---|
| LL | IHL | HL | |
| % | |||
| Wild type | 69.2 ± 7.1 | 67.9 ± 4.7 | 61.6 ± 7.5 |
| hliA− | 68.6 ± 8.0 | 41.6 ± 6.7 | 22.7 ± 2.8 |
| hliB− | 68.8 ± 3.5 | 43.8 ± 7.3 | 24.5 ± 3.0 |
| hliC− | 68.4 ± 5.3 | 42.9 ± 2.4 | 22.8 ± 5.2 |
| hliD− | 69.1 ± 5.5 | 43.4 ± 4.0 | 22.9 ± 4.5 |
| 4×hli | 69.3 ± 4.8 | 27.6 ± 12.7 | 2.8 ± 2.4 |
The protein to chlorophyll ratio (w/w) of fraction 3 was about 4.1 to 4.3 for the wild-type and all hli deletion strains when cells were grown in LL. The ratio remained unchanged for the wild type and the hliC and hliD single mutants after they were treated by intermediate HL. However, it was about 5.0 to 5.5 for the hliA and hliB single mutants, and the quadruple mutant that received intermediate HL treatment. These results suggest that hli deletion affects the stability of the trimeric PSI complex under conditions of excess excitation.
77K Fluorescence Spectra of Suc Gradient Fractions
Separation of photosynthetic pigment complexes by Suc gradient ultracentrifugation allowed us to examine the 77K fluorescence spectra of pigment complexes in more detail. Figure 5A shows typical fluorescence emission spectra of fraction 2 (PSII and PSI monomer mixture). When normalized at F685, the mutants exhibited a higher F720 and a higher F695 (Fig. 5A). The higher F695 seen in the mutant samples suggests that the PSII complexes might also be affected, directly or indirectly, by the deletion of HLIPs, even though we did not detect any HLIPs in any of our purified PSII samples. This increase in F695 in mutants does not appear to affect their recovery from photoinhibition of PSII under moderate excess excitation (Fig. 1). The F720:F685 of the mutants ranges from 0.45 to 0.5, which is much higher than the ratio in wild-type samples (about 0.33). This suggests that the monomeric PSI:total PSII ratio is higher in the mutants than in wild-type cells.
Figure 5.
77K fluorescence of Suc gradient fractions 2 (A) and 3 (B) of the Synechocystis hli deletion mutants. Thylakoid membranes from cells treated by intermediate HL for 12 h were isolated, solubilized, and fractionated by Suc gradient. The fluorescence emission spectra of the Suc gradient fractions were recorded with an excitation wavelength of 430 nm at 77K. All samples correspond to 15 μg of chlorophyll/mL. The spectra of fraction 2 (PSII and monomeric PSI) were normalized at 685 nm; the spectra of fraction 3 (PSI trimer) were normalized at 650 nm. WT, Wild type; HliA, hliA deletion mutant; HliB, hliB deletion mutant; HliC, hliC deletion mutant; HliD, hliD deletion mutant; 4×hli, the quadruple hli deletion mutant.
The 77K fluorescence spectra of the PSI trimer (fraction 3) are shown in Figure 5B. Compared with wild-type samples, the PSI trimers from mutants exhibit a much lower F720 on per chlorophyll or per protein basis. Therefore, deletion of hli genes affects not only the amounts, but also the fluorescence yield, of PSI trimers.
Two-Dimensional PAGE Analysis of HLIP Complexes
To investigate components with which HLIPs interact, we tagged each HLIP with a His9-tag at its C terminus directly on the Synechocystis genome (rather than using an expression plasmid; He et al., 2001). This allows isolation of HLIPs and the proteins associated with them by immobilized metal (cobalt) affinity chromatography (IMAC) under nondenaturing conditions (see “Materials and Methods”). Considering that the protein complexes isolated by IMAC might contain unspecific components, which normally will not remain within specific protein complexes after they are dissolved in solutions, we further fractionated the protein complexes isolated by IMAC using BN-PAGE before they were resolved by tricine-SDS-PAGE. BN-PAGE is a separation method with a higher resolution than gel filtration or Suc density ultracentrifugation that is commonly used to analyze multiprotein complexes from 10 kD to 10 MD. We then tracked HLIPs by the specific antibodies against the protein or the poly-His epitopes. The proteins that were associated with HLIPs were identified by mass spectrometry (MS; Supplemental Table S1). As shown in Figure 6, HliA and HliB were found to be associated mainly with PSI trimers; some of the HliA and HliB were also associated with Slr1128, a polypeptide with unknown function (Fig. 6, A and B). HliC was found to be associated with PsaL and TMP14 (Fig. 6C). HliD was found to be associated with various PSI components (e.g. PsaA-B, PsaE, PsaD, PsaF), but never with intact PSI complexes (Fig. 6D). No detectable amount of HliA protein was found in HliB-containing PSI trimers, and no HliB was found in HliA-containing PSI trimers by MS or western blot (data not shown). Therefore, HliA and HliB were associated with a different pool of PSI trimers. Interestingly, HliD was found in HliC complexes, but no HliC was detected in HliD complexes (mainly partial PSI aggregation complexes). This suggests there is a limited, perhaps loose, interaction between HliC and HliD.
Figure 6.
Two-dimensional PAGE analysis of protein complexes associated with HLIPs. HLIP-associated protein complexes were isolated from Synechocystis strains in which HliA (A) or HliB (B) or HliC (C) or HliD (D) was tagged with a 9×His epitope at the C terminus, using affinity chromatography under nondenaturing conditions. The protein complexes isolated were further separated by a BN gel followed by a second dimension tricine-SDS-PAGE (12%–20%) with 4 m urea. HLIPs were tracked using specific antibodies against each individual Hli polypeptide. The proteins associated with HLIPs were identified by MS (LCQ ion trap spectrometer). E, Protein complex coimmunoprecipitated with anti-HliA antibodies. HliA complexes isolated by metal chromatography as in A was further purified by coimmunoprecipitation and resolved by a BN gel followed by tricine-SDS-PAGE.
HliA and HliB are highly similar with amino acid identities of 87%. Interestingly, the proteins copurified with these two HLIPs are also highly similar (Fig. 6, A and B). Because multiple protein complexes, among other apparent impurities, were pulled out by IMAC from either HliA- or HliB-tagged strains, we further purified HliA protein complexes (isolated by IMAC; Fig. 6A) by HliA antibody (of which we have a relatively large quantity of purified antibodies) using the Pierce ProFound coimmunoprecipitation kit. The results are shown in Figure 6E. Clearly, the PSI trimers and the Slr1128 were retained by HliA antibody conjugated to antibody coupling gel. Therefore, the interaction of HliA with PSI trimers and the Slr1128 protein is rather specific.
Growth of slr1128− Mutants under Different Light Conditions
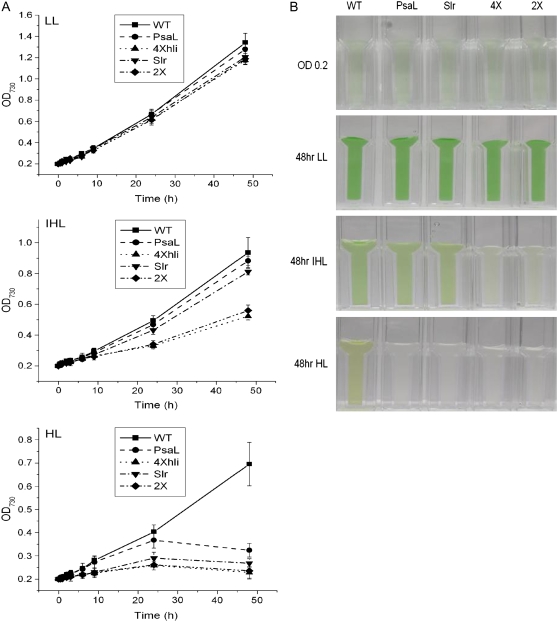
Because the 4×hli mutant lacking PSI trimers dies in HL and the psaL− mutant (lacking PSI trimers) reportedly survives in HL (Schluchter et al., 1996), we hypothesized that the Slr1128/HLIP complex might be important for protecting cells from excess excitation as well. For this reason, we inactivated the slr1128 gene from the wild type and a psaL− background (Fig. 7) and tested the growth of the homoplasmic slr1128− strains (the slr1128− single mutant and the slr1128−/psaL− double mutant) under LL, intermediate HL, and HL conditions. The growth data are shown in Figure 8. In LL, both slr1128− strains grew similarly well as compared to the wild type, suggesting that Slr1128 is not essential for Synechocystis growth under normal growth conditions. Under intermediate HL, the psaL− mutant grew at a rate similar to the wild-type growth rate, consistent with previous reports; the slr1128− mutant did not show any noticeable growth defect either. Interestingly, like the 4×hli mutant, the slr1128−/psaL− double mutant grew poorly under the same light conditions, suggesting that retaining PSI trimers and Slr1128 is important for the fitness of the cells under intermediate HL. In HL, all four mutants (slr1128−, psaL−, slr1128−/psaL−, and 4×hli) died, whereas wild-type cells grew reasonably well, suggesting PsaL, Slr1128, and HLIPs are all important for survival when excess excitations are aggravated.
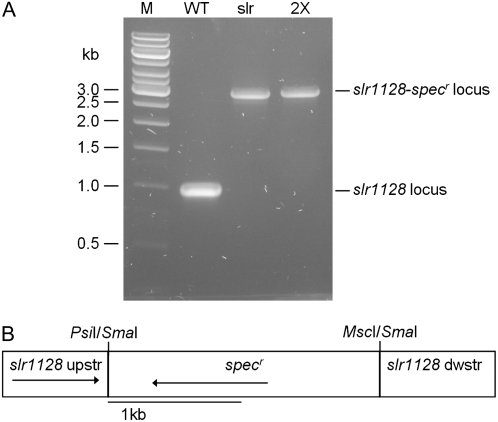
Figure 7.
Inactivation of the slr1128 gene. A, PCR analysis of the slr1128 gene in potential mutants in which a fragment of the slr1128 gene was deleted and replaced with a spectinomycin-resistant cartridge (specr). Genomic DNA for PCR analysis was isolated from a putative slr1128− mutant and a slr1128−/psaL− mutant. M, DNA size marker; WT, wild type; slr, slr1128− single mutant; 2×, slr1128−/psaL− mutant. B, Depiction of the plasmid construct used to generate the slr1128− strains. The plasmid carries a 128-bp deletion of slr1128.
Figure 8.
Growth of Synechocystis PCC 6803 strains under different light conditions. A, Growth of the wild-type (squares), psaL− (circles), 4×hli (triangles, point up), slr1128− (triangles, point down), and slr1128−/psaL− (diamonds) strains under LL, intermediate HL (IHL), and HL conditions. Cells were grown in BG-11 medium, and cell growth was monitored as a change in optical density at 730 nm. Curves were generated by averaging the data obtained from three representative experiments. B, Cultures of the wild-type (WT), psaL− (PsaL), slr1128− (Slr), 4×hli (4×), and the slr1128−/psaL− (2×) strains under LL, IHL, and HL conditions. LL-grown cells in logarithmic growth were diluted to OD730 approximately 0.2 and incubated in LL and IHL or HL for 2 d.
DISCUSSION
We have isolated Hli polypeptides and their associated proteins under nondenaturing conditions and studied the effect of hli deletion on the stability of photosystem complexes under HL conditions in Synechocystis PCC 6803. The complex isolation and localization studies were carried out using the wild-type cells and the strains in which the Hli polypeptides were individually tagged with a small epitope (9×His). No promoters or other elements essential for hli gene expression were modified nor was the PSI complex (the major chlorophyll-protein complex) deleted in these studies.
Localization of HLIP was reported previously (Andersson et al., 2003; Promnares et al., 2006; Yao et al., 2007); however, this study localized an entire family of HLIPs. One gene encoding HLIP (termed Ohp2) was found in the genome of Arabidopsis. The Ohp2 polypeptide was shown to be present in the PSI complex, but not in PSII isolated by Suc density gradient centrifugation (Andersson et al., 2003). Here, we showed that two of the four HLIPs in Synechocystis PCC 6803, the HliA and HliB proteins, were specifically associated with PSI trimers. These results, while extending the study in Arabidopsis, appear to be in conflict with two earlier reports (Promnares et al., 2006; Yao et al., 2007) that report the association of HliA and HliB with PSII when these two polypeptides were overexpressed under the control of the constitutive psbA2 promoter in LL or in PSI-less parental strain, which accumulates high levels of hli transcripts even in LL. Here, we show that HliC was associated with PsaL and TMP14, whereas HliD was associated with PSI dimers or partially dissociated PSI complexes (Fig. 6).
The PSI complex in cyanobacteria is organized in the thylakoid membrane preferentially as trimers (Tsiotis et al., 1995; Karapetyan et al., 1999). We found that PSI trimers account for 70% and approximately 60% of total cellular chlorophyll in LL and HL, respectively (Table II). The PSI trimer:monomer ratio (molar) in LL and HL samples was estimated to be about 1:1 and 2:1, respectively, based on the chlorophyll content of the corresponding bands on the BN gel (data not shown). Therefore, as in other cyanobacteria, the PSI complexes exist preferably as trimers in Synechocystis PCC 6803, and the trimer:monomer ratio was increased by high-intensity light. It has been shown that PsaL and PsaI are important in formation of PSI trimers in cyanobacteria (Chitnis and Chitnis, 1993; Sun et al., 1998). In this article, we demonstrated that HLIPs are needed for stabilization of PSI trimers in HL (Fig. 4; Table II). The mutant lacking all four hli genes lost more than 90% of PSI trimers after they were incubated in HL for 12 h. This event was precedent to a decline in cell fitness/viability, which occurred after the cells were incubated in HL for more than 24 h (He et al., 2001; Jantaro et al., 2006). This might suggest that PSI trimers were important for photoacclimation. Indeed, the psaL− mutant, lacking PSI trimers, died after it was incubated in HL for 2 d (Fig. 8). Interestingly, the psaL− mutant grew reasonably well under intermediate HL, whereas the 4×hli mutant showed retarded growth. This suggests that HLIPs also play other important roles in photoacclimation in addition to stabilization of PSI trimers. The IsiA protein, known to encircle trimeric PSI to form PSI-IsiA supercomplexes under iron-deficient conditions (Bibby et al., 2001; Kouril et al., 2005), may also be important for the stabilization of PSI trimers under HL conditions. It was reported that isiA gene transcript is inducible by HL (Havaux et al., 2005). However, this and other work documenting stress induction of isiA transcript provided no evidence that isiA gene transcription is accompanied by protein synthesis and membrane integration of IsiA (Singh and Sherman, 2007). In this study, we were able to enrich IsiA protein in HLIP-containing PSI trimers prepared from HL-treated cells. The functional relationship between HLIPs and IsiA awaits further investigation.
The PSI complex is well known for its resistance to photoinhibition under HL conditions, even though it was demonstrated that such resistance is heavily compromised at chilling temperatures (Tjus et al., 1998; Scheller and Haldrup, 2005). The association of HLIPs with PSI complexes under HL conditions indicates that the PSI complexes undergo dynamic changes in response to the light stress. The interaction of HLIPs with PsaL and TMP14, a membrane protein that was shown to interact with PsaL in plants (Zhang and Scheller, 2004), suggests that HLIPs are positioned away from high concentrations of antenna chlorophylls on the PsaA/PsaB polypeptides of PSI complex. It might also suggest that at least some of the key events leading to PSI structural and compositional changes in HL may have occurred at the interface of monomers in the center of PSI trimers. Such a change may be beneficial for photoprotection or survival as the quadruple hli mutant dies in HL.
The PSI trimers of some cyanobacteria (e.g. Spirulina platensis) contain the red chlorophyll (emitting at 760 nm at 77K) that might contribute to the dissipation of energy in PSI trimers (Shubin et al., 1992; Karapetyan et al., 1999). We did not observe the existence of the red chlorophyll species in Synechocystis PCC 6803 in PSI trimers either from the wild type or HLIP deletion mutants. Rather interestingly, we found that the intensity of F720 of PSI trimers isolated from the 4×hli strain was much lower on per chlorophyll or per protein basis (Fig. 5B). In addition, the PSI activity of the HLIP-deficient mutant is much lower than that of the wild type (Fig. 1). These findings indicate that association of HLIPs has stimulated chlorophyll fluorescence in PSI trimers and helped maintain PSI electron transport capability in HL. Because the IsiA protein is presented in the isolated HLIP-containing PSI trimers (Fig. 6), it might affect PSI trimers in a similar way as HLIPs; however, it is well established or accepted that deletion of isiA mainly affects chlorophyll fluorescence at 685 nm rather than at 720 nm (Bibby et al., 2001). In contrast to higher plants, cyanobacteria are highly enriched with PSI, and the PSI:PSII ratio is generally between 3 and 5.5 (Rakhimberdieva et al., 2001). The phenomenon is not well understood. However, it might favor PSI-driven cyclic electron transport and/or contribute to the dissipation of excess excitation (Karapetyan et al., 1999). Our results are compatible with this concept. We do not favor the idea of HLIPs as possible assemblers of PSI trimers because deletion of HLIPs does not affect PSI trimers in LL.
Another interesting finding made in this article is the consistent presence of Slr1128 in HliA and HliB complex preparations. However, only a fraction of this protein is present in the HLIP-containing PSI trimers and the rest is well separated from the trimeric PSI complexes (Fig. 6). Therefore, it appears to be tightly associated with HliA and/or HliB and rather weakly with PSI trimers (if there is any direct interaction between Slr1128 and PSI trimers). This hypothetical protein is highly conserved in cyanobacteria and the amino acid identity of the protein in various species of cyanobacteria is larger than 80%, in general. It also appears to be present in plants (with amino acid identity of 59%–66%). The protein has a transmembrane domain at its N terminus. It is likely an integral membrane protein because it was also identified as a thylakoid membrane protein by other researchers (Herranen et al., 2004). The protein also exhibits high similarity (around 35% in identity) to human and animal stomatins (thought to regulate an associated ion channel) and to a bacterial protease (HflC), but the similarity of Slr1128 to stomatins or HflC is not as high as it is to the counterparts in other cyanobacteria or plants.
We tested the hypothesis that the Slr1128/HLIP complex is important for protecting cells from excess excitation by targeted mutagenesis. We inactivated the slr1128 gene from the wild type and a psaL− strain (Fig. 7) and examined the growth of the slr1128− single mutant and the slr1128−/psaL− double mutant under LL, intermediate HL, and HL conditions. Rather excitingly, we observed that the slr1128−/psaL− double mutant resembles the 4×hli mutant in growth characteristics under different light conditions. Particularly, under intermediate HL, the slr1128−/psaL− mutant and 4×hli grew at reduced rates compared to the single mutants or wild-type cells (Fig. 8). This suggests that stabilization of PSI trimers by HLIP and the association of Slr1128 with HliA and HliB are needed for maintaining cell fitness under moderate excess excitation. Under the HL we used, even the slr1128− and the psaL− single mutants died, whereas the wild-type cells survived after incubation for 2 d. This demonstrated that PsaL, Slr1128, and HLIPs are all important for survival when excess excitation is aggravated.
The PSI complexes are the major sink of iron (Keren et al., 2004) and the iron availability might affect the structure of PSI and other photosynthetic aspects profoundly (Burnap et al., 1993; Bibby et al., 2001; Boekema et al., 2001). In a genetic analysis of the 4×hli mutant by suppressor screen, the regulatory protein PfsR was discovered. Deletion of the gene from the 4×hli mutant restored viability to the parental strain under HL conditions as a result of change in iron homeostasis (Jantaro et al., 2006). This raises a question of whether or not the iron availability plays a role in stabilization of PSI complexes, including trimers. Future studies aiming to determine the relationship among iron homeostasis, HLIPs, photosynthetic complexes, and stress responses will contribute to the understanding of HLIP function and the general survival strategies that cyanobacteria employ under HL conditions.
The work presented here demonstrates the requirement of HLIPs for maintaining PSI trimers during exposure to HL. Our finding that HliA and HliB are physically associated with PSI trimers, whereas HliD and HliC are associated with PSI components, further supports that HLIPs stabilize PSI trimers in HL. Furthermore, we demonstrated that a previously uncharacterized protein, Slr1128 (with which HliA and HliB interact), is important for maintaining cell fitness under conditions of excess excitation. Because the mutant lacking both PSI trimers (by deletion of PsaL) and Slr1128 exhibits strikingly similar growth characteristics as the 4×hli mutant under different light intensities, ranging from LL (40 μmol of photon m−2 s−1) to HL (400 μmol of photon m−2 s−1), we hypothesize that stabilization of PSI trimers and interaction with Slr1128 are the major functions of HLIPs under HL conditions. A more detailed biochemical and genetic analysis of HLIP-associated protein complexes should clearly establish the roles of PsaL, Slr1128, and HLIPs in maintaining the viability of cells in HL.
MATERIALS AND METHODS
Growth Conditions and HL Treatment
Synechocystis cells were cultivated in BG-11 medium with 10 mm TES, pH 8.2, at 30°C. The culture was bubbled with air under LL conditions (40 μmol of photon m−2 s−1); intermediate HL conditions (200 μmol of photon m−2 s−1 as indicated); or HL conditions (400 μmol of photon m−2 s−1). For HL or intermediate HL experiments, the cells reaching midlogarithmic growth phase (OD730 approximately 0.8) were diluted with fresh medium to OD730 approximately 0.1 and exposed to light at 200 μmol of photon m−2 s−1 or higher for various lengths of time at 30°C.
Thylakoid Membrane Preparation and Fractionation of Membrane Protein Complexes
Thylakoid membranes were prepared as described by Shen and Vermaas (1994) with some modifications. Briefly, cell pellets derived from cells grown to midlogarithmic phase were resuspended in ice-cold thylakoid buffer (50 mm MOPS, pH 7.0, 0.4 mm Suc, 10 mm NaCl, 1 mm freshly made phenylmethylsulfonyl fluoride [PMSF]). An equal volume of glass beads prewetted by thylakoid buffer was added to the cell suspension, and the cells were broken in a bead beater with an ice-jacketed sample chamber by six breakage cycles at full speed (30 s of bursts, followed by 5 min of chilling). The homogenate was centrifuged at 1,800g for 10 min to remove unbroken cells, cellular debris, and glass beads. The membranes in supernatant were then pelleted by centrifugation at 50,000g at 4°C for 60 min. After washing with 2 mm dodecyl maltoside to remove any remaining phycobilisomes, the membranes were washed twice and resuspended in thylakoid buffer to a chlorophyll a concentration of 1 mg/mL. The chlorophyll a concentration was estimated from the dimethylfluoride extract by the formula developed by Moran (1982).
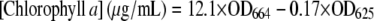 |
(1) |
To fractionate membrane protein complexes, 150 μL of 10% dodecyl maltoside was added to the thylakoid membrane to achieve a detergent to chlorophyll ratio of 15:1. The membrane was solubilized at 4°C for 30 min before it was loaded onto a 10% to 30% (w/w) step Suc gradient and centrifuged at 160,000g for 16 h at 4°C. Pigmented fractions were collected and stored at −80°C until use.
Isolation of HLIP-Associated Complexes
HLIP-associated complexes were isolated from the Synechocystis strains carrying His9-tagged HLIPs using metal chromatography under native conditions (Bricker et al., 1998). Thylakoid membranes (1 mg chlorophyll/mL) were resuspended in buffer A (50 mm MES-NaOH, pH 6.0, 10 mm MgCl2, 5 mm CaCl2, 25% glycerol, 1 mm PMSF). Then, one-tenth volume of 10% dodecyl maltoside was added to the thylakoid membranes to obtain a final concentration of 1%. After incubation on ice for 30 min, solubilized membranes were loaded onto Ni-Co metal affinity column (Talon) pre-equilibrated with buffer B (buffer A + 0.04% dodecyl maltoside). The column was then washed with 3× bed-volume of buffer B. Protein complexes were eluted from the column by 3× bed-volume of buffer C (buffer B + 50 mm His) and concentrated by precipitation with 25% PEG-8000 in 50 mm MES-NaOH, pH 6.0. The whole procedure was performed at 4°C and in dim light or in darkness, whenever possible.
Coimmunoprecipitation Assay
Coimmunoprecipitation (pull-down assay) was performed by using the ProFound coimmunoprecipitation kit (Pierce Biotechnology) in accordance with the supplier's instructions to further purify the HliA-complexes isolated by metal chromatography. Briefly, HliA antibodies were mixed with an amine-reactive gel and direct covalent immobilization of the antibodies was attained by using sodium cyanoborohydride. The HliA complexes isolated by metal chromatography were then added to the immobilized gel and incubated at 16°C for 3 h before the gel was washed and the coimmunoprecipitation complex was eluted.
BN-PAGE, Tricine-PAGE, and Immunoblotting
BN-PAGE was performed as described (Schagger and von Jagow, 1991). For electrophoresis in the second dimension, a lane of the BN gel was excised and denatured in 1.5× SDS sample buffer (50 mm Tris-HCl, pH 6.8, 3% SDS, 150 mm dithiothreitol, 0.01% bromphenol blue) for 30 min at room temperature. The gel slice was then laid onto a 12% to 20% tricine-SDS gel with 4 m urea as described (Schagger, 2006). The proteins were visualized by silver staining (Rabilloud et al., 1988). Polypeptides resolved by tricine- or SDS-PAGE were electrotransferred onto the polyvinylidene difluoride membrane. After the blocking step, the membranes were incubated with polyclonal primary antibodies followed by a horseradish peroxidase-conjugated secondary antibody. The reactive bands were detected by enhanced chemiluminescence detection reagents (GE Healthcare).
MS Analysis
Nano-liquid chromatography-tandem MS (MS/MS; Midwest Bio Services; Proteomic Services, NextGen Sciences; National Center for Toxicology) was utilized for the analysis of in-gel digests of protein bands excised from tricine-PAGE. The peptide mixture from tryptic digest was concentrated on a peptide trap column and washed to remove salts and other impurities. Desalted peptides were then separated on a microcapillary C18 reverse-phase chromatography column. Full MS and MS/MS spectra were acquired by the LCQ Deca XP Plus ion trap mass spectrometer. The sequences of the peptides were inferred by matching the MS/MS spectra to protein sequence databases using TURBOSEQUEST software.
DNA Manipulation and Mutant Construction
To inactivate slr1128, we amplified a 1,571-bp DNA fragment (corresponding to 600 bp upstream of the slr1128 start codon to 6 bp downstream of the stop codon) from Synechocystis PCC 6803 genomic DNA. The primers used were 5′-GGTgCATgcTCCCTTAACAATGGTG-3′ and 5′-AACaGatctAACTGCCCGATGGCGG-3′; the lowercase letters indicate insertions added for creation of SphI and BglII sites. The DNA fragment covering the entire slr1128 gene was cloned into pQE-70 vector. A 128-bp coding sequence of slr1128 was then deleted by digestion with restriction enzymes (PsiI and MscI) and replaced with a 2-kb spectinomycin resistance cartridge. The resulting plasmid, pQE-70-slr1128-specr, was used to transform the Synechocystis PCC 6803 wild-type cells and a strain bearing psaL deletion generated previously (Jantaro et al., 2006). The transformants were selected by screening for resistance to 20 mg/L spectinomycin in BG-11 medium. Transformants were restreaked onto the same medium, and segregation of the inactivated slr1128 gene was monitored by PCR, using genomic DNA from the transformants. The two primers used were 5′-AAACCATGGAAGCCTTTTTTCTCTTC-3′ and 5′-AACAGATCTAACTGCCCGATGGCGG-3′ that recognize sequences around start and stop codons of the slr1128 gene, respectively. The expected fragment sizes of the wild-type slr1128 and the interrupted slr1128 loci are approximately 1 and 3 kb, respectively. Homoplasmic mutants were obtained and the mutants generated by transformation of the wild-type and the psaL− strains were designated as slr1128− and slr1128−/psaL−, respectively.
Estimation of Electron Transport Rates
Electron transport rates of PSI or PSII were estimated by measuring O2 consumption/evolution using a Clark-type electrode. The light intensity used was 500 μmol of photons m−2 s−1 white light. Thylakoid membranes were adjusted to a chlorophyll content of 15 μg/mL for all measurements. The PSI reaction mixture contains 40 μm MV, 5 mm NH4Cl, 2 mm ascorbic acid, 0.1 mm 2,6-dichlorophenolindophenol (DCPIP), 2 mm NaN3, 40 μm 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), 40 mm tricine (pH 7.5), and 100 mm Suc. The reaction permits measurement of the electron transport rates from DCIP/ascorbic acid via PSI to MV; one oxygen molecule is consumed for each electron transport event. The PSII reaction mixture contains 5 mm NH4Cl, 4 mm K3FeCN, 1 mm phenyl-p-benzoquinone, 40 mm tricine (pH 7.5), and 100 mm Suc, which measures the electron transport rate from H2O via PSII to phenyl-p-benzoquinone; one oxygen molecule is produced for every four electrons transported. O2 evolution/consumption was followed for 3 min, and the rate was calculated accordingly.
Chlorophyll Fluorescence Analysis and 77K Fluorescence Spectroscopy
Chlorophyll fluorescence analysis was carried out with a dual-modulation kinetic fluorometer (model FL-3000; Photon Systems Instruments) as described (Jantaro et al., 2006). Fluorescence emission spectra at 77K were recorded with excitation and emission bandwidths of 5 and 1.5 nm, respectively, using a Cary Eclipse fluorescence spectrophotometer (Varian). The excitation wavelength used was 430 nm. Thylakoid membranes were adjusted to a chlorophyll concentration of 15 μg/mL.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Proteins identified by MS.
Supplementary Material
Acknowledgments
We thank Dr. G. Thompson, Dr. N. Ali, and Dr. S. Grace for assistance during the course of the work. We also thank J. Camp for editorial assistance.
This work was supported by the National Science Foundation (grant no. MCB0447788).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Qingfang He (qfhe@ualr.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adamska I, Ohad I, Kloppstech K (1992) Synthesis of the early light-inducible protein is controlled by blue light and related to light stress. Proc Natl Acad Sci USA 89 2610–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson U, Heddad M, Adamska I (2003) Light stress-induced one-helix protein of the chlorophyll a/b-binding family associated with photosystem I. Plant Physiol 132 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K (1994) Production and action of active oxygen species in photosynthetic tissues. In CH Foyer, PM Mullineaux, eds, Causes of Photooxidative Stress and Amelioration of Defense Systems in Plants. CRC Press, Boca Raton, FL, pp 77–104
- Bhaya D, Dufresne A, Vaulot D, Grossman A (2002) Analysis of the hli gene family in marine and freshwater cyanobacteria. FEMS Microbiol Lett 215 209–219 [DOI] [PubMed] [Google Scholar]
- Bibby TS, Nield J, Barber J (2001) Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature 412 743–745 [DOI] [PubMed] [Google Scholar]
- Boekema EJ, Hifney A, Yakushevska AE, Piotrowski M, Keegstra W, Berry S, Michel KP, Pistorius EK, Kruip J (2001) A giant chlorophyll-protein complex induced by iron deficiency in cyanobacteria. Nature 412 745–748 [DOI] [PubMed] [Google Scholar]
- Bowler C, van Camp W, van Montagu M, Inze D (1994) Superoxide dismutase in plants. Crit Rev Plant Sci 13 199–218 [Google Scholar]
- Bricker TM, Morvant J, Masri N, Sutton HM, Frankel LK (1998) Isolation of a highly active photosystem II preparation from Synechocystis 6803 using a histidine-tagged mutant of CP 47. Biochim Biophys Acta 1409 50–57 [DOI] [PubMed] [Google Scholar]
- Burnap RL, Troyan T, Sherman LA (1993) The highly abundant chlorophyll-protein complex of iron-deficient Synechococcus sp. PCC7942 (CP43′) is encoded by the isiA gene. Plant Physiol 103 893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis VP, Chitnis PR (1993) PsaL subunit is required for the formation of photosystem I trimers in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett 336 330–334 [DOI] [PubMed] [Google Scholar]
- Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, Delong EF, Chisholm SW (2006) Genomic islands and the ecology and evolution of Prochlorococcus. Science 311 1768–1770 [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B, Adams WW III (1996) The role of xanthophyll cycle carotenoids in the protection of photosynthesis. Trends Plant Sci 1 21–26 [Google Scholar]
- Demmig-Adams B, Adams WW (1992) Photoprotection and other responses of plants to high light stress. Annu Rev Plant Physiol 43 599–626 [Google Scholar]
- Di Mascio P, Devasagayam TP, Kaiser S, Sies H (1990) Carotenoids, tocopherols and thiols as biological singlet molecular oxygen quenchers. Biochem Soc Trans 18 1054–1056 [DOI] [PubMed] [Google Scholar]
- Di Mascio P, Kaiser SP, Devasagayam TP, Sies H (1991) Biological significance of active oxygen species: in vitro studies on singlet oxygen-induced DNA damage and on the singlet oxygen quenching ability of carotenoids, tocopherols and thiols. Adv Exp Med Biol 283 71–77 [DOI] [PubMed] [Google Scholar]
- Dolganov NA, Bhaya D, Grossman AR (1995) Cyanobacterial protein with similarity to the chlorophyll a/b binding proteins of higher plants: evolution and regulation. Proc Natl Acad Sci USA 92 636–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durnford DG, Falkowski PG (1997) Chloroplast redox regulation of nuclear gene transcription during photoacclimation. Photosynth Res 53 229–241 [Google Scholar]
- Foyer CH, Fletcher JM (2001) Plant antioxidants: colour me healthy. Biologist (London) 48 115–120 [PubMed] [Google Scholar]
- Fryer MJ, Ball L, Oxborough K, Karpinski S, Mullineaux PM, Baker NR (2003) Control of Ascorbate Peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J 33 691–705 [DOI] [PubMed] [Google Scholar]
- Funk C, Vermaas W (1999) A cyanobacterial gene family coding for single-helix proteins resembling part of the light-harvesting proteins from higher plants. Biochemistry 38 9397–9404 [DOI] [PubMed] [Google Scholar]
- Green BR, Pichersky E, Kloppstech K (1991) Chlorophyll a/b-binding proteins: an extended family. Trends Biochem Sci 16 181–186 [DOI] [PubMed] [Google Scholar]
- Grimm B, Kruse E, Kloppstech K (1989) Transiently expressed early light-inducible thylakoid proteins share transmembrane domains with light-harvesting chlorophyll binding proteins. Plant Mol Biol 13 583–593 [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC (1999) Free Radicals in Biology and Medicine, Ed 3. Oxford University Press, Oxford
- Havaux M, Guedeney G, Hagemann M, Yeremenko N, Matthijs HC, Jeanjean R (2005) The chlorophyll-binding protein IsiA is inducible by high light and protects the cyanobacterium Synechocystis PCC6803 from photooxidative stress. FEBS Lett 579 2289–2293 [DOI] [PubMed] [Google Scholar]
- Havaux M, Guedeney G, He Q, Grossman AR (2003) Elimination of high-light-inducible polypeptides related to eukaryotic chlorophyll a/b-binding proteins results in aberrant photoacclimation in Synechocystis PCC6803. Biochim Biophys Acta 1557 21–33 [DOI] [PubMed] [Google Scholar]
- He Q, Dolganov N, Bjorkman O, Grossman AR (2001) The high light-inducible polypeptides in Synechocystis PCC6803. Expression and function in high light. J Biol Chem 276 306–314 [DOI] [PubMed] [Google Scholar]
- Heddad M, Adamska I (2000) Light stress-regulated two-helix proteins in Arabidopsis thaliana related to the chlorophyll a/b-binding gene family. Proc Natl Acad Sci USA 97 3741–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranen M, Battchikova N, Zhang P, Graf A, Sirpio S, Paakkarinen V, Aro EM (2004) Towards functional proteomics of membrane protein complexes in Synechocystis sp. PCC 6803. Plant Physiol 134 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertwig B, Streb P, Feierabend J (1992) Light dependence of catalase synthesis and degradation in leaves and the influence of interfering stress conditions. Plant Physiol 100 1547–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horling F, Lamkemeyer P, Konig J, Finkemeier I, Kandlbinder A, Baier M, Dietz KJ (2003) Divergent light-, ascorbate-, and oxidative stress-dependent regulation of expression of the peroxiredoxin gene family in Arabidopsis. Plant Physiol 131 317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao HY, He Q, Van Waasbergen LG, Grossman AR (2004) Control of photosynthetic and high-light-responsive genes by the histidine kinase DspA: negative and positive regulation and interactions between signal transduction pathways. J Bacteriol 186 3882–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J, Lawlor DW (1993) In vivo photosynthetic electron transport does not limit photosynthetic capacity in phosphate-deficient sunflower and maize leaves. Plant Cell Environ 16 785–795 [Google Scholar]
- Jansson S, Andersson J, Kim SJ, Jackowski G (2000) An Arabidopsis thaliana protein homologous to cyanobacterial high-light-inducible proteins. Plant Mol Biol 42 345–351 [DOI] [PubMed] [Google Scholar]
- Jantaro S, Ali Q, Lone S, He Q (2006) Suppression of the lethality of high light to a quadruple HLI mutant by the inactivation of the regulatory protein PfsR in Synechocystis PCC 6803. J Biol Chem 281 30865–30874 [DOI] [PubMed] [Google Scholar]
- Karapetyan NV, Holzwarth AR, Rogner M (1999) The photosystem I trimer of cyanobacteria: molecular organization, excitation dynamics and physiological significance. FEBS Lett 460 395–400 [DOI] [PubMed] [Google Scholar]
- Keren N, Aurora R, Pakrasi HB (2004) Critical roles of bacterioferritins in iron storage and proliferation of cyanobacteria. Plant Physiol 135 1666–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Sandusky P, Bowlby NR, Aebersold R, Green BR, Vlahakis S, Yocum CF, Pichersky E (1992) Characterization of a spinach psbS cDNA encoding the 22 kDa protein of photosystem II. FEBS Lett 314 67–71 [DOI] [PubMed] [Google Scholar]
- Kouril R, Arteni AA, Lax J, Yeremenko N, D'Haene S, Rogner M, Matthijs HC, Dekker JP, Boekema EJ (2005) Structure and functional role of supercomplexes of IsiA and Photosystem I in cyanobacterial photosynthesis. FEBS Lett 579 3253–3257 [DOI] [PubMed] [Google Scholar]
- Levy H, Tal T, Shaish A, Zamir A (1993) Cbr, an algal homolog of plant early light-induced proteins, is a putative zeaxanthin binding protein. J Biol Chem 268 20892–20896 [PubMed] [Google Scholar]
- Li XP, Bjorkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403 391–395 [DOI] [PubMed] [Google Scholar]
- Mikami K, Kanesaki Y, Suzuki I, Murata N (2002) The histidine kinase Hik33 perceives osmotic stress and cold stress in Synechocystis sp PCC 6803. Mol Microbiol 46 905–915 [DOI] [PubMed] [Google Scholar]
- Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7 405–410 [DOI] [PubMed] [Google Scholar]
- Moran R (1982) Formulae for determination of chlorophyllous pigments extracted with N,N-dimethylformamide. Plant Physiol 69 1376–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami A, Kim SJ, Fujita Y (1997) Changes in photosystem stoichiometry in response to environmental conditions for cell growth observed with the cyanophyte Synechocystis PCC 6714. Plant Cell Physiol 38 392–397 [DOI] [PubMed] [Google Scholar]
- Niyogi KK (1999) Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol 50 333–359 [DOI] [PubMed] [Google Scholar]
- Ort DR, Baker NR (2002) A photoprotective role for O(2) as an alternative electron sink in photosynthesis? Curr Opin Plant Biol 5 193–198 [DOI] [PubMed] [Google Scholar]
- Peltier G, Schmidt GW (1991) Chlororespiration: an adaptation to nitrogen deficiency in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 88 4791–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl-Treves R, Galun E (1991) The tomato Cu,Zn superoxide dismutase genes are developmentally regulated and respond to light and stress. Plant Mol Biol 17 745–760 [DOI] [PubMed] [Google Scholar]
- Promnares K, Komenda J, Bumba L, Nebesarova J, Vacha F, Tichy M (2006) Cyanobacterial small chlorophyll-binding protein ScpD (HliB) is located on the periphery of photosystem II in the vicinity of PsbH and CP47 subunits. J Biol Chem 281 32705–32713 [DOI] [PubMed] [Google Scholar]
- Rabilloud T, Carpentier G, Tarroux P (1988) Improvement and simplification of low-background silver staining of proteins by using sodium dithionite. Electrophoresis 9 288–291 [DOI] [PubMed] [Google Scholar]
- Rakhimberdieva MG, Boichenko VA, Karapetyan NV, Stadnichuk IN (2001) Interaction of phycobilisomes with photosystem II dimers and photosystem I monomers and trimers in the cyanobacterium Spirulina platensis. Biochemistry 40 15780–15788 [DOI] [PubMed] [Google Scholar]
- Redinbaugh MG, Sabre M, Scandalios JG (1990) Expression of the maize Cat3 catalase gene is under the influence of a circadian rhythm. Proc Natl Acad Sci USA 87 6853–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H (2006) Tricine-SDS-PAGE. Nat Protocols 1 16–22 [DOI] [PubMed] [Google Scholar]
- Schagger H, von Jagow G (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199 223–231 [DOI] [PubMed] [Google Scholar]
- Scheller HV, Haldrup A (2005) Photoinhibition of photosystem I. Planta 221 5–8 [DOI] [PubMed] [Google Scholar]
- Schluchter WM, Shen G, Zhao J, Bryant DA (1996) Characterization of psaI and psaL mutants of Synechococcus sp. strain PCC 7002: a new model for state transitions in cyanobacteria. Photochem Photobiol 64 53–66 [DOI] [PubMed] [Google Scholar]
- Shen G, Vermaas WF (1994) Chlorophyll in a Synechocystis sp. PCC 6803 mutant without photosystem I and photosystem II core complexes. Evidence for peripheral antenna chlorophylls in cyanobacteria. J Biol Chem 269 13904–13910 [PubMed] [Google Scholar]
- Shubin VV, Bezsmertnaya IN, Karapetyan NV (1992) Isolation from Spirulina membranes of two photosystem I-type complexes, one of which contains chlorophyll responsible for the 77 K fluorescence band at 760 nm. FEBS Lett 309 340–342 [DOI] [PubMed] [Google Scholar]
- Singh AK, Sherman LA (2007) Reflections on the function of IsiA, a cyanobacterial stress-inducible, Chl-binding protein. Photosynth Res 93 17–25 [DOI] [PubMed] [Google Scholar]
- Sun J, Ke A, Jin P, Chitnis VP, Chitnis PR (1998) Isolation and functional study of photosystem I subunits in the cyanobacterium Synechocystis sp. PCC 6803. Methods Enzymol 297 124–139 [DOI] [PubMed] [Google Scholar]
- Suzuki I, Kanesaki Y, Mikami K, Kanehisa M, Murata N (2001) Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol Microbiol 40 235–244 [DOI] [PubMed] [Google Scholar]
- Teramoto H, Itoh T, Ono TA (2004) High-intensity-light-dependent and transient expression of new genes encoding distant relatives of light-harvesting chlorophyll-a/b proteins in Chlamydomonas reinhardtii. Plant Cell Physiol 45 1221–1232 [DOI] [PubMed] [Google Scholar]
- Tjus SE, Moller BL, Scheller HV (1998) Photosystem I is an early target of photoinhibition in barley illuminated at chilling temperatures. Plant Physiol 116 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang EW, Bowler C, Herouart D, Van Camp W, Villarroel R, Genetello C, Van Montagu M, Inze D (1991) Differential regulation of superoxide dismutases in plants exposed to environmental stress. Plant Cell 3 783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiotis G, Haase W, Engel A, Michel H (1995) Isolation and structural characterization of trimeric cyanobacterial photosystem I complex with the help of recombinant antibody fragments. Eur J Biochem 231 823–830 [DOI] [PubMed] [Google Scholar]
- Wedel N, Klein R, Ljungberg U, Andersson B, Herrmann RG (1992) The single-copy gene psbS codes for a phylogenetically intriguing 22 kDa polypeptide of photosystem II. FEBS Lett 314 61–66 [DOI] [PubMed] [Google Scholar]
- Xu H, Vavilin D, Funk C, Vermaas W (2004) Multiple deletions of small Cab-like proteins in the cyanobacterium Synechocystis sp. PCC 6803: consequences for pigment biosynthesis and accumulation. J Biol Chem 279 27971–27979 [DOI] [PubMed] [Google Scholar]
- Yabuta Y, Motoki T, Yoshimura K, Takeda T, Ishikawa T, Shigeoka S (2002) Thylakoid membrane-bound ascorbate peroxidase is a limiting factor of antioxidative systems under photo-oxidative stress. Plant J 32 915–925 [DOI] [PubMed] [Google Scholar]
- Yao D, Kieselbach T, Komenda J, Promnares K, Prieto MA, Tichy M, Vermaas W, Funk C (2007) Localization of the small CAB-like proteins in photosystem II. J Biol Chem 282 267–276 [DOI] [PubMed] [Google Scholar]
- Zhang S, Scheller HV (2004) Photoinhibition of photosystem I at chilling temperature and subsequent recovery in Arabidopsis thaliana. Plant Cell Physiol 45 1595–1602 [DOI] [PubMed] [Google Scholar]
- Zhong HH, Young JC, Pease EA, Hangarter RP, McClung CR (1994) Interactions between light and the circadian clock in the regulation of CAT2 expression in Arabidopsis. Plant Physiol 104 889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.